Abstract
We consider the problem of learning to simplify medical texts. This is important because most reliable, up-to-date information in biomedicine is dense with jargon and thus practically inaccessible to the lay audience. Furthermore, manual simplification does not scale to the rapidly growing body of biomedical literature, motivating the need for automated approaches. Unfortunately, there are no large-scale resources available for this task. In this work we introduce a new corpus of parallel texts in English comprising technical and lay summaries of all published evidence pertaining to different clinical topics. We then propose a new metric based on likelihood scores from a masked language model pretrained on scientific texts. We show that this automated measure better differentiates between technical and lay summaries than existing heuristics. We introduce and evaluate baseline encoder-decoder Transformer models for simplification and propose a novel augmentation to these in which we explicitly penalize the decoder for producing ‘jargon’ terms; we find that this yields improvements over baselines in terms of readability.
1. Introduction
The need for accessible medical information has never been greater. A Pew Research survey of American’s online health habits in 2013 revealed that “one in three American adults have gone online to figure out a medical condition” (Fox and Duggan, 2013). Given the rise of medical misinformation on the internet (Ioannidis et al., 2017), accessibility has become an increasingly urgent issue (World Health Organization, 2013; Armstrong and Naylor, 2019). However, sources that provide accurate and up-to-date information, including scientific papers and systematic reviews (Chalmers et al., 1995), are often effectively inaccessible to most readers because they are highly technical and laden with terminology (Damay et al., 2006).
One potential solution to this problem is text simplification, i.e., editing documents such that they are accessible to a wider audience, while preserving the key information that they contain. Although manual simplification is too expensive to feasibly apply at scale, automatic text simplification (Siddharthan, 2014; Alva-Manchego et al., 2020) provides a potential means of rendering a large volume of specialist knowledge more accessible.
Large-scale data-driven simplification systems have mostly been trained on Wikipedia (Zhu et al., 2010; Woodsend and Lapata, 2011; Coster and Kauchak, 2011) and news (Xu et al., 2015), and focus on sentence simplification (Wubben et al., 2012; Wang et al., 2016; Xu et al., 2016; Zhang and Lapata, 2017; Kriz et al., 2019; Dong et al., 2019; Alva-Manchego et al., 2020); on the other hand, medical text simplification is resource poor. Recent work has involved constructing sentence-aligned data automatically using monolingual text alignment methods (Adduru et al., 2018; Van den Bercken et al., 2019), but this process is noisy and constrains the task to sentence-level simplification.
In this work we explore new data and modern conditional text generation models (Lewis et al., 2020) to simplify medical documents. We introduce a dataset of paired (technical, simplified) texts derived from the Cochrane Database of Systematic Reviews, which is comprised of evidence syntheses on a wide range of clinical topics. Critically, each review includes a plain-language summary (PLS) written by the authors. PLS are written directly from the full reviews with their own structure and guidelines; they are not simplified versions of the corresponding technical abstracts of the reviews, nor are they summaries of the abstracts.
However, we observe that portions of the PLS can be considered simplifications of analogous sections in the abstracts, that is, they contain roughly the same content but involve simplification operations such as paraphrasing, word/sentence deletion, and summarization. We heuristically derive 4459 such pairs of sections (or paragraphs) of technical–plain English bitexts. We provide an excerpt of the dataset we have constructed in Table 1.
Table 1:
Sample excerpts from a technical abstract (top) and corresponding plain-language summary (bottom) from the Cochrane Library.
| Technical abstract: Analysis showed a higher rate of weight gain in the high-volume feeds group: mean difference 6.20 g/kg/d (95% confidence interval 2.71 to 9.69). There was no increase in the risk of feed intolerance or necrotising enterocolitis with high-volume feeds, but 95% confidence intervals around these estimates were wide. |
| Plain-language summary: Very low birth weight infants who receive more milk than standard volumes gain weight more quickly during their hospital stay. We found no evidence suggesting that giving infants high volumes of milk causes feeding or gut problems, but this finding is not certain. |
This data allows us to explore characteristics of simplified versions of technical medical texts. We show that the differences in traditional readability metrics, such as Flesch-Kincaid (Kincaid et al., 1975) and Automated Readability Index (Senter and Smith, 1967), are small. Instead, the differences are better captured using large-scale pretrained masked language models, and this reveals that there is more to the language difference than the shallow cues such as sentence and word lengths that traditional readability metrics focus on.
We present baseline methods for automatic text simplification over this data and perform analyses that highlight the challenges of this important simplification task. We find that when naively fine-tuned for the task, existing encoder-decoder models such as BART (Lewis et al., 2020) tend to prefer deletion over paraphrasing or explaining, and are prone to generating technical words. We propose a new approach to try and mitigate the latter issue by imposing a variant of unlikelihood loss (Welleck et al., 2019) that explicitly penalizes the decoder for production of ‘technical’ tokens. We show that this yields improvements in terms of readability with only a minor tradeoff with content quality.
In sum, this work takes a step towards paragraph-level simplification of medical texts by: (1) introducing a sizable new dataset, (2) proposing and validating a new masked language model (MLM)-based metric for scoring the technicality of texts, (3) analyzing and understanding the style of plain language in this important domain, and (4) presenting baselines that exploit a variant of unlikelihood training to explicitly penalize models for producing jargon. We release our code and data at https://github.com/AshOlogn/Paragraph-level-Simplification-of-Medical-Texts.
2. Related work
Recent efforts on data-driven text simplification methods have tended to rely on two resources: the Wikipedia-Simple Wikipedia aligned corpus (Zhu et al., 2010; Woodsend and Lapata, 2011; Coster and Kauchak, 2011) and the Newsela simplification corpus (Xu et al., 2015). Yet, there is an urgent need to simplify medical texts due to health literacy levels (World Health Organization, 2013). However, due to a lack of resources with which to train model-based simplification systems in this domain, past work has tended to focus on lexical simplification (Damay et al., 2006; Kandula et al., 2010; Abrahamsson et al., 2014; Mukherjee et al., 2017). Recently, Adduru et al. (2018) and Van den Bercken et al. (2019) introduced sentence-aligned corpora at the scale of thousands of sentence pairs. In contrast to our corpus, these datasets were automatically derived using paraphrase mining or monolingual alignment processes. Furthermore, as these are exclusively sentence corpora, they limit the set of potential approaches to just those that operate over sentences. Grabar and Cardon (2018) created a simplification corpus for medical texts in French, in which a small subset of the text pairs are manually sentence-aligned, resulting in 663 sentence pairs, 112 of which are also from Cochrane.
With respect to modeling, recent work has focused on sentence simplification, treating it as a monolingual machine translation task (Wubben et al., 2012; Wang et al., 2016; Xu et al., 2016) using encoder-decoder models (Zhang and Lapata, 2017; Kriz et al., 2019; Dong et al., 2019). In the medical domain, existing systems tend to adopt lexical and syntactic simplification (Damay et al., 2006; Kandula et al., 2010; Llanos et al., 2016). Research on document simplification has been sparse; to the best of our knowledge, the few prior works on this in English have focused on analysis (Petersen and Ostendorf, 2007), sentence deletion (Woodsend and Lapata, 2011; Zhong et al., 2020), and localized explanation generation (Srikanth and Li, 2020). This work proposes and evaluates an encoder-decoder model for paragraph-level simplification.
