Abstract
Background
Circular RNAs (circRNAs) are involved in the tumorigenesis and progression of lung adenocarcinoma (LUAD). This study aimed to determine the role of circ_0072088 in LUAD.
Methods
The existence and expression of circ_0072088 in human LUAD tissues and cell lines were determined through Sanger sequencing, quantitative reverse transcription‐polymerase chain reaction, and fluorescence in situ hybridization (FISH). Subsequently, the biological role of circ_0072088 was examined using loss‐of‐function assays in H1299 cells. Moreover, circ_0072088/miR‐1261/PIK3CA pathway‐mediated biological effects in H1299 were verified using bioinformatic prediction and experiments, including interaction analysis (FISH, luciferase reporter, and RNA‐pulldown assays), and tumor biological function test (CCK8 and colony formation, wound healing, and transwell assays). Finally, miR‐1261 and PIK3CA expression and LUAD patient survival were further analyzed using FISH, immunohistochemical staining, and the Kaplan–Meier plotter database, respectively.
Results
First, an increase in circ_0072088 was confirmed in human LUAD tissues. Thereafter, it was mainly localized in the cytoplasm and was found to enhance cell proliferation, migration, and invasion of H1299 cells. Mechanistically, circ_0072088 directly downregulated miR‐1261 expression, whereas increased PIK3CA gene expression was associated with poor overall survival of LUAD patients. The activation of the circ_0072088/miR‐1261/PIK3CA regulatory pathway may play a significant role in the tumorigenesis and progression of LUAD.
Conclusions
Circ_0072088‐dependent regulation of miR‐1261/PIK3CA is important for cell proliferation, migration, and invasion during the tumorigenesis and progression of LUAD, warranting the need to consider the circ_0072088/miR‐1261/PIK3CA regulatory pathway as a potential therapeutic target in patients with lung adenocarcinoma.
Keywords: bioinformatics, cell lines, circRNAs, lung adenocarcinoma, tumorigenesis
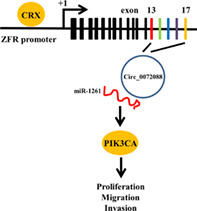
Schematic diagram of circ_0072088/miR‐1261/PIK3CA pathway accelerates lung adenocarcinoma progression.
INTRODUCTION
Lung cancer is the second most commonly diagnosed cancer and the leading cause of cancer death in 2020, with an estimated 2.2 million new cancer cases and 1.8 million deaths. 1 This disease is mainly divided into small cell lung cancer and non‐small cell lung cancer. Accordingly, lung adenocarcinoma (LUAD), the incidence of which has been increasing in recent years, accounts for ~50% of NSCLC cases. 2 Over the past decade, the 5‐year survival rate of LUAD patients has increased owing to the development of targeted therapies and immunotherapy. 3 , 4 Far from ideal, however, a considerable number of patients are still diagnosed with advanced disease or relapse and even metastasis soon after treatment, leading to an increasing demand for improved treatment modalities. Therefore, further exploration of the underlying mechanisms and key molecules multifariously involved with the major signaling pathways in tumor progression is imperative.
Circular RNAs (circRNAs), which have a closed ring structure and are hardly degraded by RNase, have attracted considerable attention in recent years. 5 , 6 Studies have shown that circRNAs, which mainly function as molecular sponge by absorbing microRNAs and affecting gene expression, are involved in tumorigenesis and deterioration. 7 , 8 , 9 Recently, new evidence has shown that circRNAs participate in the initiation and development of LUAD. 10 , 11 , 12 Through bioinformatic analysis and literature reviews, 13 , 14 , 15 the consistent upregulation of circ 0072088 expression in LUAD has persuaded us to further examine its role. Paradoxically, some reports have shown that the downregulation of circ 0072088 in hepatic carcinoma cells derived exosome is associated with tumorigenesis, 16 similarly, its decrease has been reported to be associated with metastasis of colorectal cancer, 17 suggesting that, it is necessary to further explore the expression and function of circ 0072088 in the mechanisms underlying LUAD.
Numerous studies have indicated that microRNAs (miRNAs) affect the regulation of various cancers, including lung cancer and even LUAD, by binding to mRNAs. The downregulation of a novel miR‐1261 has been found to be involved in the progression of hepatocellular carcinoma. 18 However, the impact of miR‐1261 on the initiation and deterioration of LUAD remains unclear. Phosphatidylinositol‐4,5‐bisphosphate 3‐kinase catalytic subunit alpha (PIK3CA), a well‐known oncogene, has been implicated in various cancers, such as head and neck squamous cell carcinoma, 19 breast, 20 , 21 and stomach cancers, 22 playing a crucial role in cell proliferation, migration, and invasion; however, it remains unknown whether PIK3CA participates in the invasion and metastasis of LUAD. Consequently, we hypothesized the existence of a circ_0072088‐miR‐1261‐PIK3CA signaling pathway and whether it could affect the biological functions of LUAD. The present study mainly examined the expression and location of circ_0072088 in LUAD tissues and cells, as well as its biological effects in LUAD. Our results indicated that upregulation of circ_0072088 in LUAD promoted cell proliferation, migration, and invasion by upregulating PIK3CA via functioning as a competitive endogenous RNA (ceRNA) for miR‐1261.
