Abstract
Understanding the build-up of immunity with successive severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants and the epidemiological conditions that favor rapidly expanding epidemics will help facilitate future pandemic control. We analyzed high-resolution infection and serology data from two longitudinal household cohorts in South Africa to reveal high cumulative infection rates and durable cross-protective immunity conferred by prior infection in the pre-Omicron era. Building on the history of past exposures to different SARS-CoV-2 variants and vaccination in the more representative urban cohort given South Africa’s high urbanization rate, we used mathematical models to explore the fitness advantage of the Omicron variant and its epidemic trajectory. Modelling suggests the Omicron wave likely infected a large fraction (44% - 81%) of the population, leaving a complex landscape of population immunity primed and boosted with antigenically distinct variants. We project that future SARS-CoV-2 resurgences are likely under a range of scenarios of viral characteristics, population contacts, and residual cross-protection.
This study characterizes SARS-CoV-2 shedding, transmission, and the immunologic landscape after four epidemic waves in two South African population cohorts.
INTRODUCTION
A key question for the long-term control of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is how sequential exposures to different variants of the virus shape population immunity and thereby modulate subsequent epidemic cycles and disease burden. Few studies have characterized the protection conferred by infection over long time periods, particularly in low- and middle-income settings where vaccine access is limited and high SARS-CoV-2 infection rates have been reported ( 1 – 4 ). Here we used data from two prospectively followed cohorts in South Africa to estimate the strength of cross-protective immunity conferred by infection with successive SARS-CoV-2 variants. We relied on the cohort data to reconstruct the landscape of population immunity prior to the emergence of the Omicron variant, and modeled the trajectory, scale, and long-term consequences of the Omicron epidemic in this population.
South Africa experienced three distinct SARS-CoV-2 epidemic waves prior to the emergence of the Omicron variant, with the first wave (June 2020 – December 2020) dominated by the ancestral SARS-CoV-2 strain carrying the D614G mutation (referred to as D614G hereafter) ( 5 ), the second wave (December 2020 – May 2021) dominated by the Beta (B.1.351) variant ( 6 ), and the third wave (May 2021 – October 2021) dominated by the Delta (B.1.617.2) variant ( 7 ). In South Africa, the Omicron variant was first identified in the province of Gauteng Province in November 2021 and swiftly spread nationally and globally, causing rapid growth in case counts relative to prior waves ( 7 , 8 ). Similar patterns of rapid growth despite high pre-existing immunity from infection and vaccination have also been reported in numerous countries across the world, with Omicron replacing Delta in multiple global locations, even when prevalence of Delta was high ( 9 , 10 ).
The apparent fitness advantage of the Omicron variant over Delta could be driven by immune evasion, increased intrinsic transmissibility, or a combination of both. The immune evasion hypothesis is supported by an increased reinfection risk coinciding with the rise of the Omicron variant ( 7 , 8 , 11 , 12 ). Further support for this hypothesis comes from in vitro analyses of sera from convalescent patients (infected with pre-Omicron variants) and vaccinated individuals, which show reduced neutralization titers against Omicron compared to earlier variants ( 13 – 16 ). Similarly, data from multiple settings have shown decreased vaccine effectiveness against Omicron ( 17 , 18 ). Separately, epidemiological and experimental data point to reduced clinical severity of Omicron ( 17 , 19 ), possibly due to increased tropism for the upper respiratory tract rather than the lung, which could also promote higher transmission relative to pre-Omicron variants. As the Omicron wave subsides, the relative contribution of these factors to Omicron’s spread remains elusive, in part due to uncertainty in the extent of population immunity before the rise of Omicron. Further, Omicron’s rapid spread poses immense pressure on SARS-CoV-2 testing capabilities, and its relatively benign course in most people make it difficult to assess the full scope of the epidemic.
To understand the long-term dynamics of SARS-CoV-2, we leveraged data from two longitudinal household cohorts followed over a 13-month period, from July 2020 to August 2021 in rural and urban areas of South Africa (Prospective Household study of SARS-CoV-2, Influenza and Respiratory Syncytial virus community burden, Transmission dynamics and viral interaction in South Africa, PHIRST-C, where “C” stands for coronavirus disease 2019 (COVID-19), previously described in ( 4 )). We relied on the results of densely sampled respiratory and serologic specimens testing from 222 households to model the kinetics of viral shedding, transmission dynamics among household members, and cross-protection between successive variants circulating prior to the emergence of Omicron. We used population-level models calibrated against data from these prospective cohorts and surveillance efforts to clarify long-term patterns of immunity acquisition, the impact of immune evasion, and future epidemic trajectories for SARS-CoV-2 in the aftermath of the Omicron wave.
