SUMMARY
Antibiotic susceptibilities of large cohorts of Enterobacteriaceae isolated from urine collected in the community are scarce. We report the susceptibilities of Enterobacteriaceae isolated from urine of non-selected community populations in a metropolitan area (Leeds and Bradford, UK) over 2 years. Isolates (n = 6614) were identified as follows: Escherichia coli (n = 5436), Klebsiella spp. (n = 525), Proteus mirabilis (n = 305), and 15 other species (n = 290); 58 isolates were unidentified. Ampicillin resistance was observed in 53% E. coli and 28% P. mirabilis; ⩾34% E. coli and P. mirabilis were non-susceptible to trimethoprim compared to 20% Klebsiella spp.; nitrofurantoin resistance was observed in 3% E. coli and 15% Klebsiella spp. The occurrence of extended-spectrum β-lactamases (ESBL) was low (6%), as was non-susceptibility to carbapenems, cefipime and tigecycline (<2%). Further surveillance is required to monitor this level of resistance and additional clinical studies are needed to understand the impact on the outcome of current empirical prescribing decisions.
Key words: Antibiotic resistance, Enterobacteriaceae, surveillance, urinary tract infections (UTIs)
Urinary tract infections (UTIs) are common, affecting both sexes, with highest prevalence in females between the ages of 15 and 29 years [1]. Escherichia coli are the most common cause of UTIs, followed by other Enterobacteriaceae, streptococci, staphylococci and yeasts [1]. Enterobacteriaceae have a range of intrinsic and acquired resistance mechanisms spanning multiple antibiotic classes. In particular, genes encoding extended-spectrum β-lactamases (ESBLs), such as TEM, SHV and CTX-M, are common in Enterobacteriaceae. Genes encoding ESBLs are carried by mobile genetic elements, commonly plasmids, and as such, may be associated with the carriage of resistance genes for other classes of antibiotic (e.g. aminoglycosides and fluoroquinolones), making treatment of infection difficult [2]. In the advent and spread of ESBL-producing Enterobacteriaceae with co-resistance to fluoroquinolones and aminglycosides, carbapenem agents are commonly used. Worryingly, resistance to carbapenem agents in Enterobacteriaceae, particularly Klebsiella spp., has steadily been increasing in recent years [3]. Antibiotic resistance in Enterobacteriaceae can change and spread rapidly. For example, in England and Wales in the early 2000s, there was an unexpected increase in the number of E. coli isolated from patients in the community that were resistant to extended-spectrum cephalosporins [4]. These isolates were found to be carrying CTX-M genes, which have spread rapidly within Enterobacteriaceae and are now the most common cause of resistance to extended-spectrum cephalosporins and monobactams [5].
The antibiotic susceptibilities of organisms isolated from urine are often reported from selected patient groups, usually hospital in-patients, and/or collections of bacteria that are known to be resistant [6, 7]. National and international surveillance schemes include Enterobacteriaceae; however, these mostly concentrate on the susceptibility of isolates identified from blood cultures or sterile sites [8, 9]. Data relating to the antibiotic susceptibilities of large cohorts of Enterobacteriaceae isolated from urine collected in the community setting are scarce. A standardized approach to prospective surveillance is required to obtain comparable results from multiple centres. In 2010 the Health Protection Agency (HPA) initiated a national surveillance scheme to address this lack of data, comprising multiple centres around England (Birmingham, Leeds, London, Manchester, Southampton). The work presented here reports 2 years of susceptibility testing of Enterobacteriaceae isolated from urine samples collected from community patients of the Leeds and Bradford metropolitan area, which has a population of approximately 2·3 million.
All urine samples sent for routine microbiology testing between February 2010 and March 2012 from General Practitioner (GP) surgeries in the Leeds and Bradford area were included in the surveillance (n = 215354). Samples were collected in 20-ml sterile universal tubes containing boric acid (Thermo Scientific, UK) and 6–8 ml were loaded onto an automated urine analyser (UF100™, Sysmex Corp., Japan), to enumerate red and white blood cells, squamous epithelial cells, and bacterial numbers. Positive samples (white cell count >40/μl and or >8000 bacterial cells/μl) were then inoculated onto various culture media, including chromogenic agar (Mast Group Ltd, England), using a multipoint inoculator and after 18–24 h incubation, bacterial growth was read using an automated plate reader (Mast Group Ltd). Indole-positive colonies on the chromogenic agar were presumed to be E. coli and were progressed to susceptibility testing without further identification. Colonies with blue/green or brown colouration were subjected to further identification and susceptibility testing. Each week about 10% of all isolates were subcultured from the chromogenic agar onto cystine-lactose-electrolyte deficient (CLED) agar as a check for purity.