METHODS
Human tissue harvest
The tumor and adjacent nontumor biopsy specimens were obtained from 20 patients with LUAD in the Fourth Hospital of Hebei Medical University between August 2018 and January 2020. All patients were well informed. One half of each LUAD and adjacent nontumor tissue specimen was fixed overnight in 4% paraformaldehyde and embed in paraffin, the other half was snap‐frozen in liquid nitrogen, stored at −80°C, and subsequently used for RNA extraction. This study was approved by the Institutional Ethical Committee of Hebei Medical University (Shijiazhuang, China).
Cell culture
The human lung cancer cell lines H1299, H1975, H520, H827, and 1650 were obtained from the Chinese Academy of Medical Sciences (Beijing, China). The cells were cultured in RPMI‐1640 media (Thermo Fisher) containing 10% fetal bovine serum (FBS; Gibco), 1% penicillin and streptomycin at 37°C in a humidified atmosphere of 5% CO2.
Fuorescence in situ hybridization (FISH)
The FAM probes for circ_0072088, miR‐1261 and U6 were synthesized and produced by RiboBio (Invitrogen). The specific probe was designed by using the junction of circRNA, the specific position 22‐34 nt direct position, and its specificity was confirmed through bioinformatics, and the mutation site control was added as a negative control. U6 were used as positive controls. Nuclei were stained with DAPI (Invitrogen). An RNA FISH Kit (Genepharma) was used to detect the expression and localization of circ_0072088 and miR‐1261 in LUAD tissues and cells. The fluorescence images were captured using a confocal microscope (Leica).
Immunohistochemical staining
Immunohistochemical staining for PIK3CA expression in the LUAD and adjacent nontumor tissue was performed by the SP method as previously described. 23 Primary antihuman polyclonal antibody PIK3CA was obstained from Proteintech group (67071‐1‐Ig). The percentage of brown particles in five random medium‐film fields was quantified by IPP 6.0 image analysis software.
Cell transfection
The human small interfering RNAs targeting circ_0072088 (si‐circ_0072088) and siRNA negative control (si‐NC), miRNA‐1261 mimics (miR‐1261 mimic) and miRNA‐NC (NC), as well as knockdown of miR‐1261 by transfection of miR‐1261 ASO were designed and provided by Genepharma. Cultured H1261 cells were transfected with miRNAs or siRNAs by Lipofectamine 3000 (Invitrogen) following the manufacturer's protocol.
Cell counting kit‐8 (CCK‐8)
Following appropriate treatment, the viability of H1299 cells cultured in 96‐well plates was measured using the CCK‐8, as previously described. 23 The cells were incubated for different time (0, 24, 48, 72, 96 h) and then the viability of cells was evaluated by measuring the absorbance at 450 nm.
Colony formation
The treated H1299 cells (about 200 cells per well) were seeded into six‐well plates. After 2 weeks incubation, hematoxylin staining was performed to check the colonies (<50 cells excluded in colonies).
Wound healing assay
The treated H1299 cells (5 × 105 cells) were seeded into six‐well plates until they reached about 90% confluence, and a wound scratch was performed using a 200 μl pipette tip. The detached cells were removed using PBS and were then photographed at 0 and 24 h (n = 3 for each experiment). The migratory abilities were quantified by measuring the percentage of cells in the scratched regions.
Transwell assay
Cell migration and invasion ability was examined using 24‐wells, with or without matrigel‐coated chambers (BD Biosciences), as previously described. 24 In brief, the treated H1299 cell suspension (1 × 105 cells / ml) in 0.5 ml medium containing 10% FBS was added to the upper chamber, and 600 μl of medium (no cells) was injected into the lower chamber. After 24 h incubation, nonmigratory cells on the upper membrane surface were removed and the invaded cells on the lower membrane surface were fixed with 4% paraformaldehyde and stained by 0.1% crystal violet. The migrated cells were enumerated using a light microscope (×400) in five random fields in each filter.
Reverse transcription‐quantitative polymerase chain reaction (qRT‐PCR)
Total RNA was extracted from treated cells and LUAD tissues as well as control using the TRizol reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Total RNA (1 μg) was subjected to reverse transcription and qPCR using cDNA synthesis and PCR kits according to the manufacturer's protocol (Takara Biotechnology). Primers used for PCR in circ_0072088 were: 5′‐ACGCATTCTTCGAGACCTCT‐3′(sense) and 5′‐TGCCTGTAACTCCTCTTCAGT‐3′(antisense). U6: 5′‐GCTTCGGCAGCACATATACTAAAAT‐3′(sense) and 5′‐ CGCTTCACGAATTTGCGTGTCAT‐3′ (antisense). PCR amplification procedure: 94°C, 5 min; (94°C, 30 s; 55°C, 30 s; 72°C, 1 min) ×30; 72°C, 5 min. Relative amount of transcripts were normalized with U6 or GAPDH and calculated using the 2−ΔΔCt formula. All PCRs were performed in triplicate.