RESULTS
Overview of SARS-CoV-2 epidemiology in study sites
The PHIRST-C cohort captured the dynamics of three waves of SARS-CoV-2 infections in a rural site and an urban site located in two provinces of South Africa. In total, 1,200 individuals living in 222 randomly selected households were enrolled and followed up twice a week for SARS-CoV-2 real-time reverse transcription polymerase chain reaction (rRT-PCR) testing and symptom monitoring, and blood draws were obtained every 2 months for SARS-CoV-2 serologic tests. Throughout the study, we used a broad measurement of prior and ongoing infections including both serologic and virologic evidence, irrespective of symptoms. In Fig. 1A-B, we show the weekly SARS-CoV-2 epidemic curves in the district where each study site was located ( 20 ). Population vaccination started in June 2021 in South Africa and the fraction of the cohort population fully vaccinated remained below 10% at the conclusion of the study in September 2021 (Fig. 1A-B). Both Pfizer/BioNTech’s BNT162b2 and J&J/Janssen’s Ad26.COV2.S vaccines are used in South Africa (fig. S1). We did not evaluate vaccine effectiveness in this study and focused on protection conferred by prior SARS-CoV-2 infection. However, we did consider the impact of vaccination in projections of the Omicron wave and post-Omicron future.
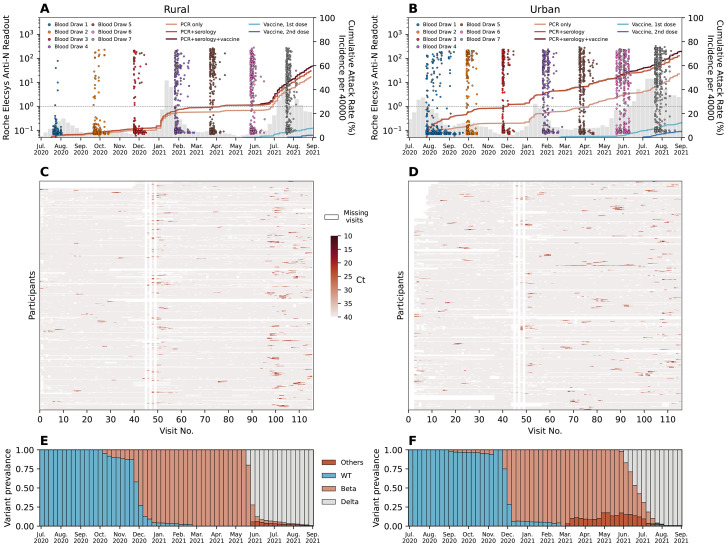
Fig. 1. PHIRST-C study June 2020 – September 2021, description of the epidemiology of SARS-CoV-2 in the two study sites, along with serology and rRT-PCR data.
(A-B) Dots in different colors represent the timing and the readouts (axis on the left) of Roche Elecsys Anti-SARS-CoV-2 assay of serum specimens collected from 4 different blood draws of the rural cohort. The dash line is the positive cutoff of the Roche Elecsys Anti-SARS-CoV-2 assay, above which a specimen is considered sero-positive. The red lines (from light to dark) are the cumulative SARS-CoV-2 variant exposures (axis on the right) over time, captured by positive rRT-PCR of mid-turbinate nasal swab samples only; by either positive serum antibody or positive mid-turbinate nasal swabs by rRT-PCR, and by either positive serum antibody or positive mid-turbinate nasal swabs by rRT-PCR or at least one dose of vaccine. The light and dark blue lines are the cumulative fraction of population receiving a 1st and 2nd dose of vaccine. The grey bars are the weekly SARS-CoV-2 incidence per 40,000 population (sharing the same axis on the right) captured by the surveillance system of Ehlanzeni District in Mpumalanga Province, where the rural site is located. (B) Same as (A) but for the urban site of Klerksdorp in the Dr Kenneth Kaunda District, North West Province. (C-D) rRT-PCR test results for all mid-turbinate nasal specimens collected from individuals in the rural (C) or urban (D) cohort over 80 visits during the 13-month study period. Color white indicates missing specimens; color red indicates the Ct value of the rRT-PCR test, the darker the red color, the lower the Ct value. (E-F) The bi-monthly relative prevalence of D614G mutation, Beta, Delta, and other variants over time at the rural (E) or urban (F) site.
