Abstract
Ovarian cancer (OC) is the most fatal gynecological malignancy because of its early asymptomatic nature and acquired resistance to chemotherapy. Small extracellular vesicles (sEVs) are a heterogeneous group of biological vesicles with a diameter <200 nm released by cells under physiological or pathological conditions. sEVs-derived non-coding RNAs (ncRNAs) are the essential effectors in the biological environment. sEVs-ncRNAs have critical roles in tumor progression via regulating mRNA expression of target cells to affect cell signaling. In addition, the status of parental cells can be disclosed via analyzing the composition of sEVs-ncRNAs, and their “cargoes” with specific changes can be used as key biomarkers for the diagnosis and prognosis of OC. Accumulating evidence has demonstrated that sEVs-ncRNAs are involved in multiple key processes that mediate the development of metastasis and chemotherapeutic resistance in OC: epithelial–mesenchymal transition; tumorigenicity of mesenchymal stem cells; immune evasion; angiogenesis. The nanomedicine delivery system based on engineering sEVs is expected to be a novel therapeutic strategy for OC. Insights into the biological roles of sEVs-ncRNAs in the invasion, metastasis, immune regulation, and chemoresistance of OC will contribute to discovery of novel biomarkers and molecular targets for early detection and innovative therapy. In this review, we highlight recent advances and applications of sEVs-ncRNAs in OC diagnosis and treatment. We also outline current challenges and knowledge gaps.
Keywords: ovarian cancer, small extracellular vesicles, non-coding RNA, biomarker, therapy, drug delivery
Introduction
Ovarian cancer (OC) is the seventh most common cancer in women with a 5-year survival of <50% (Siegel et al., 2021). Often, OC is referred to as the “silent killer”, and its pathological and molecular features are highly heterogeneous (Torre et al., 2018). Over 70% of OC cases are diagnosed at stage III or IV with widespread abdominal metastasis. The 5-year survival of patients with stage-I OC can exceed 90%. The 5-year survival of patients with stage-III–IV disease after aggressive debulking surgery and platinum-based chemotherapy is only 30% (Matulonis et al., 2016; Kuroki and Guntupalli, 2020). High-grade serous ovarian cancer (HGSOC) accounts for 70–80% of OC deaths, because most of them relapse and, ultimately, develop resistance to chemotherapy (Yang et al., 2011). HGSOC is characterized by ubiquitous TP53 mutation (96%), significant focal DNA copy number aberrations (e.g., CCNE1, MYC, MECOM, RB1, and NF1), and homologous recombination deficiency (HRD) in about half of cases (Network Cancer Genome Atlas Research, 2011). In addition, approximately 13% of HGSOC cases can be attributed to germline breast cancer gene 1 (gBRCA1) or gBRCA2 mutation (Pal et al., 2005).
The high mortality of OC is associated with the difficulty of obtaining an early diagnosis and chemoresistance-related relapse. Due to the limited expression of CA125 in early-stage OC and its correlation with non-malignant diseases (e.g., endometriosis and pelvic tuberculosis), its positive predictive value is only 40% (Menon et al., 2009; Menon et al., 2021). An effective and accurate screening strategy for OC is not available. The first-line maintenance therapy based on poly-ADP ribose polymerase inhibitor (PARPi) has become an indispensable part of OC clinical management. Regardless of gBRCA mutations or HRD status, PARPi can prolong the median progression-free survival of patients with recurrent OC to some extent (Mirza et al., 2016; Franzese et al., 2019). Unfortunately, most OC patients with platinum-resistant develop resistance to PARPi eventually. Consequently, there is an urgent need to: 1) further clarify the mechanisms of OC with widespread metastasis and chemoresistance; 2) develop novel and reliable biomarkers and molecular targets for an early diagnosis and treatment of OC.
Small extracellular vesicles (sEVs) are submicron-sized vesicles derived from cell endosomes. They have become an alternative source of biomarkers for various diseases (Wong et al., 2019; Guo et al., 2020; Alharbi et al., 2021). sEVs are found in multiple biological fluids (e.g., blood, saliva, urine, and cerebrospinal fluid) and enrich RNA species that represent specific biotypes of source cells, especially non-coding RNAs (ncRNAs) (Möller and Lobb, 2020). The latter are RNAs that do not encode specific proteins but instead act as essential regulators of physiological processes in developmental and disease environments. It has been estimated that ncRNAs regulate the translation of ∼60% of protein-coding genes (Anastasiadou et al., 2018). Increasing evidence suggests that sEVs participate closely in the communication between cancer cells and the tumor microenvironment (TME) by selectively delivering ncRNA “cargo” to alter the genes expression and phenotypic responses of recipient cells (Hoshino et al., 2015; O’Brien et al., 2020; Grunberg et al., 2021). As pathological effectors, sEVs-ncRNAs have critical roles in a variety of cancer hallmarks in OC: oncogenic reprogramming (Kanlikilicer et al., 2016); tumor stem cells maintenance (Cao et al., 2021), DNA damage repair (Chen et al., 2021), and chemotherapy resistance (Alharbi et al., 2021). Owing to their natural origin, sEVs possess excellent biocompatibility, low immunogenicity, enhanced stability, and low toxicity. sEVs have the potential to be novel nanocarriers for drugs compared with traditional nanoscale drug carriers (e.g., liposomes and synthetic nanoparticles) (Zhang et al., 2020d; Herrmann et al., 2021; Jang et al., 2021). Given these biochemical advantages, elucidating the relationship between OC and delivery of sEVs-ncRNAs may aid the development of biomarkers for an early diagnosis and the establishment of novel molecular-targeted therapies.
Due to research priorities in recent years, we will mainly focus on a subtype of sEVs named “exosomes”. However, most experimental methods do not include a sufficiently detailed description of the exosome isolation process to demonstrate their intracellular origins and biogenesis, which may contain heterogeneous EVs populations of different biological origins. Therefore, sEVs are employed to designate EVs <200 nm in diameter, and the separate term of exosome will be employed only in specific descriptions or original publications.
In this review, we summarize the latest development of sEVs-derived ncRNAs as OC biomarkers and therapeutic targets in diagnosis, surveillance, and treatment. We highlight the roles of ncRNAs delivery via sEVs in the pathogenesis and progression of OC, the potential translational applications of engineering sEVs in OC treatment, as well as the challenges and possible knowledge gaps that will be faced.
Biogenesis and “Packaging” of sEVs
“EV” is a generic term for particles with no functional nucleus naturally released from all cells in eukaryotes and prokaryotes. This definition is endorsed by the International Society for Extracellular Vesicles (ISEV) (Lotvall et al., 2014; Thery et al., 2018). The intercellular transfer of EVs has garnered extensive attention as a novel third mechanism of intercellular communication (in addition to direct cell-to-cell contact and secretion of growth factors and cytokines) (Andaloussi et al., 2013; Tkach and Thery, 2016; Raposo and Stahl, 2019). Activated vesicles were first discovered in 1983 during the in vitro culture of sheep reticulocytes, and were named “exosomes” by Johnstone et al. (1987). Exosomes were originally thought to be cellular “debris”: a way for cells to excrete waste. In recent years, the EV-research community has reached a consensus to categorize EVs broadly into three main types depending on their biogenesis mechanisms (Colombo et al., 2014). Apoptotic bodies are the largest EVs (diameter = 800–5,000 nm) and are formed from budding cells undergoing apoptosis. Mid-sized EVs (e.g., microvesicles, microparticles, and ectosomes) are produced by direct outward budding of the plasma membrane, and range in diameter from 100 to 1,000 nm. Exosomes are the smallest EVs (40–160 nm in diameter). They are exocytosed intraluminal vesicles (ILVs) released by the fusion of multivesicular bodies (MVBs) with the cell membrane (Kalluri and LeBleu, 2020).
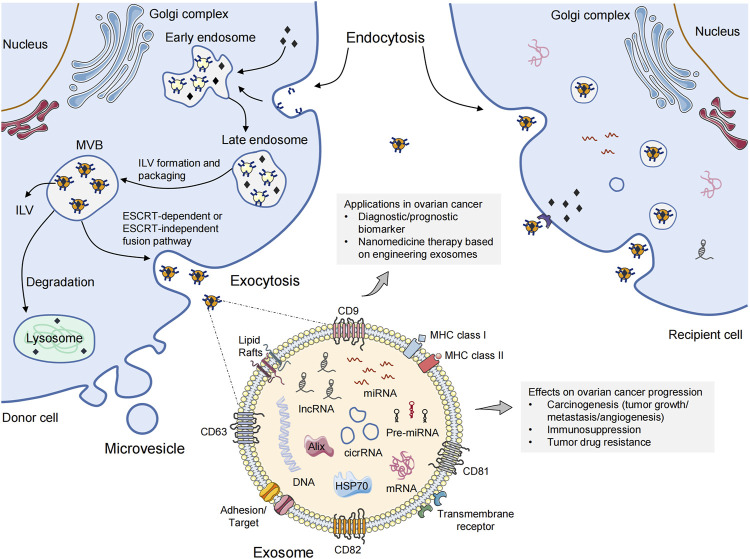
The endosomal sorting complex required for transport (ESCRT) pathway is the most well-known pathway in exosomal biogenesis and cargo classification (Maas et al., 2017; van Niel et al., 2018). This pathway mainly involves four core complexes (ESCRT 0, I, II, and III) that identify ubiquitinated membrane proteins and induce membrane budding, which leads to the formation of ILVs and MVBs (Morita et al., 2007). Briefly, ESCRT-0 recognizes and sorts ubiquitinated endosomal membrane proteins. Further binding to ESCRT-I and ESCRT-II causes inward budding of the endosomal membrane to allow capture of different cargoes (Katzmann et al., 2001). Then, the ESCRT-III complex undergoes deubiquitination and cleaves the budding vesicles (Chiaruttini et al., 2015). This process is coordinated by multiple ancillary proteins in the ESCRT pathway, including ALG2-interacting protein X (ALIX), tumor susceptibility gene 101 (TSG101), and L domain-containing proteins (Christ et al., 2017). These budding vesicles form early endosomes and eventually mature into late endosomes and MVBs. Most MVBs are degraded via fusion with lysosomes. Only a few MVBs containing CD63 and lysosomal-associated membrane protein 1 (LAMP1) fuse with the plasma membrane to release exosomes exocytotically, which is facilitated by Rab guanosine triphosphatase (GTPase) 27a and 27b (Ostrowski et al., 2010). In addition, cells can also generate ILVs and MVBs independent of the ESCRT pathway through lipids, ceramides, tetraspanins, or heat-shock proteins (HSPs) (Shao et al., 2018). These specific membrane transport and fusion processes lead to the presence of typically marker proteins in exosomes. Such marker proteins for exosome recognition include CD9, CD81, CD63, TSG101, ALIX, and HSP70 (Pegtel and Gould, 2019). Exosomes also contain unique cell function-related proteins. For example, major histocompatibility complex (MHC) class-I or class-II molecules are abundant in exosomes derived from antigen-presenting cells (APCs) (Andre et al., 2002). Exosomes secreted by epithelial tumor cells carry epithelial cell adhesion molecule (EpCAM) (Takao et al., 2018). Exosomes released from breast cancer (Ciravolo et al., 2012) and gastric cancer (Baran et al., 2010) cells express human epidermal receptor family proteins. Importantly, the distinct nucleic-acid, protein, and lipid contents of exosomes are not merely a reflection of donor cells. Studies have shown that the expression profile of exosomal cargoes is different from that of the original cells to some extent, which indicates a highly controlled sorting process (Zaborowski et al., 2015). The interaction between exosomes and recipient cells is dependent upon specific glycans, lipids, proteins, and total negative charge on their membrane surface. The cellular-uptake pathways of exosomes mainly include receptor-mediated endocytosis, clathrin interaction, micropinocytosis, direct fusion, and interaction between “lipid rafts”. The biogenesis and cellular-uptake mechanisms of EVs are presented in Figure 1.
FIGURE 1.
The biogenesis and cellular-uptake of extracellular vesicles. Microvesicles are released directly from the plasma membrane via outward budding. The biogenesis of exosomes follows the endocytic-exocytic pathway, inward budding from the plasma membrane to form early endosomes. Late endosomal membrane invagination results in the loading of cytosolic proteins/RNAs, which further forms the exosomal precursor–intraluminal vesicles (ILVs) in multivesicular bodies (MVBs). Exosomes are then secreted into the extracellular space after fusion with the plasma membrane via an ESCRT-dependent or ESCRT-independent pathway. Exosomes can be taken up by recipient cells through endocytosis, direct fusion, or binding to surface proteins, causing the transfer of oncogenic cargoes, and changing the biological behaviors of recipient cells. A multitude of non-coding RNA species (e.g., miRNAs, circRNAs, lncRNAs) are contained in exosome. The roles of exosomes in ovarian cancer are also shown. ESCRT, endosomal sorting complex required for transport; MHC, major histocompatibility complex; miRNA, microRNA; circRNA, circular RNA; lncRNA, long non-coding RNA.