3. Technical abstracts vs. plain-language summaries
We compiled a dataset of technical abstracts of biomedical systematic reviews and corresponding PLS from the Cochrane Database of Systematic Reviews, which comprises thousands of evidence synopses (where authors provide an overview of all published evidence relevant to a particular clinical question or topic). The PLS are written by review authors; Cochrane’s PLS standards (Cochrane, 2013) recommend that “the PLS should be written in plain English which can be understood by most readers without a university education”. PLS are not parallel with every sentence in the abstract; on the contrary, they are structured heterogeneously (Kadic et al., 2016).
3.1. Data compilation
To derive the dataset we scraped the online interface to the database for articles containing PLS, extracting the raw text of the technical abstracts and PLS for those that we identified. In this way we obtained 7820 pairs after removing problematic links (e.g., HTTP 404 errors). We also excluded reviews with atypical formatting that would have required extensive manual inspection.
On average, PLS are shorter than abstracts (Table 2, ‘raw’). They contain sections different from those in the abstracts, emphasize different content, and sometimes contain information not in the abstract. We divided documents into those that are split into sections with subheadings and those without (henceforth “long-form” summaries); 56% of the data are long-form. For the sectioned PLS, headers are quite different from those found in the abstracts. The latter adhere to one of the 2 following formats:
Table 2:
Means and standard deviations of original abstract and PLS lengths (tokens), and our compiled data before & after filtering out texts with more than 1024 tokens.
| Compiled data | |||
|---|---|---|---|
| Raw | Before-filter | After-filter | |
|
| |||
| Abstract | 815 ± 331 | 551 ± 272 | 501 ± 211 |
| PLS | 394 ± 216 | 284 ± 156 | 264 ± 136 |
Background, Objectives, Search Methods, Selection Criteria, Data Collection and Analysis, Main Results, Authors’ Conclusions
Background, Objectives, Methods, Main Results, Authors’ Conclusions
In contrast, PLS contain a variety of headings, with the most common ones shown below:
background, study characteristics, key results, review question, quality of the evidence, search date, quality of evidence, conclusions
Others include questions such as What was the aim of this review? And How up-to-date was the review?
Manual inspection revealed that the results, discussion, and conclusion sections of abstracts and summaries tended to occur in parallel. This motivated us to extract aligned subsets of abstracts and summaries to compose our dataset. More specifically, we determined the approximate location of the section describing studies and results in each text and kept everything from that point forward.
Therefore, in the abstracts we kept the text from the Main Results section onward. For the sectioned PLS we kept every section after and including the first that contained one of the following substrings: find, found, evidence, tell us, study characteristic. For the long-form PLS, we found the first paragraph containing any of the following words within the first couple sentences and included that and subsequent paragraphs: journal, study, studies, trial. We keep one-paragraph PLS in their entirety. We also exclude instances where the PLS and abstracts are drastically different in length, by keeping only instances where the length ratio between the two falls between 0.2 and 1.3. Our final dataset comprises 4459 pairs of technical abstracts and PLS, all containing ≤1024 tokens (so that they can be fed into the BART model in their entirety).
3.2. Characterizing readability differences Readability metrics.
Designing metrics that reliably capture readability remains an open topic of research. In recent years, a host of metrics have been developed that use a wide variety of linguistic features to assess readability in a supervised manner. For example, Kate et al. (2010) developed a metric based on syntactical, semantic, and language model-based features, and Vajjala and Lučić (2018) developed a new readability corpus, on which they trained support vector machines to predict text readability. For this medical text simplification task, however, we considered a couple established heuristics-based readability metrics due to clear domain differences between our Cochrane corpus and those used to train supervised readability metrics: the Flesch-Kincaid score (Kincaid et al., 1975) and the automated readability index (ARI) (Senter and Smith, 1967), which estimate the educational maturity (grade-level) required to comprehend a text. These metrics rely on a combination of shallow cues, most notably lengths of words, sentences, and documents.
Table 3 reports the mean grade levels of abstracts and PLS calculated via the above metrics. There are small but statistically significant (p < 0.01, paired t-test) differences between the abstract and PLS distributions, especially for Flesch-Kincaid. For instance, the maximum difference in mean minimum grades (1.5) is achieved by Flesch-Kincaid, and the number is only 0.6 with ARI. By contrast, a 3–5 grade level difference was shown on the Wikipedia and Britannica simplification datasets (Li and Nenkova, 2015). The high gradelevel suggested by standard readability metrics confirms prior studies highlighting that these ‘plain language’ summaries of medical systematic reviews remain at higher reading levels than those of average US adults (Karačić et al., 2019).
Table 3:
Means and standard deviations of different readability scores calculated over abstracts and PLS.
| Metric | Abstracts | PLS |
|---|---|---|
|
| ||
| Flesch-Kincaid | 14.4 ± 2.3 | 12.9 ± 2.4 |
| ARI | 15.5 ± 2.8 | 14.9 ± 3.0 |
Masked language models.
Despite the small differences in readability metrics, PLS do qualitatively seem easier to understand (see Table 1 for an example). This suggests that existing measures are incomplete. We propose adopting modern masked language models — namely BERT (Devlin et al., 2019) — as another means of scoring the ‘technicality’ of text. In particular, when such models are trained on specialized or technical language (e.g., scientific articles) we would expect the likelihoods subsequently assigned to ‘jargon’ tokens to be relatively high compared to a model trained over general lay corpora, as in the original BERT model (Devlin et al., 2019).
Capitalizing on this intuition, we consider two large-scale pre-trained masked language models: (1) BERT (Devlin et al., 2019) trained on BooksCorpus (Zhu et al., 2015) and English Wikipedia; and (2) SciBERT (Beltagy et al., 2019), trained on a sample of 1.14 million technical papers from Semantic Scholar (Ammar et al., 2018) (mostly biomedical and computer science articles). Inspired by the original training objective for these models, we compute a probability score for a document by splitting it into sentences, masking 10 subsets of 15% of the tokens in each sentence (exempting CLS and SEP), computing the likelihoods of the original tokens in the distributions output by the model in each masked position, and averaging these probabilities over all the masked subsets and sentences in the document. The details are shown in Algorithm 1.
Algorithm 1.
Used to compute a probability score for a text document D given a masked language model M. The output of the model returned by a call to Forward is a matrix where each row maps to a distribution over all the tokens in the vocabulary. The Append function adds a value to the end of a list.
| procedure Masked-Prob(D, M) |
| sents ← Sentence-Split(D) |
| P ← Initialize empty list |
| for i = 1 … |sents| do |
| T ← Tokenize(sents[i]) |
| for j = 1 … 10 do |
| A ← sample 15% from 1… |T| |
| T′ ← T |
| for all a ∈ A do |
| T′[a] ← [MASK] |
| outputs ← Forward(M, T′) |
| for all a ∈ A do |
| prob ← outputs[a][T[a]] |
| Append(P, prob) |
| return mean(P) |
Figure 1 depicts the distributions of probabilities output by general BERT and SciBERT for the abstracts and PLS in our dataset. Both masked LMs induce distributions over instances from the respective sets that are clearly different. For example, SciBERT (which yields sharper differences) outputs higher likelihoods for tokens comprising the technical abstracts than for those in the plain language versions, as we might expect given that this is pretrained on technical literature. A paired t-test confirms that these observed differences between the abstracts and PLS distributions are statistically significant (with p < 0.01).