RNA binding protein immunoprecipitation assay
RNA binding protein immunoprecipitation (RIP) assay was employed for the detection of enrichment of circ_0072088 decoyed by miR‐1261 according to the manufacturer's protocol (Millipore) and our previous report. Briefly, incubation with anti‐Ago2 antibody (ab32381, 1:50, Abcam) or anti‐IgG (ab109489, 1:100, Abcam Inc.) was performed and coupled with magnetic beads in transfected cell lysate overnight at 4°C, followed by qRT‐PCR detection for coprecipitated RNA after proteinase K treatment.
Luciferase report assay
The fragments of circ_0072088 and PIK3CA containing miR‐1261 binding sites or the corresponding mutated binding sites were synthesized by Shanghai Sangon Biotech. These fragments were subcloned into the luciferase plasmid pGL‐3 (Takara Biotechnology). H1299 cells were seeded into 24‐well plates (1 × 105 cells / well) and incubated at 37 C overnight. Cells were transfected with wild‐type circ_0072088 (WT circ_0072088), mutant circ_0072088 (MUT circ_0072088), WT PIK3CA or MUT PIK3CA constructs in the presence of miR‐1261 mimics or NC by cotransfection with Lipofectamine 3000. Relative luciferase activity was measured using the dual‐luciferase reporter assay system (Promega). Renilla luciferase activity was normalized to the activity of Firefly luciferase.
Statistical analysis
All data are presented as the mean ± standard deviation. SPSS 23.0 software (SPSS, Inc.) was used for statistical analyses including Student's t‐test and ANOVA post‐hoc tests (Bonferroni) for cell viability, cell number, band density, gene expression. *p < 0.05 was considered to indicate a statistically significant difference.
RESULTS
Circ_0072088 is overexpressed in LUAD tissues
According to previous studies, 13 , 14 , 15 the production of circ_0072088 was presented in FIGURE 1a, containing five exons from exon 13 to exon 17 (693 nt) from the ZFR gene flanked by long introns on either side. Subsequently, the distinct products of the expected sizes were amplified using outward‐facing primers in LUAD tissues and were identified by sequencing. Moreover, we confirmed circ_0072088 upregulation in LUAD tissues relative to its matched paracancerous tissues (n = 20) through FISH probe for circ_0072088 detection (Figure FIGURE 1b). Furthermore, qRT‐PCR results showed differential expression of the circ_0072088 in five human lung cancer cell lines, with the highest expression observed in the H1299 cell line (Figure FIGURE 1c). Given that H1299 cells had the highest level of circ_0072088, it was selected for subsequent study. Meanwhile, cytoplasm localization of circ_0072088 in H1299 cells was further verified by FISH staining (Figure FIGURE 1d). These results indicated the upregulation and cytoplasm localization of circ_0072088 in LUAD cells.
FIGURE 1.
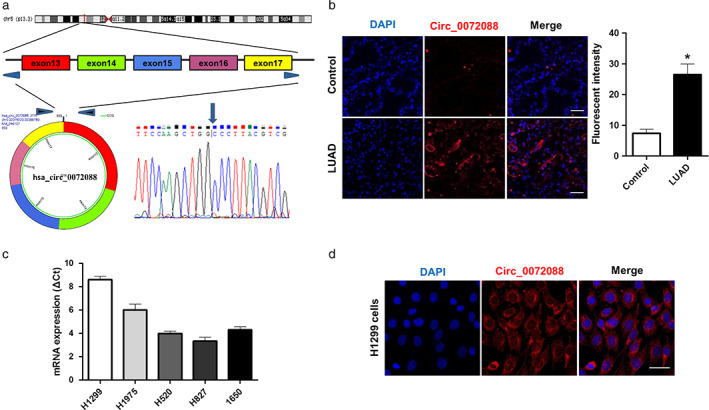
Cytoplasmic localization and up‐regulation of hsa_Circ_0072088 was identified in lung adenocarcinoma cells and tissues. (a) Schematic diagram of circ_0072088 formation and Sanger sequencing for identifying circ_0072088. (b) Representative photographs of RNA fluorescence in site hybridization (FISH) staining for Circ_0072088 expression (red) in lung adenocarcinoma (LUAD) tumors and their adjacent normal tissues (n = 20). Nuclei were stained with DAPI (blue). Scale bars = 50 μm. Right: statistical analysis of fluorescence intensity. *p < 0.05. (c) Distributions of the circ_0072088 expression levels were assessed in five various lung cancer cell lines (H1299, H1975, H520, H872, 1650). (d) The cytoplasmic localization of circ_0072088 in H1299 cells was determined by RNA FISH
Effect of Circ_0072088 in biological behaviors of H1299 cells
To subsequently illustrate the biological function of circ_0072088 in LUAD cells, the expression of circ_0072088 was knocked down by transfecting H1299 cells with human circ_0072088‐specific siRNA, after which they were compared with those transfected with nonspecific siRNA (Figure 2a). CCK‐8 assays showed that the viability of H1299 cells transfected with si‐circ0072088 was significantly lower than those transfected with si‐NC (Figure 2b). The clone formation assays also indicated similar results, which showed that the si‐circ_0072088 group had significantly less numbers of cells compared to the si‐NC group (Figure 2c). Furthermore, we performed scratch and transwell assays to determine the role of circ_0072088 in LUAD metastasis. As shown in Figure 2d, the migration of the cells was significantly inhibited after circ_0072088 knockdown. Meanwhile, transwell migration and matrigel invasion assays of the transfected cells indicated significant suppression of migration and invasion of H1299 cells in the si‐circ_0072088 group (Figure 2e). Collectively, these results suggested that upregulated circ_0072088 contributed to proliferation, migration, and invasion of H1299 cells.