Assigning EVs to specific biogenerative pathways is extraordinarily difficult unless they are captured via the real-time imaging system during release (Gould and Raposo, 2013). Therefore, if reliable markers of subcellular origin cannot be established, ISEV recommends using size-based classification terms for EV subtypes, such as “sEVs” and “medium/large EVs” (Thery et al., 2018). Additionally, technical problems hinder the extraction and purification of sEVs to some extent (Tian et al., 2020). Protein aggregates, cell debris, viruses, and liposomes must be removed to purify sEVs completely. There are five main methods for sEVs isolation: differential ultracentrifugation (Thery et al., 2006); sucrose and iodixanol density ultracentrifugation (Kowal et al., 2016); size-exclusion chromatography (Hong et al., 2016); polyethylene-glycol precipitation (Weng et al., 2016); and immunoaffinity capture (Cai et al., 2018). In the future, in-depth understanding of the biogenesis and packaging mechanisms of sEVs may have vital roles in optimization of isolation methods.
Functional Delivery of ncRNAs by sEVs
Non-coding nucleic acid sequences (which do not encode proteins) account for about 98% of the human genome. In general, the biological complexity of organisms is positively correlated with these non-protein-coding sequences, which are widely transcribed into a large number of ncRNAs (Esteller, 2011; Slack and Chinnaiyan, 2019). During the biogenesis of sEVs, cellular-vesiculation mechanisms package multiple ncRNAs into different EVs subclasses to form a substantial pool of extracellular ncRNAs (Fabbiano et al., 2020). With the development of high-throughput sequencing, a multitude of ncRNA species contained in sEVs have been identified, including some highly conserved (e.g., microRNAs [miRNAs], circular RNAs [circRNAs]), as well as others lacking conservation across species (e.g., long non-coding RNAs [lncRNAs]) (Pegtel and Gould, 2019). Agilent Bioanalyzer 2,100 analysis shows that sEVs have a higher content of small RNAs (miRNA) than large EVs, and no prominent ribosomal RNA peaks (Crescitelli et al., 2020).
Small size, high abundance, and targeted localization aid sEVs-mediated intercellular transport of specific ncRNAs (Das et al., 2019). The lipid bilayer of sEVs can protect ncRNAs from RNase degradation during transport (Min et al., 2019). Once sEVs-ncRNAs escape the degradation pathway and are delivered to recipient cells, they can elicit the corresponding functional response. There may be a functional correlation between sEVs biogenesis and miRNA-regulated mRNA silencing (Murphy et al., 2019; Garcia-Martin et al., 2022). Gibbings et al. (2009) have found that GW182, an important component of miRNA-containing RNA-induced silencing complex (miRISC), is enriched in monocyte-derived exosomes. Moreover, depletion of some ESCRT-related components (ALIX, vacuolar protein-sorting-associated protein 36 [VPS36], and hepatocyte growth factor-regulated tyrosine kinase substrate [HRS]) reduce miRNA activity. Hence, interference with the integrity of ESCRT limits miRNA function, possibly via altering MVBs uptake of GW182. Mittelbrunn et al. (2011) have discovered a novel mechanism of cellular communication: CD63 + exosomes transfer miRNAs unidirectionally between cells using immunological synapses. They also have found that T cells-, B cells-, and dendritic cells-derived exosomes contained miRNA repertoires that are different from their parent cells. Targeting neutral sphingomyelinase-2 can inhibit exosomes production and destroy the transfer of miRNAs to APCs. Other EV-modification signals (e.g., ubiquitination, uridylation, phosphorylation, and sumoylation) also affect the splicing and translation of RNA, and miRNA biogenesis (Mukherjee et al., 2016; Montaldo et al., 2021). The effect of these mechanisms on RNA packaging of sEVs depends largely on particular RNA species, and how cells regulate RNA-cargo loading of sEVs is not known. Thus, a deeper understanding of the complex network of sEVs-ncRNAs synergies may provide a unique opportunity to design better diagnostic methods and therapeutic interventions for OC.
sEVs-ncRNAs in Oncogenic Alterations of OC
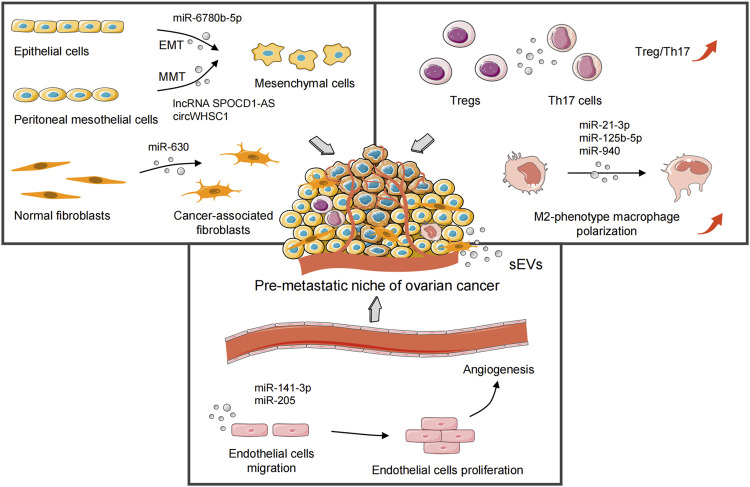
In general, OC is characterized by widespread metastatic growth in the peritoneal cavity and accumulation of malignant ascites (Feng W. et al., 2019). Studies have shown an abundance of exosomes in the ascites of OC patients. They promote OC progression via clearing the interstitial barrier and remodeling the peritoneal environment (Graves et al., 2004; Yamamoto et al., 2018). Oncogenic ncRNAs can be transmitted horizontally to recipient cells in local and distant microenvironments via sEVs. Then, sEVs mediate formation of the pre-metastatic niche of OC by local stromal remodeling (Wang C. et al., 2021), immunosuppression and evasion (Zhou et al., 2018), and angiogenesis (He et al., 2019) (Figure 2). This sEVs-mediated pre-metastatic microenvironment is coordinated through reciprocal interplay with multiple components in the TME, including the extracellular matrix (chemokines and proinflammatory cytokines) and stromal cells (peritoneal mesothelial cells [PMCs], fibroblasts, macrophages, and endothelial cells) (Hoshino et al., 2015). Furthermore, the release of cancer-derived and TME-derived sEVs is increased under various types of microenvironmental crosstalk in inflammation (Li G. et al., 2021), oxygen tension (Dorayappan et al., 2018; Lian et al., 2020), and the therapeutic pressures from irradiation (Zhang F. et al., 2021) or chemotherapy (Zang et al., 2021), and the biomolecular properties of sEVs-ncRNAs are changed accordingly. Table 1 summarizes the relevant studies of sEVs-ncRNAs in OC carcinogenesis.
FIGURE 2.
sEVs-derived oncogenic ncRNAs mediate formation of the pre-metastatic niche of ovarian cancer by local stromal remodeling, immunosuppression and evasion, and angiogenesis. sEVs, small extracellular vesicles; EMT, epithelial–mesenchymal transition; MMT, mesothelial–mesenchymal transition; Tregs, regulatory T cells; Th17 cells, T helper 17 cells.
TABLE 1.
Summary of sEVs-ncRNAs involved in OC carcinogenesis.
| sEVs-ncRNAs | Source | Recipient Cells | Roles/Mechanisms | References |
|---|---|---|---|---|
| miR-6780b-5p | Ascites | OC cells | Induces epithelial–mesenchymal transition of OC cells | Cai et al. (2021) |
| miR-330-3p | Plasma cells | OC cells | Promotes core epithelial-mesenchymal transition programs | Yang et al. (2021b) |
| lncRNA SPOCD1-AS | OC cells | Peritoneal mesothelial cells | Remodels mesothelial cells via interacting with G3BP1 | Wang et al. (2021a) |
| circWHSC1 | OC cells | Peritoneal mesothelial cells | Sponges miR-145 and miR-1182 to promote proliferation and invasion of OC cells | Zong et al. (2019) |
| circPUM1 | OC cells | Peritoneal mesothelial cells | Sponges miR-615-5p and miR-6753-5p to inhibit apoptosis via up-regulating NF-κB and MMP2 | Guan et al. (2019) |
| miR-630 | OC cells | Normal fibroblasts | Facilitates CAFs activation | Cui et al. (2021) |
| miR-221-3p | M2-phenotype macrophages | OC cells | Promotes proliferation and G1/S transition of OC cells | Li and Tang, (2020) |
| miR-21-3p, miR-125b-5p, miR-181d-5p | ||||
| miR-940 | Hypoxic OC cells | Macrophages | Induce polarization of M2-phenotype macrophages | (Chen et al., 2017; Chen et al., 2018) |
| miR-29a-3p | ||||
| miR-21-5p | TAMs | CD4+ T cells | Suppress STAT3 and result in the imbalance of Treg/Th17 cells | Zhou et al. (2018) |
| miR-141-3p | OC cells | HUVECs | Promotes endothelial cells migration and angiogenesis | Masoumi-Dehghi et al. (2020) |
| miR-205 | OC cells | HUVECs | Increases microvessel density | He et al. (2019) |
OC, ovarian cancer; CAFs, cancer-associated fibroblasts; TAMs, tumor-associated macrophages; HUVECs, human umbilical vein endothelial cells.
OC is different from other neoplasms in that it prefers to invade the abdominal cavity through ascites. This strategy makes OC particularly adept at involving the omentum and various abdominal organs (Lheureux et al., 2019). Exosomal miR-21 derived from OC ascites regulates programmed cell death four to facilitate oncogenic transformation in distant target cells without direct colonization by tumor cells (Cappellesso et al., 2014). Ascites-derived exosomal miR-6780b-5p promotes the epithelial–mesenchymal transition (EMT) of OC cells, a key step in the invasion and metastasis (Cai et al., 2021). In addition, similar to epithelial cells that acquire fibroblast-like motility and phenotype in EMT, “mesothelial–mesenchymal transition” (MMT) refers to PMCs being converted into cancer-associated fibroblasts (CAFs) with a mesenchymal phenotype, which leads to tissue fibrosis and peritoneal adhesions (Si et al., 2019; Yang Z. G. et al., 2021). The lncRNA SPOCD1-AS is derived from the sEVs of OC. It remodels PMCs to induce the MMT phenotype by interacting with Ras-GTPase-activating protein-binding protein 1 (G3BP1), which promotes peritoneal implantation and dissemination of OC cells (Wang C. et al., 2021). circWHSC1 shows high expression in OC cells. It acts on the peritoneal mesothelium in the form of exosomes to induce tumor metastasis through sponging miR-145 and miR-1182 (Zong et al., 2019). Exosomal circPUM1 promotes peritoneal metastasis of OC via increasing NF-κB and MMP-2 expression in PMCs (Guan et al., 2019). In addition, OC cells-secreted exosomal miR-630 can induce normal fibroblasts transformation into CAFs by remodeling the extracellular matrix, and trigger tumor progression via targeting Kruppel-like factor 6 (Cui et al., 2021).
Tumor-associated macrophages (TAMs), regulatory T cells (Tregs), and T helper 17 (Th17) cells in the immune microenvironment are also crucial for OC progression (Gordon and Martinez, 2010). M2-phenotype TAMs remodel the pre-metastatic TME by suppressing the adaptive immune response (Li and Tang, 2020; Zhao et al., 2022). The Treg/Th17 ratio in the peritoneum of patients with metastatic OC is significantly higher than that in benign ovarian tumors and a benign peritoneum (Zhou et al., 2018). Moreover, Szajnik et al. (2010) have shown that tumor-derived exosomes in OC patients enhance the anti-apoptotic ability of Tregs, which may promote immune evasion of tumor cells. Hypoxia-induced overexpression of miR-21-3p, miR-125b-5p (Chen et al., 2018), and miR-940 (Chen et al., 2017) in OC cells-derived exosomes stimulate the polarization of M2-phenotype macrophages. Interestingly, Hu et al. (2017)have observed that exosomes secreted from tumor necrosis factor-like weak inducer of apoptosis (TWEAK)-stimulated macrophages can be internalized by OC cells and block metastasis thanks to miR-7 inhibiting EGFR/AKT/ERK pathway.