In the rural site, baseline enrollment visits started prior to the peak of the first epidemic wave. The seroprevalence of anti-SARS-CoV-2 nucleocapsid antibodies was 1.1% (5/445) at enrollment, increased to 7.3% (42/574) after the first wave (third blood draw), to 25.4% (151/595) after the second wave (fifth blood draw), and reached 39.1% (227/581) around the peak of the third wave (seventh blood draw). The timing and individual results of the serological assay are visualized in Fig. 1A. During the study period (July 2020 to August 2021), 50.9% (327/643) of individuals tested positive by rRT-PCR for at least one infection episode. The cumulative infection rate (confirmed by either a rRT-PCR or a serological test) was 59.7% (384/643) by the end of the study in the rural cohort. In contrast, in the urban site, enrollment started near the time of the peak of the first wave and SARS-CoV-2 seroprevalence was higher at enrollment (14.3%, 73/511), increased to 27.0% (143/530) after the first wave, to 40.3% (207/514) after the second wave, and reached 55.7% (279/501) around the peak of the third wave (see Fig. 1B for bi-monthly results). During the study period, 53.1% (296/557) of participants tested positive by rRT-PCR for at least one infection episode. The cumulative infection rate (confirmed by either a rRT-PCR or a serological test) was 69.4% (387/557) in the urban cohort by the end of the study.
In total across both sites, we observed 669 rRT-PCR-confirmed infection episodes, including 599/669 (89.5%) primary infections and 70/669 (9.5%) reinfections. The weekly incidence of SARS-CoV-2 infections within each cohort (fig. S2) matched the epidemic trajectory at the district level (Fig. 1A-B), except for a less pronounced third wave in the urban cohort compared to the district. Lineage-specific rRT-PCR and sequencing data revealed that 14.3% (96/669) of infections were D614G, 33.2% (222/669) were Beta, 44.1% (295/669) were Delta, 2.7% (18/669) were other lineages including Alpha and C.1.2 variants, and 5.7% (38/669) were inconclusive. Figure 1E-F shows the relative prevalence of different lineages over time for the rural and urban site, respectively (details in Materials and Methods Section 2.2).
Kinetics of viral RNA shedding.
To study the risk of infection and re-infection in the cohort and better understand acquisition of immunity before the rise of Omicron, we first built a time-varying model that captured the dynamics of viral RNA shedding for each individual in the cohort, adjusted for host characteristics and variant types. Household exposure depends on the degree of viral shedding among household members; to obtain a correlate of shedding intensity, we used the serial Ct values of nasal swab specimens collected twice-weekly and tested for SARS-CoV-2 using rRT-PCR. We considered Ct value of the rRT-PCR test of a specimen as a proxy for the RNA shedding intensity. We used the serial rRT-PCR test results to model the shedding kinetics of SARS-CoV-2 infection episodes, following prior work ( 21 , 22 ). To account for the potential role of adaptive immunity in limiting transmission in the later phases of infection, we allowed for different transmission risks during the viral RNA proliferation stage (before peak shedding) and the viral RNA clearance stage (after peak shedding), with shedding increases and decreases assumed to following linear curves on the scale of Ct values. Because the nasal swab sampling period ended on August 28, 2021, around the peak of the Delta wave in both sites, we limited our analysis to infection episodes with first positive PCR specimen 30 days prior to the end of sampling to avoid censoring bias.