FIGURE 2.
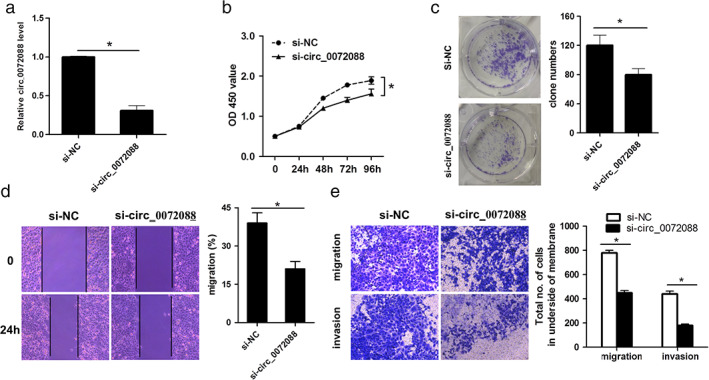
Effect of Circ_0072088 knockdown on proliferation, migration, and invasion of H1299 cells. (a) After transfection with si‐circ_0072088 and negative control (si‐NC) for 36 h in H1299 cells, the efficiency of si‐circ_0072088 knockdown was confirmed by qRT‐PCR detection. (b) CCK‐8 assay in H1299 cells with or without circ_0072088 knockdown by transfection with si‐circ_0072088 or non‐special short interfering siRNA as a control (si‐NC) for different time. (c) Representative photomicrographs of colony formation assays using H1299 cells treated as described above. Right: statistical analysis of cell number. (d) Representative photomicrographs of a scratch assay performed with transfected cells after scratching and 24 h later. Right: Quantification of cells migrating into the scratch gap. (e) Representative photomicrographs of transwell migration and matrigel invasion assay in transfected cells. Right: statistical analysis of cells migrated to the lower side of the membrane. All data represent mean ± SD (n = 3). *p < 0.05 versus si‐NC
Circ_0072088 acts as a sponge of miR‐1261 in H1299 cells
Given that the circ_0072088 is mainly located in cytoplasm, it may function as a sponge to absorb miRNAs that inhibit downstream genes. After searching circRNA interactome databases (http://circinteractome.nia.nih.gov), the target miRNAs (miR‐1261 and miR‐620) with high predicted score was selected for further exploration (Figure 3a). Moreover, the downregulation of miR‐1261 expression was further confirmed by qRT‐PCR (Figure 3b) and FISH staining (Figure 3c) in 20 LUAD tissues relative to its matched paracancerous tissues. The FISH staining results showed that circ_0072088 was colocalized with miR‐1261 in the cytoplasm of H1299 cells (Figure 3d). Moreover, RIP assay further showed the interactions between circ_0072088 and miR‐1261 in H1299 cells (Figure 3e). To confirm the direct interaction between them, wild‐type (WT) and mutant (MUT) circ_0072088 luciferase reporter plasmids (pLG3 vector) were constructed according to the predicted interacting site and were then cotransfected with miR‐1261 mimics or negative control (NC) into H1299 cells. As shown in Figure 3f, the relative luciferase activity was dramatically decreased in H1299 cells cotransfected with WT circ_0072088 luciferase reporter and miR‐1261 mimics, whereas this alteration of luciferase activity disappeared with cotransfection of MUT circ_0072088 and miR‐1261 mimics. These results indicated that circ_0072088 functioned as a sponge to absorb miR‐1261, which would later be involved in initiation and progression of LUAD.
FIGURE 3.
