Abstract
This study aimed to explore the prognostic impact of spatial distribution of tumor‐infiltrating lymphocytes (TILs) quantified by deep learning (DL) approaches based on digitalized whole‐slide images stained with hematoxylin and eosin in patients with colorectal cancer (CRC). The prognostic impact of spatial distributions of TILs in patients with CRC was explored in the Yonsei cohort (n = 180) and validated in The Cancer Genome Atlas (TCGA) cohort (n = 268). Two experienced pathologists manually measured TILs at the most invasive margin (IM) as 0–3 by the Klintrup–Mäkinen (KM) grading method and this was compared to DL approaches. Inter‐rater agreement for TILs was measured using Cohen's kappa coefficient. On multivariate analysis of spatial TIL features derived by DL approaches and clinicopathological variables including tumor stage, microsatellite instability, and KRAS mutation, TIL densities within 200 μm of the IM (f_im200) remained the most significant prognostic factor for progression‐free survival (PFS) (hazard ratio [HR] 0.004 [95% confidence interval, CI, 0.0001–0.15], p = 0.0028) in the Yonsei cohort. On multivariate analysis using the TCGA dataset, f_im200 retained prognostic significance for PFS (HR 0.031 [95% CI 0.001–0.645], p = 0.024). Inter‐rater agreement of manual KM grading was insignificant in the Yonsei (κ = 0.109) and the TCGA (κ = 0.121) cohorts. The survival analysis based on KM grading showed statistically significant different PFS in the TCGA cohort, but not the Yonsei cohort. Automatic quantification of TILs at the IM based on DL approaches shows prognostic utility to predict PFS, and could provide robust and reproducible TIL density measurement in patients with CRC.
Keywords: colorectal cancer, tumor‐infiltrating lymphocytes, deep learning, prognosis, whole‐slide image
Introduction
Standard treatment of colorectal cancer (CRC) includes curative intent surgical resection followed by postoperative selective chemotherapy with or without radiotherapy [1]. In patients with unresectable CRC, chemotherapy is the main treatment option to sustain the survival durations or to convert patients into resectable status. Postoperative chemotherapy has its own role to decrease recurrence, but its adoption is solely dependent on postoperative staging under current guidelines [1]. Personalized treatment is demanding, because CRC patients with the same stage of disease may have different survival outcomes.
The presence of tumor‐infiltrating lymphocytes (TILs) is increasingly recognized as an important biomarker in multiple cancer types [2, 3, 4, 5]. Previous studies using immunohistochemical (IHC) staining of various T‐cell markers suggested that densities of TILs in the tumor microenvironment are associated with survival outcomes in patients with CRC [2]. In particular, the Immunoscore that quantifies TIL densities and spatial distributions has shown a high prognostic value in patients with CRC, which could provide a more precise stratification of patient prognosis [6]. However, application of the Immunoscore requires IHC staining and expert interpretation, which could incur substantial cost and requires specialized facilities [2]. Few studies measuring TILs using hematoxylin and eosin (H&E)‐stained slides also demonstrated its positive role as a prognostic indicator in patients with CRC [4, 7, 8, 9, 10, 11]. Nevertheless, standardizing methods to quantify TILs are known to be labor‐intensive and pathologist‐dependent [7]. Although assessing TILs is considered to be clinically important, TILs have had limited use as a prognostic biomarker due to the additional requirements of IHC staining as well as substantial lack of standardization of measurement [6, 12].
With recent advances in digital pathology and artificial intelligence (AI) (i.e. deep learning [DL] algorithms), there has been increasing interest in developing automated methods for TIL quantification and analysis from pathology slides. Saltz et al used DL approaches to detect TILs in H&E‐stained whole‐slide images (WSIs), and showed that spatial TIL distribution reflected by clustering indexes was linked to patient survival across different tumor types [13]. Corredor et al performed computerized analysis on H&E‐stained tissue microarray slides, which identified TIL spatial distribution and its co‐localization with cancer cell nuclei to predict likelihood of recurrence in early‐stage non‐small cell lung cancer [12]. Bankhead et al showed that the TIL densities of tumor regions quantified by the QuPath® software [14] in H&E‐stained pathology images could be used to predict overall survival (OS) in patients with melanoma [15]. Yoo et al analyzed IHC‐stained CRC pathology slides by using the QuPath® software. The lymphocyte (e.g. CD3, CD8) densities inside the tumor core and invasive margins (IMs) were used to classify CRC patients into clinicopathologically relevant subgroups [16]. AbdulJabbar et al quantified lymphocytic infiltration variability between lung cancer regions in pathology slides, which was shown to correlate with patient disease‐free survival [17]. Although these recent studies suggest that TIL‐related variables extracted by computational pathology could potentially predict patient clinical outcomes across different cancer types, the quantitative analysis of TILs according to spatial distribution using H&E‐stained WSIs has been investigated to only a limited extent in patients with CRC. To the best of our knowledge, only one study has investigated quantitative TIL measurement in CRC pathology slides, but it required additional IHC staining and manual interactions with the analyzing software which hindered its wide application in clinical settings [16].
In this study, we aimed to investigate whether automated quantification of spatial distribution of TILs in tumor IMs based on DL approaches utilizing H&E‐stained WSIs could predict progression‐free survival (PFS) outcomes in patients with CRC. In addition, we also sought to compare the Deep Learning based TIL density measurement (DeepTIL) in terms of its ability to predict prognosis in these patients against the manual scoring of TILs at the deepest invasive area by pathologists. Overall, the DeepTIL tools developed are aimed at providing effective and efficient assessments of TIL distributions inside the H&E‐stained WSI, and assisting in CRC patient prognosis via fully automatic analysis.
Materials and methods
Datasets
Two independent cohorts of H&E‐stained WSIs from 448 colorectal cancer patients were included in this study: the Yonsei (n = 180) and The Cancer Genome Atlas (TCGA) (n = 268) cohorts. The Yonsei cohort consists of 180 diagnostic WSIs and corresponding clinical information from stage II or III colon cancer patients who underwent curative resection followed by FOLFOX (5‐fluorouracil, leucovorin, and oxaliplatin) chemotherapy between September 2005 and January 2014. The Yonsei cohort was collected from Gangnam Severance Hospital, Yonsei University College of Medicine, Republic of Korea.
The whole TCGA CRC cohort with corresponding clinical information and diagnostic WSIs was downloaded from TCGA data portal (https://portal.gdc.cancer.gov/). After visual evaluation by two pathologists, 268 CRC patients whose WSIs have reasonably good quality and enough clinical information were used for analysis in this study.
The WSIs from the Yonsei dataset (.mrxs format slides) were generated using Pannoramic® 250 Flash III scanner (3DHISTECH, Budapest, Hungary) with the pixel resolution of 0.2428 μm/pixel. The TCGA pathology slides were generated and uploaded by many different institutions, where images (.svs format slide) were scanned by using the Aperio Scanscope Q5 CS scanner (Leica Biosystems, Wetzlar, Germany) with the pixel resolution of 0.2527 μm/pixel.