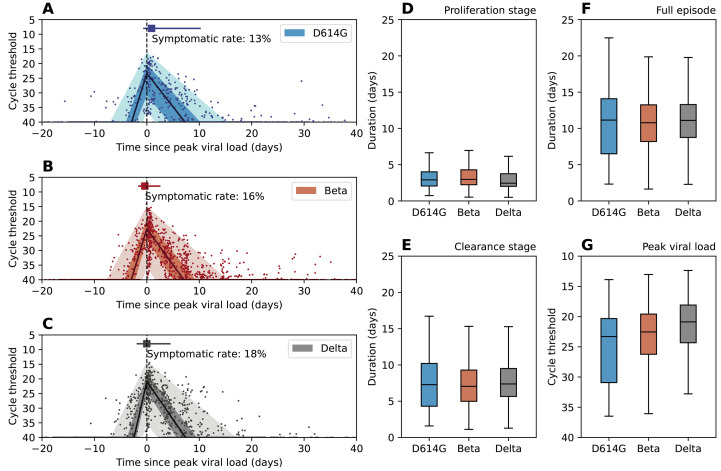
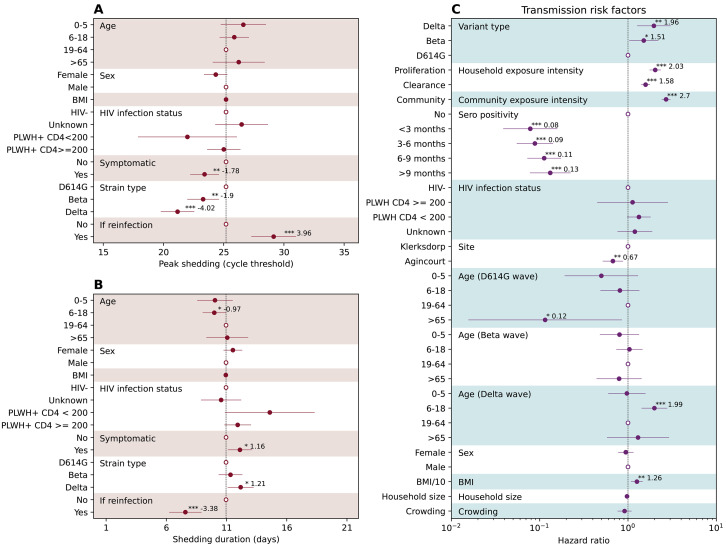
Figure 2 A-C shows the RNA shedding kinetics of the D614G, Beta, and Delta variants respectively. All three variants had similar shedding kinetics characterized by a short proliferation stage (Fig. 2D, median and interquartile range (IQR) for D614G: 3.2 (2.1 – 4.0) days, Beta: 3.3 (2.2 – 4.3) days, Delta: 3.1 (2.0 – 3.8) days) and a longer clearance stage (Fig. 2E, median and IQR for D614G: 7.4 (4.3 – 10.2) days, Beta: 7.5 (5.0 – 9.3) days, Delta: 8.0 (5.7 – 9.5) days). Symptomatic rates among infection episodes were low across all variants, at 13% for D614G, 16% for Beta, and 18% for Delta. The timing of symptom onset coincided with the timing of peak viral shedding, suggesting substantial shedding had already occurred prior to symptom presentation. After adjusting for age, sex, body mass index (BMI), and HIV infection status, symptomatic infections had a significantly lower trough Ct value (peak shedding intensity, P < 0.05) than asymptomatic infections (Fig. 3A). The Beta variant’s trough Ct value was lower than D614G, whereas Delta’s was the lowest among the three. We also found that prior infection significantly reduced peak shedding (P < 0.001) by 4.0 Ct (95% CI 2.1 – 5.8) and shedding duration (P < 0.001) by 3.4 days (95% CI 2.0 – 4.7) upon reinfection (Fig. 3A-B).
Fig. 2. SARS-CoV-2 shedding patterns by D614G, Beta, and Delta variants.
(A-C) Characterization of the RNA shedding kinetics for D614G (A), Beta (B), and Delta (C) infections. The solid dots are longitudinal Ct values observation for each infection episode, aligned based on the estimated timing of trough Ct. The solid line is the population median of all individual fits, the dark shade is the interquartile range, and the light shade is the 95% confidence interval. Dashed vertical line indicate the timing of peak viral load. The square marker and the horizontal line indicate the median time and interquartile range of symptom onset for symptomatic infections. We also reported the fraction of symptomatic infections among all infections (symptomatic rate) for each variant. (D-G) Distribution of the estimated duration of viral RNA proliferation (D), viral RNA clearance (E), full duration (proliferation stage + clearance stage) of rRT-PCR positivity (F), and distribution of the estimated trough Ct (G) for D614G, Beta, and Delta variants. Boxplots show median, interquartile range, minimum and maximum of the distribution.
Fig. 3. Drivers of SARS-CoV-2 peak shedding, shedding duration, and risk factors associated with SASR-CoV-2 infection.