Vascular endothelial growth factor receptor 1 (VEGFR1)+ hematopoietic progenitor cells have initiated angiogenesis before tumor colonization to a metastatic site (Ribatti et al., 2007). This antecedent angiogenic microenvironment enables the pre-metastatic niche to meet the nutritional requirements for subsequent rapid metastasis and growth of tumor cells (Azarin et al., 2015). Furthermore, OC-derived exosomes advance angiogenesis by triggering migration of vascular endothelial cells, which contributes to tumor cells colonization into the pre-metastasis niche (Tang et al., 2018). Zhang X. et al. (2021) have found that prokineticin receptor-1(PKR1)+ exosomes also facilitate migration of vascular endothelial cells and tube formation by STAT3 phosphorylation. Masoumi-Dehghi et al. (2020) have suggested that epithelial ovarian cancer cells-derived sEVs can activate the intracellular reactive oxygen species (ROS)-dependent NF-κB signaling of endothelial cells. sEVs-miR-141-3p inhibits the suppressors of cytokine signaling-5 (SOCS-5) expression, thereby leading to up-regulation of VEGFR-2. Another study has found that OC cells-secreted exosomal miR-205 promotes metastatic progression and microvessel density by regulating the PTEN-AKT pathway (He et al., 2019).
sEVs-ncRNAs in Drug Resistance of OC
A platinum agent combined with paclitaxel is the first-line chemotherapy regimen for OC patients (Jelovac and Armstrong, 2011). Platinum binds with double-stranded DNA to form platinum–DNA adducts, which interfere with DNA replication and RNA transcription and, eventually, initiate apoptosis. Paclitaxel is an antimitotic agent. It binds specifically to intracellular β-tubulin, and induces tubulin polymerization to hyper-stable microtubules, thereby suppressing mitosis to trigger apoptosis. PARPi kills OC cells in HRD status through a “synthetic lethal” effect (Kurian et al., 2021). However, multidrug resistance during chemotherapy has become a major obstacle in OC treatment (Agarwal and Kaye, 2003; Patch et al., 2015). The acquisition of drug resistance involves many factors: defects in DNA damage repair mechanisms; loss of BRCA1 promoter methylation; multiple independent reversions of gBRCA mutations; breakage of tumor suppressor genes; recurrent promoter fusion related to overexpression of multidrug-resistant protein-1 (MDR1) (Sarkadi et al., 2006; Christie and Bowtell, 2017; Norouzi-Barough et al., 2018; McMullen et al., 2020). sEVs have been shown to be important mediators for the development of resistance to anticancer drugs (Challagundla et al., 2015; Yang Q. et al., 2021). Sousa et al. (2015) have revealed that sEVs derived from tumor drug-resistant cells can confer resistant phenotypes to drug-sensitive cells via transferring miRNAs, lncRNAs, and drug-efflux pumps. In addition to tumor cells, tumor stromal cells-derived sEVs also contribute to transmitting the resistant phenotype (Zheng et al., 2017; Jia et al., 2021).
Au Yeung et al. (2016)have identified a new pathway causing paclitaxel resistance in OC cells. Exosomes secreted from cancer-associated adipose cells or CAFs transfer miR-21 into cancer cells, thereby increasing the apoptotic threshold of paclitaxel treated OC cells via down-regulating expression of apoptotic protease-activating factor-1 (APAF1). Kanlikilicer et al. (2018) have discovered that the massive release of exosomal miR-1246 from OC cells is absorbed by M2-phenotype macrophages in TME. MiR-1246 inhibitor treatment reduces expression of P-glycoprotein protein, thereby enhancing paclitaxel sensitivity of OC cells. The endogenous exosomal miR-433 inhibits the apoptosis of OC cells via down-regulating expression of phosphorylated retinoblastoma (p-Rb) and cyclin-dependent kinase-6 (CDK6), so as to induce paclitaxel resistance (Weiner-Gorzel et al., 2015). Pink et al. (2015) have found that exosomes from cisplatin-resistant OC cells can raise resistance in sensitive cells. Their data suggest that the passenger strand, miR-21-3p, directly facilitates cisplatin resistance of OC cells via targeting neuron navigator-3 (NAV3). Similarly, lncRNA urothelial carcinoma-associated-1 (UCA1) is upregulated in serum exosomes from OC patients with cisplatin-resistant. The results have shown that lncRNA UCA1 promotes cisplatin resistance of OC cells through the miR-143/FOS-like antigen 2 (FOSL2) pathway, which may be a novel target of OC therapy (Li et al., 2019). Interestingly, exosomal miR-30a-5p enhances apoptosis and sensitivity to cisplatin by regulating SRY- (SOX9) expression in OC cells (Liu et al., 2020). Feng Y. et al. (2019) have analyzed the differential expression profile of miRNAs between drug-resistant exosomes and primitive OC cells through the Gene Expression Omnibus database. They have suggested that miR-922, miR-574-3p, and miR-30a-5p may induce drug resistance by modulating a Cullin 2-mediated hypoxia-inducible factor-1 (HIF-1) pathway, whereas miR-183-5p promotes drug resistance via regulating expression of methyl-CpG-binding protein 2 (MECP2). A recent study (Zhang et al., 2021c) has indicated that sEVs-derived miRNAs can regulate oxaliplatin-induced peripheral neuropathy and chemotherapy efficacy in OC treatment. Table 2 summarizes the gene targets and mechanisms of sEVs-ncRNAs associated with resistance to OC chemotherapy.
TABLE 2.
Summary of sEVs-ncRNAs involved in OC chemoresistance.
| sEVs-ncRNAs | Targets | Drugs | Roles/Mechanisms | |
|---|---|---|---|---|
| miR-21 | APAF1 | Paclitaxel | Suppresses apoptosis and confers chemoresistance on OC cells | Au Yeung et al. (2016) |
| miR-1246 | Caveolin-1 | Paclitaxel | Regulates the Caveolin-1/P-gp/M2-phenotype macrophage axis to induce drug resistance | Kanlikilicer et al. (2018) |
| miR-433 | CDK6 | Paclitaxel | Drives senescence of OC cells to induce chemoresistance | Weiner-Gorzel et al. (2015) |
| miR-21-3p | NAV3 | Cisplatin | Contributes to cisplatin resistance via down-regulation of NAV3 | Pink et al. (2015) |
| lncRNA UCA1 | FOSL2 | Cisplatin | Regulates the miR-143/FOSL2 signaling to confer chemoresistance on OC cells | Li et al. (2019) |
| lncRNA NEAT1 | SOX3 | Cisplatin | Sponges miR-491-5p to inhibit OC apoptosis | Jia et al. (2021) |
| miR-429 | CASR | Cisplatin | Enhances proliferation of OC cells by targeting CASR/STAT3 pathway | Li et al. (2021b) |
| miR-21-5p | PDHA1 | Cisplatin | Promotes glycolysis and OC cells viability | Zhuang et al. (2021) |
| miR-21-3p | ||||
| miR-21-5p | ||||
| miR-891-5p | MYC, CNBP | Carboplatin | Activate glycolysis and increase expression of DNA repair proteins | Alharbi et al. (2020) |
| miR-223 | PTEN | Cisplatin | Promotes drug resistance and malignant phenotypes of OC cells via the PTEN-PI3K/AKT pathway | Zhu et al. (2019) |
| miR-4315 | Bim | Anti-PD1 | Induces apoptosis-resistance phenomenon via down-regulation of Bim | Guyon et al. (2020) |
| miR-30a-5p | SOX9 | Cisplatin | Enhances OC apoptosis and cellular sensitivity to cisplatin | Liu et al. (2020) |
| miR-146a | LAMC2 | Docetaxel | Inhibits OC cells growth and increases chemosensitivity by targeting the LAMC2-mediated PI3K/Akt axis | Qiu et al. (2020) |
OC, ovarian cancer; P-gp, P-glycoprotein protein; PD-1, programmed cell death protein 1 .
sEVs-ncRNAs as Diagnostic and Prognostic Biomarkers of OC
The prospect of significantly improved OC survival is based on an early diagnosis. Serum CA125 and human epididymis protein (HE4) are the most widely used biomarkers in the clinical diagnosis of OC. However, because of their low sensitivity and specificity, CA125 and HE4 are usually employed to monitor the progression or recurrence of OC rather than early detection. A randomized study has illustrated that the false-positive results of serum CA125 combined with transvaginal ultrasound screening lead to unnecessary surgical procedures and serious complications (Partridge et al., 2013). It has been demonstrated that the ncRNA composition in sEVs is a “snapshot” of parental cells status, in which cargoes with specific changes could be used as key biomarkers of diseases (Royo and Falcon-Perez, 2012). sEVs-derived ncRNAs as noninvasive biomarkers in liquid biopsies have three main advantages. First, they are quite stable (not degraded by RNase) (Möller and Lobb, 2020). Second, the vesicle surface displays the antigenic markers of original cells, which allows enrichment of vesicles from specific tissue sources (e.g., capture of OC-derived sEVs in plasma using EpCAM antibody) (Nagrath et al., 2007). Third, the content of sEV-ncRNAs is related to tumor stage and treatment outcome (Yokoi et al., 2018).
Taylor and Gercel-Taylor (2008) have separated tumor cells-derived exosomes from the plasma of OC patients using an improved immunoaffinity magnetic bead-sorting system with anti-EpCAM. Their microarray data has suggested that the “signature” of circulating tumor-derived exosomal miRNAs mirror and exhibit a strong correlation with the profile of tumor tissue-derived miRNAs in the same OC patients (exhibiting correlations from 0.71 to 0.90). Hence, circulating tumor-derived exosomal miRNAs can be used as candidate biomarkers for OC diagnosis. Another study has illustrated that there are a large number of abnormally expressed miRNAs (miR-106a-5p, let-7d-5p, miR-93-5p, miR-122-5p, miR-185-5p, miR-99b-5p) in plasma and exosomes of OC patients compared with healthy controls (Zhang et al., 2019). Moreover, the concentration of serum exosomal miR-373, miR-200a, miR-200b, and miR-200c in OC patients have been found to be significantly higher than those in healthy women and to be associated with tumor stage (Meng et al., 2016). Interestingly, expression of exosomal miR-34a in the serum of early-stage OC patients is significantly higher than that in advanced OC patients, and is negatively correlated with the risk of lymph-node metastasis and OC recurrence (Maeda et al., 2020).
sEVs-derived miRNAs can also be employed to distinguish the different histological types of OC. Kobayashi et al. (2018) have found a significantly increased expression of miR-1290 in the serum of HGSOC patients, which can distinguish HGSOC from patients with other histological types of OC. Importantly, miRNA expression of circulating exosomes does not always parallel that of the original tumor tissue. Kim et al. (2019) have discovered that miR-145 (identified as a down-regulated biomarker in OC tissue) is highly expressed in the serum exosomes of OC patients. Moreover, low expression of exosomal miR-484 in serum has been associated significantly with aggressive clinical variables and shorter overall survival and progression-free survival (Zhang W. et al., 2020). Wang et al. (2020) have detected the exosomal circRNAs expression profile in serum between OC patients and healthy subjects. Serum exosomal circ-0001068 expression in OC patients is up-regulated significantly compared with that in healthy subjects. OC cells-derived circ-0001068 is delivered into T cells as a comepting endogenous RNA of miR-28-5p, resulting in increased expression of programmed cell death protein 1 (PD1) and T-cell exhaustion. They have suggested that circ-0001068 may be a novel noninvasive diagnostic biomarker and therapeutic target for OC.
Engineering sEVs Targeted Delivery of ncRNAs in OC Therapy
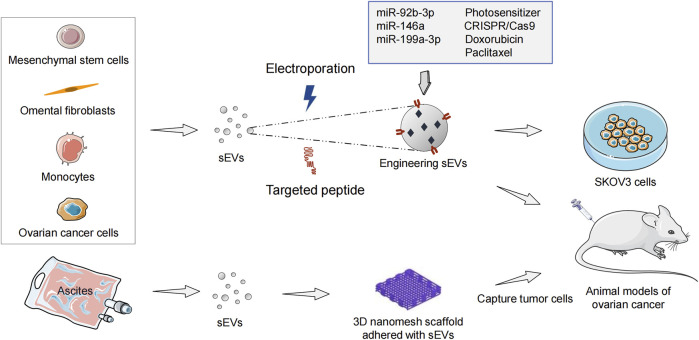
Extensive efforts are being devoted to developing therapeutic strategies that can effectively bypass the physical and biological barriers within the peritoneum in metastatic OC patients. A new direction of OC therapy is to develop various targeted delivery systems based on drug conjugates and nanomedicines (Wang Z. et al., 2021). As biogenic and bioactive nanovectors for cancer therapy, sEVs include the following advantages. First, therapeutic biomaterials can be loaded into sEVs. Second, they can be absorbed by recipient cells, thereby altering cellular processes. Third, sEVs have high histocompatibility and can reduce immune clearance in drug delivery (Kamerkar et al., 2017; Al-Dossary et al., 2021). As drug-delivery vesicles, sEVs can penetrate through anatomical barriers and have been shown to deliver miRNAs (Jc Bose et al., 2018), siRNAs (Kamerkar et al., 2017), CRISPR/Cas9 (Yao et al., 2021), and chemotherapeutic drugs (Qiu et al., 2019) safely and effectively in animal models (Figure 3).