Identification and susceptibility tests were performed on the VITEK® 2 (bioMérieux, France) system with a 0·5 McFarland suspension of the isolate under test. GN-ID cards were used for identification and AST-N054 (February–March 2010) and AST-N144 cards (March 2010 onwards) were used for antimicrobial susceptibility testing. Susceptibility to the following antibiotics was tested: amikacin (AST-N054 only), ampicillin, amoxicillin/clavulanic acid, aztreonam (AST-N054 only), cefalexin, cefepime, cefotaxime, ceftazidime, cefoxitin, cefuroxime, ciprofloxacin, ertapenem, fosfomycin (AST-N144 only), gentamicin, meropenem, nalidixic acid, nitrofurantoin, piperacillin, piperacillin/tazobactam, temocillin (AST-N144 only), tobramycin (AST-N054 only) and trimethoprim. European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints were used by VITEK 2 for the interpretation of susceptibility results.
Between February 2010 and March 2012, 6614 isolates were tested and identified as follows: E. coli (n = 5436), Klebsiella spp. (K. pneumoniae, n = 434; K. oxytoca, n = 91), Proteus mirabilis (n = 305), plus 15 other species of Enterobacteriaceae (n = 308), comprising the genera Enterobacter (n = 108), Citrobacter (n = 97), Morganella (n = 28), Serratia (n = 22), Raoultella (n = 12), other Proteus (n = 6), other Escherichia (n = 3), Kluyvera (n = 3), Shigella (n = 3), Pantoea (n = 2), Providencia (n = 2) and a single isolate of each of the following: Cedeca davisae, Delftia acidovorans, Hafnia alvei and Salmonella enteritidis. Fifty-eight isolates could not be identified using the VITEK 2 analyser. Figure 1 shows the average proportion of E. coli, Klebsiella spp., and P. mirabilis with non-susceptibility to the antibiotics tested. Results of antibiotic susceptibilities presented per quarterly period are provided as Supplementary material (available online). The analysis excluded cefoxitin, which is not used for treatment of UTIs, carbapenems, which had levels of non-susceptibility <1%, and piperacillin/tazobactam following advice from the manufacturer of the AST cards (bioMérieux, France).