Patients who had postoperative WSIs and available clinicopathological and follow‐up data were included. For the TCGA dataset, patients with very poor quality slides, absence of survival status or duration, and patients whose survival time was denoted as 0 months were excluded. When the pathologists evaluated the manual scoring of TILs, they found that some slides did not include the IMs and these patients were excluded at the stage of measuring the agreement rating and further comparison processing between human scoring versus AI‐based scoring (supplementary material, Figure S1).
For each patient, the following outcomes were collected if available: sex, age (years), American Society of Anesthesiologists classification, body mass index (BMI) (kg/m2), carcinoembryonic antigen (ng/ml), tumor location, complications, histological grade, lymphovascular invasion (LVI), total retrieved lymph node numbers, stage, microsatellite instability (MSI), and KRAS mutation status.
This study was conducted after approval from the Institutional review board of the Gangnam Severance Hospital, Yonsei University College of Medicine (Seoul, Republic of Korea) (approval no. 3‐2020‐0076). The need for informed consent was waived for this retrospective study.
Development of DeepTILs using H&E‐stained WSIs
The developmental process of DeepTILs was composed of four main modules: tumor detector, TIL detector, automatic quantification of TILs, and statistical and survival analyses (Figure 1). The details are described in the following sections.
Figure 1.
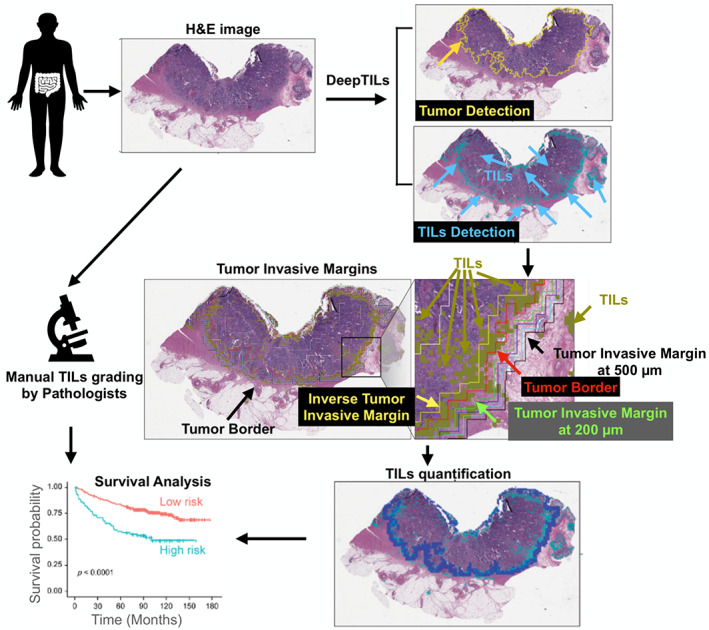
Overview of the approach taken in this study. Tumor region and TILs are first detected in the H&E‐stained WSI based on DL approaches. We then quantify TILs across and within tumors. The spatial distributions of TILs densities at tumor IMs and tumor core are used to identify patient subgroups with distinct PFS outcome.
Tumor detector
To develop an automatic tumor detector, we performed transfer learning on Resnet18 DL model that was originally trained on ImageNet dataset [18]. We trained the Resnet18 on a public dataset containing 11,977 image patches (256 μm edge length per image) of H&E‐stained histological samples of human CRC [18, 19, 20]. Regions in these images were manually annotated into three classes: tumor tissue, loose non‐tumor tissue (i.e. adipose tissue and mucus), and dense non‐tumor tissue (i.e. stroma and muscle). To train and test tumor detector, we randomly divided the whole dataset into training (80%), validation (10%), and testing (10%) sets. We then trained the Resnet18 model to distinguish tumor and non‐tumor image patches. Image augmentations including random horizontal and vertical flipping and color jittering (i.e. alteration of image contrast, brightness, and saturation) were applied along with training. We trained the model by freezing different percentiles of trainable layers, and using different parameter configurations in terms of optimizer, batch size, and learning rate (supplementary material, Table S1). Overall, we trained and tested 24 different tumor detection models. These DL models were trained on a local GPU server (CentOS Linux7 system) with the Intel Xeon 8353H CPU (128 G RAM) and two RTX 3090 GPUs (24 G Memory). The PyTorch (torch: 1.7.1; torchvision: 0.8.2) and scikit‐image (v 0.17.2) libraries were used to implement these models. Testing accuracies of the 24 models with different configurations are described in supplementary material, Figure S2. The tumor detector, which was trained by fine‐tuning all trainable layers of Resnet18 and using Adam optimizer with a learning rate of 0.0001 and batch size of 64, provided the best test accuracy (100%). This best performing tumor detector was applied to predict tumor regions for all WSIs from the Yonsei and TCGA cohorts. Specially, the WSI was divided into a set of non‐overlapping image tiles (256 μm × 256 μm per tile), which were predicted with the probabilities of belonging to tumor tiles by using our best tumor detector. The predicted probabilities of different image tiles were then stitched together according to tile locations, such that the WSI‐level tumor predictions were obtained. Finally, the typical threshold 0.5 was applied on the WSI‐level prediction map to obtain the tumor detection results that are indicated by red pixels in supplementary material, Figure S3A (tumor detection).
TIL detector
By following a similar procedure to that for tumor detection, we trained and applied DL models to identify TIL regions in the WSI. As TIL identification is more challenging, we retrained three different DL architectures including Resent18, Resent34, and Shufflenet [21] on a public dataset, where 43,440 image tiles were adopted [13]. The whole dataset was randomly divided into three parts: training (80%), validation (10%), and testing (10%) sets. Image augmentations including random flipping and color jittering were applied along with training, which was performed in the same manner as training tumor detectors. We trained the models by freezing different percentiles of trainable layers, and using different parameter configurations in terms of optimizer, batch size, and learning rate (supplementary material, Table S2). Overall, we trained and tested 144 different TIL detectors, and testing accuracies of 144 models with different configurations are described in supplementary material, Figure S4. The TIL detection model, which was trained by fine‐tuning all trainable layers of Resnet18 and using Adam optimizer with a learning rate of 0.0001 and batch size of 4, provided the best test accuracy (80.06%). Several TIL prediction examples randomly selected from the testing set are provided in supplementary material, Figure S5. Supplementary material, Figure S6 shows visualized features learned by the TIL detector, where the reconstructed color and gray‐scale images are generated by using the guided backpropagation method. The best TIL detection model was used to identify TIL regions for all WSIs from the Yonsei and TCGA cohorts. Specially, the WSI was divided into a set of non‐overlapping image patches (112 μm × 112 μm/patch) which were predicted with the probabilities of belonging to TILs. The WSI‐level TIL prediction was finally obtained by stitching tile‐level predictions. The threshold of 0.5 was used to determine regions containing TILs in the WSI‐level prediction map. Supplementary material, Figure S3B (TIL detection) illustrates a TIL detection example (yellow pixels).