Figure 1:
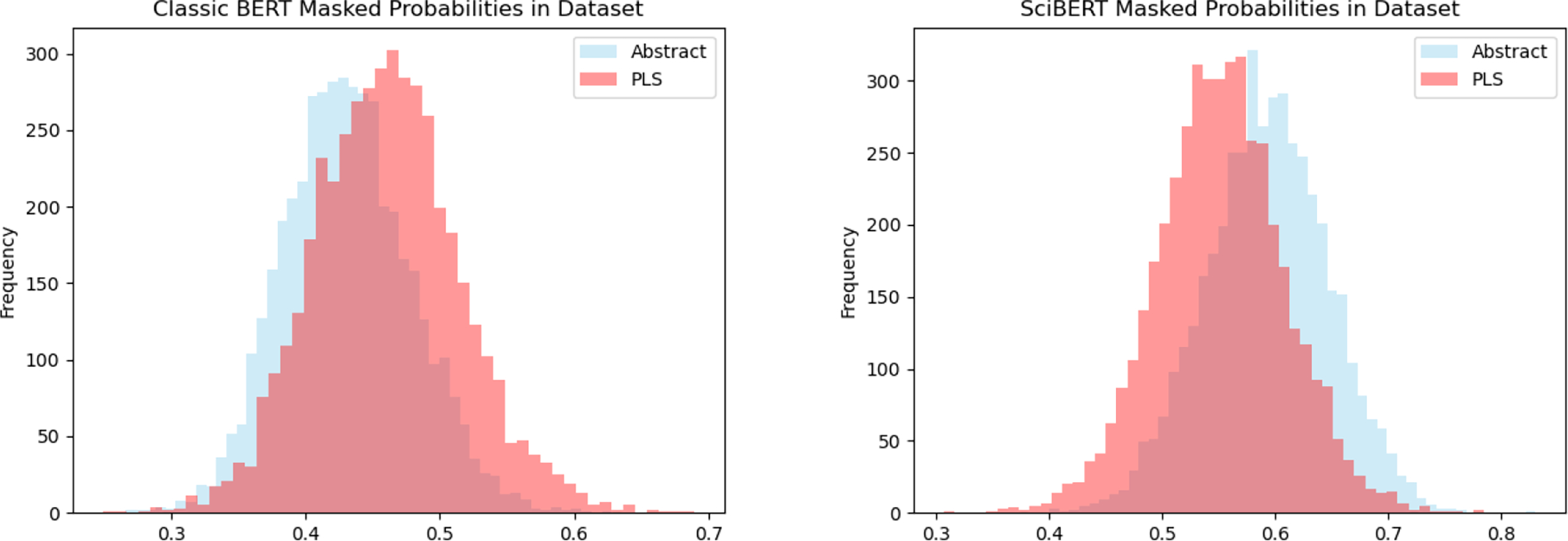
BERT (left) vs SciBERT (right) probabilities of technical abstracts (blue) and PLS (red).
Which metric discriminates better?
To better determine how well the proposed masked probability outputs discriminate between technical abstracts and PLS, we plot receiver operating characteristic (ROC) curves for the outputs of BERT, SciBERT, Flesch-Kincaid and ARI, coding technical and PLS abstracts as 0 and 1, respectively. The SciBERT curve has a higher AUC score (0.70) than the general BERT curve (0.66), indicating that it is better at discriminating between plain language and technical abstracts.For this reason, we use the SciBERT masked probabilities when analyzing the texts generated by our models.
The AUC score for SciBERT is also higher than that for Flesch-Kincaid, indicating that simplicity in PLS can be better captured by probabilistic means than by surface-level linguistic cues, and that it is more appropriately viewed as a stylistic difference rather than one of readability.This echoes the arguments made by early investigators of readability metrics that these measures do not replace more subtle linguistic characteristics, e.g., style (Klare, 1963; Chall, 1958).
3.3. Lexical analysis
We next investigate lexical differences between technical abstracts and PLS. In prior work, Gledhill et al. 2019 performed extensive lexical analysis on this corpus by comparing the relative frequencies of different part-of-speech n-grams found in the abstracts and PLS. Here, we analyze the weights from a logistic regression model that classifies whether a text is a technical abstract or a PLS (coding the latter as y = 1); the weights learned by the model can be conveniently incorporated into the loss function we use to train our simplification model (Section 4.2).
We represent texts as normalized bag-of-words frequency vectors (with a feature for each token in the BART vocabulary). We performed 5-fold cross validation on the data and observed an average accuracy of 92.7%, which indicated that even this relatively simple model is capable of accurately distinguishing technical abstracts from PLS. We also evaluated this model on the train-validation split described in Section 4.3. The model achieves a very high AUC score of 0.99, indicating that it almost perfectly separates abstracts from PLS.
To better understand which kinds of tokens are most associated with technical abstracts and PLS, we examined the tokens with the highest-magnitude learned weights in the model, with the most negative weights corresponding to tokens indicative of technical abstracts and the most positive ones being indicative of PLS. These notable tokens are displayed in Table 4. From this table it is clear that numerical tokens and those related to statistical analysis, like bias and CI (confidence interval) are most indicative of abstracts. The tokens indicative of PLS are less illuminating and merely reflect common phrases include in PLS, such as In this review and We searched scientific databases.
Table 4:
The tokens with the most negative and most positive weights in a logistic regression model trained to distinguish technical abstracts from PLS.
| Token | Weight | Token | Weight |
|---|---|---|---|
|
| |||
| 0 | −7.262 | people | 4.681 |
| . | −6.126 | review | 4.551 |
| % | −5.379 | We | 4.461 |
| CI | −4.986 | This | 3.413 |
| ; | −4.821 | that | 2.943 |
| 95 | −4.593 | The | 2.836 |
| significant | −4.273 | side | 2.722 |
| R | −3.726 | who | 2.671 |
| 1 | −3.685 | blood | 2.515 |
| There | −3.477 | found | 2.514 |
| bias | −3.303 | searched | 2.407 |
| criteria | −3.263 | The | 2.114 |
| outcome | −3.247 | results | 2.098 |
| ( | −3.195 | their | 2.022 |
| inclusion | −3.148 | current | 1.984 |
In Section 4, we use this model as a discriminator along with our transformer encoder-decoder model during training to penalize the generation of tokens that are indicative of technical abstracts.
4. Baseline models for simplification
4.1. Pretrained BART
Our baseline simplification model is BART (Lewis et al., 2020), an encoder-decoder architecture in which both components are transformers (Vaswani et al., 2017). The decoder is auto-regressive, making it a natural fit for generation tasks. BART has been shown to achieve strong performance on text summarization, specifically on the CNN/Daily Mail (Hermann et al., 2015) and XSum (Narayan et al., 2018) datasets.
We initialize the weights in BART to those estimated via fine-tuning on the XSum (Narayan et al., 2018) dataset as provided by HuggingFace’s Model Hub (Wolf et al., 2019). We then fine-tune these models on our corpus.1
In the decoding step, we use nucleus sampling (Holtzman et al., 2019): at each step of token generation the next token is sampled from a probability distribution constructed by removing the ‘tail’ of probability mass from BART’s output distribution and then renormalizing. This strategy mitigates the awkward repetition typical of greedy methods like beam search while still avoiding incoherence by truncating the unlikely tail in the original model distribution.