Fig. 1.
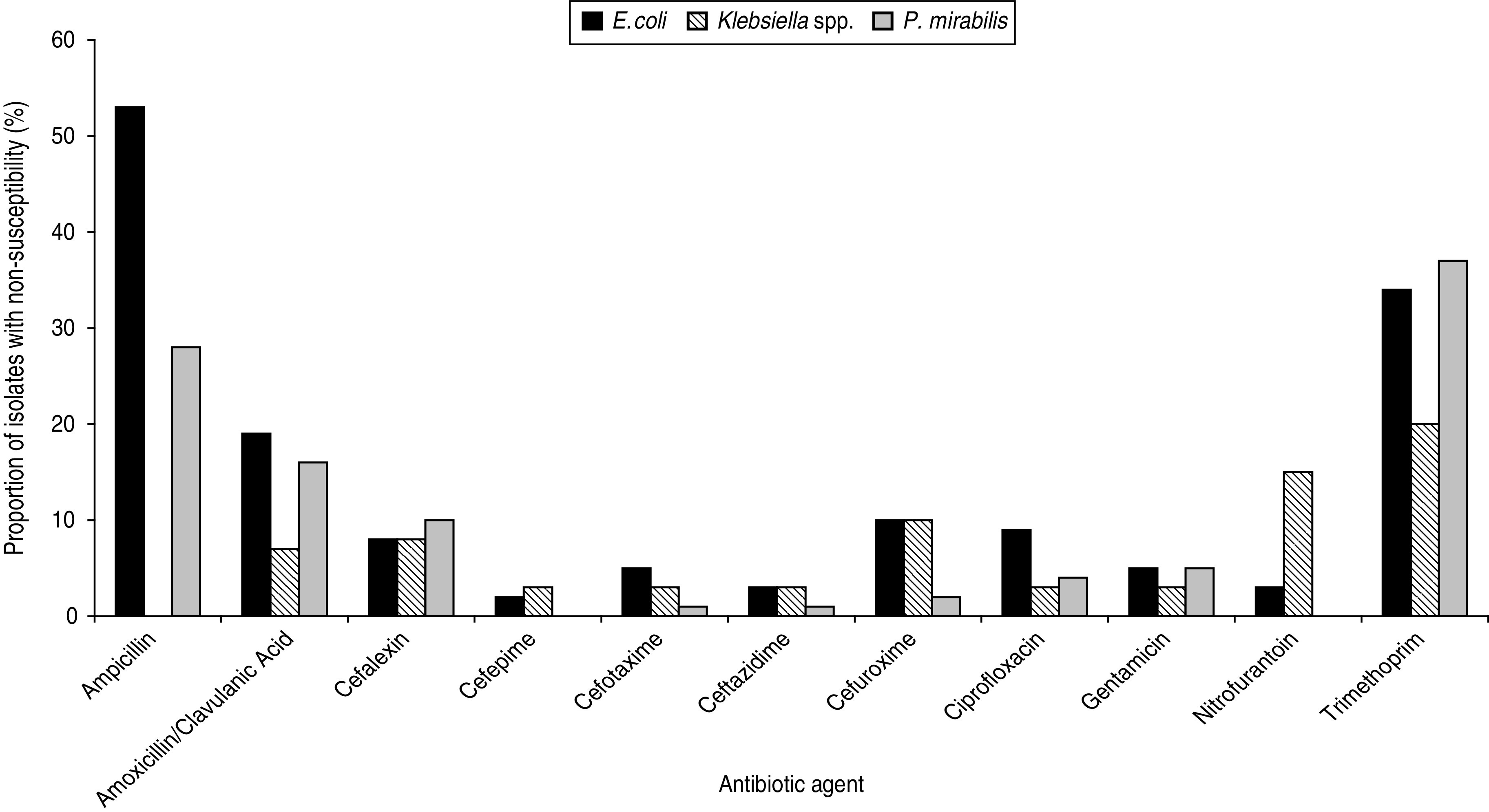
Average quarterly values of antibiotic susceptibilities of Escherichia coli (n = 5436), Klebsiella spp. (n = 525), and Proteus mirabilis (n = 305), February 2010 to March 2012.
Of the Enterobacteriaceae isolates, 53% E. coli and 28% P. mirabilis were resistant to ampicillin; 34% E. coli and 37% P. mirabilis were non-susceptible to trimethoprim compared to 20% of Klebsiella spp. Nitrofurantoin resistance was observed in 3% E. coli and 15% Klebsiella spp. Non-susceptibility to third-generation cephalosporins, ciprofloxacin and gentamicin in all three groups of Enterobacteriaceae was <10%. Non-susceptibility to ertapenem and meropenem was low (<1%) for E. coli, Klebsiella spp. and P. mirabilis, as was non-susceptibility to cefipime (<2%). The susceptibility of E. coli or Klebsiella spp. remained unchanged during the study period (Supplementary Figs S1 and S2); however, notable changes in antibiotic susceptibility were observed in P. mirabilis (Supplementary Fig. S3). Ampicillin resistance in P. mirabilis increased from 20% (February–March 2010) to 43% (April–June 2011) but returned to 21% by January–March 2012 (Supplementary Fig. S3). Resistance to amoxicillin/clavulanic acid in P. mirabilis also increased (5% February–March 2010 vs. 18% January–March 2012) (Supplementary Fig. S2). According to the Advanced Expert System (AES) of the VITEK 2 analyser, 5% (n = 279) E. coli were inferred to have an ESBL phenotype compared to 15% (n = 81) Klebsiella spp. and 2% (n = 5) P. mirabilis. Eighty-one (1·5%) E. coli isolates were inferred to have an AmpC phenotype (either increased chromosomal AmpC cephalosporinase or an acquired AmpC). An inhibitor-resistant TEM β-lactamase phenotype was inferred in 2·5% (n = 139) E. coli, 0·6% (n = 3) Klebsiella and 2·4% (n = 7) P. mirabilis. Fifty-three percent of E. coli inferred as having an ESBL phenotype were also resistant to ciprofloxacin compared to 7% E. coli without an ESBL phenotype.
