Abstract
Cleavage of the Cry2Aa1 protoxin (molecular mass, 63 kDa) from Bacillus thuringiensis by midgut juice of gypsy moth (Lymantria dispar) larvae resulted in two major protein fragments: a 58-kDa fragment which was highly toxic to the insect and a 49-kDa fragment which was not toxic. In the midgut juice, the protoxin was processed into a 58-kDa toxin within 1 min, but after digestion for 1 h, the 58-kDa fragment was further cleaved within domain I, resulting in the protease-resistant 49-kDa fragment. Both the 58-kDa and nontoxic 49-kDa fragments were also found in vivo when 125I-labeled toxin was fed to the insects. N-terminal sequencing revealed that the protease cleavage sites are at the C termini of Tyr49 and Leu144 for the active fragment and the smaller fragment, respectively. To prevent the production of the nontoxic fragment during midgut processing, five mutant proteins were constructed by replacing Leu144 of the toxin with Asp (L144D), Ala (L144A), Gly (L144G), His (L144H), or Val (L144V) by using a pair of complementary mutagenic oligonucleotides in PCR. All of the mutant proteins were highly resistant to the midgut proteases and chymotrypsin. Digestion of the mutant proteins by insect midgut extract and chymotrypsin produced only the active 58-kDa fragment, except that L144H was partially cleaved at residue 144.
During sporulation, Bacillus thuringiensis, a group of gram-positive bacteria, produces crystalline (Cry) proteins which are toxic against a range of insect groups. Recently, over 120 cry genes have been reported and a new nomenclature system classifying Cry proteins based on the amino acid homology of the proteins has been proposed (5). The mechanism of action of most Cry proteins consists of three major steps: solubilization and activation of protoxin in the insect midgut (34), binding of the activated fragment to midgut receptor (2, 13), and insertion of the toxin into the midgut apical membrane, which causes destruction of membrane potential (9, 12).
The molecular sizes of most Cry proteins are around 130 to 140 kDa (14). After digestion by trypsin-like enzymes in the midgut (23, 34), the active fragment is produced. In general, the protease-resistant core protein is in the range of 60 to 70 kDa (1, 14), covering three domains of the active toxin (10, 19). Deletion of the cry genes beyond the active sequences completely abolished the toxic activity of the gene products (33, 35).
Proteolytic processing of Cry protein by midgut proteases is reported to generate active toxins of varying potency and specificity (11). However, digestion of Cry1A by diamondback moth (Plutella xylostella) midgut extract generated a core that lacks α-helix 1 in domain I of Cry1A (27). Similarly, digestion of Cry3A with chymotrypsin caused a nick at the region between α-helix 3 and α-helix 4 of domain I (3). The protease cleavage inside domain I was also found in Cry9Ca1, and more-stable protein was produced after removal of the trypsin-cleaved residue (16).
Cry2Aa1, a 633-amino-acid toxin with molecular mass of 63 kDa, was originally described by Yamamoto as a dipteran- and lepidopteran-active protein (6, 36, 37). Even though the sequence of this Cry protein shows rather limited homology to those of the other Cry proteins, its structure has recently been determined and observed to be similar to those of Cry1Aa and Cry3A, consisting of three distinct domains (24). English et al. (7) reported the distinct binding and ion channels formed by Cry2Aa1 in Helicoverpa zea, indicating a mode of action unique among the Cry proteins.
Here, we report that Cry2Aa1 is rapidly cleaved at a single position in domain I by midgut enzymes of gypsy moth (Lymantria dispar). The cleavage product is not toxic to the insect. Several mutant proteins were constructed by removing the amino acid targeted by the proteases. All of these mutants were able to release the active fragment, which is more resistant to protease digestion than the wild-type toxin.
MATERIALS AND METHODS
All enzymes and reagents were purchased from Boehringer Mannheim Biochemicals (BMB) unless otherwise stated.
Bacterial strains and plasmids.
Escherichia coli BL21 and plasmids pDL103 (20) and pOS4201 (8), which carry the cry2Aa1 and cry1Aa1 genes, respectively, were obtained from our laboratory stocks and from the Bacillus Genetic Stock Center, The Ohio State University.
Gut juice preparation.
Fourth-instar larvae of L. dispar were dissected, and the midguts were recovered. The midguts were centrifuged at 15,000 × g for 30 min, and the supernatant was collected and used as the midgut extract.
Protein purification.