FIGURE 3.
Engineering sEVs targeted delivery of ncRNAs and anticancer drugs in ovarian cancer treatment. sEVs, small extracellular vesicles; ncRNA, non-coding RNA.
Pinto et al. (2021) have found sEVs from mesenchymal stem cells (MSCs) to be immune-active photosensitizers (PS) vectors that can be employed to treat OC patients with peritoneal metastasis. Intraperitoneal injection of MSCs-derived sEVs-PS in model mice significantly enhances tumoral targeting compared with liposomal (Foslip) and the free drug. The mean ratio of PS in tumors/organs is 40 in the sEVs-PS group, 5.5 in the Foslip group, and 1.5 in the free PS group. sEVs-PS mediated photodynamic therapy promotes 55% of tumoral nodules necrosis (sEVs-PS versus Foslip [p < 0.0001]) and enhances CD8+ effector T cells infiltration to significantly prolong mice survival. Fuente et al. (2015) have embedded exosomes purified from ascites of OC patients into 3D scaffolds (metastatic trap [M-Trap]) to capture tumor cells disseminated in the peritoneal cavity, thereby interfering with the natural process of peritoneal metastasis. After the M-Trap device is applied to the murine model of OC with peritoneal metastasis, the mean survival increases from 117.5 to 309.4 days (p < 0.001). Overall, these findings support the importance of sEVs in the treatment of OC with peritoneal metastasis.
Numerous studies indicate that inhibiting the function of overexpressed oncogenic miRNA (oncomiR) is a potent anticancer therapeutic strategy. MiR-21 is a robust oncomiR in most cancers (Abels et al., 2019) as well as OC (Cappellesso et al., 2014). Antisense miR-21 therapy can enhance apoptosis and chemosensitivity in OC cells (Chan et al., 2014; Raniolo et al., 2021). Targeted therapy of engineering sEVs based on antisense miR-21 has achieved good efficacy in animal models of breast cancer (Jc Bose et al., 2018), and colorectal cancer (Liang et al., 2020). Wang J. et al. (2021) have investigated the potential of peptide-engineered exosomes overexpressing miR-92b-3p in anti-angiogenic treatment of OC. Their data has shown that DSPE-PEG2K-RGD modified exosomes packaged with miR-92b-3p can produce combined antitumor and antiangiogenic effects with Apatinib in intraperitoneal tumorigenesis model of nude mice. Qiu et al. (2020) have found that human umbilical cord MSCs-derived exosomes carrying miR-146a increase the sensitivity of OC cells to docetaxel and taxane by targeting the laminin gamma-2 (LAMC2)-mediated PI3K/Akt axis. In another xenograft study, miR199a-3p encapsulated in omental fibroblasts-derived exosomes significantly inhibits the peritoneal dissemination of OC cells in a mice model (Kobayashi et al., 2020).
CRISPR/Cas9 is a therapeutic genome editing technology, which has great application prospects in OC. Tumor-derived exosomes carrying CRISPR/Cas9 via electroporation disrupt PARP-1 in SKOV3 cells, resulting in apoptosis and enhancing chemosensitivity to cisplatin (Kim et al., 2017). As a carrier with low immunogenicity, exosomes achieve efficient CRISPR/cas9-mediated genome editing in vivo. However, the biggest challenges for sEVs-based clinical treatment are large-scale production, isolation, purification, and modification. Pisano et al. (2020) have developed an exosome production platform based on monocytes. Doxorubicin (DOX) is injected into exosomes as a model drug to treat SKOV3 cells. Monocytes-derived immune exosomes have shown higher production yields (2.5-fold increase), encapsulation efficiency, and drug release rate than natural exosomes. In addition, Hadla et al. (2016) have demonstrated that exosomes not only increase the therapeutic index of DOX in OC mouse models, but also avoid the cardiotoxic side effects caused by drug accumulation in myocardial endothelial cells. Paclitaxel-loaded MSCs-derived exosomes (MSCs-exos) significantly reduce the number of SKOV3 cells through apoptotic and necrotic disintegration (Melzer et al., 2019). Compared with the equivalent cytotoxic obtained via paclitaxel in vitro, the concentration of paclitaxel in MSCs-exos is reduced 7.6-fold, which indicates a higher specificity and tumor targeting of MSCs-exos.
The sEVs delivery system loaded with siRNAs or anticancer drugs is a promising approach for the treatment of OC patients with peritoneal metastasis. However, the efficacious drugs to eliminate harmful sEVs in OC patients are not available. Five efficacious exosome inhibitors have been identified through high-throughput drug screening: ketoconazole, triadimenol, tipifarnib, climbazole, and neticonazole. Their efficacy in vivo needs further validation (Datta et al., 2018). Furthermore, it has been found that an inhibitor of the ceramide-biosynthesis regulator, GW4869, can damage the exosomal secretion of 293T cells (Essandoh et al., 2015). Those findings indicate that the application of sEV inhibitors in OC treatment is worthy of further exploration.
Conclusion and Outlook
Many experimental studies have revealed the importance of sEVs-derived ncRNAs in OC progression and resistance to platinum-based chemotherapy. Hence, sEVs-ncRNAs from biological fluids (e.g., plasma or ascites) can indicate tumor status and an adaptative pathologic response to drug exposure in OC patients. Compared with the difficulty of obtaining tumor tissue, detection of sEVs-ncRNAs in plasma can aid the early diagnosis of OC, monitor tumor progression, and assess drug resistance in real-time. Furthermore, sEVs-ncRNAs, as quasi-fragments derived from tumor cells, allow for the integration of multiple molecular biomarkers to establish the diagnostic model. Studies have indicated that the diagnostic performance of the sEVs miRNAs model appears to be equivalent to or even better than the circulating miRNAs model. However, most currently available OC biomarkers based on sEVs-ncRNAs require further prospective validation. Previous studies are not directly comparable in terms of study design, investigation population, methods of sEVs isolation, and biopsy materials. Therefore, reproducible results must be achieved in large-scale studies based on patient stratification and standardized isolation methods for sEVs-ncRNAs.
sEVs that have been optimized naturally over a long period can provide unique advantages over synthetic nanoparticles. Engineering sEVs-based nanotechnology has made great strides in OC treatment, but this has not translated to longer survival for OC patients. Thus, there is a need to use sEVs-RNAi or immunotherapy (or a combination of the two options) to target OC tumors (especially in the peritoneal cavity) rather than relying on a passive therapeutic mechanism. Additionally, to become clinically usable therapeutic vectors, sEVs must face the challenges of highly controlled purification and separation, industrial-scale production, loading efficiency, and long-term efficacy. On the other hand, achieving translational application of sEVs requires direct quantitative comparisons with clinical therapy systems of OC, such as liposomal-based drug therapy. More importantly, the natural content contained in sEVs will be jointly delivered to target cells and may play a biological role. Their harmful side-effects remain to be determined, and the safety and efficacy of sEVs-based drug delivery in vivo need further evaluation. With the development and optimization of research methods, sEVs-ncRNAs are expected to promote the formulation of novel diagnosis and treatment strategies for OC.
Author Contributions
ML designed the outline of the article and wrote the initial draft. JT designed the outline of the article and revised the manuscript. XZ gave some helpful suggestions. All authors reviewed the manuscript and approved the final version.
Funding
This work was supported by Grants from the General Project of Natural Science Foundation of Hunan Province (No. 2020JJ4051); the Promotion Project of Health Suitability Program in Health Department of Hunan Province (No. WZ2020-15); Science and Technology Innovation Program of Hunan Province (No. 2020SK51101); General Project in Health Department of Hunan Province (No. 202205015388); Hunan Cancer Hospital Climbing Fund (No. ZX2020004); Capacity Building Project of Central Subsidy Medical and Health Institutions (No. 20201127-1001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abels E. R., Maas S. L. N., Nieland L., Wei Z., Cheah P. S., Tai E., et al. (2019). Glioblastoma-Associated Microglia Reprogramming Is Mediated by Functional Transfer of Extracellular miR-21. Cell Rep. 28, 3105–3119. 10.1016/j.celrep.2019.08.036 PubMed Abstract | 10.1016/j.celrep.2019.08.036 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal R., Kaye S. B. (2003). Ovarian Cancer: Strategies for Overcoming Resistance to Chemotherapy. Nat. Rev. Cancer 3, 502–516. 10.1038/nrc1123 PubMed Abstract | 10.1038/nrc1123 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Al-Dossary A. A., Tawfik E. A., Isichei A. C., Sun X., Li J., Alshehri A. A., et al. (2021). Engineered EV-Mimetic Nanoparticles as Therapeutic Delivery Vehicles for High-Grade Serous Ovarian Cancer. Cancers 13, 3075. 10.3390/cancers13123075 PubMed Abstract | 10.3390/cancers13123075 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alharbi M., Sharma S., Guanzon D., Lai A., Zuñiga F., Shiddiky M. J. A., et al. (2020). miRNa Signature in Small Extracellular Vesicles and Their Association with Platinum Resistance and Cancer Recurrence in Ovarian Cancer. Nanomedicine Nanotechnol. Biol. Med. 28, 102207. 10.1016/j.nano.2020.102207 10.1016/j.nano.2020.102207 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Alharbi M., Lai A., Sharma S., Kalita-de Croft P., Godbole N., Campos A., et al. (2021). Extracellular Vesicle Transmission of Chemoresistance to Ovarian Cancer Cells Is Associated with Hypoxia-Induced Expression of Glycolytic Pathway Proteins, and Prediction of Epithelial Ovarian Cancer Disease Recurrence. Cancers 13, 3388. 10.3390/cancers13143388 PubMed Abstract | 10.3390/cancers13143388 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiadou E., Jacob L. S., Slack F. J. (2018). Non-coding RNA Networks in Cancer. Nat. Rev. Cancer 18, 5–18. 10.1038/nrc.2017.99 PubMed Abstract | 10.1038/nrc.2017.99 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- EL Andaloussi S., Mäger I., Breakefield X. O., Wood M. J. A. (2013). Extracellular Vesicles: Biology and Emerging Therapeutic Opportunities. Nat. Rev. Drug Discov. 12, 347–357. 10.1038/nrd3978 PubMed Abstract | 10.1038/nrd3978 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Andre F., Schartz N. E., Movassagh M., Flament C., Pautier P., Morice P., et al. (2002). Malignant Effusions and Immunogenic Tumour-Derived Exosomes. Lancet 360, 295–305. 10.1016/s0140-6736(02)09552-1 PubMed Abstract | 10.1016/s0140-6736(02)09552-1 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Au Yeung C. L., Co N.-N., Tsuruga T., Yeung T.-L., Kwan S.-Y., Leung C. S., et al. (2016). Exosomal Transfer of Stroma-Derived miR21 Confers Paclitaxel Resistance in Ovarian Cancer Cells through Targeting APAF1. Nat. Commun. 7, 11150. 10.1038/ncomms11150 PubMed Abstract | 10.1038/ncomms11150 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azarin S. M., Yi J., Gower R. M., Aguado B. A., Sullivan M. E., Goodman A. G., et al. (2015). In Vivo capture and Label-free Detection of Early Metastatic Cells. Nat. Commun. 6, 8094. 10.1038/ncomms9094 PubMed Abstract | 10.1038/ncomms9094 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran J., Baj-Krzyworzeka M., Weglarczyk K., Szatanek R., Zembala M., Barbasz J., et al. (2010). Circulating Tumour-Derived Microvesicles in Plasma of Gastric Cancer Patients. Cancer Immunol. Immunother. 59, 841–850. 10.1007/s00262-009-0808-2 PubMed Abstract | 10.1007/s00262-009-0808-2 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., Luo B., Jiang P., Zhou X., Lan F., Yi Q., et al. (2018). Immuno-modified Superparamagnetic Nanoparticles via Host-Guest Interactions for High-Purity Capture and Mild Release of Exosomes. Nanoscale 10, 14280–14289. 10.1039/c8nr02871k PubMed Abstract | 10.1039/c8nr02871k | Google Scholar [DOI] [PubMed] [Google Scholar]