Twenty-eight isolates had an ertapenem minimum inhibitoty concentration (MIC) >1 mg/l and/or a meropenem MIC >4 mg/l and were interpreted as non-susceptible to carbapenem agents (E. coli, n = 16; K. pneumoniae, n = 5; Enterobacter spp., n = 4 and one of each of the following P. mirabilis, S. fonticola, C. freundii). According to AES, 11 of these isolates had an inconsistent phenotype and were excluded from analysis. Low-level ertapenem (MIC values between 2 and 4 mg/ml) resistance was observed in the isolates of P. mirabilis and Enterobacter spp., which is an expected phenotype probably due to the presence of AmpC and impermeability changes [3]. The isolates of E. coli and K. pneumoniae were identified by VITEK 2 to have ESBLs plus impermeability changes. Confirmation of resistance mechanisms or further characterization of isolates was not done.
This study has tested the susceptibility of a large number of Enterobacteriaceae identified from the urine samples of a non-selected population, over a 2-year period using an automated, standardized method. Due to differences in study population, methodology and/or panels of antibiotic agents used for testing, comparison of antibiotic susceptibility data from uropathogens is not straightforward. In addition, sampling bias is likely given that only the patients with the most serious and persistent UTIs will have a urine sample referred for microbiology investigation. Three other pieces of work from around the UK are available which provide a suitable comparison for the present study [10–12]. Rates of non-susceptibility of E. coli and Klebsiella spp. found here are comparable with these studies for the antibiotics tested (ampicillin, amoxicillin/clavulanic acid, trimethoprim, ciprofloxacin, nitrofurantoin); however, an important difference is noted in the prevalence of amoxicillin/clavulanic acid resistance in P. mirabilis. Farrell et al. [11] reported P. mirabilis collected between 1999 and 2000 with 3·2% resistance to amoxicillin/clavulanic acid, which is in marked contrast to the present study and similar surveillance in Wales which recorded ∼16% resistance for this drug species combination [12]. No such increase in amoxicillin/clavulanic acid resistance was identified in isolates of E. coli but it should be noted that difference in susceptibility between these studies may be due to the method or breakpoints used by each centre.
Surveillance of the antibiotic susceptibilities of Enterobacteriaceae isolated from community urine samples is important in order to monitor for changes in resistance to the commonly used empirical antibiotics. Many patients who present to GPs with a suspected uncomplicated UTI are treated with empirical therapy. In the UK, this is most commonly trimethoprim or nitrofurantoin and urine samples are not routinely sent for microbiological investigation [13]. This study identified that >30% E. coli and P. mirabilis were non-susceptible to trimethoprim, whereas rates of resistance to nitrofurantoin were less frequent (3% E. coli and 15% Klebsiella spp., respectively). What is not known is how closely these non-susceptibility rates correlate with clinical failure rates of empirical antimicrobial therapy which result in patients requiring further visits to their GP, additional second-line therapies and further investigation. While the prevalence of patients with ESBL- and AmpC-producing E. coli may appear low (5% and 1·5%, respectively), UTIs with this organism are so common that the absolute figure of community-based patients experiencing UTI with one of these strains across the UK will be high. These strains are often resistant to other agents (e.g. fluoroquinolones), which limits the number of therapeutic options available to GPs. Failure to successfully treat a UTI provides an opportunity for bacteria to persist and cause a more serious infection, such as a bacteraemia. Could this failure to treat be contributing to the marked increase in reports of E. coli bacteraemia in England observed in recent years?
Changes to prescribing policies in the primary-care setting have occurred over the past few years. For instance, the use of ciprofloxacin and third-generation cephalosporins has been discouraged in order to reduce the rates of Clostridium difficile infection [14]. This study showed that rates of resistance to ciprofloxacin and third-generation cephalosporins remain low (<10%) and although rates of non-susceptibility to trimethoprim (34%, 37% and 20% for E. coli, P. mirabilis and Klebsiella spp., respectively) and nitrofurantoin (3% and 15% for E. coli and Klebsiella spp., respectively) are higher, there was no sustained increase in resistance to these commonly used empirical agents during the study period.