(A) The association between peak shedding (trough Ct) and age, sex BMI, HIV infection status, symptom presentation, variant type, and prior infection history, based on Gaussian multiple regression. Regression coefficients along with 95%CIs are reported as solid dots and horizontal lines relative to the value of the regression intercept. The hollow dots are reference class for each of the categorical variable. (B) Same as (A) but for shedding duration. (C) Piecewise exponential hazard model on risk factors associated with infection acquisition. Hazard ratios (HR) along with 95%CIs are reported as solid dots and horizontal lines. The hollow dots are reference class for each of the categorical variable. Protection is measured as . * p<0.05; ** p<0.01; *** p < 0.001 based on t test (A-B) and z-test (C). Abbreviations: HIV- (HIV-uninfected individuals), PLWH+ CD4 <200 (Persons living with HIV, CD4+ T cell count under 200 cells/ml), PLWH+ CD4 >=200 (Persons living with HIV, CD4+ T cell count equal or above 200 cells/ml).
The population of the PHIRST-C cohort had a high prevalence of HIV, 13% in the rural site and 16% in the urban site, reflecting the burden of HIV infections in South Africa (Table 1). However, in this cohort, most (93.8%) persons living with HIV (PLWH) had CD4+ T cell counts ≥ 200 cells/ml (Table 1), and they did not differ from HIV-uninfected individuals in terms of SARS-CoV-2 shedding (Fig. 3A-B).
Table 1: PHIRST-C study June 2020 – August 2021, characteristics of the population and SARS-CoV-2 infections at two study sites, South Africa.
*PLWH: persons living with HIV
| Rural | Urban | |
| Characteristics | No. (%) | No. (%) |
| All | 643 (100) | 557 (100) |
| Age group, in years | ||
| 0-5 | 99 (15) | 55 (10) |
| 6-18 | 299 (47) | 211 (38) |
| 19-65 | 218 (34) | 260 (47) |
| >65 | 27 (4) | 31 (5) |
| Sex | ||
| Male | 234 (36) | 249 (45) |
| Female | 409 (64) | 308 (55) |
| Household size | ||
| <4 | 44 (7) | 62 (11) |
| 4-6 | 282 (44) | 296 (53) |
| 7-10 | 257 (40) | 141 (25) |
| >10 | 60 (9) | 58 (10) |
| BMI | ||
| Underweight | 259 (40) | 140 (25) |
| Healthy weight | 206 (32) | 192 (35) |
| Overweight | 93 (14) | 103 (18) |
| Obesity | 85 (13) | 120 (22) |
| Unknown | 0 (0) | 2 (0) |
| HIV status | ||
| Negative | 520 (81) | 451 (81) |
| PLWH*: CD4 < 200 | 5 (1) | 9 (2) |
| PLWH*: CD4 ≥200 | 75 (12) | 78 (14) |
| Unknown | 43 (7) | 19 (3) |
| Vaccination status | ||
| None | 593 (92) | 488 (88) |
| J&J | 12 (2) | 9 (2) |
| Pfizer first dose only | 27 (4) | 32 (6) |
| Pfizer first and second doses | 11 (2) | 28 (5) |
Infection risk and protection against reinfection.