4.2. Unlikelihood training
As an additional mechanism to encourage simple terminology in the PLS generated by our model, we propose a new method in which we explicitly penalize the model for producing seemingly technical words via unlikelihood training (Welleck et al., 2019; Li et al., 2020). The idea is to add a term to the objective that encourages the model to decrease the probability mass assigned to some set of tokens . This is realized by adding a term to the (log) loss: , where x is the technical abstract input to the encoder, y<t is the prefix of the target summary y input to the decoder at time t, and pθ(sj|y<t, x) is the probability assigned to token sj in the distribution output by BART (with model parameters θ) at time t. This expression is referred to as Unlikelihood Loss (UL). The UL term is weighted by a positive constant α and added to the typical log-likelihood objective.
We construct by collecting tokens with negative weights from a bag-of-words logistic regression model trained to classify whether a document is simple (1) or complex (0), for which negative tokens are indicative of complex language. We then softmax the absolute values of these weights so that they sum to 1 and the tokens most indicative of technical abstracts (i.e., those with the most negative weights initially) contribute the most to this sum. We consider three variants of this procedure. (1) We classify whether a document is a PLS or an abstract (Section 3.3). (2) We use external data, namely the Newsela corpus (Xu et al., 2015), and train a model to distinguish between documents of reading levels 0 and 3.2 (3) We train two different models for the previous tasks and then sum the weight vectors before applying a softmax to derive token penalties.
Let wj denote the learned logistic regression weight for token . The final weight used in the unlikelihood loss function is:
| (1) |
where T is the temperature of the softmax.
A modification we make to the unlikelihood loss function is that we only apply the loss for a given token sj if the probability distribution output for the token at position t indicates that sj should be output, that is, if where denotes BART’s token vocabulary. Denoting an indicator function for this event by , our final unlikelihood loss term is:
| (2) |
4.3. Experimental setup
Data.
We split our dataset of 4459 abstract-PLS pairs so that 3568 reviews are in the training set, 411 in the validation set, and 480 in the test set. We experimented with hyperparameters by manually inspecting a subset of the validation set and report results on the entire test set.
Hyperparameters.
For nucleus sampling, we use a top-p value of 0.9. In the unlikelihood training procedure, we experimented with different values of α in our total loss function (1, 10, 103, 106) on the validation set and different temperatures T in the softmax step (1, 2, 5, 10). Based on manual examination of the generated texts in the validation set, we determined that (T = 2, α = 100) yields the most coherent and high-quality simplifications, so we only report results for this case. All models are fine-tuned on our dataset for 1 epoch with a batch size of 1 and a learning rate that starts at 3e-5 and decreases linearly to 0 over the course of training. For optimizer, we used AdamW with ε = 1e-8 (Kingma and Ba, 2015; Loshchilov and Hutter, 2019).
5. Results
In this section we comment on the generated texts’ readability, quality of summarization and simplification, stylistic fidelity with the PLS, and overall coherence and simplicity based on human examination. In the results tables, we indicate whether lower or higher scores for the metrics reported are better with ↓ and ↑ symbols, respectively.
5.1. Readability scores
Table 5 reports the mean readability scores achieved under different training settings. Results generated via models trained with the proposed UL objective achieve significantly lower Flesch-Kincaid scores than those achieved by both the technical abstracts and reference PLS, whereas the model trained without UL produced texts with a higher reading level than the PLS. Rather surprisingly, the UL-Newsela and UL-both settings, both of which use the Newsela dataset to produce unlikelihood weights, did not yield a decrease in estimated grade levels. We suspect that this could be attributed to the difference in domains, that is, the tokens contributed by the Newsela classifier are not generated frequently enough to have a noticeable impact during unlikelihood training.
Table 5:
Flesch-Kincaid, ARI, and SciBERT masked probability scores for generated PLS. Differences wbetween abstracts and generated PLS are statistically significant; so are differences in FK and ARI between UL models and No-UL (p < 0.01, paired t-test).
| FK↓ | ARI↓ | SciBERT↓ | |
|---|---|---|---|
|
| |||
| Abstracts | 14.42 | 15.58 | 0.57 |
| PLS | 13.11 | 15.08 | 0.53 |
|
| |||
| No UL | 13.44 | 15.09 | 0.55 |
| UL-Cochrane | 11.97 | 13.73 | 0.55 |
| UL-Newsela | 12.51 | 14.15 | 0.54 |
| UL-Both | 12.26 | 14.04 | 0.54 |
These results suggest that: (1) BART is capable of performing simplification of medical texts such that outputs enjoy reduced reading levels compared to those of the technical abstracts; (2) The proposed use of UL to explicitly penalize the model for outputting jargon allows for the generation of text with even greater readability than the reference PLS. The reading levels of even the simplified outputs, however, are at the late-high school/early college levels. This could reflect the relatively small differences in readability scores between abstracts and PLS in general (Section 3.2).
5.2. Style
In Section 3.2 we showed that SciBERT masked probability scores are more useful as a discriminator between technical abstracts and PLS than the standard readability metrics, which use surface-level cues like word and sentence counts. Experiments by Jawahar et al. (2019) suggest that BERT-style masked language models encode a wide array of syntactic and semantic features of language, which they then employs for downstream tasks. For this reason, we use SciBERT masked probability scores as our notion of style, with lower scores corresponding to simpler, less technical language. To explore the extent to which the generated summaries stylistically resemble the PLS, we computed the average of the SciBERT masked probability scores of the generated texts for each model. The results are shown in Table 5 along with the readability scores.
We see that every model produces text with significantly lower probability scores than the abstracts, which suggests that they successfully convert input abstracts into less-technical summaries. Though the average scores are higher than that of the PLS, this difference is not statistically significant, so we can consider the outputs of the models to be stylistically on par with the target PLS.
5.3. Content
We report SARI (Xu et al., 2016), a standard edit-based metric for text simplification, and BLEU (Papineni et al., 2002), a precision-based method for machine translation that is also often reported for simplification systems. Xu et al. (2016) showed that SARI correlates better with human evaluation for simplification tasks, focusing more on simplicity, while BLEU is stronger with respect to meaning and grammar. Finally we report the F1 versions of ROUGE-1, ROUGE-2, and ROUGE-L (Lin, 2004), which are the standard metrics typically used for summarization tasks.
Table 6 shows the mean ROUGE, BLEU, and SARI scores. While UL models yielded small but significantly better SARI scores, the opposite is true for the ROUGE and BLEU measures. Despite the lack of clear patterns in these scores, there are clear qualitative differences between the different models’ outputs, which are expounded upon in Section 5.4.
Table 6:
ROUGE, BLEU, and SARI scores for generated PLS. All differences between No-UL and UL models, except for (BLEU, UL-Newsela), are statistically significant (p < 0.01, paired t-test).
| R1↑ | R2↑ | RL↑ | BLEU↑ | SARI↑ | |
|---|---|---|---|---|---|
|
| |||||
| No UL | 0.40 | 0.15 | 0.37 | 0.44 | 0.38 |
| UL-Cochrane | 0.38 | 0.14 | 0.36 | 0.39 | 0.40 |
| UL-Newsela | 0.39 | 0.15 | 0.37 | 0.43 | 0.39 |
| UL-Both | 0.38 | 0.14 | 0.37 | 0.40 | 0.39 |
Extractive vs. abstractive?
Although not reflected in the automatic evaluation metrics above, the increase in readability of UL models led us to suspect that UL models are more abstractive than extractive, namely, they contain more paraphrases. To determine the degree to which the outputs directly copy content from the technical abstracts, we computed the fraction of n-grams in the output PLS that also occur in the abstract (without considering repetition). These results are shown in Table 7.