E. coli harboring the cry2Aa1 gene was cultured in Luria-Bertani broth containing 100 μg of ampicillin per ml at 37°C for 72 h. Inclusion bodies were purified from the bacteria as previously described (20). The bacterial cells from a 500-ml culture were resuspended in 100 ml of lysis buffer (15% sucrose, 50 mM EDTA, 50 mM Tris [pH 8.0]) containing 10 μg of lysozyme per ml at 37°C for 2 h, followed by sonication and washing three times in 100 ml of 2% Triton X-100–0.5 M NaCl, five times in 100 ml of 0.5 M NaCl, and three times in distilled water. The crystal protein was solubilized in 100 mM Na3PO4, pH 12, at 37°C for 1 h. The soluble protein samples were then dialyzed in 50 mM Na2CO3, pH 10.5. Protein concentration was determined by the bicinchoninic acid (Pierce) method with a gel densitometer (Kodak Digital Science Electrophoresis Documentation and Analysis System).
Protein digestion.
Midgut extract digestion reaction was performed at room temperature as indicated. The reaction was stopped by the addition of Complete (a protease inhibitor [final concentration, 1×]; BMB), 2 μM E-64 (a papain inhibitor), and 0.4 N NaOH. Sodium dodecyl sulfate (SDS)-loading buffer was added, and the samples were boiled. The protein samples were neutralized with equal molar concentrations of HCl and then separated by SDS-10% polyacrylamide gel electrophoresis (SDS-PAGE) (15). Chymotrypsin digestion was performed by using a mass of the enzyme that was 20% that of the protein. The reaction was stopped by adding Complete (Roche; final concentration, 1×) and boiling the samples in SDS-loading buffer. The molecular masses of the digested products were determined by Kodak Digital Science 1D Image Analysis Software.
Autoradiography and in vivo cleavage of Cry2Aa1.
The protoxin of Cry2Aa1 was iodinated with IODO-BEAD (Pierce) as previously described (18). Twenty-five micrograms of the toxin was iodinated with 1 mCi of [125I]NaI. Each overnight-starved fourth-instar larva of gypsy moth was fed a diet contaminated with 1 μg of 125I-labeled protein per cm2. After feeding for 1 h, the insect was longitudinally dissected. The peritrophic membrane and contents were gently removed from the midgut membrane. The midgut tissue was washed in 50 ml of binding buffer (150 mM NaCl, 8 mM Na2HPO4, 2 mM KH2PO4 [pH 7.4]) for 10 min twice. The peritrophic membrane, with its contents, and the midgut were separately placed on a piece of Whatman filter paper and then vacuum dried before being subjected to autoradiography for 48 h. To determine the midgut-processed fragments in vivo, another group of the insects were separately treated with 125I-labeled toxin as described above. After dissecting, the midgut contents in the peritrophic membrane were centrifuged at 15,000 × g for 30 min. The supernatant was diluted with distilled water (1:10) before Complete, E-64, and NaOH were added as described above. After boiling in SDS-loading buffer, the protein samples were neutralized with HCl and analyzed by gel electrophoresis. The bound toxin on the midgut tissue was extracted by boiling the whole gut tissue in 200 μl of a solution containing 1× Complete, 2 μM E-64, and 1× SDS-loading buffer. The samples were centrifuged at 15,000 × g for 15 min, and the supernatant was analyzed by gel electrophoresis. The gel was vacuum dried and autoradiographed for 48 h.
Site-directed mutagenesis.
Double-primer site-directed mutagenesis was performed by using double-stranded pDL103 as a template DNA. The method was a modification of the QuikChange site-directed mutagenesis method (Stratagene). Two complementary oligonucleotides were used as the mutagenic primers, 5′-CAAAACCCTGTTCCTCACTCAATAACTTCTTCG-3′ to replace Leu144 by His and 5′-CGAAGAAGTTATTGAGNCAGGAACAGGGTTTTG-3′ to replace Leu144 by Ala, Asp, Gly, and Val (the underlined portions indicate the codons that were altered). The mutagenic double-stranded DNA was amplified by PCR (25). Pwo was used as a DNA polymerase enzyme in the reaction. The temperatures and times for annealing, extension, and denaturing were 48°C for 1 min, 68°C for 10 min, and 95°C for 30 s, respectively. The reactions were performed for 15 cycles. PCR products were treated with DpnI for 1 h to digest methylated parental DNA template before transforming E. coli BL21.
N-terminal sequencing of the protein fragments.
The protein samples from the midgut juice digestion reaction were separated on an SDS-10% PAGE gel and then transferred by electrophoresis to an Immobilon-P polyvinylidene difluoride membrane (Bio-Rad). The peptide fragments were analyzed by Edman degradation with an Applied Biosystems model 477A pulsed liquid-sequencer equipped with an on-line high-performance liquid chromatograph for PTH amino acid analysis at the USDA Forest Service facility (Delaware, Ohio).
Bioassays.