Automatic quantification of TILs from H&E‐stained WSIs
Based on predicted tumor and TIL regions by DL models, we quantified the density of TILs inside IM and tumor regions, respectively. If more than one tumor region was detected, the largest tumor region was selected. We used image morphological dilation operation with a square structuring element to detect the tumor IM automatically. Note that by adjusting the physical size of structuring elements, we could obtain different widths of IM layers. For instance, in order to detect the 200‐μm IM layer, we first used the square structuring element with an edge length of 400 μm to perform the morphological dilation. The 200‐μm IM layer was then obtained by removing tumor pixels in the dilated binary mask. The schematic explanation of tumor IMs is shown in supplementary material, Figure S7, and the example overlapped with automatically extracted IM boundaries is illustrated in Figure 1. The red contour indicates the tumor border, while the IM layers with the distance of 200, 300, 400, and 500 μm from the tumor border are indicated by different color lines. The TIL densities were computed and denoted as ‘f_im200’, ‘f_im300’, ‘f_im400’, and ‘f_im500’ at each IM layer, respectively. For example, ‘f_im200’ represents the ratio between the number of TIL pixels inside the 200‐μm IM layer and the whole number of pixels at the 200‐μm IM layer. In addition to TIL densities at IM layers, we further quantified TIL densities at tumor regions. The TIL density at the whole tumor region was computed as ‘f_wt’. The area that occupies 25% of the central part of the whole tumor region was defined as the tumor core and the TIL density in the tumor core was denoted as ‘f_tc’. We applied morphological erosion operation to obtain the 200‐μm inverse IM layer and computed the corresponding TIL density as ‘f_inv200’. Finally, in the whole tumor region, the area of inverse 200‐μm IM layer was subtracted and the remaining area was defined as TC2, and the TIL density of this area was defined as ‘f_tc2’ (for further descriptions, please see supplementary material, Table S3 and Figure S7).
Manual scoring of TILs by pathologists using the KM recommendation
Two board‐certified pathologists, who had 9 and 10 years of experience, respectively, graded each patient's TILs using the Klintrup–Mäkinen (KM) recommendation (KM grading) [10]. The overall inflammatory reaction of the deepest area of the IM of the tumor was assessed by using a 4° scale based on visual examination of H&E‐stained pathology slides (supplementary material, Figure S8). A score 0 denoted no increase in inflammatory cells, 1 denoted a mild and patchy increase in inflammatory cells, 2 denoted a moderate and band‐like inflammatory infiltrate with some destruction of cancer cell islands, and 3 denoted a marked and florid cuplike inflammatory infiltrate with frequent destruction of cancer cell islands [8, 9]. Patients were dichotomized as KM‐low (KM gradings 0 and 1) and KM‐high (KM gradings 2 and 3), and survival outcome were compared between these two groups.
Statistical analysis
All statistical analyses were performed using R version 3.6.3 (R‐project, Institute for Statistics and Mathematics, Vienna, Austria). Patients' clinicopathological characteristics were compared using chi‐square and t‐tests for categorical and continuous variables, respectively. Correlation between TIL counts and clinical variables were assessed using the Mann–Whitney U‐test for dichotomous values, Kruskal–Wallis tests for more than three‐group comparisons, and Spearman correlations for continuous parameters.
The kappa statistic was used to assess inter‐rater agreement with respect to scoring and was interpreted according to the guidelines of Landis and Koch [22]. We used the following definition to interpret the kappa coefficients: a kappa (κ) value of ≤0.20 indicated insignificant agreement, values of 0.21–0.40 indicated median agreement, values of 0.41–0.60 indicated moderate agreement, values of 0.61–0.80 indicated substantial agreement, and values of 0.81–1.00 indicated almost perfect agreement.
PFS was calculated from the date of surgery until the date of recurrence detection in the Yonsei dataset. Patients alive at the last follow‐up or dead were censored. The Kaplan–Meier method was used to construct survival curves and the log‐rank test was used to compare survival rates between groups. Cox proportional hazards models were used to estimate the hazard ratios (HRs) and 95% confidence interval (CI). All variables with p < 0.1 on univariate analysis were entered for multivariate analysis with backward stepwise selection of variables. A two‐sided p < 0.05 was considered statistically significant.
Results
Clinicopathological characteristics of the patients
Among the patients included in the study, 29 of 180 patients (16.1%) in the Yonsei cohort and 74 of 268 patients (27.6%) in the TCGA cohort had recurrences. The median follow‐up period for patients was 89 months (interquartile range [IQR], 71–122 months) for the Yonsei cohort and 19.6 months (IQR, 12.4–32.9 months) for the TCGA cohort. Details of clinicopathological features for the included patients are presented in Table 1.
Table 1.
Patient characteristics of the Yonsei and TCGA datasets
| Variables | Yonsei (n = 180), N (%) | TCGA (n = 268), N (%) | p | |
|---|---|---|---|---|
| Sex | Female | 74 (41.1) | 130 (48.5) | |
| Male | 106 (58.9) | 138 (51.5) | 0.149 | |
| Age (years) | <70 | 28 (15.6) | 178 (66.4) | |
| ≥70 | 152 (84.4) | 90 (33.6) | <0.001 | |
| BMI (kg/m2) | <25 | 134 (74.4) | 54 (20.1) | |
| ≥25 | 46 (25.6) | 150 (56) | ||
| No data | — | 64 (23.9) | <0.001 | |
| CEA (ng/ml) | <5 | 113 (62.8) | — | |
| ≥5 | 67 (37.2) | — | NA | |
| Tumor location | Right colon | 62 (34.4) | 134 (50) | |
| Left colon | 118 (65.6) | 105 (39.2) | ||
| Rectum | — | 19 (7.1) | ||
| No data | — | 10 (3.7) | NA | |
| Complications | No | 145 (80.6) | — | |
| Yes | 35 (19.4) | — | NA | |
| Histological grade | G1 and G2 | 149 (82.8) | — | |
| G3, etc. | 31 (17.2) | — | NA | |
| LVI | Absent | 109 (60.6) | 102 (38.1) | |
| Present | 66 (36.7) | 144 (53.7) | ||
| No data | 5 (2.8) | 22 (8.2) | <0.001 | |
| LN numbers | <12 | 10 (5.6) | 35 (13.1) | |
| ≥12 | 170 (94.4) | 212 (79.1) | ||
| No data | — | 21 (7.8) | <0.001 | |
| Stage | I | — | 39 (14.6) | |
| II | 27 (15) | 105 (39.2) | ||
| III | 153 (85) | 94 (35.1) | ||
| IV | — | 30 (11.2) | <0.001 | |
| MSI | MSS/MSI‐low | 83 (46.1) | 159 (59.3) | |
| MSI‐high | 13 (7.2) | 49 (18.3) | ||
| No data | 84 (46.7) | 60 (22.4) | <0.001 | |
| f_wt | Continuous | 0.1 (0.1) | 0.1 (0.1) | 0.619 |
| f_im200 | Continuous | 0.2 (0.1) | 0.1 (0.1) | <0.001 |
CEA, carcinoembryonic antigen; LN, lymph node; MSS, microsatellite stable; NA, not available.