Table 7:
% of n-grams in reference/generated PLS that are also in the abstracts.
| N=1 | N=2 | N=3 | N=4 | |
|---|---|---|---|---|
|
| ||||
| PLS | 0.56 | 0.29 | 0.19 | 0.14 |
|
| ||||
| No-UL | 0.95 | 0.89 | 0.84 | 0.79 |
| UL-Cochrane | 0.84 | 0.67 | 0.57 | 0.49 |
| UL-Newsela | 0.92 | 0.81 | 0.73 | 0.66 |
| UL-Both | 0.89 | 0.76 | 0.67 | 0.59 |
We observe that the introduction of UL clearly decreases n-gram overlap, and the difference becomes more marked as n increases. The use of Cochrane weights (those from the logistic regression model trained to discriminate between technical abstracts and PLS) likely reduces n-gram overlap because the tokens most penalized in UL training are those used to represent numerical data, e.g., statistics and confidence intervals. Penalizing these tokens discourages the regurgitation of numerical details from the technical abstract. The use of Newsela weights does not have the same effect, again likely due to the domain difference between the tokens penalized during unlikelihood training and those generated by the model. None of the model settings, however, achieve n-gram overlap scores nearly as low as the reference PLS, indicating that the generated summaries remain considerably more extractive than human-written PLS.
5.4. Manual examination and analysis
We manually examined the outputs generated by our models on a random sample of 40 technical abstracts from the test split of our dataset. While reading these outputs, we made special note of text length, readability and coherence, the presence of hallucinated information not found in the corresponding abstract, and artifacts such as repetition and misspelled words.
Our examination demonstrated that the generated texts were all significantly shorter than their respective abstracts and also shorter than the reference PLS. Furthermore, the models trained with Cochrane weights (‘UL-Cochrane’ and ‘UL-Both’) produced shorter texts on average than the models trained without UL or with Newsela weights. This observation is supported by the results in Table 9, which displays the average number of tokens and sentences in the summaries generated under different training settings.
Table 9:
Lengths of generated PLS.
| # Tokens | # Sentences | |
|---|---|---|
|
| ||
| Abstracts | 492.04 | 14.03 |
| PLS | 254.60 | 9.59 |
|
| ||
| No UL | 228.27 | 8.34 |
| UL-Cochrane | 163.79 | 7.10 |
| UL-Newsela | 201.01 | 8.45 |
| UL-Both | 173.88 | 7.75 |
One explanation for why UL with Cochrane weights produces shorter summaries is that training with these weights discourages the copying of statistics from the original abstract, a phenomenon exemplified in Appendix A, Table 10. Another trend that we noticed was that higher α values produce shorter, more readable summaries at the expense of information completeness. Training with a high α also increases the likelihood of hallucination, misspelling, and repetition. These drawbacks greatly impacted coherence for α ≥ 1000. These observations suggest a tradeoff between completness of information and conciseness as α is varied in the training process.
The most common hallucination found in all settings, and especially with high α, was the inclusion of a statement of the form The evidence is current to [month] [year]. The reason for this is that many PLS contain such a statement of currency not found in the technical abstracts, so models learn to include such a statement even if it cannot be factually deduced from the abstract. Another observation is that most commonly misspelled words are those of medications and diseases. Table 8 provides examples of the various kinds of artifacts found in the generated PLS. The presence of these artifacts suggest that in practice, generated texts should be reviewed before being used.
Table 8:
Example of artifacts found in generated PLS.
| Hallucination: The evidence is up-to-date as of February 2016. We found seven studies, involving 1839 participants, that compared home-based treatment with hospital-based care for venous thromboembolism. |
| Misspelling: The review authors provided no information on other important outcomes, including gastro-oesophageal reflux, aspiration pneumonia, necrotise enterulitis… |
| Repetition: However, we were not able to combine their results because of the small number and small number of people in the included studies. |
6. Conclusions
In this work we considered the important task of medical text simplification. We derived a new resource for this task made up of technical abstracts summarizing medical evidence paired with plain language versions of the same; we have made this data publicly available to facilitate further research.3 We proposed a new masked language model (MLM)-based measure of the technicality of text, which quantifies technicality by calculating the likelihood of tokens in the input text with respect to a transformer-based MLM trained on a technical corpus. We demonstrated that this metric better discriminated technical abstracts from PLS than more traditional notions of readability.
We proposed models for automated simplification based on BART (Lewis et al., 2020), extending the training objective by incorporating an explicit penalty for production of ‘jargon’ terms. We found that this method can improve model outputs (i.e., can increase simplicity and the abstractiveness of summaries) according to the metrics considered.
7. Ethical Considerations
This paper presents a dataset from the Cochrane library; this comprises only the freely available portion of the information on Cochrane (abstracts that are readily available to all). No annotators other than the authors of this paper are involved in the manual inspection of this data. In addition, the Cochrane data in itself, and our collection and inspection of it, does not involve any personally identifiable information.
The baseline models presented involves simplifying medical texts. Inconsistencies (e.g., hallucinations) of the generated PLS with respect to the original review is an artifact discussed in Section 5.4. This can lead to misinformed readers. Therefore, the outputs of the proposed systems should always be manually examined before being used.
Figure 2:
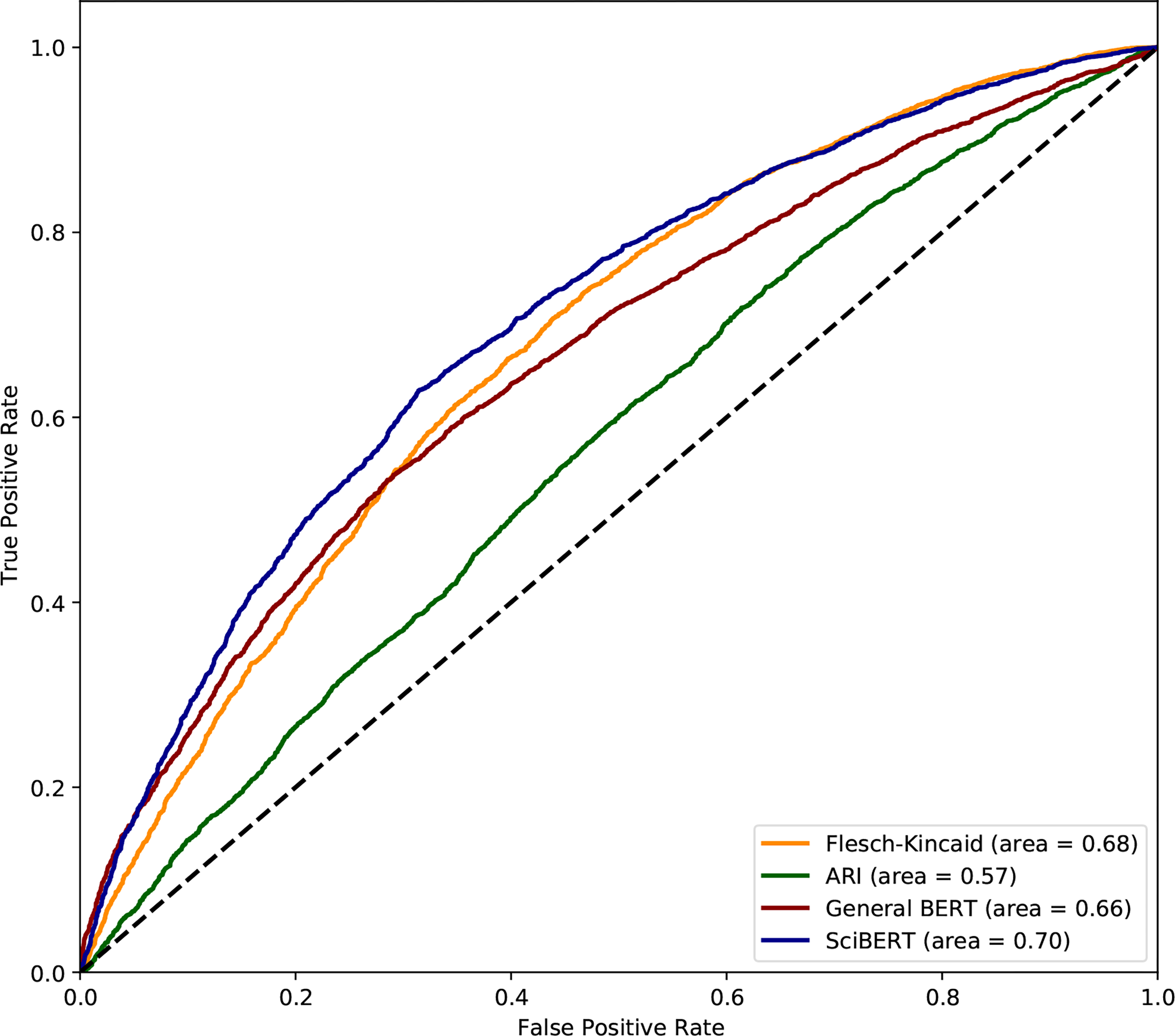
ROC Curves for Readability Metrics.