Gypsy moth diet (F9631B; Bioserv) was used in all experiments. The protein samples were treated as indicated in Results, and the digestion products were examined by SDS-PAGE just prior to bioassay experiments. The 58-kDa fragment was prepared by digestion of the protoxin in the diluted (1:50 [vol/vol]) gut extract for 10 min, and the reaction was stopped by adding Complete and was stored at 4°C. The 49-kDa fragment was prepared by the same procedure, except that digestion was for 16 h. The proteins were added to the surface of the diet in Multiwell-24 plates. The 50% lethal concentration (LC50) was determined after 5 days of intoxication of the insect neonates. The 50% growth inhibition dose (ID50) was determined by using second-instar larvae. The weight of each larva was measured before transfer to the toxin-contaminated diet and 5 days after intoxication. The growth inhibition effect was considered positive when the larvae failed to gain weight. The data was analyzed by the Probit method (32).
RESULTS
Digestion of Cry proteins by midgut extract.
Proteolysis of Cry proteins after adding loading buffer has been reported by Choma and Kaplan (4). At high concentrations of L. dispar gut extract (greater than 1:10 [vol/vol]), we also observed degradation of the protein after adding protein-loading buffer. The degradation might be from the digestion of SDS-induced denatured toxin proteins by SDS-resistant proteases in the insect midgut juice. Therefore, both protein inhibitors and NaOH were added before mixing the protein samples with protein-loading buffer.
Within 1 min, at a 1:10 (vol/vol) ratio of L. dispar gut extract to protein solution, at room temperature, Cry2Aa1 protoxin was digested to a 58-kDa fragment. After 1 h, the toxin was further digested to a 49-kDa fragment (Fig. 1). The sequences from the amino termini of the Cry2Aa1 protoxin, the 58-kDa fragment, and the 49-kDa fragment are shown in Table 1. According to the protein sequence of Cry2Aa1, the first amino acid of the 58-kDa fragment was Val50 and that of the smaller fragment was Ser145.
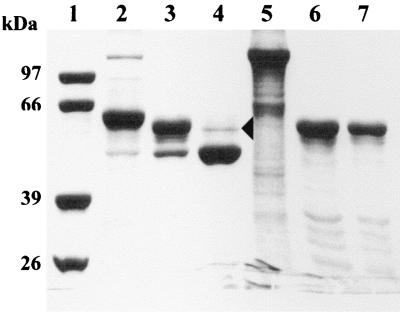
FIG. 1.
SDS–10% polyacrylamide gel electrophoresis of the products of Cry2Aa1 and Cry1Aa1 processed by gypsy moth larval midgut extract at different periods of incubation. The protein solutions were mixed with the gut extract at a ratio of 10:1 (vol/vol) and incubated at room temperature. The reactions were stopped by adding protease inhibitors, as described in Methods and Materials. Lanes: 1, molecular size markers (phosphorylase b, 97 kDa; bovine serum albumin, 66 kDa; aldolase, 39 kDa; triosephosphate isomerase, 26 kDa); 2, Cry2Aa1 protoxin; 3, Cry2Aa1 digested for 1 min; 4, Cry2Aa1 digested for 1 h; 5, Cry1Aa1 protoxin; 6, Cry1Aa1 digested for 1 h; 7, Cry1Aa1 digested for 16 h. The arrowhead indicates a trace amount of the 58-kDa fragment.
TABLE 1.
Toxicity against L. dispar larvae of Cry2Aa1 protoxin and protein fragments from the processing of larval midgut extract
| Protein | N-terminal sequence | LC50a (ng/cm2) | ID50b (ng/cm2) |
|---|---|---|---|
| 63-kDa Cry2Aa1 protoxin | MNNVLNSGRT | 5.6 (2.1–8.7) | 26.9 (17.2–38.2) |
| 58-kDa fragment | VAPVVGTVSSFL | 4.7 (1.3–7.5) | 30.4 (19.1–48.5) |
| 49-kDa fragment | SITSSVNTMQ | NDc | NDc |
LC50s are for insect neonates. Confidence intervals (95%) are given in parentheses.
ID50s are for second-instar larvae. Confidence intervals (95%) are given in parentheses.
ND, not detectable (no death or growth inhibition was observed for toxin concentrations in the range of 10 to 5,000 ng/cm2).
For comparison, the Cry1Aa1 protoxin was processed into a 60-kDa fragment within 1 min. Within this short period of digestion smaller protein fragments were also found in the gel (results not shown). Further digestion for 1 h reduced the presence of the small fragments (Fig. 1, lane 6). The 60-kDa fragment was stable and remains present in the solution after digestion in the midgut extract for 16 h.
Cleavage of Cry2Aa1 in vivo.