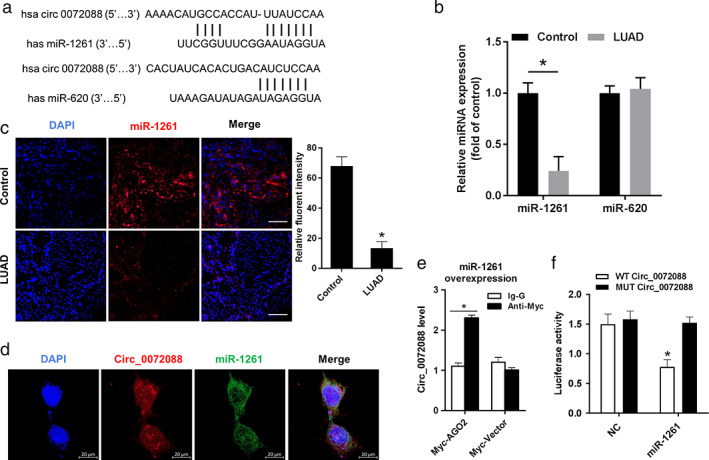
Circ_0072088 acts as a sponge of miR‐1261 in H1299 cells. (a) Schematic diagram for the potential binding sites of miR‐1261 and miR‐620 in circ_0072088 were predicted by CircInteractome databases. (b) qRT‐PCR analysis for the relative expression of miR‐1261 in lung adenocarcinoma (LUAD) tumors and their adjacent normal tissues (n = 20). (c) Representative photographs of RNA FISH staining for miR‐1261 (red) in lung adenocarcinoma (LUAD) tumors and their adjacent normal tissues (n = 20). Nuclei were stained with DAPI (blue). Scale bars = 50 μm. Right: statistical analysis of fuorescence intensity. *p < 0.05. (d) RNA‐FISH assay for colocalization between circ_0072088 (red) and miR‐1261 (green) in H1299 cells. Scale bars = 50 μm. (e) RNA‐binding protein immunoprecipitation (RIP) assays for identification of circ_0072088 interact with miR‐1261 in H1299. (f) Luciferase reporter assay for demonstration of the interaction between circ_0072088 and miR‐1261 in H1299 cells with transfection of WT‐circ_0072088 or MUT‐circ_0072088. All data represent mean ± SEM (n = 3). *p < 0.05 versus IgG or circ_0072088‐WT
Circ_0072088 promotes cell proliferation, migration, and invasion of H1299 cells by suppressing miR‐1261 expression
Circ_0072088 was required for cell proliferation, migration, and invasion in H1299 cells and directly interacted with miR‐1261. Thus, rescue experiments in H1299 cells were performed to further confirm the role of the circ_0072088/miR‐1261 axis in the biological function of LUAD cells. Accordingly, CCK‐8 (Figure 4a), colony formation (Figure 4b), scratch (Figure 4c, d), and transwell experiments (Figure 4e, f) showed that miR‐1261 suppression by transfection of antisense oligonucleotide (ASO) reversed circ_0072088 inhibition and induced the proliferation, migration, and invasion in H1299 cells (Figures 4b–f), suggesting that circ_0072088 could maintain the carcinogenicity of LUAD cells by antagonizing the tumor‐suppressive role of miR‐1261 in LUAD.
FIGURE 4.
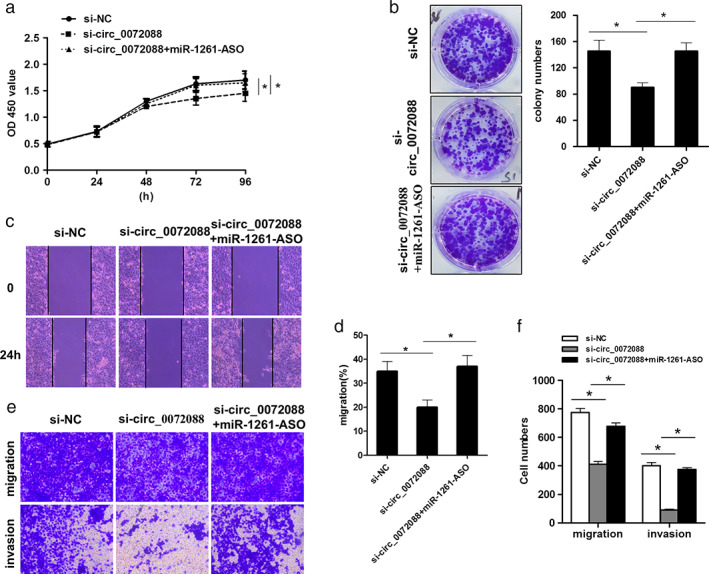
miR‐1261 mediated Circ_0072088‐induced proliferation, migration, and invasion in H1299 cells. (a–f) Effects of miR‐1261 inhibition by transfection with miR‐1261‐ASO on circ_0072088 induced the proliferation, migration, and invasion of H1299 cells by CCK8 assay (a), clone formation (b), a scratch assay (c and d), and transwell assay (e and f), respectively. All data represent mean ± SD (n = 3). *p < 0.05 versus si‐NC or si‐circ_0072088
Circ_0072088 upregulated PIK3CA, which has been associated with poor overall survival of LUAD, by suppressing miR‐1261 expression
To further determine the potential mechanism by which the circ_0072088/miR‐1261 axis exerts its cancer‐promoting role in LUAD, the downstream genes were explored. Through TargetScan and miRBD databases, 23 common predicted targets of miR‐1261 were identified (Figure 5a). KEGG analysis exhibited several critical cancer‐associated pathways, such as the mTOR signaling pathway, cAMP signaling pathway, JAK–STAT signaling pathway, and PI3K‐Akt signaling pathway (Figure 5b). Intriguingly, among them, PIK3CA could be a target of miR‐1261 (Figure 5c), as well as a hub for many signaling pathways in LUAD cells. As expected, the relative luciferase activity was significantly decreased in H1299 cells cotransfected with WT‐PIK3CA and miR‐1261 mimics, whereas the change was abrogated in cells cotransfected with MUT‐PIK3CA and miR‐1261 mimics (Figure 5d). These results revealed that PIK3CA was a target of miR‐1261 in LUAD cells. Lastly, to confirm the clinical relevance of our findings, PIK3CA expression was measured by qRT‐PCR and immunochemistry staining in tissues from patients with LUAD and control. Accordingly, the results showed a marked increase in transcriptional (Figure 5e) and protein levels of PIK3CA (Figure 5h). In line with this, bioinformatic assay results also showed that the high expression of PIK3CA was associated with poor overall survival in LUAD patients according to Kaplan–Meier plotter database (Figure 5f) and Human protein atlas database (Figure 5g). These results indicated that circ_0072088 increased PIK3CA expression, which has been associated with the poor overall survival of LUAD patients, by suppressing miR‐1261 expression.