Acknowledgments
This work was supported in part by the National Institutes of Health (NIH), grant R01-LM012086, and the National Science Foundation (NSF), grant IIS-1850153. We acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing HPC resources that have contributed to the research results reported within this paper.
Appendix A: Example outputs
Table 10:
A full example of technical abstract, reference PLS and model outputs.
| Technical abstract: We included a total of 40 studies in the review, with more than 140,000 women aged between 20 and 70 years old. Many studies were at low risk of bias. There were a sufficient number of included studies with adequate methodology to perform the following test comparisons: hybrid capture 2 (HC2) (1 pg/mL threshold) versus conventional cytology (CC) (atypical squamous cells of undetermined significance (ASCUS)+ and low-grade squamous intraepithelial lesions (LSIL)+ thresholds) or liquid-based cytology (LBC) (ASCUS+ and LSIL+ thresholds), other high-risk HPV tests versus conventional cytology (ASCUS+ and LSIL+ thresholds) or LBC (ASCUS+ and LSIL+ thresholds). For CIN 2+, pooled sensitivity estimates for HC2, CC and LBC (ASCUS+) were 89.9%, 62.5% and 72.9%, respectively, and pooled specificity estimates were 89.9%, 96.6%, and 90.3%, respectively. The results did not differ by age of women (less than or greater than 30 years old), or in studies with verification bias. Accuracy of HC2 was, however, greater in European countries compared to other countries. The results for the sensitivity of the tests were heterogeneous ranging from 52% to 94% for LBC, and 61% to 100% for HC2. Overall, the quality of the evidence for the sensitivity of the tests was moderate, and high for the specificity. The relative sensitivity of HC2 versus CC for CIN 2+ was 1.52 (95% CI: 1.24 to 1.86) and the relative specificity 0.94 (95% CI: 0.92 to 0.96), and versus LBC for CIN 2+ was 1.18 (95% CI: 1.10 to 1.26) and the relative specificity 0.96 (95% CI: 0.95 to 0.97). The relative sensitivity of HC2 versus CC for CIN 3+ was 1.46 (95% CI: 1.12 to 1.91) and the relative specificity 0.95 (95% CI: 0.93 to 0.97). The relative sensitivity of HC2 versus LBC for CIN 3+ was 1.17 (95% CI: 1.07 to 1.28) and the relative specificity 0.96 (95% CI: 0.95 to 0.97). Whilst HPV tests are less likely to miss cases of CIN 2+ and CIN 3+, these tests do lead to more unnecessary referrals. However, a negative HPV test is more reassuring than a negative cytological test, as the cytological test has a greater chance of being falsely negative, which could lead to delays in receiving the appropriate treatment. Evidence from prospective longitudinal studies is needed to establish the relative clinical implications of these tests. |
| Plain-language summary: We searched for all relevant studies up to November 2015. Forty studies compared the HPV test to the Pap test on over 140,000 women between 20 to 70 years old who attended for their routine cervical screening. The studies examined which test can detect precancerous cervical changes which are called cervical intraepithelial neoplasias (CIN 2 and CIN 3). There were enough studies with enough women in them to allow us to draw conclusions. However, some of the results from the studies were different from each other. For example, tests were more accurate in studies in Europe than in Asia or Central or South America. Overall, the quality of the evidence was moderate to high. A perfect test would correctly say if a woman has precancerous changes or if a woman does not. But most tests are not perfect. This review found that for every 1000 women screened, around 20 women will have precancerous changes. The HPV test will correctly identify 18 of these women (but will miss 2 women). The Pap test will identify 15 of the women (but will miss 5 women). The women who are missed could develop cervical cancer. For every 1000 women screened, there will be 980 women who will not have precancerous changes. The HPV test will correctly identify 881 women (but 99 women will be incorrectly told that they have a lesion). The Pap test will correctly identify 885 women (but 95 will be incorrectly told that they have a lesion). Women who are incorrectly told that they have a lesion may have their cervix examined or may receive surgery unnecessarily. |
| No UL: We included 40 studies in the review, with more than 140,000 women aged between 20 and 70 years old. Many studies were at low risk of bias. There were a sufficient number of included studies with adequate methodology to perform the following test comparisons: hybrid capture 2 (HC2) (1 pg/mL threshold) versus conventional cytology (atypical squamous cells of undetermined significance (ASCUS)+ and low-grade squamous intraepithelial lesions (LSIL)+ thresholds) or liquid-based cytology or other high-risk HPV tests versus conventional or liquid cytology thresholds. For CIN 2+, pooled sensitivity estimates for HC2, CC and LBC were 89.9%, 96.6%, and 90.3%, respectively. The results did not differ by age of women (less than or greater than 30 years old), or in studies with verification bias. The accuracy of HC2 was, however, greater in European countries compared to other countries. Overall, the quality of the evidence for the sensitivity of the tests was moderate, and high for the specificity. Whilst HPV tests are less likely to miss cases of CIN2+ and CIN 3+, these tests do lead to more unnecessary referrals. However, a negative HPV test is more reassuring than a negative cytological test, as the cytology test has a greater chance of being falsely negative, which could lead to delays in receiving the appropriate treatment. Evidence from prospective longitudinal studies is needed to establish the relative clinical implications of these tests. |
| UL, Cochrane Weights, Temp=2, α=100: We found 40 studies involving more than 140,000 women aged between 20 and 70 years old. The studies compared different types of HPV tests with each other, or with conventional cytology. The results showed that the tests were more accurate than conventional cytological tests. However, the results did not differ by age of women (less than or greater than 30 years old), or in studies with verification bias. Overall, the quality of the evidence for the sensitivity of the tests was moderate, and the specificity was high for the tests. The accuracy of the test was, however, greater in European countries compared to other countries. Whilst HPV tests are less likely to miss cases, they do lead to more unnecessary referrals, as the cytological test has a greater chance of being falsely negative, which could lead to delays in receiving the appropriate treatment. Evidence from prospective longitudinal studies is needed to establish the relative clinical implications of these tests. |
Footnotes
We also considered starting from a checkpoint corresponding to training over CNN/Daily News but preliminary manual examination of model outputs suggested starting from XSum yielded higher quality outputs.