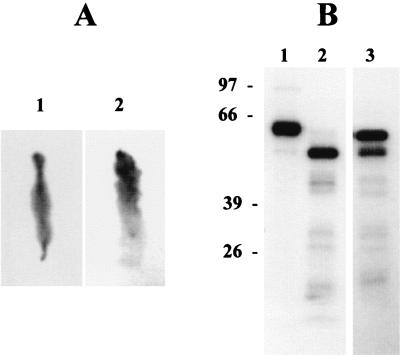
To investigate whether the 58-kDa and 49-kDa fragments were produced in vivo, 125I-labeled Cry2Aa1 was applied to the diet. After the larvae fed on toxin-contaminated diet for 1 h, we found diffusion of the toxin throughout the food tract (Fig. 2A, image 1), and the toxin was also found on the midgut membrane (Fig. 2A, image 2). More bound toxin was detected in the anterior and the middle regions of the midgut. After intoxication for 16 h, the toxin was equally detected on all midgut regions (results not shown). The 49-kDa fragment was found in the midgut fluid (Fig. 2B, lane 2), while the 58-kDa fragment and small amounts of 49-kDa fragment were found on the midgut membrane (Fig. 2B, lane 3).
FIG. 2.
Autoradiography of L. dispar larval midgut and digested Cry2Aa1 after 1 h of feeding on diet contaminated with 125I-labeled Cry2Aa1. (A) The peritrophic membrane with its contents (image 1) and midgut tissue after removal of peritrophic membrane (image 2). Note that the anterior regions of both the peritrophic sac and the midgut tissue were arranged upward. (B) Lanes: 1, 125I-labeled Cry2Aa1 before it was applied to the insect; 2, 125I-labeled Cry2Aa1 extracted from the midgut fluid; 3, 125I-labeled Cry2Aa1 extracted from the midgut membrane of the same insect as in lane 2.
Toxicity of the digested Cry2Aa1 to L. dispar larvae.
The LC50 of Cry2Aa1 protoxin against the insect neonate larvae was 5.6 ng/cm2, and similarly, the LC50 of the 58-kDa fragment was 4.7 ng/cm2 (Table 1). The toxicities of these two proteins were not significantly different. The results were confirmed by measuring the ID50s for second-instar larvae. The ID50s of the protoxin and the 58-kDa fragment were 26.9 and 30.4 ng/cm2, respectively. In contrast to the protoxin and the 58-kDa fragment, the 49-kDa fragment was not toxic to the insect in treatment concentrations of up to 5,000 ng/cm2. This 49-kDa fragment was insoluble and found in the precipitation form (results not shown). It was easily separated from solution by centrifugation. However, we still found a trace amount of 58-kDa protein coprecipitated with this fragment (Fig. 1, lane 4). This contaminating active fragment might be the protein that caused growth inhibition and lethality when a higher concentration of the 49-kDa fragment was applied to the insects.
Mutagenesis.
Site-direct mutagenesis was used to replace Leu144 of Cry2Aa1 with different amino acids. The reverse primer was designed to have His at this position because we expected that the residue in this position might have the same properties as His161 in the loop between α3 and α4 of Cry3A. The “spin” codon designed in the forward primer allowed other small amino acids, Ala, Asp, Gly, and Val, to replace Leu144 so that the stability and toxicity of the new mutant proteins could be studied. With these two oligonucleotides, five mutant proteins, Leu144Asp (L144D), Leu144Ala (L144A), Leu144Gly (L144G), Leu144His (L144H), and Leu144Val (L144V), were produced. All mutations yielded highly expressed protoxins, and these proteins were more stable than the wild-type protoxin (Fig. 3).
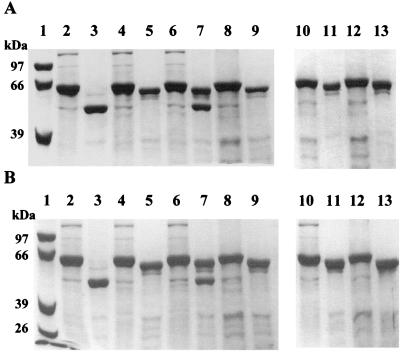
FIG. 3.
Digestion of Cry2Aa1 protoxin and the mutant proteins by gut extract (A) and chymotrypsin (B). Samples (10 μg) of the protein were incubated with midgut extract at a 10:1 ratio (vol/vol) of protein solution to gut extract or with chymotrypsin (20% by mass). Lanes 1, molecular size markers. Even-numbered lanes show protoxins, as follows: lanes 2, Cry2Aa1; lanes 4, L144D; lanes 6, L144H; lanes 8, L144V; lanes 10, L144A; and lanes 12, L144G. Odd-numbered lanes show digestion products of the proteins after 1 h of incubation, as follows: lanes 3, Cry2Aa1 wild-type protein; lanes 5, L144D; lanes 7, L144H; lanes 9, L144V; lanes 11, L144A; and lanes 13, L144G.
Protease resistance of the mutant proteins.
Digestion of the L144D, L144V, L144A, and L144G protoxins with a 10:1 dilution (vol/vol) of protein solution and gut juice produced only a 58-kDa fragment. Stability of these mutants was also found with gut juice digestion for 16 h (results not shown). Digestion of L144H yielded two protein fragments, 58 kDa and 49 kDa, respectively (Fig. 3A, lane 7). The remaining 58-kDa fragment indicates a higher degree of protease resistance in the L144H protein. Digestion of these proteins with chymotrypsin produced the same results as midgut extract digestion (Fig. 3B).