Prognostic evaluation of automated TIL features by DeepTILs from the Yonsei dataset
The composition of TILs according to spatial distribution is illustrated in supplementary material, Table S4. The median TIL density of each spatial distribution ranged from 0.0447 to 0.2002. Univariate Cox proportional hazards model of PFS in the Yonsei cohort revealed that ‘f_im200’, ‘f_im300’, ‘f_im400’, ‘f_im500’, and ‘f_inv200’ are significant prognostic factors (supplementary material, Table S5). In multivariate analysis using features derived from various IMs, ‘f_im200’ remained as an independent significant factor (supplementary material, Table S6A). For features derived from inner tumor area, ‘f_wt’ and ‘f_inv200’ were identified as prognostic factors (supplementary material, Table S6B). When all features were considered, ‘f_wt’ and ‘f_im200’ remained statistically significant prognostic factors (supplementary material, Table S6C). Thus, these two variables were included in further analysis by adjusting with clinicopathological variables.
In the Yonsei cohort, univariate analysis revealed that LVI (p = 0.009) and ‘f_im200’ (p = 0.002) were significantly associated with PFS. In the multivariate analysis, ‘f_im200’ was an independent significant prognostic factor (HR 0.004, 95% CI 0.0001–0.15, p = 0.0028) (Table 2).
Table 2.
Univariate and multivariate analyses associated with PFS in the Yonsei dataset (n = 180)
| Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|
| Variables | HR (95% CI) | p | HR (95% CI) | p | |
| Sex | Female | Ref | |||
| Male | 0.83 (0.40–1.72) | 0.622 | |||
| Age (years) | <70 | Ref | |||
| ≥70 | 1.21 (0.42–3.48) | 0.719 | |||
| BMI (kg/m2) | <25 | Ref | |||
| ≥25 | 0.56 (0.21–1.48) | 0.245 | |||
| CEA (ng/ml) | <5 | Ref | |||
| ≥5 | 0.59 (0.26–1.33) | 0.205 | |||
| Tumor location | Right colon | Ref | |||
| Left colon | 1.72 (0.73–4.03) | 0.21 | |||
| Complications | No | Ref | |||
| Yes | 1.11 (0.45–2.73) | 0.812 | |||
| Histological grade | G1 and G2 | Ref | |||
| G3, etc. | 1.03 (0.39–2.72) | 0.939 | |||
| LVI | Absent | Ref | Ref | ||
| Present | 2.71 (1.27–5.80) | 0.009 | 2.60 (1.21–5.55) | 0.013 | |
| No data | 1.81 (0.23–14.03) | 0.569 | 1.36 (0.17–10.57) | 0.766 | |
| LN numbers | <12 | Ref | |||
| ≥12 | 1.51 (0.20–11.12) | 0.684 | |||
| Stage | II | Ref | |||
| III | 2.55 (0.60–10.73) | 0.201 | |||
| MSI | MSS/MSI‐low | Ref | |||
| MSI‐high | 1.18 (0.26–5.30) | 0.822 | |||
| No data | 1.29 (0.60–2.77) | 0.502 | |||
| f_wt | Continuous | 0.095 (0.0007–12.16) | 0.342 | ||
| f_im200 | Continuous | 0.003 (0.0001–0.14) | 0.002 | 0.004 (0.0001–0.15) | 0.0028 |
CEA, carcinoembryonic antigen; LN, lymph node; MSS, microsatellite stable; Ref, reference.
Validation of automated TIL features in the TCGA dataset
The median TIL densities of spatial distribution of ‘f_wt’ and ‘f_im200’ were 0.0808 and 0.1085, respectively (supplementary material, Table S7). Among the 268 patients from the included TCGA dataset, age, stage, ‘f_wt’, and ‘f_im200’ were significant prognostic factors in the univariate analysis. Factors with p < 0.1 were entered into the multivariate analysis. Multivariate analysis revealed that ‘f_im200’ retained prognostic significance for PFS (HR 0.031 [95% CI 0.001–0.645], p = 0.024) along with age (HR 1.994 [95% CI 1.233–3.222], p = 0.004) and stage (I and II versus IV, HR 3.342 [95% CI 1.804–6.191], p = 0.0001) (Table 3).
Table 3.
Univariate and multivariable analyses of factors associated with PFS in the TCGA dataset (n = 268)
| Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|
| Variables | HR (95% CI) | p | HR (95% CI) | p | |
| Sex | Female | Ref | |||
| Male | 1.592 (0.992–2.555) | 0.053 | |||
| Age | <70 | Ref | Ref | ||
| ≥70 | 1.708 (1.079–2.705) | 0.022 | 1.994 (1.233–3.222) | 0.004 | |
| BMI (kg/m2) | <25 | Ref | |||
| ≥25 | 1.454 (0.782–2.701) | 0.236 | |||
| No data | 0.896 (0.426–1.887) | 0.774 | |||
| CEA (ng/ml) | <5 | NA | |||
| ≥5 | NA | ||||
| Tumor location | Right colon | Ref | |||
| Left colon | 0.830 (0.504–1.365) | 0.464 | |||
| Rectum | 1.017 (0.428–2.417) | 0.969 | |||
| No data | 1.662 (0.589–4.690) | 0.337 | |||
| Complications | No | NA | |||
| Yes | NA | ||||
| Histological grade | G1 and G2 | NA | |||
| G3, etc. | NA | ||||
| LVI | Absent | Ref | |||
| Present | 1.620 (1.008–2.603) | 0.046 | |||
| No data | 1.338 (0.522–3.424) | 0.543 | |||
| LN numbers | <12 | Ref | Ref | ||
| ≥12 | 0.629 (0.337–1.175) | 0.146 | 0.571 (0.302–1.078) | 0.084 | |
| No data | 0.323 (0.090–1.164) | 0.084 | 0.270 (0.072–1.017) | 0.053 | |
| Stage | I and II | Ref | Ref | ||
| III | 1.598 (0.945–2.702) | 0.080 | 1.557 (0.910–2.662) | 0.105 | |
| IV | 4.062 (2.241–7.361) | <0.001 | 3.342 (1.804–6.191) | 0.0001 | |
| MSI | MSS/MSI‐low | Ref | |||
| MSI‐high | 0.873 (0.457–1.667) | 0.682 | |||
| No data | 1.485 (0.881–2.503) | 0.137 | |||
| f_wt | Continuous | 0.007 (0.0002–0.2793) | 0.0079 | ||
| f_im200 | Continuous | 0.0103 (0.0005–0.1999) | 0.0024 | 0.031 (0.001–0.645) | 0.024 |
CEA, carcinoembryonic antigen; LN, lymph node; MSS, microsatellite stable; NA, not available; Ref, reference.