Five-fold evaluation showed that the model achieved > 90% accuracy. We also experimented with the Simple Wikipedia/Wikipedia dataset (Zhu et al., 2010), but this model was not effective in early experiments.
We emphasize that the data here comprises only text derived from publicly accessible abstracts.
Contributor Information
Ashwin Devaraj, The University of Texas at Austin.
Byron C. Wallace, Northeastern University
Iain J. Marshall, King’s College London
Junyi Jessy Li, The University of Texas at Austin.
References
- Abrahamsson Emil, Forni Timothy, Skeppstedt Maria, and Kvist Maria. 2014. Medical text simplification using synonym replacement: Adapting assessment of word difficulty to a compounding language. In Proceedings of the 3rd Workshop on Predicting and Improving Text Readability for Target Reader Populations (PITR), pages 57–65. [Google Scholar]
- Adduru Viraj, Sadid A Hasan Joey Liu, Ling Yuan, Vivek V Datla Ashequl Qadir, and Farri Oladimeji. 2018. Towards dataset creation and establishing baselines for sentence-level neural clinical paraphrase generation and simplification. In KHD@ IJCAI. [Google Scholar]
- Alva-Manchego Fernando, Scarton Carolina, and Specia Lucia. 2020. Data-driven sentence simplification: Survey and benchmark. Computational Linguistics, 46(1):135–187. [Google Scholar]
- Ammar Waleed, Groeneveld Dirk, Bhagavatula Chandra, Beltagy Iz, Crawford Miles, Downey Doug, Dunkelberger Jason, Elgohary Ahmed, Feldman Sergey, Ha Vu, et al. 2018. Construction of the literature graph in semantic scholar. In Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 3 (Industry Papers), pages 84–91. [Google Scholar]
- Armstrong Paul W and Naylor C David. 2019. Counteracting health misinformation: a role for medical journals? Jama, 321(19):1863–1864. [DOI] [PubMed] [Google Scholar]
- Beltagy Iz, Lo Kyle, and Cohan Arman. 2019. SciBERT: A pretrained language model for scientific text. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), pages 3606–3611. [Google Scholar]
- Chall Jeanne Sternlicht. 1958. Readability: An appraisal of research and application. 34. Ohio State University. [Google Scholar]
- Chalmers Iain, Altman Douglas G, et al. 1995. Systematic reviews. BMJ Publishing London. [Google Scholar]
- Cochrane. 2013. Standards for the reporting of plain language summaries in new cochrane intervention reviews (pleacs). [Google Scholar]
- Coster William and Kauchak David. 2011. Simple English Wikipedia: a new text simplification task. In Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies, pages 665–669. [Google Scholar]
- Damay Jerwin Jan S, Lojico Gerard Jaime D, Lu Kimberly Amanda L, Tarantan D, and Ong E. 2006. SIMTEXT: Text simplification of medical literature. In Proceedings of the 3rd National Natural Language Processing Symposium-Building Language Tools and Resources, pages 34–38. [Google Scholar]
- Devlin Jacob, Chang Ming-Wei, Lee Kenton, and Toutanova Kristina. 2019. Bert: Pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers), pages 4171–4186. [Google Scholar]
- Dong Yue, Li Zichao, Rezagholizadeh Mehdi, and Cheung Jackie Chi Kit. 2019. EditNTS: An neural programmer-interpreter model for sentence simplification through explicit editing. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, pages 3393–3402. [Google Scholar]
- Fox Susannah and Duggan Maeve. 2013. Health online 2013. https://www.pewresearch.org/internet/2013/01/15/health-online-2013/. Online; accessed April 2, 2021.
- Gledhill Chris, Martikainen Hanna, Mestivier Alexandra, and Zimina Maria. 2019. Towards a linguistic definition of ‘simplified medical english’: Applying textometric analysis to cochrane medical abstracts and their plain language versions. LCM - La Collana / The Series, 9788879169196:91–114. [Google Scholar]
- Grabar Natalia and Cardon Rémi. 2018. CLEAR – simple corpus for medical French. In Proceedings of the 1st Workshop on Automatic Text Adaptation (ATA), pages 3–9. [Google Scholar]
- Hermann Karl Moritz, Kocisky Tomas, Grefenstette Edward, Espeholt Lasse, Kay Will, Suleyman Mustafa, and Blunsom Phil. 2015. Teaching machines to read and comprehend. In Cortes C, Lawrence ND, Lee DD, Sugiyama M, and Garnett R, editors, Advances in Neural Information Processing Systems 28, pages 1693–1701. Curran Associates, Inc. [Google Scholar]
- Holtzman Ari, Buys Jan, Du Li, Forbes Maxwell, and Choi Yejin. 2019. The curious case of neural text degeneration. In International Conference on Learning Representations. [Google Scholar]
- Ioannidis John PA, Stuart Michael E, Brownlee Shannon, and Strite Sheri A. 2017. How to survive the medical misinformation mess. European journal of clinical investigation, 47(11):795–802. [DOI] [PubMed] [Google Scholar]
- Jawahar Ganesh, Sagot Benoît, and Seddah Djamé. 2019. What does BERT learn about the structure of language? In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, pages 3651–3657. [Google Scholar]
- Kadic Antonia Jelicic, Fidahic Mahir, Vujcic Milan, Saric Frano, Propadalo Ivana, Marelja Ivana, Dosenovic Svjetlana, and Puljak Livia. 2016. Cochrane plain language summaries are highly heterogeneous with low adherence to the standards. BMC medical research methodology, 16(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandula Sasikiran, Curtis Dorothy, and Zeng-Treitler Qing. 2010. A semantic and syntactic text simplification tool for health content. In AMIA annual symposium proceedings, pages 366–370. [PMC free article] [PubMed] [Google Scholar]
- Karačić Jasna Dondio Pierpaolo, Buljan Ivan, Hren Darko, and Marušić Ana. 2019. Languages for different health information readers: multitrait-multimethod content analysis of cochrane systematic reviews textual summary formats. BMC medical research methodology, 19(1):75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kate Rohit, Luo Xiaoqiang, Patwardhan Siddharth, Franz Martin, Florian Radu, Mooney Raymond, Roukos Salim, and Welty Chris. 2010. Learning to predict readability using diverse linguistic features. In Proceedings of the 23rd International Conference on Computational Linguistics (Coling 2010), pages 546–554. [Google Scholar]
- Kincaid J Peter, Fishburne Robert P Jr, Rogers Richard L, and Chissom Brad S. 1975. Derivation of new readability formulas (automated readability index, fog count and flesch reading ease formula) for navy enlisted personnel. Technical report, Naval Technical Training Command Millington TN Research Branch. [Google Scholar]
- Kingma Diederik and Ba Jimmy. 2015. Adam: A method for stochastic optimization. In International Conference on Learning Representations. [Google Scholar]
- Klare George Roger. 1963. Measurement of readability.