Toxicity of the mutant proteins.
Results from a bioassay of the mutant proteins treated with the midgut extract and chymotrypsin against second-instar larvae of gypsy moth are shown in Table 2. Even though all mutant proteins were more resistant to protease cleavage, compared to the wild-type protein, they did not show higher toxicity. We tested the toxicity of the protoxin and confirmed the data by treating the proteins with either gut extract or chymotrypsin before applying them to the insect diets. The LC50s of all the mutant proteins were the same as that of the wild type, between 2.8 and 8.8 ng/cm2.
TABLE 2.
Toxicity of Cry2Aa1 and the mutant proteins against L. dispar larvaea
| Protein | LC50 (ng/cm2)
|
||
|---|---|---|---|
| No digestion | Digestion with:
|
||
| Gut juice | Chymotrypsin | ||
| Cry2Aa1 | 5.6 (2.1–8.7) | NDb | ND |
| L144A | 3.1 (0.9–5.8) | 5.2 (1.1–9.5) | 5.8 (4.2–7.1) |
| L144D | 6.4 (3.5–9.1) | 4.2 (0.5–7.7) | 6.9 (2.9–10.5) |
| L144G | 4.8 (1.3–8.1) | 8.0 (5.2–11.4) | 7.7 (2.1–12.8) |
| L144H | 8.8 (4.5–12.3) | 18.4 (9.7–25.3) | 19.6 (10.4–29.1) |
| L144V | 6.3 (4.7–7.4) | 2.8 (0.7–5.2) | 7.1 (5.0–9.7) |
The proteins were digested with midgut extract (1:50 [vol/vol]) or chymotrypsin (20% [wt/wt]) for 1 h before the treatment was administered to the insect larvae. Confidence intervals (95%) are given in parentheses.
ND, not detectable (no mortality was observed for protein concentrations in the range of 5 to 5,000 ng/cm2).
DISCUSSION
It is commonly believed that the products from the proteolytic cleavage of the Cry protoxins in the insect larval gut are the active fragments that play the major roles in binding to the receptors on the midgut columnar epithelial cells (13, 18, 30, 31) and irreversible insertion into the cell membrane, which destroys the electric potential on the apical midgut membrane (9, 12). Furthermore, the proteolytic activation of the protoxins was also reported as an important factor for the mechanism of resistance of the insects to B. thuringiensis toxins (28). Since it was observed that the intact Cry3A and chymotrypsin-treated protein exhibited different affinity binding (22), it is important to investigate which portion of the polypeptides from the proteolytic process plays a role in toxicity.
Structural analysis of the trypsinized Cry1Aa and Cry3A revealed three domains in both Cry proteins of which seven helices are the main secondary structure in domain I, and β-sheets and loops are the main secondary structures in both domain II and domain III (10, 19). This led to the hypothesis that all Cry proteins may exhibit three-domain structures (10). The protease-resistant core polypeptide starts at Ile29 of Cry1Aa, Cry1Ab, and Cry1Ac and at Asp58 of Cry3A. Both residues are located just before α1 on domain I. Further removal of codons from the N terminus of the peptide leads to the loss of toxicity of the proteins (33).
The three-dimensional structure of Cry2Aa1 has been determined (24). Compared to the structures of Cry1Aa and Cry3A, there is one more α-helix in domain I of Cry2Aa1, designated α0, based on the nomenclature of Cry3A structure given by Li et al. (19). Tyr49 is on the loop between α0 and α1. Removal of α0 from the mutant protoxin by midgut protease or chymotrypsin generates the protease-resistant core that is comparable to the trypsinized Cry3A and Cry1Aa that starts from α1. Another cleavage site in Cry2Aa1 is Leu144 that is on the loop before α4. The cleavage after Leu144 was expected as an activity of chymotrypsin-like proteases in the insect gut extract. This cleavage was found during digestion of Cry2Aa1 with chymotrypsin (Fig. 3B, lane 3). Unlike Nicolls et al. (26), who reported the N-terminal sequences of these protease-resistant cores using limited proteolysis by chymotrypsin, we prepared the protein fragments by using insect midgut extract. The obtained sequences were the same for both digestions, indicating the prevalence of the enzyme in the insect midgut.