- Cai J., Gong L., Li G., Guo J., Yi X., Wang Z. (2021). Exosomes in Ovarian Cancer Ascites Promote Epithelial-Mesenchymal Transition of Ovarian Cancer Cells by Delivery of miR-6780b-5p. Cell Death Dis. 12, 210. 10.1038/s41419-021-03490-5 PubMed Abstract | 10.1038/s41419-021-03490-5 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J.-Y., Wang B., Tang T.-T., Wen Y., Li Z.-L., Feng S.-T., et al. (2021). Exosomal miR-125b-5p Deriving from Mesenchymal Stem Cells Promotes Tubular Repair by Suppression of P53 in Ischemic Acute Kidney Injury. Theranostics 11, 5248–5266. 10.7150/thno.54550 PubMed Abstract | 10.7150/thno.54550 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellesso R., Tinazzi A., Giurici T., Simonato F., Guzzardo V., Ventura L., et al. (2014). Programmed Cell Death 4 and microRNA 21 Inverse Expression Is Maintained in Cells and Exosomes from Ovarian Serous Carcinoma Effusions. Cancer Cytopathol. 122, 685–693. 10.1002/cncy.21442 PubMed Abstract | 10.1002/cncy.21442 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Challagundla K. B., Wise P. M., Neviani P., Chava H., Murtadha M., Xu T., et al. (2015). Exosome-mediated Transfer of microRNAs within the Tumor Microenvironment and Neuroblastoma Resistance to Chemotherapy. J. Natl. Cancer Inst. 107. djv135. 10.1093/jnci/djv135 PubMed Abstract | 10.1093/jnci/djv135 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. K., Blansit K., Kiet T., Sherman A., Wong G., Earle C., et al. (2014). The Inhibition of miR-21 Promotes Apoptosis and Chemosensitivity in Ovarian Cancer. Gynecol. Oncol. 132, 739–744. 10.1016/j.ygyno.2014.01.034 PubMed Abstract | 10.1016/j.ygyno.2014.01.034 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Chen X., Ying X., Wang X., Wu X., Zhu Q., Wang X. (2017). Exosomes Derived from Hypoxic Epithelial Ovarian Cancer Deliver microRNA-940 to Induce Macrophage M2 Polarization. Oncol. Rep. 38, 522–528. 10.3892/or.2017.5697 PubMed Abstract | 10.3892/or.2017.5697 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Chen X., Zhou J., Li X., Wang X., Lin Y., Wang X. (2018). Exosomes Derived from Hypoxic Epithelial Ovarian Cancer Cells Deliver microRNAs to Macrophages and Elicit a Tumor-Promoted Phenotype. Cancer Lett. 435, 80–91. 10.1016/j.canlet.2018.08.001 PubMed Abstract | 10.1016/j.canlet.2018.08.001 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Chen F., Xu B., Li J., Yang X., Gu J., Yao X., et al. (2021). Hypoxic Tumour Cell-Derived Exosomal miR-340-5p Promotes Radioresistance of Oesophageal Squamous Cell Carcinoma via KLF10. J. Exp. Clin. Cancer Res. 40, 38. 10.1186/s13046-021-01834-9 PubMed Abstract | 10.1186/s13046-021-01834-9 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaruttini N., Redondo-Morata L., Colom A., Humbert F., Lenz M., Scheuring S., et al. (2015). Relaxation of Loaded ESCRT-III Spiral Springs Drives Membrane Deformation. Cell 163, 866–879. 10.1016/j.cell.2015.10.017 PubMed Abstract | 10.1016/j.cell.2015.10.017 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ L., Raiborg C., Wenzel E. M., Campsteijn C., Stenmark H. (2017). Cellular Functions and Molecular Mechanisms of the ESCRT Membrane-Scission Machinery. Trends Biochem. Sci. 42, 42–56. 10.1016/j.tibs.2016.08.016 PubMed Abstract | 10.1016/j.tibs.2016.08.016 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Christie E. L., Bowtell D. D. L. (2017). Acquired Chemotherapy Resistance in Ovarian Cancer. Ann. Oncol. 28, viii13–viii15. 10.1093/annonc/mdx446 PubMed Abstract | 10.1093/annonc/mdx446 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Ciravolo V., Huber V., Ghedini G. C., Venturelli E., Bianchi F., Campiglio M., et al. (2012). Potential Role of HER2-Overexpressing Exosomes in Countering Trastuzumab-Based Therapy. J. Cell. Physiol. 227, 658–667. 10.1002/jcp.22773 PubMed Abstract | 10.1002/jcp.22773 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Colombo M., Raposo G., Théry C. (2014). Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Biol. 30, 255–289. 10.1146/annurev-cellbio-101512-122326 PubMed Abstract | 10.1146/annurev-cellbio-101512-122326 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Crescitelli R., Lässer C., Jang S. C., Cvjetkovic A., Malmhäll C., Karimi N., et al. (2020). Subpopulations of Extracellular Vesicles from Human Metastatic Melanoma Tissue Identified by Quantitative Proteomics after Optimized Isolation. J. Extracell. Vesicles 9, 1722433. 10.1080/20013078.2020.1722433 PubMed Abstract | 10.1080/20013078.2020.1722433 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Wang D., Xie M. (2021). Tumor-Derived Extracellular Vesicles Promote Activation of Carcinoma-Associated Fibroblasts and Facilitate Invasion and Metastasis of Ovarian Cancer by Carrying miR-630. Front. Cell Dev. Biol. 9, 652322. 10.3389/fcell.2021.652322 PubMed Abstract | 10.3389/fcell.2021.652322 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Das S., Extracellular R. C. C., Ansel K. M., Bitzer M., Breakefield X. O., Charest A., et al. (2019). The Extracellular RNA Communication Consortium: Establishing Foundational Knowledge and Technologies for Extracellular RNA Research. Cell 177, 231–242. 10.1016/j.cell.2019.03.023 PubMed Abstract | 10.1016/j.cell.2019.03.023 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A., Kim H., McGee L., Johnson A. E., Talwar S., Marugan J., et al. (2018). High-throughput Screening Identified Selective Inhibitors of Exosome Biogenesis and Secretion: A Drug Repurposing Strategy for Advanced Cancer. Sci. Rep. 8, 8161. 10.1038/s41598-018-26411-7 PubMed Abstract | 10.1038/s41598-018-26411-7 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorayappan K. D. P., Wanner R., Wallbillich J. J., Saini U., Zingarelli R., Suarez A. A., et al. (2018). Hypoxia-induced Exosomes Contribute to a More Aggressive and Chemoresistant Ovarian Cancer Phenotype: a Novel Mechanism Linking STAT3/Rab Proteins. Oncogene 37, 3806–3821. 10.1038/s41388-018-0189-0 PubMed Abstract | 10.1038/s41388-018-0189-0 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essandoh K., Yang L., Wang X., Huang W., Qin D., Hao J., et al. (2015). Blockade of Exosome Generation with GW4869 Dampens the Sepsis-Induced Inflammation and Cardiac Dysfunction. Biochimica Biophysica Acta (BBA) - Mol. Basis Dis. 1852, 2362–2371. 10.1016/j.bbadis.2015.08.010 10.1016/j.bbadis.2015.08.010 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. (2011). Non-coding RNAs in Human Disease. Nat. Rev. Genet. 12, 861–874. 10.1038/nrg3074 PubMed Abstract | 10.1038/nrg3074 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Fabbiano F., Corsi J., Gurrieri E., Trevisan C., Notarangelo M., D'Agostino V. G. (2020). RNA Packaging into Extracellular Vesicles: An Orchestra of RNA‐binding Proteins? J. Extracell. Vesicles 10, e12043. 10.1002/jev2.12043 PubMed Abstract | 10.1002/jev2.12043 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Dean D. C., Hornicek F. J., Shi H., Duan Z. (2019a). Exosomes Promote Pre-metastatic Niche Formation in Ovarian Cancer. Mol. Cancer 18, 124. 10.1186/s12943-019-1049-4 PubMed Abstract | 10.1186/s12943-019-1049-4 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Hang W., Sang Z., Li S., Xu W., Miao Y., et al. (2019b). Identification of Exosomal and Non-exosomal microRNAs A-ssociated with the D-rug R-esistance of O-varian C-ancer. Mol. Med. Rep. 19, 3376–3392. 10.3892/mmr.2019.10008 PubMed Abstract | 10.3892/mmr.2019.10008 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzese E., Centonze S., Diana A., Carlino F., Guerrera L. P., Di Napoli M., et al. (2019). PARP Inhibitors in Ovarian Cancer. Cancer Treat. Rev. 73, 1–9. 10.1016/j.ctrv.2018.12.002 PubMed Abstract | 10.1016/j.ctrv.2018.12.002 | Google Scholar [DOI] [PubMed] [Google Scholar]
- de la Fuente A., Alonso-Alconada L., Costa C., Cueva J., Garcia-Caballero T., Lopez-Lopez R., et al. (2015). M-trap: Exosome-Based Capture of Tumor Cells as a New Technology in Peritoneal Metastasis. JNCI.J. 107, djv184. 10.1093/jnci/djv184 10.1093/jnci/djv184 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martin R., Wang G., Brandão B. B., Zanotto T. M., Shah S., Kumar Patel S., et al. (2022). MicroRNA Sequence Codes for Small Extracellular Vesicle Release and Cellular Retention. Nature 601, 446–451. 10.1038/s41586-021-04234-3 PubMed Abstract | 10.1038/s41586-021-04234-3 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings D. J., Ciaudo C., Erhardt M., Voinnet O. (2009). Multivesicular Bodies Associate with Components of miRNA Effector Complexes and Modulate miRNA Activity. Nat. Cell Biol. 11, 1143–1149. 10.1038/ncb1929 PubMed Abstract | 10.1038/ncb1929 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Gordon S., Martinez F. O. (2010). Alternative Activation of Macrophages: Mechanism and Functions. Immunity 32, 593–604. 10.1016/j.immuni.2010.05.007 PubMed Abstract | 10.1016/j.immuni.2010.05.007 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Gould S. J., Raposo G. (2013). As We Wait: Coping with an Imperfect Nomenclature for Extracellular Vesicles. J. Extracell. Vesicles 2, 20389. 10.3402/jev.v2i0.20389 PubMed Abstract | 10.3402/jev.v2i0.20389 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves L. E., Ariztia E. V., Navari J. R., Matzel H. J., Stack M. S., Fishman D. A. (2004). Proinvasive Properties of Ovarian Cancer Ascites-Derived Membrane Vesicles. Cancer Res. 64, 7045–7049. 10.1158/0008-5472.can-04-1800 PubMed Abstract | 10.1158/0008-5472.can-04-1800 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Grunberg N., Pevsner-Fischer M., Goshen-Lago T., Diment J., Stein Y., Lavon H., et al. (2021). Cancer-Associated Fibroblasts Promote Aggressive Gastric Cancer Phenotypes via Heat Shock Factor 1-Mediated Secretion of Extracellular Vesicles. Cancer Res. 81, 1639–1653. 10.1158/0008-5472.CAN-20-2756 PubMed Abstract | 10.1158/0008-5472.CAN-20-2756 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X., Zong Z.-h., Liu Y., Chen S., Wang L.-l., Zhao Y. (2019). circPUM1 Promotes Tumorigenesis and Progression of Ovarian Cancer by Sponging miR-615-5p and miR-6753-5p. Mol. Ther. - Nucleic Acids 18, 882–892. 10.1016/j.omtn.2019.09.032 PubMed Abstract | 10.1016/j.omtn.2019.09.032 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Lv X., Ru Y., Zhou F., Wang N., Xi H., et al. (2020). Circulating Exosomal Gastric Cancer-Associated Long Noncoding RNA1 as a Biomarker for Early Detection and Monitoring Progression of Gastric Cancer. JAMA Surg. 155, 572. 10.1001/jamasurg.2020.1133 PubMed Abstract | 10.1001/jamasurg.2020.1133 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon N., Garnier D., Briand J., Nadaradjane A., Bougras-Cartron G., Raimbourg J., et al. (2020). Anti-PD1 Therapy Induces Lymphocyte-Derived Exosomal miRNA-4315 Release Inhibiting Bim-Mediated Apoptosis of Tumor Cells. Cell Death Dis. 11, 1048. 10.1038/s41419-020-03224-z PubMed Abstract | 10.1038/s41419-020-03224-z | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadla M., Palazzolo S., Corona G., Caligiuri I., Canzonieri V., Toffoli G., et al. (2016). Exosomes Increase the Therapeutic Index of Doxorubicin in Breast and Ovarian Cancer Mouse Models. Nanomedicine 11, 2431–2441. 10.2217/nnm-2016-0154 PubMed Abstract | 10.2217/nnm-2016-0154 | Google Scholar [DOI] [PubMed] [Google Scholar]