Despite the large numbers of Enterobacteriaceae tested here, the sample represents only 10% of the total Enterobacteriaceae identified from urine samples; testing more isolates may increase the number of isolates with resistant phenotypes. However, urine sampling in the community setting; is known to be biased towards cases where resistant uropathogens are more likely [15]. It was not possible to compare the susceptibility profiles of bacteria collected from different demographic (i.e. male/female or age related) or clinical groups as information about the patients was not collected. Nevertheless, our data represent a contemporaneous picture of the antibiotic susceptibilities of Enterobacteriaceae collected from urine samples taken from community patients in a large metropolitan area in England over a 2-year period. It is reassuring that no sustained changes in antimicrobial susceptibility were observed during the 2-year study period, which is admittedly relatively short in ecological terms; however, further surveillance is required to monitor for the emergence of resistance in Enterobacteriaceae. As E. coli is a common cause of UTI, emergence of resistance would make treating such infections, particularly in the community, a serious challenge. Additional clinical studies are required to understand the impact that a change in susceptibility may have on the outcome of current empirical prescribing decisions.
Supplementary Material
Supplementary information supplied by authors.
ACKNOWLEDGEMENTS
We thank all the staff in the urine section of the Department of Microbiology, Leeds General Infirmary. The surveillance was funded by the Health Protection Agency.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268813000988.
click here to view supplementary material
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Foxman B. The epidemiology of urinary tract infection. Nature Reviews Urology 2010; 7: 653–660. [DOI] [PubMed] [Google Scholar]
- 2.Woodford N, et al. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrobial Agents and Chemotherapy 2009; 53: 4472–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Health Protection Agency. Department of Health Advisory Committee on Antimicrobial Resistance and Hospital Associated Infection (ARHAI). Advice on carbapenemase producers: recognition, infection control and treatment (http://www.hpa.org.uk/Topics/InfectiousDiseases/InfectionsAZ/CarbapenemResistance/GuidanceOnCarbapenamProducers/). Accessed 24 April 2013.
- 4.Woodford N, et al. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum beta-lactamases in the UK. Journal of Antimicrobial Chemotherapy 2004; 54: 735–743. [DOI] [PubMed] [Google Scholar]
- 5.Rossolini GM, D'Andrea MM, Mugnaioli C. The spread of CTX-M-type extended-spectrum beta-lactamases. Clinical Microbiology and Infection 2008; 14 (Suppl. 1): 33–41. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez J, et al. Prevalence and risk factors of infections by multiresistant bacteria in cirrhosis: a prospective study. Hepatology 2012; 55: 1551–1561. [DOI] [PubMed] [Google Scholar]
- 7.Roux D, et al. Diversity of extended-spectrum beta-lactamase-producing Enterobacteriaceae on hospital admission. Scandinavian Journal of Infectious Diseases 2012; 44: 231–236. [DOI] [PubMed] [Google Scholar]
- 8.European Centre for Disease Prevention and Control. European Antimicrobial Resistance Surveillance System (EARSS-Net) (http://www.rivm.nl/earss/). Accessed 26 November 2012.
- 9.Livermore DM, et al. Non-susceptibility trends among Enterobacteriaceae from bacteraemias in the UK and Ireland, 2001–06. Journal of Antimicrobial Chemotherapy 2008; 62 (Suppl. 2): ii41–ii54. [DOI] [PubMed] [Google Scholar]
- 10.Brabazon E, et al. Epidemiology and resistance patterns in urinary pathogens from long-term care facilities and GP populations. Irish Medical Journal 2012; 105: 177–180. [PubMed] [Google Scholar]
- 11.Farrell DJ, et al. A UK multicentre study of the antimicrobial susceptibility of bacterial pathogens causing urinary tract infection. Journal of Infection 2003; 46: 94–100. [DOI] [PubMed] [Google Scholar]
- 12.Heginbothom M, Howe R. Antibacterial resistance and usage in Wales, 2005–2010. 2011. (http://www.wales.nhs.uk/news/21018). Accessed 13 December 2012.
- 13.Health Protection Agency. Management of infection guidance for primary care for consultation and local adaptation (http://www.hpa.org.uk/webc/HPAwebFile/HPAweb_C/1194947333801). Accessed 24 April 2013.
- 14.Health Protection Agency. Clostridium difficile infection: how to deal with the problem (http://www.hpa.org.uk/webc/HPAwebFile/HPAweb_C/1232006607827). Accessed 24 April 2013.
- 15.Hillier S, et al. When do general practitioners request urine specimens for microbiology analysis? The applicability of antibiotic resistance surveillance based on routinely collected data. Journal of Antimicrobial Chemotherapy 2006; 58: 1303–1306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information supplied by authors.
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268813000988.
click here to view supplementary material