Carroll et al. (3) reported that cabbage white butterfly (Pieris brassicae) gut extract and chymotrypsin cleaved Cry3A at the N termini of Arg158 and His161, respectively. These residues are aligned around the loop region between α3 and α4. However, because the small component is still associated with the major component, the nicked toxin is soluble and as active as the protoxin. Different results were found in experiments with Cry9Ca1 (16). Cry9Ca1 contains a protease cleavage site at Arg164, but trypsinization of this protein generated a nontoxic 55-kDa fragment without the small additional N-terminal fragment. In contrast to Cry3A and Cry9Ca1, Cry2Aa1 cleaved by gut protease or by chymotrypsin at the carboxyl terminus of Leu144 tended to form precipitation. We digested Cry2Aa1 in different buffers with pH values ranging from 7 to 12, but the precipitation was still found. The insoluble form might be an aggregation of Cry2Aa1 pieces created when domain I loses helices α0 to α3. SDS-PAGE analysis of the precipitate showed that almost all of it was the nontoxic 49-kDa fragment, contaminated by a trace amount of the 58-kDa fragment (Fig. 1, lane 4).
More evidence that the 58-kDa peptide is the active fragment against L. dispar is the absence of the 49-kDa fragment from proteolytic processing of our mutant proteins. Leu144 was replaced so that the cleavage site was eliminated. These proteins were highly resistant to both chymotrypsin and gut juice digestion (Fig. 3). They were stable even in digestion by midgut juice for 16 h (results not shown). Initially, we expected that these more resistant proteins might show higher toxicity against L. dispar. But the results from Table 2 indicate that their toxicities were not significantly different from that of the wild-type protein.
Taking together the change in protease resistance of the mutants compared to that of the wild-type toxin and a degree of toxicity similar among all of these protoxins, we propose that the process of solubilization, proteolytic processing, and binding (or insertion) of Cry2Aa1 must happen more rapidly in vivo than in vitro. We further propose that the cleavage to produce the 58-kDa toxin in vivo occurs more rapidly than that which produces the 49-kDa protein. The evidence for this is obvious as more amounts of the 58-kDa fragment were found on the midgut membrane (Fig. 2B, lane 3), indicating that this fragment is bound to the midgut membrane and protected by the membrane components before being cleaved at Leu144. Another possible mechanism is that binding of the 55-kDa fragment to the receptor renders a conformational change in such a way that the second cleavage site, Leu144, is away from the active site of the proteases.
The rapid toxicity of Cry2Aa1 against H. zea larvae was previously observed by English et al. (7), who reported that Cry2Aa1-fed larvae of this insect stopped feeding about 1 min after intoxication and showed morbidity within 4 min. Experiments performed with the other Cry proteins also showed that the apical cell membrane electrical potential response was detected within a few minutes after adding the toxins (21, 29). The results from our experiments also imply that the toxin binds and inserts very rapidly in the insect midgut membrane before proteolytic cleavage by chymotrypsin-like protease of the midgut enzymes.
In vivo and in vitro cleavage at Leu144 of Cry2Aa1 might produce different forms of the processed fragments. Digestion at Leu144 in the midgut environment, which is more viscous and contains many more biochemical carriers, might only create a nick on the protein molecule, leaving the active fragment. The nick fragment of wild-type Cry2Aa1 is as toxic as the 58-kDa fragment produced by the mutants. This might explain why both mutant proteins and wild-type Cry2Aa1 showed the same degree of toxicity. On the other hand, nicking at Leu144 during in vitro digestion might cause dissociation of α1 to α3 helices from the rest of the protein molecule.
Making chymotrypsin-resistant Cry proteins by site-directed mutagenesis is an alternative method that can facilitate the production of the active fragment that is used in an investigation of the mode of action of these toxins. In addition, this strategy can be used in production of more stable proteins. Protease cleavage generating an inactive protein is one of those limitations. Furthermore, we expect this technique to be used to stabilize other labile Cry proteins like Cry20Aa1, which loses its mosquitocidal activity due to degradation (17).
ACKNOWLEDGMENTS
We thank Gary Bernon (Animal and Plant Health Inspection Service [APHIS], USDA, OTIS ANGB, MA) for kindly supplying L. dispar eggs.
This work was supported by a National Institutes of Health grant to D.H.D. (RØ1AI29Ø92).