- He L., Zhu W., Chen Q., Yuan Y., Wang Y., Wang J., et al. (2019). Ovarian Cancer Cell-Secreted Exosomal miR-205 Promotes Metastasis by Inducing Angiogenesis. Theranostics 9, 8206–8220. 10.7150/thno.37455 PubMed Abstract | 10.7150/thno.37455 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann I. K., Wood M. J. A., Fuhrmann G. (2021). Extracellular Vesicles as a Next-Generation Drug Delivery Platform. Nat. Nanotechnol. 16, 748–759. 10.1038/s41565-021-00931-2 PubMed Abstract | 10.1038/s41565-021-00931-2 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Hong C.-S., Funk S., Muller L., Boyiadzis M., Whiteside T. L. (2016). Isolation of Biologically Active and Morphologically Intact Exosomes from Plasma of Patients with Cancer. J. Extracell. Vesicles 5, 29289. 10.3402/jev.v5.29289 PubMed Abstract | 10.3402/jev.v5.29289 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A., Costa-Silva B., Shen T.-L., Rodrigues G., Hashimoto A., Tesic Mark M., et al. (2015). Tumour Exosome Integrins Determine Organotropic Metastasis. Nature 527, 329–335. 10.1038/nature15756 PubMed Abstract | 10.1038/nature15756 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Li D., Wu A., Qiu X., Di W., Huang L., et al. (2017). TWEAK-stimulated Macrophages Inhibit Metastasis of Epithelial Ovarian Cancer via Exosomal Shuttling of microRNA. Cancer Lett. 393, 60–67. 10.1016/j.canlet.2017.02.009 PubMed Abstract | 10.1016/j.canlet.2017.02.009 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Jang Y., Kim H., Yoon S., Lee H., Hwang J., Jung J., et al. (2021). Exosome-based Photoacoustic Imaging Guided Photodynamic and Immunotherapy for the Treatment of Pancreatic Cancer. J. Control. Release 330, 293–304. 10.1016/j.jconrel.2020.12.039 PubMed Abstract | 10.1016/j.jconrel.2020.12.039 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Jc Bose R., Uday Kumar S., Zeng Y., Afjei R., Robinson E., Lau K., et al. (2018). Tumor Cell-Derived Extracellular Vesicle-Coated Nanocarriers: An Efficient Theranostic Platform for the Cancer-specific Delivery of Anti-miR-21 and Imaging Agents. ACS Nano 12, 10817–10832. 10.1021/acsnano.8b02587 PubMed Abstract | 10.1021/acsnano.8b02587 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelovac D., Armstrong D. K. (2011). Recent Progress in the Diagnosis and Treatment of Ovarian Cancer. CA A Cancer J. Clin. 61, 183–203. 10.3322/caac.20113 PubMed Abstract | 10.3322/caac.20113 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X., Wei L., Zhang Z. (2021). NEAT1 Overexpression Indicates a Poor Prognosis and Induces Chemotherapy Resistance via the miR-491-5p/SOX3 Signaling Pathway in Ovarian Cancer. Front. Genet. 12, 616220. 10.3389/fgene.2021.616220 PubMed Abstract | 10.3389/fgene.2021.616220 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Johnstone R. M., Adam M., Hammond J. R., Orr L., Turbide C. (1987). Vesicle Formation during Reticulocyte Maturation. Association of Plasma Membrane Activities with Released Vesicles (Exosomes). J. Biol. Chem. 262, 9412–9420. 10.1016/s0021-9258(18)48095-7 PubMed Abstract | 10.1016/s0021-9258(18)48095-7 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Kalluri R., LeBleu V. S. (2020). The Biology , Function , and Biomedical Applications of Exosomes. Science 367. 10.1126/science.aau6977 PubMed Abstract | 10.1126/science.aau6977 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerkar S., LeBleu V. S., Sugimoto H., Yang S., Ruivo C. F., Melo S. A., et al. (2017). Exosomes Facilitate Therapeutic Targeting of Oncogenic KRAS in Pancreatic Cancer. Nature 546, 498–503. 10.1038/nature22341 PubMed Abstract | 10.1038/nature22341 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanlikilicer P., Rashed M. H., Bayraktar R., Mitra R., Ivan C., Aslan B., et al. (2016). Ubiquitous Release of Exosomal Tumor Suppressor miR-6126 from Ovarian Cancer Cells. Cancer Res. 76, 7194–7207. 10.1158/0008-5472.CAN-16-0714 PubMed Abstract | 10.1158/0008-5472.CAN-16-0714 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanlikilicer P., Bayraktar R., Denizli M., Rashed M. H., Ivan C., Aslan B., et al. (2018). Exosomal miRNA Confers Chemo Resistance via Targeting Cav1/p-gp/M2-type Macrophage axis in Ovarian Cancer. EBioMedicine 38, 100–112. 10.1016/j.ebiom.2018.11.004 PubMed Abstract | 10.1016/j.ebiom.2018.11.004 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann D. J., Babst M., Emr S. D. (2001). Ubiquitin-Dependent Sorting into the Multivesicular Body Pathway Requires the Function of a Conserved Endosomal Protein Sorting Complex, ESCRT-I. Cell 106, 145–155. 10.1016/s0092-8674(01)00434-2 PubMed Abstract | 10.1016/s0092-8674(01)00434-2 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Kim S. M., Yang Y., Oh S. J., Hong Y., Seo M., Jang M. (2017). Cancer-derived Exosomes as a Delivery Platform of CRISPR/Cas9 Confer Cancer Cell Tropism-dependent Targeting. J. Control. Release 266, 8–16. 10.1016/j.jconrel.2017.09.013 PubMed Abstract | 10.1016/j.jconrel.2017.09.013 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Kim S., Choi M. C., Jeong J.-Y., Hwang S., Jung S. G., Joo W. D., et al. (2019). Serum Exosomal miRNA-145 and miRNA-200c as Promising Biomarkers for Preoperative Diagnosis of Ovarian Carcinomas. J. Cancer 10, 1958–1967. 10.7150/jca.30231 PubMed Abstract | 10.7150/jca.30231 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Sawada K., Nakamura K., Yoshimura A., Miyamoto M., Shimizu A., et al. (2018). Exosomal miR-1290 Is a Potential Biomarker of High-Grade Serous Ovarian Carcinoma and Can Discriminate Patients from Those with Malignancies of Other Histological Types. J. Ovarian Res. 11, 81. 10.1186/s13048-018-0458-0 PubMed Abstract | 10.1186/s13048-018-0458-0 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Sawada K., Miyamoto M., Shimizu A., Yamamoto M., Kinose Y., et al. (2020). Exploring the Potential of Engineered Exosomes as Delivery Systems for Tumor-Suppressor microRNA Replacement Therapy in Ovarian Cancer. Biochem. Biophysical Res. Commun. 527, 153–161. 10.1016/j.bbrc.2020.04.076 10.1016/j.bbrc.2020.04.076 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Kowal J., Arras G., Colombo M., Jouve M., Morath J. P., Primdal-Bengtson B., et al. (2016). Proteomic Comparison Defines Novel Markers to Characterize Heterogeneous Populations of Extracellular Vesicle Subtypes. Proc. Natl. Acad. Sci. U.S.A. 113, E968–E977. 10.1073/pnas.1521230113 PubMed Abstract | 10.1073/pnas.1521230113 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian A. W., Ward K. C., Abrahamse P., Bondarenko I., Hamilton A. S., Deapen D., et al. (2021). Time Trends in Receipt of Germline Genetic Testing and Results for Women Diagnosed with Breast Cancer or Ovarian Cancer, 2012-2019. J. Clin. Oncol. 39, 1631–1640. 10.1200/JCO.2010.1200/JCO.20.02785 PubMed Abstract | 10.1200/JCO.2010.1200/JCO.20.02785 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroki L., Guntupalli S. R. (2020). Treatment of Epithelial Ovarian Cancer. Bmj 371, m3773. 10.1136/bmj.m3773 PubMed Abstract | 10.1136/bmj.m3773 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Lheureux S., Braunstein M., Oza A. M. (2019). Epithelial Ovarian Cancer: Evolution of Management in the Era of Precision Medicine. CA A Cancer J. Clin. 69, 280–304. 10.3322/caac.21559 PubMed Abstract | 10.3322/caac.21559 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Li X., Tang M. (2020). Exosomes Released from M2 Macrophages Transfer miR‐221‐3p Contributed to EOC Progression through Targeting CDKN1B. Cancer Med. 9, 5976–5988. 10.1002/cam4.3252 PubMed Abstract | 10.1002/cam4.3252 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Niu H., Qin Q., Yang S., Wang Q., Yu C., et al. (2019). lncRNA UCA1 Mediates Resistance to Cisplatin by Regulating the miR-143/fosl2-Signaling Pathway in Ovarian Cancer. Mol. Ther. - Nucleic Acids 17, 92–101. 10.1016/j.omtn.2019.05.007 PubMed Abstract | 10.1016/j.omtn.2019.05.007 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Wang B., Ding X., Zhang X., Tang J., Lin H. (2021a). Plasma Extracellular Vesicle Delivery of miR-210-3p by Targeting ATG7 to Promote Sepsis-Induced Acute Lung Injury by Regulating Autophagy and Activating Inflammation. Exp. Mol. Med. 53, 1180–1191. 10.1038/s12276-021-00651-6 PubMed Abstract | 10.1038/s12276-021-00651-6 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Lin L., Liu Q., Gao W., Chen L., Sha C., et al. (2021b). Exosomal Transfer of miR-429 Confers Chemoresistance in Epithelial Ovarian Cancer. Am. J. Cancer Res. 11, 2124–2141. PubMed Abstract | Google Scholar [PMC free article] [PubMed] [Google Scholar]
- Lian X. Y., Zhang H., Liu Q., Lu X., Zhou P., He S. Q., et al. (2020). Ovarian Cancer‐excreted Exosomal miR‐199a‐5p Suppresses Tumor Metastasis by Targeting Hypoxia‐inducible Factor‐2α in Hypoxia Microenvironment. Cancer Commun. 40, 380–385. 10.1002/cac2.12034 10.1002/cac2.12034 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Zhu Y., Ali D. J., Tian T., Xu H., Si K., et al. (2020). Engineered Exosomes for Targeted Co-delivery of miR-21 Inhibitor and Chemotherapeutics to Reverse Drug Resistance in Colon Cancer. J. Nanobiotechnol. 18, 10. 10.1186/s12951-019-0563-2 PubMed Abstract | 10.1186/s12951-019-0563-2 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Zhang Y., Sun P., Wang C. (2020). DDP-resistant Ovarian Cancer Cells-Derived Exosomal microRNA-30a-5p Reduces the Resistance of Ovarian Cancer Cells to DDP. Open Biol. 10, 190173. 10.1098/rsob.190173 PubMed Abstract | 10.1098/rsob.190173 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lötvall J., Hill A. F., Hochberg F., Buzás E. I., Di Vizio D., Gardiner C., et al. (2014). Minimal Experimental Requirements for Definition of Extracellular Vesicles and Their Functions: a Position Statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 3, 26913. 10.3402/jev.v3.26913 PubMed Abstract | 10.3402/jev.v3.26913 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S. L. N., Breakefield X. O., Weaver A. M. (2017). Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 27, 172–188. 10.1016/j.tcb.2016.11.003 PubMed Abstract | 10.1016/j.tcb.2016.11.003 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K., Sasaki H., Ueda S., Miyamoto S., Terada S., Konishi H., et al. (2020). Serum Exosomal microRNA-34a as a Potential Biomarker in Epithelial Ovarian Cancer. J. Ovarian Res. 13, 47. 10.1186/s13048-020-00648-1 PubMed Abstract | 10.1186/s13048-020-00648-1 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoumi-Dehghi S., Babashah S., Sadeghizadeh M. (2020). microRNA-141-3p-containing Small Extracellular Vesicles Derived from Epithelial Ovarian Cancer Cells Promote Endothelial Cell Angiogenesis through Activating the JAK/STAT3 and NF-Κb Signaling Pathways. J. Cell Commun. Signal. 14, 233–244. 10.1007/s12079-020-00548-5 PubMed Abstract | 10.1007/s12079-020-00548-5 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matulonis U. A., Sood A. K., Fallowfield L., Howitt B. E., Sehouli J., Karlan B. Y. (2016). Ovarian Cancer. Nat. Rev. Dis. Prim. 2, 16061. 