Reconstruction of the variant-specific shedding kinetics of each infected individual allowed us not only to infer the timing of their infections, but also to evaluate the daily intensity of exposure to SARS-CoV-2 within their households. We used a piecewise exponential hazard model (Materials and Methods Section 2.5) to explore how variant type and prior infection history affect the risk of SARS-CoV-2 infection and reinfection, after adjustment for time-varying SARS-CoV-2 exposure and host factors. We regressed the daily infection risk of each individual on covariates including variant type, time since prior SARS-CoV-2 infection, age (allowing for variant-specific age patterns), sex, BMI, HIV infection status, household size, crowding, study site, household exposures to SARS-CoV-2, and community SARS-CoV-2 infection prevalence within the cohort. Household exposure intensity was measured as the combined shedding intensity of all actively infected household members (Materials and Methods Section 2.5). We found that the risk of acquiring infection increased with household exposure intensity, with a stronger effect in the proliferation than the clearance phase (Fig. 3C). A one-unit increase in the household exposure intensity during the proliferation stage led to a 103% (95% CI 76% – 136%) increase in the hazard of infection, whereas a one-unit increase in the clearance stage led to a 58% (95% CI 41% – 77%) increase in the hazard of infection. Compared to D614G, we found that infectiousness was highest for the Delta variant followed by the Beta variant, after adjusting for household and community exposure intensity, among other risk factors (hazard ratio against D614G: Delta 1.96, 95% CI 1.27 – 3.05, Beta 1.51, 95% CI 1.03 – 2.21). The difference between Delta and Beta’s infectiousness was not statistically different (P > 0.05), with overlapping confidence intervals in their hazard ratios. We found that prior infection provided durable protection against reinfection throughout the study period. Compared to seronegative individuals, prior infection was 92% (95% CI 84 – 96%) protective against reinfection for the first 3 months and decreased marginally to 87% (95% CI 78 – 92%) after 9 months (Fig. 3C). Individuals older than 65 years were significantly less affected (P<0.05) during the D614G wave whereas children and adolescents aged 6 – 18 years were significantly more affected (P<0.05) during the Delta wave (Fig. 3C). In addition, higher BMI (P < 0.01) and residing in an urban setting (P<0.01) were independently associated with increased risk of SARS-CoV-2 infection. We did not find significant associations between SARS-CoV-2 infection risk and sex, household size, HIV infection status or crowding (Fig. 3C).
Projecting the Omicron wave and post-Omicron futures in PHIRST-C’s urban site.
The study cohorts provide estimates of the duration and degree of cross-protective immunity between SARS-CoV-2 variants predating Omicron, with evidence of persistence of clinical protection beyond a year for these variants. Building on these estimates, we used mathematical models to explore a range of plausible scenarios compatible with the observed transmission dynamics of the Omicron epidemic wave in South Africa. Specifically, we explored how Omicron’s potential differences (relative to Delta) in infectivity, immune evasion, and severity could shape the scale and severity of the Omicron epidemic wave and the likelihood of recurrences of SARS-CoV-2 outbreaks post-Omicron. We focused the analysis on the study’s urban cohort, which was sampled from the city of Klerksdorp in Dr Kenneth Kaunda Health District located on a national transport route, and the population was well-mixed with the district population.
We built and calibrated a transmission model to infer the COVID-19 epidemic trajectory as the Omicron variant took over and eventually replaced the Delta variant, and project potential resurgences in the aftermath of Omicron. To briefly summarize, we utilized PHIRST-C’s urban site data and district-level SARS-CoV-2 surveillance information to reconstruct SARS-CoV-2’s antigen exposure(s) history in this population, considering infections and vaccinations through time and until August 2021. We then built a variant-specific transmission model to capture the declining phase of the Delta wave after the end of the PHIRST-C study and infer the population-level transmission rate during that time. Next, we added a second strain to our model to account for Omicron’s dynamics, with free parameters representing the relative infectiousness of Omicron vs Delta (ratio of basic reproduction numbers , Omicron’s degree of immune evasion against infection among individuals infected by prior variants , and Omicron’s degree of immune evasion against transmission given reinfection (with higher values of indicating higher degrees of immune evasion, where corresponds to no evasion while whereas indicates 100% evasion, detailed in Materials and Methods Section 4). We explored the parameter space and initial conditions of Omicron’s introduction and selected epidemic trajectories that were compatible with the observed growth advantage of Omicron over Delta and the peak timing of the Omicron wave – the two most reliable epidemiologic observations during the Omicron epidemic. We then evaluated Omicron’s possible epidemic trajectories for the remaining parameters conditional on the observed growth rate and peak timing. Lastly, we selected a reference scenario that was most compatible with independent evidence on the degree of Omicron’s immune evasion and projected the likelihood of resurgence by different variants.