REFERENCES
- 1.Aronson A I, Beckman W, Dunn P. Bacillus thuringiensis and related insect pathogens. Microbiol Rev. 1986;50:1–24. doi: 10.1128/mr.50.1.1-24.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bravo A, Hendrickx K, Jensens S, Peferoen M. Immunocytochemical analysis of specific binding of Bacillus thuringiensis insecticidal crystal proteins to lepidopteran and coleopteran midgut membranes. J Invertebr Pathol. 1992;60:247–253. [Google Scholar]
- 3.Carroll J, Convents D, Van Damme J, Boets A, Van Rie J, Ellar D J. Intramolecular proteolytic cleavage of Bacillus thuringiensis Cry3A δ-endotoxin may facilitate its coleopteran toxicity. J Invertebr Pathol. 1997;70:41–49. doi: 10.1006/jipa.1997.4656. [DOI] [PubMed] [Google Scholar]
- 4.Choma C T, Kaplan H. Folding and unfolding of the protoxin from Bacillus thuringiensis: evidence that the toxic moiety is present in an active conformation. Biochemistry. 1990;29:10971–10977. doi: 10.1021/bi00501a015. [DOI] [PubMed] [Google Scholar]
- 5.Crickmore N, Zeigler D R, Feitelson J, Schnepf E, Van Rie J, Lereclus D, Baum J, Dean D H. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:807–813. doi: 10.1128/mmbr.62.3.807-813.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donovan W P, Dankocsik C C, Gilbert M P, Gawron-Burke W C, Groat R R, Carton B C. Amino acid sequence and entomocidal activity of the P2 crystal protein. An insect toxin from Bacillus thuringiensis var. kurstaki. J Biol Chem. 1988;263:561–567. [PubMed] [Google Scholar]
- 7.English L, Robbins H L, Von Tersch M A, Kulesza C A, Ave D, Coyle D, Jany C S, Slatin S L. Mode of action of CryIIA: a Bacillus thuringiensis delta-endotoxin. Insect Biochem Mol Biol. 1994;24:1025–1035. [Google Scholar]
- 8.Ge A Z, Shivarova N I, Dean D H. Location of the Bombyx mori specificity domain on a Bacillus thuringiensis δ-endotoxin protein. Proc Natl Acad Sci USA. 1989;86:4037–4041. doi: 10.1073/pnas.86.11.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giordana B, Tasca M, Villa M, Chiantore C, Hanozet G M, Parenti P. Bacillus thuringiensis subsp. aizawai δ-endotoxin inhibits the K+/amino acid cotransporters of lepidopteran larval midgut. Comp Biochem Physiol C. 1993;106:403–407. [Google Scholar]
- 10.Grochulski P, Masson L, Borisova S, Pusztai-Carey M, Schwartz J-L, Brousseau R, Cygler M. Bacillus thuringiensis CryIA(a) insecticidal toxin: crystal structure and channel formation. J Mol Biol. 1995;254:447–464. doi: 10.1006/jmbi.1995.0630. [DOI] [PubMed] [Google Scholar]
- 11.Haider M Z, Knowles B H, Ellar D J. Specificity of Bacillus thuringiensis var. colmeri insecticidal δ-endotoxin determined by differential proteolytic processing of the protoxin by larval gut proteases. Eur J Biochem. 1986;156:531–540. doi: 10.1111/j.1432-1033.1986.tb09612.x. [DOI] [PubMed] [Google Scholar]
- 12.Harvey W R, Wolfersberger M G. Mechanism of inhibition of active potassium transport in isolated midgut of Manduca sexta by Bacillus thuringiensis endotoxin. J Exp Biol. 1979;83:293–304. doi: 10.1242/jeb.83.1.293. [DOI] [PubMed] [Google Scholar]
- 13.Hofmann C, Vanderberggen H, Höfte H, Van Rie J, Jansens S, Van Mellaert H. Specificity of Bacillus thuringiensis δ-endotoxin is correlated with the presence of high-affinity binding sites in the brush border membrane of target insect midguts. Proc Natl Acad Sci USA. 1988;85:7844–7848. doi: 10.1073/pnas.85.21.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Höfte H, Whiteley H R. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol Rev. 1989;53:242–255. doi: 10.1128/mr.53.2.242-255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Lambert B, Buysse L, Decock C, Jansens S, Piens C, Saey B, Seurinck J, Van Audenhove K, Van Rie J, Van Vliet A, Peferoen M. A Bacillus thuringiensis insecticidal crystal protein with a high activity against members of the family Noctuidae. Appl Environ Microbiol. 1996;62:80–86. doi: 10.1128/aem.62.1.80-86.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee H-K, Gill S S. Molecular cloning and characterization of a novel mosquitocidal protein gene from Bacillus thuringiensis subsp. fukuokaensis. Appl Environ Microbiol. 1997;63:4664–4670. doi: 10.1128/aem.63.12.4664-4670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee M K, Milne R E, Ge A Z, Dean D H. Location of Bombyx mori receptor binding on a Bacillus thuringiensis δ-endotoxin. J Biol Chem. 1992;267:3115–3121. [PubMed] [Google Scholar]
- 19.Li J, Carroll J, Ellar D J. Crystal structures of insecticidal δ-endotoxin from Bacillus thuringiensis at 2.5 Å resolution. Nature. 1991;353:815–821. doi: 10.1038/353815a0. [DOI] [PubMed] [Google Scholar]
- 20.Liang Y, Dean D H. Location of a lepidopteran specificity region in insecticidal crystal protein CryIIA from Bacillus thuringiensis. Mol Microbiol. 1994;13:569–575. doi: 10.1111/j.1365-2958.1994.tb00451.x. [DOI] [PubMed] [Google Scholar]
- 21.Liebig B, Stetson D L, Dean D H. Quantification of the effect of Bacillus thuringiensis toxins on short-circuit current in the midgut of Bombyx mori. J Insect Physiol. 1995;41:17–22. [Google Scholar]
- 22.Martínez-Ramírez A C, Real M D. Proteolytic processing of Bacillus thuringiensis CryIIIA toxin and specific binding to brush-border membrane vesicles of Leptinotarsa decemlineata (Colorado potato beetle) Pestic Biochem Physiol. 1996;54:115–122. [Google Scholar]
- 23.Milne R, Kaplan H. Purification and characterization of trypsin-like digestive enzyme from spruce budworm Choristoneura fumiferana responsible for the activation of delta endotoxin from Bacillus thuringiensis. Insect Biochem Mol Biol. 1993;23:663–673. doi: 10.1016/0965-1748(93)90040-y. [DOI] [PubMed] [Google Scholar]
- 24.Morse, R. J., G. Powell, V. Ramalingam, M. Audtho, D. H. Dean, T. Yamamoto, and R. M. Stroud. Unexpected binding epitope and specificity determinants of insecticidal δ-endotoxins revealed by structure of the Cry2Aa protoxin. Submitted for publication.