10.1038/nrdp.2016.61 PubMed Abstract | 10.1038/nrdp.2016.61 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen M., Karakasis K., Madariaga A., Oza A. M. (2020). Overcoming Platinum and PARP-Inhibitor Resistance in Ovarian Cancer. Cancers 12, 1607. 10.3390/cancers12061607 PubMed Abstract | 10.3390/cancers12061607 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer C., Rehn V., Yang Y., Bähre H., von der Ohe J., Hass R. (2019). Taxol-Loaded MSC-Derived Exosomes Provide a Therapeutic Vehicle to Target Metastatic Breast Cancer and Other Carcinoma Cells. Cancers 11, 798. 10.3390/cancers11060798 PubMed Abstract | 10.3390/cancers11060798 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Müller V., Milde-Langosch K., Trillsch F., Pantel K., Schwarzenbach H. (2016). Diagnostic and Prognostic Relevance of Circulating Exosomal miR-373, miR-200a, miR-200b and miR-200c in Patients with Epithelial Ovarian Cancer. Oncotarget 7, 16923–16935. 10.18632/oncotarget.7850 PubMed Abstract | 10.18632/oncotarget.7850 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon U., Gentry-Maharaj A., Hallett R., Ryan A., Burnell M., Sharma A., et al. (2009). Sensitivity and Specificity of Multimodal and Ultrasound Screening for Ovarian Cancer, and Stage Distribution of Detected Cancers: Results of the Prevalence Screen of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS). Lancet Oncol. 10, 327–340. 10.1016/s1470-2045(09)70026-9 PubMed Abstract | 10.1016/s1470-2045(09)70026-9 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Menon U., Gentry-Maharaj A., Burnell M., Singh N., Ryan A., Karpinskyj C., et al. (2021). Ovarian Cancer Population Screening and Mortality after Long-Term Follow-Up in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a Randomised Controlled Trial. Lancet 397, 2182–2193. 10.1016/s0140-6736(21)00731-5 PubMed Abstract | 10.1016/s0140-6736(21)00731-5 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min L., Zhu S., Chen L., Liu X., Wei R., Zhao L., et al. (2019). Evaluation of Circulating Small Extracellular Vesicles Derived miRNAs as Biomarkers of Early Colon Cancer: a Comparison with Plasma Total miRNAs. J. Extracell. Vesicles 8, 1643670. 10.1080/20013078.2019.1643670 PubMed Abstract | 10.1080/20013078.2019.1643670 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza M. R., Monk B. J., Herrstedt J., Oza A. M., Mahner S., Redondo A., et al. (2016). Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N. Engl. J. Med. 375, 2154–2164. 10.1056/NEJMoa1611310 PubMed Abstract | 10.1056/NEJMoa1611310 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Mittelbrunn M., Gutiérrez-Vázquez C., Villarroya-Beltri C., González S., Sánchez-Cabo F., González M. Á., et al. (2011). Unidirectional Transfer of microRNA-Loaded Exosomes from T Cells to Antigen-Presenting Cells. Nat. Commun. 2, 282. 10.1038/ncomms1285 PubMed Abstract | 10.1038/ncomms1285 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller A., Lobb R. J. (2020). The Evolving Translational Potential of Small Extracellular Vesicles in Cancer. Nat. Rev. Cancer 20, 697–709. 10.1038/s41568-020-00299-w PubMed Abstract | 10.1038/s41568-020-00299-w | Google Scholar [DOI] [PubMed] [Google Scholar]
- Montaldo C., Terri M., Riccioni V., Battistelli C., Bordoni V., D’Offizi G., et al. (2021). Fibrogenic Signals Persist in DAA-Treated HCV Patients after Sustained Virological Response. J. Hepatology 75, 1301–1311. 10.1016/j.jhep.2021.07.003 10.1016/j.jhep.2021.07.003 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Morita E., Sandrin V., Chung H.-Y., Morham S. G., Gygi S. P., Rodesch C. K., et al. (2007). Human ESCRT and ALIX Proteins Interact with Proteins of the Midbody and Function in Cytokinesis. EMBO J. 26, 4215–4227. 10.1038/sj.emboj.7601850 PubMed Abstract | 10.1038/sj.emboj.7601850 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K., Ghoshal B., Ghosh S., Chakrabarty Y., Shwetha S., Das S., et al. (2016). Reversible HuR‐micro RNA Binding Controls Extracellular Export of miR‐122 and Augments Stress Response. EMBO Rep. 17, 1184–1203. 10.15252/embr.201541930 PubMed Abstract | 10.15252/embr.201541930 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D. E., de Jong O. G., Brouwer M., Wood M. J., Lavieu G., Schiffelers R. M., et al. (2019). Extracellular Vesicle-Based Therapeutics: Natural versus Engineered Targeting and Trafficking. Exp. Mol. Med. 51, 1–12. 10.1038/s12276-019-0223-5 10.1038/s12276-019-0223-5 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagrath S., Sequist L. V., Maheswaran S., Bell D. W., Irimia D., Ulkus L., et al. (2007). Isolation of Rare Circulating Tumour Cells in Cancer Patients by Microchip Technology. Nature 450, 1235–1239. 10.1038/nature06385 PubMed Abstract | 10.1038/nature06385 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network Cancer Genome Atlas Research (2011). Integrated Genomic Analyses of Ovarian Carcinoma. Nature 474, 609–615. 10.1038/nature10166 PubMed Abstract | 10.1038/nature10166 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norouzi-Barough L., Sarookhani M. R., Sharifi M., Moghbelinejad S., Jangjoo S., Salehi R. (2018). Molecular Mechanisms of Drug Resistance in Ovarian Cancer. J. Cell. Physiol. 233, 4546–4562. 10.1002/jcp.26289 PubMed Abstract | 10.1002/jcp.26289 | Google Scholar [DOI] [PubMed] [Google Scholar]
- O’Brien K., Breyne K., Ughetto S., Laurent L. C., Breakefield X. O. (2020). RNA Delivery by Extracellular Vesicles in Mammalian Cells and its Applications. Nat. Rev. Mol. Cell Biol. 21, 585–606. 10.1038/s41580-020-0251-y PubMed Abstract | 10.1038/s41580-020-0251-y | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski M., Carmo N. B., Krumeich S., Fanget I., Raposo G., Savina A., et al. (2010). Rab27a and Rab27b Control Different Steps of the Exosome Secretion Pathway. Nat. Cell Biol. 12, 19–30. 10.1038/ncb2000 PubMed Abstract | 10.1038/ncb2000 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Pal T., Permuth-Wey J., Betts J. A., Krischer J. P., Fiorica J., Arango H., et al. (2005). BRCA1andBRCA2mutations Account for a Large Proportion of Ovarian Carcinoma Cases. Cancer 104, 2807–2816. 10.1002/cncr.21536 PubMed Abstract | 10.1002/cncr.21536 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Partridge E. E., Greenlee R. T., Riley T. L., Commins J., Ragard L., Xu J.-L., et al. (2013). Assessing the Risk of Ovarian Malignancy in Asymptomatic Women with Abnormal CA 125 and Transvaginal Ultrasound Scans in the Prostate, Lung, Colorectal, and Ovarian Screening Trial. Obstetrics Gynecol. 121, 25–31. 10.1097/aog.0b013e3182755e14 PubMed Abstract | 10.1097/aog.0b013e3182755e14 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patch A.-M., Christie E. L., Etemadmoghadam D., Garsed D. W., George J., Fereday S., et al. (2015). Whole-genome Characterization of Chemoresistant Ovarian Cancer. Nature 521, 489–494. 10.1038/nature14410 PubMed Abstract | 10.1038/nature14410 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Pegtel D. M., Gould S. J. (2019). Exosomes. Annu. Rev. Biochem. 88, 487–514. 10.1146/annurev-biochem-013118-111902 PubMed Abstract | 10.1146/annurev-biochem-013118-111902 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Pink R. C., Samuel P., Massa D., Caley D. P., Brooks S. A., Carter D. R. F. (2015). The Passenger Strand, miR-21-3p, Plays a Role in Mediating Cisplatin Resistance in Ovarian Cancer Cells. Gynecol. Oncol. 137, 143–151. 10.1016/j.ygyno.2014.12.042 PubMed Abstract | 10.1016/j.ygyno.2014.12.042 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Pinto A., Marangon I., Méreaux J., Nicolás-Boluda A., Lavieu G., Wilhelm C., et al. (2021). Immune Reprogramming Precision Photodynamic Therapy of Peritoneal Metastasis by Scalable Stem-Cell-Derived Extracellular Vesicles. ACS Nano 15, 3251–3263. 10.1021/acsnano.0c09938 PubMed Abstract | 10.1021/acsnano.0c09938 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Pisano S., Pierini I., Gu J., Gazze A., Francis L. W., Gonzalez D., et al. (2020). Immune (Cell) Derived Exosome Mimetics (IDEM) as a Treatment for Ovarian Cancer. Front. Cell Dev. Biol. 8. 10.3389/fcell.2020.553576 PubMed Abstract | 10.3389/fcell.2020.553576 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Li Z., Han X., Zhen L., Luo C., Liu M., et al. (2019). Tumor-derived Nanovesicles Promote Lung Distribution of the Therapeutic Nanovector through Repression of Kupffer Cell-Mediated Phagocytosis. Theranostics 9, 2618–2636. 10.7150/thno.32363 PubMed Abstract | 10.7150/thno.32363 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L., Wang J., Chen M., Chen F., Tu W. (2020). Exosomal microRNA-146a Derived from Mesenchymal Stem Cells Increases the Sensitivity of Ovarian Cancer Cells to Docetaxel and Taxane via a LAMC2-Mediated PI3K/Akt axis. Int. J. Mol. Med. 46, 609–620. 10.3892/ijmm.2020.4634 PubMed Abstract | 10.3892/ijmm.2020.4634 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raniolo S., Unida V., Vindigni G., Stolfi C., Iacovelli F., Desideri A., et al. (2021). Combined and Selective miR-21 Silencing and Doxorubicin Delivery in Cancer Cells Using Tailored DNA Nanostructures. Cell Death Dis. 12, 7. 10.1038/s41419-020-03339-3 PubMed Abstract | 10.1038/s41419-020-03339-3 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G., Stahl P. D. (2019). Extracellular Vesicles: a New Communication Paradigm? Nat. Rev. Mol. Cell Biol. 20, 509–510. 10.1038/s41580-019-0158-7 PubMed Abstract | 10.1038/s41580-019-0158-7 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Ribatti D., Conconi M. T., Nussdorfer G. G. (2007). Nonclassic Endogenous Novel Regulators of Angiogenesis. Pharmacol. Rev. 59, 185–205. 10.1124/pr.59.2.3 PubMed Abstract | 10.1124/pr.59.2.3 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Royo F., Falcon-Perez J. M. (2012). Liver Extracellular Vesicles in Health and Disease. J. Extracell. Vesicles 1. 10.3402/jev.v1i0.18825 10.3402/jev.v1i0.18825 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkadi B., Homolya L., Szakacs G., Varadi A. (2006). Human Multidrug Resistance ABCB and ABCG Transporters: Participation in a Chemoimmunity Defense System. Physiol. Rev. 86, 1179–1236. 10.1152/physrev.00037.2005 PubMed Abstract | 10.1152/physrev.00037.2005 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Shao H., Im H., Castro C. M., Breakefield X., Weissleder R., Lee H. (2018). New Technologies for Analysis of Extracellular Vesicles. Chem. Rev. 118, 1917–1950. 10.1021/acs.chemrev.7b00534 PubMed Abstract | 10.1021/acs.chemrev.7b00534 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si M., Wang Q., Li Y., Lin H., Luo D., Zhao W., et al. (2019). Inhibition of Hyperglycolysis in Mesothelial Cells Prevents Peritoneal Fibrosis. Sci. Transl. Med. 11, eaav5341. 10.1126/scitranslmed.aav5341 PubMed Abstract | 10.1126/scitranslmed.aav5341 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D., Fuchs H. E., Jemal A. (2021). Cancer Statistics, 2021. CA Cancer J. Clin. 71, 7–33. 10.3322/caac.21654 PubMed Abstract | 10.3322/caac.21654 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Slack F. J., Chinnaiyan A. M. (2019). The Role of Non-coding RNAs in Oncology. Cell 179, 1033–1055. 10.1016/j.cell.2019.10.017 PubMed Abstract | 10.1016/j.cell.2019.10.017 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa D., Lima R. T., Vasconcelos M. H. (2015). Intercellular Transfer of Cancer Drug Resistance Traits by Extracellular Vesicles. Trends Mol. Med. 21, 595–608. 10.1016/j.molmed.2015.08.002 PubMed Abstract | 10.1016/j.molmed.2015.08.002 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Szajnik M., Czystowska M., Szczepanski M. J., Mandapathil M., Whiteside T. L. (2010). Tumor-derived Microvesicles Induce, Expand and Up-Regulate Biological Activities of Human Regulatory T Cells (Treg). PLoS One 5, e11469. 