FIGURE 5.
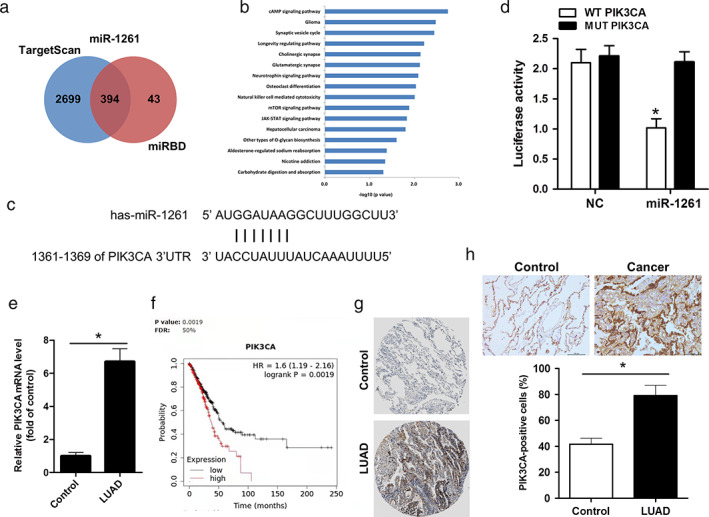
PIK3CA as a target of miR‐1261 is the hub gene of many signaling pathways in lung adenocarcinoma cells. (a) Schematic diagram of prediction for the downstream target genes of miR‐1261 using targetscan and miRBD databases. (b) KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis of 23 common potential target genes involving PIK3CA‐associated 16 top signaling pathways. (c) Schematic diagram for the potential binding sites of miR‐1261 in PIK3CA mRNA were predicted by targetscan and miRBD databases. (d) The relative luciferase activity of PIK3CA 3′‐UTR‐wild‐type (WT PIK3CA) or ‐mutant (MUT PIK3CA) luciferase vector after transfection with miR‐1261 mimics or negative control (NC) was detected in H1299 cells. All data represent mean ± SD (n = 3). *p < 0.05 versus WT PIK3CA. (e) qRT‐PCR assay for PIK3CA mRNA level in lung adenocarcinoma (LUAD) tumors and their adjacent normal tissues (n = 20). *p < 0.05 versus control. (f) Kaplan–Meier plotter database (http://kmplot.com/analysis/) was used to further validate the poor survival with high expression of PIK3CA in LUAD patients. (g) Human protein atlas database (http://www.proteinatlas. org/) was performed to validate the stronger positive staining of PIK3CA in cancer tissues than normal lung tissues. (h) Representative immunohistochemistry (IHC) staining for PIK3CA in lung adenocarcinoma (LUAD) tumors and their adjacent normal tissues (n = 20). Down: Statistic analysis of PIK3CA‐positive cells. *p < 0.05 versus control
DISCUSSION
Our study revealed that upregulation and cytoplasmic localization of circ_0072088 in LUAD tissues and cells play an oncogenic role by releasing miR‐1261‐mediated inhibition of PIK3CA, which mediates circ_0072088‐induced cell proliferation, migration, and invasion via crosstalk among several tumor‐associated signaling pathways. The novel circRNA–miRNA–mRNA regulatory framework established herein may provide a potential therapeutic target for LUAD.
Previous studies have suggested that circRNAs were either up‐ or downregulated in different cancers and are accompanied by carcinogenic or carcinostasis effects. Li et al. found that hepatocellular carcinoma (HCC) tissues had a greater expression of circ_0072088 compared to adjacent hepatocellular tissues by RT‐PCR. Moreover, the high expression of circ_0072088 was correlated with cell proliferation, migration, and invasion. 25 In contrast, Liu et al. confirmed that circ_0072088 suppressed gastric cancer cell proliferation and promoted apoptosis by regulating PTEN via sponging miR‐130a/miR‐107. 26 To clarify the role of circ_0072088 in the initiation and development of LUAD, we initially validated the existence of circ_0072088 and its increase in LUAD tissues relative to paired paracancerous normal tissues, with cytoplasmic localization in tissue and cellular levels. Moreover, circ_0072088 knockdown could suppress the proliferation, migration, and invasion in vitro. These results indicated that lung tissue‐specific upregulation of circ_0072088 functioned as an oncogene and participated in the occurrence and development of LUAD.