Inter‐rater agreement of KM grading for Yonsei and TCGA datasets by two pathologists
KM grading was manually performed by two pathologists for the Yonsei (n = 180) and TCGA (n = 249) datasets. Note that some patients (n = 19) from the TCGA dataset were excluded from the analysis due to poor slide quality and/or difficulty by both pathologists in manually finding the IMs. The agreement of KM grading for each dataset by two pathologists was evaluated using kappa statistics.
Inter‐rater agreement of each KM grading was insignificant in the Yonsei dataset (κ = 0.109) and in the TCGA dataset (κ = 0.121). When we repeated kappa statistics using KM‐low (KM gradings 0 and 1) versus KM‐high (KM gradings 2 and 3) groups, insignificant agreement was still observed in the Yonsei dataset (κ = 0.151); however, median agreement was observed in the TCGA dataset (κ = 0.404) (supplementary material, Table S8). Kappa scores for DeepTILs versus manual KM are provided in supplementary material, Table S9. The kappa score indicated insignificant and median agreement between DeepTILs and pathologic evaluation.
Comparison of patient subgrouping based on the DeepTILs and KM grading by the pathologists
We first investigated whether there were statistically significant differences between ‘f_im200’ values across KM grading patient subgroups. The median values of ‘f_im200’ according to the KM gradings 0, 1, 2, and 3 by pathologist 1 were 0.075, 0.142, 0.230, and 0.329, respectively, in the Yonsei dataset (p < 0.001). The median values of ‘f_im200’ according to the KM gradings 1, 2, and 3 by pathologist 2 were 0.055, 0.194, and 0.234, respectively, in the Yonsei dataset (p < 0.001). There was no KM grading 0 by pathologist 2 in the Yonsei dataset.
The median values of ‘f_im200’ according to KM gradings 0, 1, 2, and 3 by pathologist 1 were 0.058, 0.109, 0.142, and 0.225, respectively, in the TCGA dataset (p < 0.001). The median values of ‘f_im200’ according to KM gradings 0, 1, 2, and 3 by pathologist 2 were 0.049, 0.077, 0.123, and 0.179, respectively, in the TCGA dataset (p < 0.001) (supplementary material, Figure S9). These results indicate that patient subgroups with higher KM grading patient by the pathologists carry higher TIL densities within the 200‐μm IM layer by the DeepTILs.
We used the X‐tile program to find an optimal cut‐off value of ‘f_im200’ in the Yonsei dataset [23] and identified 0.14 as the cut‐off value producing the largest χ 2 using the Mantel–Cox test (supplementary material, Figure S10). Based on this cut‐off value, we divided patients from the Yonsei dataset into two subgroups. Specifically, patients with a ‘f_im200’ value higher than 0.14 were included in the TIL‐high subgroup, otherwise they were included in the low subgroup. The same cut‐off value (0.14) was also applied to identify TIL‐high and ‐low subgroups for patients from the TCGA dataset. A total of 71 patients (39.4%) in the Yonsei dataset and 168 patients (62.6%) in the TCGA dataset were allocated into the low DeepTIL group. There was no significant difference in clinicopathological characteristics between patients with low and high DeepTILs in the Yonsei dataset. In contrast, there were statistically significant differences in BMI, LVI, and stage between the two groups in the TCGA dataset (supplementary material, Table S10).
As 249 patients in the TCGA cohort were graded by both pathologists, those 249 patients were included for the survival analysis. When we re‐analyzed the multivariate analysis using the 249 selected patients in the TCGA dataset, f_im200 was again identified as an independent prognostic factor for PFS (HR 0.005 [95% CI 0.001–0.173], p = 0.003) (supplementary material, Table S11). When we evaluated the association between the binary classification of DeepTILs and pathologists' KM grading systems in these 249 patients, as the KM grading was increased, the rate of high grading by DeepTILs was also increased (supplementary material, Figures S11 and S12).
Prognostic utility of spatial TIL features based on DeepTILs and KM grading
We performed Kaplan–Meier survival analysis in the Yonsei cohort, and found that patients in the TIL‐high subgroup (i.e. patients with higher densities defined by the DeepTILs) showed better PFS compared with those patients belonging to the TIL‐low subgroup (log‐rank test, p = 0.0018) (Figure 2A). Interestingly, there were no significant survival differences between KM‐low and KM‐high groups assigned either by pathologist 1 (p = 0.16) or pathologist 2 (p = 0.66) (Figure 2B,C). Based on the same cut‐off value determined in the Yonsei cohort, Kaplan–Meier survival analysis was performed in the TCGA cohort. Those patients with higher densities defined by the DeepTILs showed better PFS compared with patients with lower TIL densities (log‐rank test, p = 0.0026) (Figure 2D). The KM‐high groups defined by pathologists 1 and 2 showed better PFS than the KM‐low groups (log‐rank test, p = 0.001 by pathologist 1 and p = 0.026 by pathologist 2) (Figure 2E,F).
Figure 2.
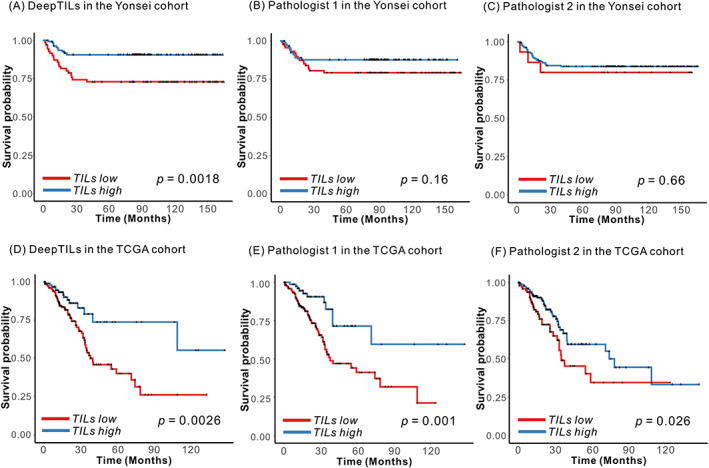
Kaplan–Meier plots for patient subgroup analysis identified by DeepTILs and KM gradings by the pathologists. (A) TIL‐high and ‐low subgroups identified by DeepTILs in the Yonsei cohort. (B, C) TIL‐high and ‐low subgroups identified by KM gradings from (B) pathologist 1 and (C) pathologist 2 in the Yonsei cohort. (D) TIL‐high and ‐low subgroups identified by DeepTILs in TCGA cohort. (E, F) TIL‐high and ‐low subgroups identified by KM gradings from (E) pathologist 1 and (F) pathologist 2 in the TCGA cohort.