10.1371/journal.pone.0011469 PubMed Abstract | 10.1371/journal.pone.0011469 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao M., Nagai Y., Ito M., Ohba T. (2018). Flow Cytometric Quantitation of EpCAM-Positive Extracellular Vesicles by Immunomagnetic Separation and Phospholipid Staining Method. Genes cells. 23, 963–973. 10.1111/gtc.12645 PubMed Abstract | 10.1111/gtc.12645 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Tang M. K. S., Yue P. Y. K., Ip P. P., Huang R. L., Lai H. C., Cheung A. N. Y., et al. (2018). Soluble E-Cadherin Promotes Tumor Angiogenesis and Localizes to Exosome Surface. Nat. Commun. 9, 2270. 10.1038/s41467-018-04695-7 PubMed Abstract | 10.1038/s41467-018-04695-7 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D. D., Gercel-Taylor C. (2008). MicroRNA Signatures of Tumor-Derived Exosomes as Diagnostic Biomarkers of Ovarian Cancer. Gynecol. Oncol. 110, 13–21. 10.1016/j.ygyno.2008.04.033 PubMed Abstract | 10.1016/j.ygyno.2008.04.033 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Thery C., Amigorena S., Raposo G., Clayton A. (2006). Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 3, 1–29. 10.1002/0471143030.cb0322s30 10.1002/0471143030.cb0322s30 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Thery C., Witwer K. W., Aikawa E., Alcaraz M. J., Anderson J. D., Andriantsitohaina R., et al. (2018). Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): a Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 7, 1535750. 10.1080/20013078.2018.1535750 PubMed Abstract | 10.1080/20013078.2018.1535750 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y., Gong M., Hu Y., Liu H., Zhang W., Zhang M., et al. (2020). Quality and Efficiency Assessment of Six Extracellular Vesicle Isolation Methods by Nano-Flow Cytometry. J. Extracell. Vesicles 9, 1697028. 10.1080/20013078.2019.1697028 PubMed Abstract | 10.1080/20013078.2019.1697028 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach M., Thery C. (2016). Communication by Extracellular Vesicles: Where We Are and where We Need to Go. Cell 164, 1226–1232. 10.1016/j.cell.2016.01.043 PubMed Abstract | 10.1016/j.cell.2016.01.043 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Torre L. A., Trabert B., DeSantis C. E., Miller K. D., Samimi G., Runowicz C. D., et al. (2018). Ovarian Cancer Statistics, 2018. CA Cancer J. Clin. 68, 284–296. 10.3322/caac.21456 PubMed Abstract | 10.3322/caac.21456 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G., D'Angelo G., Raposo G. (2018). Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 19, 213–228. 10.1038/nrm.2017.125 PubMed Abstract | 10.1038/nrm.2017.125 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Wang X., Yao Y., Jin M. (2020). Circ-0001068 Is a Novel Biomarker for Ovarian Cancer and Inducer of PD1 Expression in T Cells. Aging-US 12, 19095–19106. 10.18632/aging.103706 PubMed Abstract | 10.18632/aging.103706 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Wang J., Shen X., Li M., Yue Y., Cheng X., et al. (2021a). LncRNA SPOCD1-AS from Ovarian Cancer Extracellular Vesicles Remodels Mesothelial Cells to Promote Peritoneal Metastasis via Interacting with G3BP1. J. Exp. Clin. Cancer Res. 40, 101. 10.1186/s13046-021-01899-6 10.1186/s13046-021-01899-6 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wang C., Li Y., Li M., Zhu T., Shen Z., et al. (2021b). Potential of Peptide-Engineered Exosomes with Overexpressed miR-92b-3p in Anti-angiogenic Therapy of Ovarian Cancer. Clin. Transl. Med. 11, e425. 10.1002/ctm2.425 PubMed Abstract | 10.1002/ctm2.425 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Meng F., Zhong Z. (2021c). Emerging Targeted Drug Delivery Strategies toward Ovarian Cancer. Adv. Drug Del. Rev. 178, 113969. 10.1016/j.addr.2021.113969 10.1016/j.addr.2021.113969 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Weiner-Gorzel K., Dempsey E., Milewska M., McGoldrick A., Toh V., Walsh A., et al. (2015). Overexpression of the microRNA miR-433 Promotes Resistance to Paclitaxel through the Induction of Cellular Senescence in Ovarian Cancer Cells. Cancer Med. 4, 745–758. 10.1002/cam4.409 PubMed Abstract | 10.1002/cam4.409 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y., Sui Z., Shan Y., Hu Y., Chen Y., Zhang L., et al. (2016). Effective Isolation of Exosomes with Polyethylene Glycol from Cell Culture Supernatant for In-Depth Proteome Profiling. Analyst 141, 4640–4646. 10.1039/c6an00892e PubMed Abstract | 10.1039/c6an00892e | Google Scholar [DOI] [PubMed] [Google Scholar]
- Wong L. L., Zou R., Zhou L., Lim J. Y., Phua D. C. Y., Liu C., et al. (2019). Combining Circulating MicroRNA and NT-proBNP to Detect and Categorize Heart Failure Subtypes. J. Am. Coll. Cardiol. 73, 1300–1313. 10.1016/j.jacc.2018.11.060 PubMed Abstract | 10.1016/j.jacc.2018.11.060 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Yamamoto C. M., Oakes M. L., Murakami T., Muto M. G., Berkowitz R. S., Ng S. W. (2018). Comparison of Benign Peritoneal Fluid- and Ovarian Cancer Ascites-Derived Extracellular Vesicle RNA Biomarkers. J. Ovarian Res. 11, 20. 10.1186/s13048-018-0391-2 PubMed Abstract | 10.1186/s13048-018-0391-2 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Khan S., Sun Y., Hess K., Shmulevich I., Sood A. K., et al. (2011). Association of BRCA1 and BRCA2 Mutations with Survival, Chemotherapy Sensitivity, and Gene Mutator Phenotype in Patients with Ovarian Cancer. JAMA 306, 1557–1565. 10.1001/jama.2011.1456 PubMed Abstract | 10.1001/jama.2011.1456 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Zhao S., Shi Z., Cao L., Liu J., Pan T., et al. (2021a). Chemotherapy-elicited Exosomal miR-378a-3p and miR-378d Promote Breast Cancer Stemness and Chemoresistance via the Activation of EZH2/STAT3 Signaling. J. Exp. Clin. Cancer Res. 40, 120. 10.1186/s13046-021-01901-1 10.1186/s13046-021-01901-1 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. G., Wang W., Zhao L. J., Wang X., Gimple R. C., Xu L., et al. (2021b). Plasma Cells Shape the Mesenchymal Identity of Ovarian Cancers through Transfer of Exosome-Derived microRNAs. Sci. Adv. 7 (9), eabb0737. 10.1126/sciadv.abb0737 PubMed Abstract | 10.1126/sciadv.abb0737 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Lyu P., Yoo K., Yadav M. K., Singh R., Atala A., et al. (2021). Engineered Extracellular Vesicles as Versatile Ribonucleoprotein Delivery Vehicles for Efficient and Safe CRISPR Genome Editing. J. Extracell. Vesicles 10, e12076. 10.1002/jev2.12076 PubMed Abstract | 10.1002/jev2.12076 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi A., Matsuzaki J., Yamamoto Y., Yoneoka Y., Takahashi K., Shimizu H., et al. (2018). Integrated Extracellular microRNA Profiling for Ovarian Cancer Screening. Nat. Commun. 9, 4319. 10.1038/s41467-018-06434-4 PubMed Abstract | 10.1038/s41467-018-06434-4 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborowski M. P., Balaj L., Breakefield X. O., Lai C. P. (2015). Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 65, 783–797. 10.1093/biosci/biv084 PubMed Abstract | 10.1093/biosci/biv084 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R., Qiu X., Song Y., Wang Y. (2021). Exosomes Mediated Transfer of Circ_0000337 Contributes to Cisplatin (CDDP) Resistance of Esophageal Cancer by Regulating JAK2 via miR-377-3p. Front. Cell Dev. Biol. 9, 673237. 10.3389/fcell.2021.673237 PubMed Abstract | 10.3389/fcell.2021.673237 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Xu S., Liu X. (2019). MicroRNA Profiling of Plasma Exosomes from Patients with Ovarian Cancer Using High-Throughput Sequencing. Oncol. Lett. 17, 5601–5607. 10.3892/ol.2019.10220 PubMed Abstract | 10.3892/ol.2019.10220 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Su X., Li S., Liu Z., Wang Q., Zeng H. (2020a). Low Serum Exosomal miR-484 Expression Predicts Unfavorable Prognosis in Ovarian Cancer. Cancer Biomark. 27, 485–491. 10.3233/CBM-191123 PubMed Abstract | 10.3233/CBM-191123 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Zhang X., Xu Q., Zi Z., Liu Z., Wan C., Crisman L., et al. (2020c). Programmable Extracellular Vesicles for Macromolecule Delivery and Genome Modifications. Dev. Cell 55, 784–801. 10.1016/j.devcel.2020.11.007 PubMed Abstract | 10.1016/j.devcel.2020.11.007 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Sang Y., Chen D., Wu X., Wang X., Yang W., et al. (2021a). M2 Macrophage-Derived Exosomal Long Non-coding RNA AGAP2-AS1 Enhances Radiotherapy Immunity in Lung Cancer by Reducing microRNA-296 and Elevating NOTCH2. Cell Death Dis. 12, 467. 10.1038/s41419-021-03700-0 PubMed Abstract | 10.1038/s41419-021-03700-0 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Sheng Y., Li B., Wang Q., Liu X., Han J. (2021b). Ovarian Cancer Derived PKR1 Positive Exosomes Promote Angiogenesis by Promoting Migration and Tube Formation In Vitro . Cell biochem. Funct. 39, 308–316. 10.1002/cbf.3583 PubMed Abstract | 10.1002/cbf.3583 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Zhang Y., Li C., Qin Y., Cepparulo P., Millman M., Chopp M., et al. (2021d). Small Extracellular Vesicles Ameliorate Peripheral Neuropathy and Enhance Chemotherapy of Oxaliplatin on Ovarian Cancer. J. Extracell. Vesicles 10, e12073. 10.1002/jev2.12073 PubMed Abstract | 10.1002/jev2.12073 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Pang X., Yang Z., Wang S., Deng H., Chen X. (2022). Nanomaterials Targeting Tumor Associated Macrophages for Cancer Immunotherapy. J. Control. Release 341, 272–284. 10.1016/j.jconrel.2021.11.028 PubMed Abstract | 10.1016/j.jconrel.2021.11.028 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Zheng P., Chen L., Yuan X., Luo Q., Liu Y., Xie G., et al. (2017). Exosomal Transfer of Tumor-Associated Macrophage-Derived miR-21 Confers Cisplatin Resistance in Gastric Cancer Cells. J. Exp. Clin. Cancer Res. 36, 53. 10.1186/s13046-017-0528-y PubMed Abstract | 10.1186/s13046-017-0528-y | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li X., Wu X., Zhang T., Zhu Q., Wang X., et al. (2018). Exosomes Released from Tumor-Associated Macrophages Transfer miRNAs that Induce a Treg/Th17 Cell Imbalance in Epithelial Ovarian Cancer. Cancer Immunol. Res. 6, 1578–1592. 10.1158/2326-6066.CIR-17-0479 PubMed Abstract | 10.1158/2326-6066.CIR-17-0479 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Zhu X., Shen H., Yin X., Yang M., Wei H., Chen Q., et al. (2019). Macrophages Derived Exosomes Deliver miR-223 to Epithelial Ovarian Cancer Cells to Elicit a Chemoresistant Phenotype. J. Exp. Clin. Cancer Res. 38, 81. 10.1186/s13046-019-1095-1 PubMed Abstract | 10.1186/s13046-019-1095-1 | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang L., Zhang B., Liu X., Lin L., Wang L., Hong Z., et al. (2021). Exosomal miR-21-5p Derived from Cisplatin-Resistant SKOV3 Ovarian Cancer Cells Promotes Glycolysis and Inhibits Chemosensitivity of its Progenitor SKOV3 Cells by Targeting PDHA1. Cell Biol. Int. 45 (10), 2140–2149. 10.1002/cbin.11671 PubMed Abstract | 10.1002/cbin.11671 | Google Scholar [DOI] [PubMed] [Google Scholar]
- Zong Z. H., Du Y. P., Guan X., Chen S., Zhao Y. (2019). CircWHSC1 Promotes Ovarian Cancer Progression by Regulating MUC1 and hTERT through Sponging miR-145 and miR-1182. J. Exp. Clin. Cancer Res. 38, 437. 10.1186/s13046-019-1437-z PubMed Abstract | 10.1186/s13046-019-1437-z | Google Scholar [DOI] [PMC free article] [PubMed] [Google Scholar]