- 25.Mullis K B, Faloona F A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 26.Nicolls C N, Ahmad W, Ellar D J. Evidence for two different types of insecticidal P2 toxins with dual specificity in Bacillus thuringiensis subspecies. J Bacteriol. 1989;171:5141–5147. doi: 10.1128/jb.171.9.5141-5147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogiwara K, Indrasith L S, Asano S, Hori H. Processing of δ-endotoxin from Bacillus thuringiensis subsp. kurstaki HD-1 and HD-73 by gut juices of various insect larvae. J Invertebr Pathol. 1992;60:121–126. doi: 10.1016/0022-2011(92)90084-h. [DOI] [PubMed] [Google Scholar]
- 28.Oppert B, Kramer K J, Beeman R W, Johnson D, McGaughey W H. Proteinase-mediated insect resistance to Bacillus thuringiensis toxins. J Biol Biochem. 1997;272:23473–23476. doi: 10.1074/jbc.272.38.23473. [DOI] [PubMed] [Google Scholar]
- 29.Peyronnet O, Vachon V, Brousseau R, Baines D, Schwartz J-L, Laprade R. Effect of Bacillus thuringiensis toxins on the membrane potential of lepidopteran insect midgut cells. Appl Environ Microbiol. 1997;63:1679–1684. doi: 10.1128/aem.63.5.1679-1684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajamohan F, Alzate O, Cotrill J A, Curtiss A, Dean D H. Protein engineering of Bacillus thuringiensis δ-endotoxin: mutations at domain II of CryIAb enhance receptor affinity and toxicity toward gypsy moth larvae. Proc Natl Acad Sci USA. 1996;93:14338–14343. doi: 10.1073/pnas.93.25.14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rajamohan F, Hussain S R A, Cotrill J A, Gould F, Dean D H. Mutations at domain II, loop 3, of Bacillus thuringiensis CryIAa and CryIAb delta-endotoxins suggest loop 3 is involved in initial binding to lepidopteran midguts. J Biol Chem. 1996;271:25220–25226. doi: 10.1074/jbc.271.41.25220. [DOI] [PubMed] [Google Scholar]
- 32.Raymond M. Présentation d’un programme d’analyse log probit pour micro-ordinateur. Cah ORSTOM Ser Entomol Med Parasitol. 1985;22:117–121. [Google Scholar]
- 33.Schnepf H E, Whiteley H R. Delineation of a toxin-encoding segment of a Bacillus thuringiensis crystal protein gene. J Biol Chem. 1985;260:6273–6280. [PubMed] [Google Scholar]
- 34.Tojo A, Aizawa K. Dissolution and degradation of Bacillus thuringiensis δ-endotoxin by gut juice protease of the silkworm Bombyx mori. Appl Environ Microbiol. 1983;45:576–580. doi: 10.1128/aem.45.2.576-580.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wabiko H, Raymond K C, Bulla L A., Jr Bacillus thuringiensis entomocidal protoxin gene sequence and gene product analysis. DNA. 1986;5:305–314. doi: 10.1089/dna.1986.5.305. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto T, McLaughlin R E. Isolation of a protein from the parasporal crystal of Bacillus thuringiensis var. kurstaki toxic to mosquito larva, Aedes taeniorhynchus. Biochem Biophys Res Commun. 1981;103:414–421. doi: 10.1016/0006-291x(81)90468-x. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto T. Identification of entomocidal toxins of Bacillus thuringiensis by high performance liquid chromatography. J Gen Microbiol. 1983;129:2595–2603. [Google Scholar]