Several tumor‐associated circRNAs regulate transcription and translation processes through a common mechanism, sponging miRNAs to regulate the function of target genes. 9 Therefore, we mainly focused on the sponging function of circ_0072088 in absorbing key miR‐1261 in LUAD according to bioinformatic analysis and our results suggesting cytoplasmic localization. Only a few reports have been available on miR‐1261 downregulation in hepatocellular cancer, 18 , 27 papillary thyroid cancer, 28 and gliomas. 29 Consistently, our results also showed that miR‐1261 was a target for circ_0072088 and that it was downregulated in LUAD tissues. Moreover, reversing the effects of circ_0072088 inhibition by miR‐1261 suppression promoted proliferation, migration, and invasion, suggesting that circ_0072088 could maintain the carcinogenicity of LUAD cells by antagonizing the tumor‐suppressive role of miR‐1261 in LUAD.
Mutations in PIK3CA, which lead to a gain of function, determine a constitutive activation of the PI3K/AKT/mTOR pathway, which is among the main driving mechanisms in cancers. 30 , 31 Moreover, phosphatidylinositol kinase prompted PDK1 to phosphorylate the AKT protein Set308, leading to the activation of AKT, thereby regulating cell proliferation, migration, and invasion. 32 The results presented herein also demonstrated that miR‐1261 negatively targeted PIK3CA. Importantly, the high expression of PIK3CA, confirmed by database and immunochemistry methods, was associated with poor prognosis of LUAD patients, suggesting that circ_0072088 accelerates tumorigenesis and progression of lung adenocarcinoma by regulating PIK3CA.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGMENTS
The authors thank Professor Men‐xiang Sang, from the Fourth Hospital of HebeiMedical University for technical guidance. This study is supported by the Hebei Natural Science Foundation (No. H2021206130).
Cao F, Liu S, Li Z, Meng L, Sang M, Shan B. Activation of circ_0072088/miR‐1261/PIK3CA pathway accelerates lung adenocarcinoma progression. Thorac Cancer. 2022;13:1548–1557. 10.1111/1759-7714.14369
Funding information Hebei Natural Science Foundation, Grant/Award Number: H2021206130
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. [DOI] [PubMed] [Google Scholar]
- 2. Barta JA, Powell CA, Wisnivesky JP. Global epidemiology of lung cancer. Ann Glob Health. 2019;85(1):8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Succony L, Rassl DM, Barker AP, FM MC, Rintoul RC. Adenocarcinoma spectrum lesions of the lung: detection, pathology and treatment strategies. Cancer Treat Rev. 2021;99:102237. [DOI] [PubMed] [Google Scholar]
- 4. Xu LS, Qi Q, Zhang Y, Cui J, Liu R, Li Y. Combination of icotinib and chemotherapy as first‐line treatment for advanced lung adenocarcinoma in patients with sensitive EGFR mutations: a randomized controlled study. Lung Cancer. 2019;7(133):23–31. [DOI] [PubMed] [Google Scholar]
- 5. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8. [DOI] [PubMed] [Google Scholar]
- 6. Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol. 2020;21(8):475–90. [DOI] [PubMed] [Google Scholar]
- 7. Vo J, Marcin C, Zhang YJ, Shukla S, Xiao L, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176:869–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Papatsirou M, Artemaki PI, Karousi P, Scorilas A, Kontos CK. Circular RNAs: emerging regulators of the major signaling pathways involved in cancer progression. Cancer. 2021;13:2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8. [DOI] [PubMed] [Google Scholar]
- 10. Wang T, Liu Z, She Y, Deng J, Zhong Y, Zhao M, et al. A novel protein encoded by circASK1 ameliorates gefitinib resistance in lung adenocarcinoma by competitively activating ASK1‐dependent apoptosis. Cancer Lett. 2021;520(1):321–31. [DOI] [PubMed] [Google Scholar]
- 11. Shen HY, Shi LX, Wang L, Fang LP, Xu W, Xu JQ, et al. Hsa_circ_0001361 facilitates the progress of lung adenocarcinoma cells via targeting miR‐525‐5p/VMA21 axis. J Transl Med. 2021;19(1):389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Y, Sun R, Li R, Chen Y, Du H. Prognostic nomogram based on circular RNA‐associated competing endogenous RNA network for patients with lung adenocarcinoma. Oxid Med Cell Longev. 2021;2021:9978206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liang LM, Zhang L, Zhang JG, Bai S, Fu H. Identification of circRNA‐miRNA‐mRNA networks for exploring the fundamental mechanism in lung adenocarcinoma. Onco Targets Ther. 2020;13:2945–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu YC, Wang X, Bi LL, Huo H, Yan S, Cui Y, et al. Identification of differentially expressed circular RNAs as miRNA sponges in lung adenocarcinoma. J Oncol. 