- Kriz Reno, Sedoc João, Apidianaki Marianna, Zheng Carolina, Kumar Gaurav, Miltsakaki Eleni, and Callison-Burch Chris. 2019. Complexity-weighted loss and diverse reranking for sentence simplification. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers), pages 3137–3147. [Google Scholar]
- Lewis Mike, Liu Yinhan, Goyal Naman, Ghazvininejad Marjan, Mohamed Abdelrahman, Levy Omer, Stoyanov Veselin, and Zettlemoyer Luke. 2020. BART: Denoising sequence-to-sequence pretraining for natural language generation, translation, and comprehension. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, pages 7871–7880. [Google Scholar]
- Li Junyi Jessy and Nenkova Ani. 2015. Fast and accurate prediction of sentence specificity. In Proceedings of the AAAI Conference on Artificial Intelligence, pages 2281–2287. [Google Scholar]
- Li Margaret, Roller Stephen, Kulikov Ilia, Welleck Sean, Boureau Y-Lan, Cho Kyunghyun, and Weston Jason. 2020. Don’t say that! making inconsistent dialogue unlikely with unlikelihood training. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, pages 4715–4728. [Google Scholar]
- Lin Chin-Yew. 2004. Rouge: A package for automatic evaluation of summaries. In Text summarization branches out, pages 74–81. [Google Scholar]
- Llanos Leonardo Campillos, Bouamor Dhouha, Zweigenbaum Pierre, and Rosset Sophie. 2016. Managing linguistic and terminological variation in a medical dialogue system. In Proceedings of the Tenth International Conference on Language Resources and Evaluation (LREC’16), pages 3167–3173. [Google Scholar]
- Loshchilov Ilya and Hutter Frank. 2019. Decoupled weight decay regularization. International Conference on Learning Representations. [Google Scholar]
- Mukherjee Partha, Leroy Gondy, Kauchak David, Rajanarayanan Srinidhi, Romero Diaz Damian Y, Yuan Nicole P, Gail Pritchard T, and Colina Sonia. 2017. Negait: A new parser for medical text simplification using morphological, sentential and double negation. Journal of biomedical informatics, 69:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan Shashi, Cohen Shay B, and Lapata Mirella. 2018. Don’t give me the details, just the summary! topic-aware convolutional neural networks for extreme summarization. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, pages 1797–1807. [Google Scholar]
- Papineni Kishore, Roukos Salim, Ward Todd, and Zhu Wei-Jing. 2002. Bleu: a method for automatic evaluation of machine translation. In Proceedings of the 40th annual meeting of the Association for Computational Linguistics, pages 311–318. [Google Scholar]
- Petersen Sarah E and Ostendorf Mari. 2007. Text simplification for language learners: a corpus analysis. In Workshop on Speech and Language Technology in Education. [Google Scholar]
- Senter RJ and Smith Edgar A. 1967. Automated readability index. Technical report, CINCINNATI UNIV OH. [PubMed] [Google Scholar]
- Siddharthan Advaith. 2014. A survey of research on text simplification. ITL-International Journal of Applied Linguistics, 165(2):259–298. [Google Scholar]
- Srikanth Neha and Li Junyi Jessy. 2020. Elaborative simplification: Content addition and explanation generation in text simplification. arXiv preprint arXiv:2010.10035. [Google Scholar]
- Vajjala Sowmya and Lučić Ivana. 2018. Onestopenglish corpus: A new corpus for automatic readability assessment and text simplification. In Proceedings of the thirteenth workshop on innovative use of NLP for building educational applications, pages 297–304. [Google Scholar]
- Van den Bercken Laurens, Sips Robert-Jan, and Lofi Christoph. 2019. Evaluating neural text simplification in the medical domain. In The World Wide Web Conference, pages 3286–3292. [Google Scholar]
- Vaswani Ashish, Shazeer Noam, Parmar Niki, Uszkoreit Jakob, Jones Llion, Gomez Aidan N, Kaiser Ł ukasz, and Polosukhin Illia. 2017. Attention is all you need. In Guyon I, Luxburg UV, Bengio S, Wallach H, Fergus R, Vishwanathan S, and Garnett R, editors, Advances in Neural Information Processing Systems 30, pages 5998–6008. Curran Associates, Inc. [Google Scholar]
- Wang Tong, Chen Ping, Rochford John, and Qiang Jipeng. 2016. Text simplification using neural machine translation. In Thirtieth AAAI Conference on Artificial Intelligence. [Google Scholar]
- Welleck Sean, Kulikov Ilia, Roller Stephen, Dinan Emily, Cho Kyunghyun, and Weston Jason. 2019. Neural text generation with unlikelihood training. In International Conference on Learning Representations. [Google Scholar]
- Wolf Thomas, Debut Lysandre, Sanh Victor, Chaumond Julien, Delangue Clement, Moi Anthony, Cistac Pierric, Rault Tim, Louf Rémi, Funtowicz Morgan, Davison Joe, Shleifer Sam, Patrick von Platen Clara Ma, Jernite Yacine, Plu Julien, Xu Canwen, Teven Le Scao Sylvain Gugger, Drame Mariama, Lhoest Quentin, and Rush Alexander M.. 2019. Huggingface’s transformers: State-of-the-art natural language processing. ArXiv, abs/1910.03771. [Google Scholar]
- Woodsend Kristian and Lapata Mirella. 2011. Learning to simplify sentences with quasi-synchronous grammar and integer programming. In Proceedings of the Conference on Empirical Methods in Natural Language Processing, pages 409–420. [Google Scholar]
- World Health Organization. 2013. Health literacy: the solid facts. World Health Organization. Regional Office for Europe. [Google Scholar]
- Wubben Sander, van den Bosch Antal, and Krahmer Emiel. 2012. Sentence simplification by monolingual machine translation. In Proceedings of the 50th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), pages 1015–1024. [Google Scholar]
- Xu Wei, Callison-Burch Chris, and Napoles Courtney. 2015. Problems in current text simplification research: New data can help. Transactions of the Association for Computational Linguistics, 3:283–297. [Google Scholar]
- Xu Wei, Napoles Courtney, Pavlick Ellie, Chen Quanze, and Callison-Burch Chris. 2016. Optimizing statistical machine translation for text simplification. Transactions of the Association for Computational Linguistics, 4:401–415. [Google Scholar]
- Zhang Xingxing and Lapata Mirella. 2017. Sentence simplification with deep reinforcement learning. In Proceedings of the 2017 Conference on Empirical Methods in Natural Language Processing, pages 584–594. [Google Scholar]
- Zhong Yang, Jiang Chao, Xu Wei, and Li Junyi Jessy. 2020. Discourse level factors for sentence deletion in text simplification. In Proceedings of the AAAI Conference on Artificial Intelligence. [Google Scholar]
- Zhu Yukun, Kiros Ryan, Zemel Rich, Salakhutdinov Ruslan, Urtasun Raquel, Torralba Antonio, and Fidler Sanja. 2015. Aligning books and movies: Towards story-like visual explanations by watching movies and reading books. In Proceedings of the IEEE International Conference on Computer Vision (ICCV). [Google Scholar]
- Zhu Zhemin, Bernhard Delphine, and Gurevych Iryna. 2010. A monolingual tree-based translation model for sentence simplification. In Proceedings of the 23rd International Conference on Computational Linguistics, pages 1353–1361. [Google Scholar]