Combination of DeepTILs and pathologic grading and its prognostic effect
Lastly, we investigated whether integrating KM grading by the pathologists with TIL subgroups by DeepTILs could improve patient prognostication. Specifically, we grouped patients into four subgroups: TIL high and high, TIL high and low, TIL low and high, and TIL low and low by DeepTILs and the pathologists. We generated Kaplan–Meier plots for four subgroups and performed univariate and multivariate analyses of subgroups (Figure 3 and supplementary material, Table S12). Patients belonging to the TIL‐high subgroup by both KM grading and DeepTILs showed better PFS across datasets, while patients belonging to the TIL‐low subgroup by both approaches showed the poorest PFS. For example, on multivariate analysis using the TCGA dataset, we found that TIL‐high subgroups identified by both approaches showed statistically better PFS compared to TIL‐low subgroups identified by two approaches (e.g. TIL‐high subgroup by pathologist 1 and DeepTILs: HR 0.372 [95% CI 0.154–0.896], p = 0.027, and TIL‐high subgroup by pathologist 2 and DeepTILs: HR 0.472 [95% CI 0.230–0.968], p = 0.040) (supplementary material, Table S12C,D). Another interesting observation is that patients assigned as TIL‐low by KM grading but TIL‐high by DeepTILs showed a trend toward better PFS compared to TIL‐low subgroups by both approaches. However, patients assigned as TIL‐high by KM grading but TIL‐low by DeepTILs showed poorer PFS compared to those of TIL‐high subgroup by both approaches. This might indicate that incorporating TIL subgroups derived by DeepTILs with the pathologists' KM grading could improve patient stratification compared to the KM grading alone.
Figure 3.
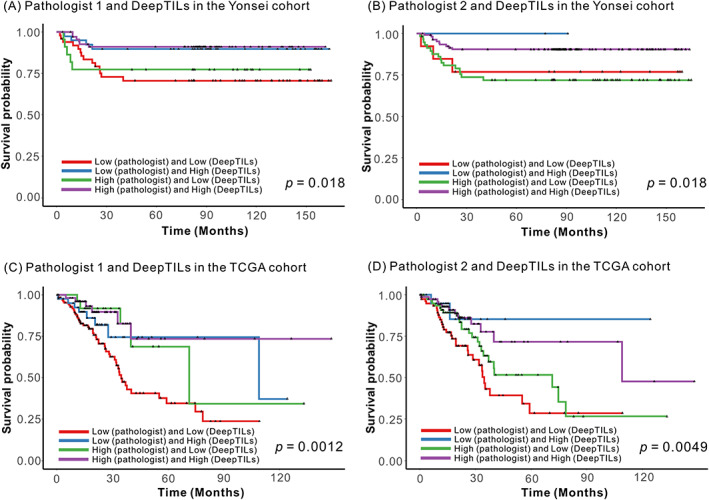
Kaplan–Meier survival analysis based on combination of subgroups derived from KM grading and DeepTILs. Patients were assigned to four subgroups; TILs high and high, TILs high and low, TILs low and high, and TILs low and low using KM grading by two pathologists and DeepTILs in the Yonsei and TCGA CRC cohorts, respectively. (A) Combination of patient subgroups by pathologist 1 and DeepTILs in the Yonsei cohort. (B) Combination of patient subgroups by pathologist 2 and DeepTILs in the Yonsei cohort. (C) Combination of patient subgroups by pathologist 1 and DeepTILs in the TCGA cohort. (D) Combination of patient subgroups by pathologist 2 and DeepTILs in the TCGA cohort.
Discussion
We have shown that the DL models based on H&E‐stained WSIs can quantify TIL densities at the IMs and across regions of surgically resected CRC. In experiments using datasets from the Yonsei and TCGA cohorts, we have shown that TIL densities quantified by DeepTILs could be used to identify subgroups of CRC patients with distinct survival differences. In particular, we performed comprehensive evaluation of TIL densities at various IMs as well as tumor core and whole tumor regions. Our subgroup analysis based on the automatic quantification of TIL densities showed that the patient subgroup with high TIL densities at the 200‐μm IM layer (i.e. ‘f_im200’) has statistically significant better PFS in the Yonsei and TCGA cohorts. However, the subgroup analysis based on pathologists' independent TIL grading only showed statistically significant different PFS outcome in the TCGA cohort, but insignificant difference in the Yonsei cohort.
Pathologists' manual TIL grading on H&E‐ or IHC‐stained slides has to manually identify tumor boundary and its IM, which is time‐consuming and labor‐intensive. In addition, inter‐rater variability in manual TIL grading by pathologists could make the universal application of TIL analysis in routine clinical practice difficult. For example, the agreements of KM scoring using a 4‐grade system between pathologists were insignificant in both the Yonsei and TCGA datasets, which indicates inter‐rater issues for manual grading. The agreement of KM scoring using a 2‐grade system (i.e. KM‐low versus ‐high subgroups) showed that kappa value was increased in the TCGA dataset, but no clear change was observed in the Yonsei dataset. This also indicates that both 2 and 4 manual KM grading systems contain inter‐rater variability.
One strength of our approaches is to reduce inter‐rater variability by evaluating TIL densities across different tumor regions and IMs with fewer biases compared to human grading. Specifically, DL approaches measured TIL density of whole tumor regions and IMs without manually selecting certain regions (i.e. manually selected representative or subsets of tumor regions and/or IMs), which can provide a less subjective measurement of TIL densities. In addition, our approach can effectively measure TIL densities throughout the tumor center and IMs, and thus could provide a comprehensive prognostic evaluation across the whole tumor and its boundary using H&E‐stained WSIs.
The pathologists revisited cases from the TCGA cohort that had different TIL‐high or ‐low grading at the tumor IM by KM grading and DeepTILs. We found that the disagreement was largely due to lack of consensus in the definition of tumor IM (supplementary material, Figure S13). For instance, most of the cases identified as belonging to the TIL‐high subgroup by pathologists had a high‐level TIL quantification within tumors by DeepTILs. However, those cases have few TILs present at the 200‐μm tumor IM, and thus they are graded as TIL‐low by DeepTILs (supplementary material, Figure S13C,D). Similarly, the TIL‐low subgroup by KM grading had few TILs found within the slide, while these cases were identified as belonging to the TIL‐high group by DeepTILs (supplementary material, Figure S13A,B) as those few TILs appeared at the 200‐μm tumor IM. We also found that some inflammatory cells in necrotic debris and/or fibrosis tissue from few cases were detected as TILs by DeepTILs (supplementary material, Figure S13A,B). Although these false positives did not significantly affect patient subgrouping in our study, the DL approaches could be further improved to reduce the false‐positive prediction by learning these patterns.