2021;2021:5193913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang ZJ, Pei HZ, Liang HS, Zhang Q, Wei L, Shi D, et al. Construction and analysis of a circRNA‐mediated ceRNA network in lung adenocarcinoma. Onco Targets Ther. 2021;14:3659–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin Y, Zheng ZH, Wang JX, Zhao Z, Peng TY. Tumor cell‐derived exosomal circ‐0072088 suppresses migration and invasion of hepatic carcinoma cells through regulating MMP‐16. Front Cell Dev Biol. 2021;9:726323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bian L, Zhi X, Ma L, Zhang J, Chen P, Sun S, et al. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR‐532e3p/FOXO4 axis. Biochem Biophys Res Commun. 2018;505(2):346–52. [DOI] [PubMed] [Google Scholar]
- 18. Lyu N, Zeng Y, Kong Y, Chen Q, Deng H, Ou S, et al. Ferroptosis is involved in the progression of hepatocellular carcinoma through the circ0097009/miR‐1261/SLC7A11 axis. Ann Transl Med. 2021;9(8):675–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Borkowska EM, Barańska M, Kowalczyk M, Pietruszewska W. Detection of PIK3CA gene mutation in head and neck squamous cell carcinoma using droplet digital PCR and RT‐qPCR. Biomolecules. 2021;11(6):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shi X, Sun Y, Zhang Y, Wang W, Xu J, Guan Y, et al. MEX3A promotes development and progression of breast cancer through regulation of PIK3CA. Exp Cell Res. 2021;404(1):112580. [DOI] [PubMed] [Google Scholar]
- 21. Gris‐Oliver A, Ibrahim YH, Rivas MA, García‐García C, Sánchez‐Guixé M, Ruiz‐Pace F, et al. PI3K activation promotes resistance to eribulin in HER2‐negative breast cancer. Br J Cancer. 2021;124(9):1581–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu Y, Xie Z, Zhao M, Lian X. Identification of PIK3CA multigene mutation patterns associated with superior prognosis in stomach cancer. BMC Cancer. 2021;21(1):368–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Meng LJ, Liu SH, Liu F, Sang M, Ju Y, Fan X, et al. ZEB1‐mediated transcriptional upregulation of circWWC3 promotes breast cancer progression through activating Ras signaling pathway. Mol Ther Nucleic Acids. 2020;22:124–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ma D, Zheng B, Suzuki T, Zhang R, Jiang C, Bai D, et al. Inhibition of KLF5–Myo9b–RhoA pathway‐mediated Podosome formation in macrophages ameliorates abdominal aortic aneurysm. Circ Res. 2017;120(5):799–815. [DOI] [PubMed] [Google Scholar]
- 25. Li LH, Xiao CJ, He K, Xiang G. Circ_0072088 promotes progression of hepatocellular carcinoma by activating JAK2/STAT3 signaling pathway via miR‐375. IUBMB Life. 2021;73(9):1153–65. [DOI] [PubMed] [Google Scholar]
- 26. Liu TL, Liu S, Xu Y, Shu R, Wang F, Chen C, et al. Circular RNA‐ZFR inhibited cell proliferation and promoted apoptosis in gastric cancer by sponging miR‐130a/miR‐107 and modulating PTEN. Cancer Res Treat. 2018;50(4):1396–417. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27. Zhang Y, Zhou H. LncRNA BCAR4 promotes liver cancer progression by upregulating ANAPC11 expression through sponging miR‐1261. Int J Mol Med. 2020;46(1):159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wei H, Pan L, Tao D, Li R. Circular RNA circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging miR‐1261 and facilitating C8orf4 expression. Biochem Biophys Res Commun. 2018;503(1):56–61. [DOI] [PubMed] [Google Scholar]
- 29. Zhang F, Mai SR, Cao FP, Cao CX, Zhang L. MiR‐1261/circ‐PTPRZ1/PAK1 pathway regulates glioma cell growth and invasion. Hum Cell. 2019;32(4):540–7. [DOI] [PubMed] [Google Scholar]
- 30. Swaney DL, Ramms DJ, Wang Z, Park J, Goto Y, Soucheray M, et al. A protein network map of head and neck cancer reveals PIK3CA mutant drug sensitivity. Science. 2021;374(6563):eabf2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pascual J, Lim JSJ, Macpherson IR, Armstrong AC, Ring A, et al. PIK3CATriplet therapy with Palbociclib, Taselisib, and Fulvestrant in ‐mutant breast cancer and doublet Palbociclib and Taselisib in pathway‐mutant solid cancers. Cancer Discov. 2021;11(1):92–107. [DOI] [PubMed] [Google Scholar]
- 32. Ali T, Badshah H, Kim TH, Kim MO. Melatonin attenuates D‐galactose‐induced memory impairment, neuroinflammation and neurodegeneration via RAGE/NF‐K B/JNK signaling pathway in aging mouse model. J Pineal Res. 2015;58:71–85. [DOI] [PubMed] [Google Scholar]