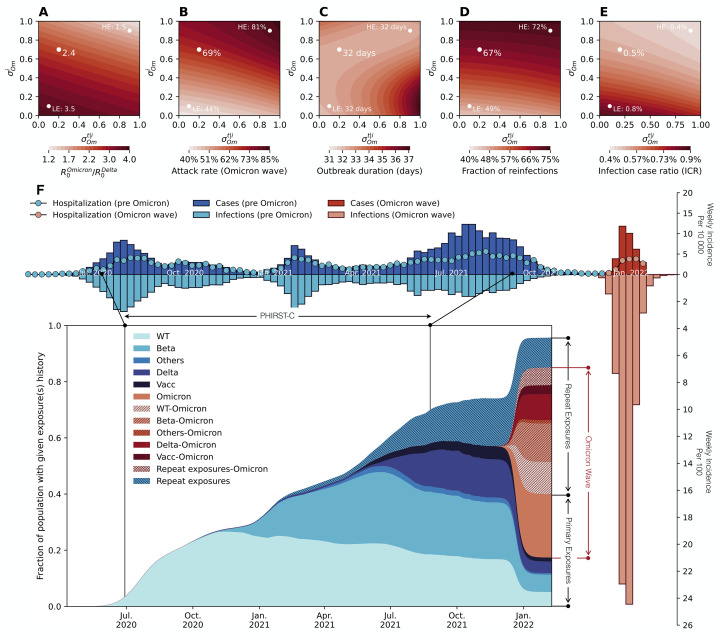
Assuming that the Delta variant was in an exponentially declining phase after week 35 of 2021 and that the Omicron variant was growing exponentially until week 48 of 2021 in the study urban district ( 20 ), we estimated that the daily growth rate was -0.063 for Delta and 0.275 for Omicron, which translated into a growth advantage of 0.338 per day for Omicron over Delta (fig. S7). The Delta and Omicron transmission models were calibrated to match their observed growth rates during this period, respectively. In Fig. 4A, we show the trade-off between the estimated ratio of basic reproduction numbers between Omicron and Delta and different degrees of evasion of protection against infection and transmission . We assumed mean intrinsic generation times of 5 and 4 days for Delta and Omicron ( 23 ). We found that across the full range of immune evasion parameters, Omicron had a higher basic reproduction number than Delta. However, there was clear compensation between immune evasion and intrinsic transmissibility: a higher degree of immune evasion would require a lower basic reproduction number for the Omicron variant to match the observed epidemic trajectory. We also found that for all parameters explored, the Omicron epidemic led to higher infection rates than prior epidemic waves, with the most optimistic scenario resulting in an infection rate above 40%. We projected that Omicron’s infection rate positively correlated with its immune-evasive property (Fig. 4B). Omicron infections were expected to accumulate within a relatively short period of time, with epidemic duration (measured as 4 times the standard deviation of the onset dates of all infections within the epidemic wave) projected to range from 31 – 37 days depending on the parameters. Notably, our model projected that a large fraction of Omicron infections (>40%) would be reinfections or vaccine breakthrough infections, with higher proportions observed for higher immune evasion parameters (Fig. 4D). Furthermore, a low rate of clinical cases was projected for Omicron, after controlling for the number of infections due to this variant. The projected infection case ratio (ICR) for Omicron ranged from 0.4% to 0.9% (Fig. 4E), much lower than the ICRs of 3.6%, 3.3%, and 9.4% estimated for the D614G, Beta, and Delta waves (table S2). In sensitivity analyses (fig. S9), we further explored the impact of Omicron mean generation times ranging from 3-6 days ( 24 ).
Fig. 4. Modelling the plausible epidemic trajectories of the SARS-CoV-2 Omicron wave in the urban site district.
(A) Phase diagram of estimated reproduction number ratio between Omicron and Delta ( ) as a function of immune escape parameters ( =escape on infection) and (escape on transmission reduction conditional on infection). Parameters shown are those that matched the observed growth advantage of Omicron over Delta and the timing of the Omicron peak in the urban district of the PHIRST-C study. (B-E) Phase diagram of the infection rate of the Omicron wave (B), epidemic duration (C), the fraction of reinfections (D), and the infection case ratio (ICR) (E) of the Omicron wave as a function of and and the corresponding in (A). In (A)-(E), white dots marks three specific scenarios including a reference scenario (RS) with and , a low immune escape scenario (LE) with and , and a high immune escape scenario (HE) with and . (F) For the reference scenario (dot in the middle), reconstruction of infection time series and exposure histories by variant are shown. Top panel y axis upwards: Weekly incidence per 10,000 individuals of SARS-CoV-2 cases reported to the District till January 2022, in the period before Omicron (dark blue bars) and during Omicron (dark red bars). Top panel y axis downwards: Weekly incidence per 100 individuals of SARS-CoV-2 infections reconstructed based on PHRIST-C data (prior to September 2021) and estimated using Delta/Omicron-specific transmission models from September 2021 to the end of the Omicron wave at the end of January 2022. Pre-Omicron infections are in light blue and Omicron infections are in light red. For the top panel, the y axis upwards and downwards have different scales (by a factor of 100). Insert panel: the prevalence of the population with specific SARS-CoV-2 antigen exposure histories. Legend abbreviations: D614G: individuals who