Recent meta‐analysis showed that manual TIL analysis of certain T‐cells (e.g. CD4, CD8, etc.) in tumor center, stroma, and at the IM based on IHC staining was associated with OS and disease‐free survival [2]. The discrepancies between our findings (i.e. no statistical PFS difference using TIL densities in the tumor center derived by DeepTILs) compared to the recent meta‐analysis could be due our approaches only utilizing H&E‐stained WSIs where we cannot take into account densities of certain types of T‐cells within the tumor regions and at IMs. In addition, our discovery cohort, the Yonsei cohort, was collected from patients who underwent surgical resection at a single institute and could pose ethnic‐specific disease outcome and/or morphological differences across ethnicities [20, 24]. We plan to collect WSIs immunostained for CD3, CD8, and FOXP3 and integrate them with matched H&E‐stained WSIs, to allow more accurate quantification of TIL density and evaluation of prognostic information of TIL features from certain types of T‐cell across the tumor center, tumor core, and at IMs. With additional larger training and testing datasets from multiple centers and international institutes, we will further validate our findings and assess whether TIL densities at the 200‐μm IM layer could serve as a robust prognostic biomarker for patients with CRC. Another limitation was that we did not incorporate morphological features present in tumor and stroma regions with TIL densities at various tumor regions and IMs to correlate with patient outcome. There are several new studies showing that H&E‐stained WSI‐based DL models utilizing morphological features from WSIs can accurately predict CRC patient survival. Incorporating such morphologic features with TIL densities could possibly further improve CRC patient prognostication. Lastly, while we showed that combining manual TIL grading by pathologists and DeepTILs could improve patient subgrouping, we did not attempt to use the DL model as an assistant for pathologists' TIL grading. A recent study showed that systematic incorporation of the DL model to assist physicians in predicting MSI status based on H&E‐stained WSI could provide labor and cost‐saving benefits [25]. Similarly, proper integration of the DL models as guidance for TIL grading could potentially provide similar benefits.
Taken together, we have developed DL approaches for TIL detection and their spatial quantification in H&E‐stained WSIs. Our analyses indicate that automatic TIL grading could identify CRC patient subgroups with distinct PFS. The analyses also indicate that the DeepTILs could address the inter‐rater disagreements of TIL evaluation by pathologists in H&E‐stained slides. Finally, we have shown that combining the DL‐based TIL subgroup with KM grading could improve patient prognostication. Our trained TIL detector has the potential to be deployed in the clinical setting, which could help pathologists in TIL identification and quantification. In future, we plan to test and validate the DL approaches for spatial TIL quantification through a larger number and diversity of datasets as well as compare the IHC‐based scoring system to help in selecting CRC patients with high or low risk.
Author contributions statement
JK, SHL and THH conceived the study, collected the data, performed the analysis, verified the results, supervised the project and wrote the manuscript. YJC and SHL collected the data, reviewed the slides, performed the analyses, verified the results and wrote the manuscript with support from JK and THH. HX developed and implemented the algorithms, performed the analysis and wrote the manuscript with support from JK, SHL and THH. JC and JRC assisted with implementation of the algorithms, performed the analysis and wrote the manuscript with support from JK, SHL and THH.
Supporting information
Figure S1. Patient inclusion in this study from the Yonsei and TCGA datasets
Figure S2. Independent testing of tumor detectors
Figure S3. Examples of tumor and TIL detections
Figure S4. Independent testing of TIL detectors
Figure S5. Examples of TIL prediction in the testing patches from the public dataset
Figure S6. Visualizing features learned by the TIL detector with the guided backpropagation method
Figure S7. Illustration of tumor IMs based on manually labeled contours
Figure S8. Two representative cases with different interobserver agreement
Figure S9. Distribution of f_im200 according to KM grading measured by pathologists 1 and 2 using the Yonsei and the TCGA datasets
Figure S10. Determining cut‐off values of ‘f_im200’ using the X‐tile program in the Yonsei dataset
Figure S11. Distribution of DeepTILs by KM 4 grading and by the two pathologists
Figure S12. Distribution of DeepTILs by KM‐low and ‐high, and by the two pathologists
Figure S13. TCGA CRC patients who were assigned to different TIL subgroups by KM grading and DeepTILs
Table S1. Grid search of parameter settings for Resnet18 based tumor detectors
Table S2. Grid search of parameter settings for finding the best TIL detector
Table S3. Descriptions of computed TIL spatial distribution variables
Table S4. Composition of TILs according to spatial distribution in the Yonsei dataset
Table S5. Univariate Cox proportional hazard ratio of PFS in the Yonsei dataset
Table S6. Multivariate analysis of factors associated with PFS using spatial TIL densities in the Yonsei dataset
Table S7. Composition of TILs according to spatial distribution in the TCGA dataset
Table S8. Comparison of kappa values between the two pathologists using validated slides in the Yonsei and TCGA datasets
Table S9. Comparison of kappa values between DeepTILs and the two pathologists using validated slides in the Yonsei and TCGA datasets
Table S10. Patient characteristics according to low and high DeepTILs in the Yonsei and TCGA datasets
Table S11. Univariate and multivariable analyses of factors associated with PFS in the TCGA dataset after selection by the pathologists
Table S12. Multivariate analysis of factors associated with PFS in the Yonsei and TCGA datasets using combination of KM grading by the pathologists and DeepTILs
Acknowledgements
The authors thank Medical Illustration & Design, part of the Medical Research Support Services of Yonsei University College of Medicine, for all artistic support related to this work.
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2020R1G1A1102555). This work was also partially supported by the Fundamental Research Funds for the Central Universities in China (No. DUT21RC(3)038) funded by Dalian University of Technology, and Youth Fund (No. 82102135) funded by National Natural Science Foundation of China (NSFC).
Conflict of interest statement: THH is the co‐founder of KURE.AI and received consulting fees and research funding from AITRICS. All other authors declare no conflicts of interest.
Contributor Information
Sung Hak Lee, Email: hakjjang@gmail.com.
Jeonghyun Kang, Email: ravic@naver.com.
Tae Hyun Hwang, Email: hwang.taehyun@mayo.edu.
Data availability statement
Source code used for this study is available at the following link: https://github.com/hwanglab/TILs_Analysis
References
Reference 26 is cited only in the supplementary material.
- 1. NCCN Guidelines® . National Comprehensive Cancer Network (NCCN) Guidelines for Treatment of Cancer by Site. 2020. [Accessed 30 June 2020]. Available from: https://www.nccn.org/professionals/physician_gls/default.aspx
- 2. Idos GE, Kwok J, Bonthala N, et al. The prognostic implications of tumor infiltrating lymphocytes in colorectal cancer: a systematic review and meta‐analysis. Sci Rep 2020; 10: 3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Orhan A, Vogelsang RP, Andersen MB, et al. The prognostic value of tumour‐infiltrating lymphocytes in pancreatic cancer: a systematic review and meta‐analysis. Eur J Cancer 2020; 132: 71–84. [DOI] [PubMed] [Google Scholar]
- 4. Cha YJ, Park EJ, Baik SH, et al. Clinical significance of tumor‐infiltrating lymphocytes and neutrophil‐to‐lymphocyte ratio in patients with stage III colon cancer who underwent surgery followed by FOLFOX chemotherapy. Sci Rep 2019; 9: 11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sinicrope FA, Graham RP. Tumor‐infiltrating lymphocytes for prognostic stratification in nonmetastatic colon cancer – are we there yet? JAMA Oncol 2021; 7: 969–970. [DOI] [PubMed] [Google Scholar]
- 6. Pagès F, Mlecnik B, Marliot F, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet 2018; 391: 2128–2139. [DOI] [PubMed] [Google Scholar]
- 7. Rozek LS, Schmit SL, Greenson JK, et al. Tumor‐infiltrating lymphocytes, Crohn's‐like lymphoid reaction, and survival from colorectal cancer. J Natl Cancer Inst 2016; 108: djw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huh JW, Lee JH, Kim HR. Prognostic significance of tumor‐infiltrating lymphocytes for patients with colorectal cancer. Arch Surg 2012; 147: 366–372. [DOI] [PubMed] [Google Scholar]
- 9. Roxburgh CS, Salmond JM, Horgan PG, et al. Comparison of the prognostic value of inflammation‐based pathologic and biochemical criteria in patients undergoing potentially curative resection for colorectal cancer. Ann Surg 2009; 249: 788–793. [DOI] [PubMed] [Google Scholar]
- 10. Klintrup K, Mäkinen JM, Kauppila S, et al. Inflammation and prognosis in colorectal cancer. Eur J Cancer 2005; 41: 2645–2654. [DOI] [PubMed] [Google Scholar]
- 11. Nagtegaal ID, Marijnen CA, Kranenbarg EK, et al. Local and distant recurrences in rectal cancer patients are predicted by the nonspecific immune response; specific immune response has only a systemic effect – a histopathological and immunohistochemical study. BMC Cancer 2001; 1: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corredor G, Wang X, Zhou Y, et al. Spatial architecture and arrangement of tumor‐infiltrating lymphocytes for predicting likelihood of recurrence in early‐stage non‐small cell lung cancer. Clin Cancer Res 2019; 25: 1526–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saltz J, Gupta R, Hou L, et al. Spatial organization and molecular correlation of tumor‐infiltrating lymphocytes using deep learning on pathology images. Cell Rep 2018; 23: 181–193.e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bankhead P, Loughrey MB, Fernández JA, et al. QuPath: open source software for digital pathology image analysis. Sci Rep 2017; 7: 16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Acs B, Ahmed FS, Gupta S, et al. An open source automated tumor infiltrating lymphocyte algorithm for prognosis in melanoma. Nat Commun 2019; 10: 5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yoo SY, Park HE, Kim JH, et al. Whole‐slide image analysis reveals quantitative landscape of tumor‐immune microenvironment in colorectal cancers. Clin Cancer Res 2020; 26: 870–881. [DOI] [PubMed] [Google Scholar]
- 17. AbdulJabbar K, Raza SEA, Rosenthal R, et al. Geospatial immune variability illuminates differential evolution of lung adenocarcinoma. Nat Med 2020; 26: 1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA. 2016; 770–778. 10.1109/CVPR.2016.90. [DOI]
- 19. Kather JN, Krisam J, Charoentong P, et al. Predicting survival from colorectal cancer histology slides using deep learning: a retrospective multicenter study. PLoS Med 2019; 16: e1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kather JN, Pearson AT, Halama N, et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med 2019; 25: 1054–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang X, Zhou X, Lin M, et al. Shufflenet: an extremely efficient convolutional neural network for mobile devices. In 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA. 2018; 6848–6856. 10.1109/CVPR.2018.00716. [DOI]
- 22. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977; 33: 159–174. [PubMed] [Google Scholar]
- 23. Camp RL, Dolled‐Filhart M, Rimm DL. X‐tile: a new bio‐informatics tool for biomarker assessment and outcome‐based cut‐point optimization. Clin Cancer Res 2004; 10: 7252–7259. [DOI] [PubMed] [Google Scholar]
- 24. Lee W, Nelson R, Mailey B, et al. Socioeconomic factors impact colon cancer outcomes in diverse patient populations. J Gastrointest Surg 2012; 16: 692–704. [DOI] [PubMed] [Google Scholar]
- 25. Yamashita R, Long J, Longacre T, et al. Deep learning model for the prediction of microsatellite instability in colorectal cancer: a diagnostic study. Lancet Oncol 2021; 22: 132–141. [DOI] [PubMed] [Google Scholar]
- 26. Springenberg JT, Dosovitskiy A, Brox T, et al. Striving for simplicity: the all convolutional net. arXiv 2014: 1412.6806 [Not peer reviewed]. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Patient inclusion in this study from the Yonsei and TCGA datasets
Figure S2. Independent testing of tumor detectors
Figure S3. Examples of tumor and TIL detections
Figure S4. Independent testing of TIL detectors
Figure S5. Examples of TIL prediction in the testing patches from the public dataset
Figure S6. Visualizing features learned by the TIL detector with the guided backpropagation method
Figure S7. Illustration of tumor IMs based on manually labeled contours
Figure S8. Two representative cases with different interobserver agreement
Figure S9. Distribution of f_im200 according to KM grading measured by pathologists 1 and 2 using the Yonsei and the TCGA datasets
Figure S10. Determining cut‐off values of ‘f_im200’ using the X‐tile program in the Yonsei dataset
Figure S11. Distribution of DeepTILs by KM 4 grading and by the two pathologists
Figure S12. Distribution of DeepTILs by KM‐low and ‐high, and by the two pathologists
Figure S13. TCGA CRC patients who were assigned to different TIL subgroups by KM grading and DeepTILs
Table S1. Grid search of parameter settings for Resnet18 based tumor detectors
Table S2. Grid search of parameter settings for finding the best TIL detector
Table S3. Descriptions of computed TIL spatial distribution variables
Table S4. Composition of TILs according to spatial distribution in the Yonsei dataset
Table S5. Univariate Cox proportional hazard ratio of PFS in the Yonsei dataset
Table S6. Multivariate analysis of factors associated with PFS using spatial TIL densities in the Yonsei dataset
Table S7. Composition of TILs according to spatial distribution in the TCGA dataset
Table S8. Comparison of kappa values between the two pathologists using validated slides in the Yonsei and TCGA datasets
Table S9. Comparison of kappa values between DeepTILs and the two pathologists using validated slides in the Yonsei and TCGA datasets
Table S10. Patient characteristics according to low and high DeepTILs in the Yonsei and TCGA datasets
Table S11. Univariate and multivariable analyses of factors associated with PFS in the TCGA dataset after selection by the pathologists
Table S12. Multivariate analysis of factors associated with PFS in the Yonsei and TCGA datasets using combination of KM grading by the pathologists and DeepTILs
Data Availability Statement
Source code used for this study is available at the following link: https://github.com/hwanglab/TILs_Analysis