only experienced one D614G infection; Beta: individuals who only experienced one Beta infection; Delta: individuals who only experienced one Delta infection; Omicron: individuals who only experienced one Omicron infection; Others: individuals who only experienced one SARS-CoV-2 infection with genotype other than the D614G, Beta, Delta and Omicron variants; Vacc: individuals who had received at least one dose of vaccines but had not yet been infected by SARS-CoV-2; Vacc-Omicron: individuals who were vaccinated first then infected by Omicron; D614G-Omicron: individuals who were first infected by D614G then infected by Omicron; Beta-Omicron: individuals who were infected by Beta first then infected by Omicron; Delta-Omicron: individuals who were infected by Delta first then infected by Omicron; Others-Omicron: individuals who were infected by a variant other than D614G, Beta, Delta and Omicron first then infected by Omicron; Repeat exposures: individuals who were exposed to SARS-CoV-2 antigens more than once (through vaccination or infection) without Omicron infection; Repeat exposures-Omicron: individuals who were exposed to SARS-CoV-2 antigens more than twice (through vaccination or infection) then infected by Omicron.
To estimate the immune footprint of the Omicron wave and project a post-Omicron future, we considered a reference scenario for Omicron’s immune evasion characteristics, guided by independent data. We set the degrees of evasion of protection against infection and transmission corresponding to a drop in protection against infection from 88% for pre-Omicron variants (Fig. 2J) to 47% for Omicron ( 17 , 25 ), and a drop in protection against transmission from 60% ( 26 ) to 52%, reflecting weak immune evasion on onward transmission ( 18 ). Under this scenario, the estimated ratio of Omicron vs. Delta’s basic reproduction number was 2.4, the infection rate was 69%, the epidemic lasted 32 days, and the fraction of reinfections and vaccine breakthroughs was 68% (Fig. 4A-D, white dot). In Fig. 4F, we visualized the observed incidence of reported SARS-CoV-2 cases in all four epidemic waves in the study district and report the reconstructed infection time series and population-level history of SARS-CoV-2 antigen exposures. We found variable degrees of under-reporting depending on the SARS-Cov-2 variant, with infection case ratio (ICR) of 3.6% (95% CI 3.4 – 3.8%) for D614G, 3.3% (95%CI 3.0 – 3.6%) for Beta, 9.4% (95%CI 8.7 – 10.2%) for Delta (table S2), and 0.5% for Omicron’s reference scenario projection (Fig. 4E). Findings for the D614G and Beta wave in agreement with previous findings ( 27 ). For the reference scenario, more than 90% of the population was projected to have been infected with one or more SARS-CoV-2 variants by the end of the Omicron wave (Fig. 4F). In particular, we estimated that 22.5% of the population would have seen Omicron as their first SARS-CoV-2 exposure, 16.8% would have been exposed to a pre-Omicron variant and this would remain their only SARS-CoV-2 exposure, and 45.7% would have experienced Omicron reinfections or vaccine breakthrough infections. In a sensitivity analysis (fig. S5), we further explored a high immune escape scenario where and and a low immune escape scenario where and . Comparing to the reference scenario, the high immune escape scenario corresponded to a more moderate increase in transmissibility (basic reproduction number 1.5 times of the Delta variant, Fig. 4A), a higher infection attack rate (81%, Fig. 4B), and a larger proportion of reinfections (72%, Fig. 4D). In contrast, the low immune escape scenario corresponded to a substantial increase in transmissibility (basic reproduction number 3.5 times of the Delta variant, Fig. 4A), a lower infection attack rate (44%, Fig. 4B), and a smaller proportion of reinfections (49%, Fig. 4D). The epidemic trajectories and build-up of population immunity for the low and high immune escape scenarios are reported in fig. S5A and S5B respectively.
Because the Omicron variant is antigenically distinct from all previously circulating variants in South Africa ( 14 , 15 , 28 ), the degree of immunity conferred by Omicron’s primary and breakthrough infections/reinfections against itself, other circulating variants, and new variants is a key determinant for the likelihood of SARS-CoV-2 resurgence ( 29 , 30 ). To study the post-Omicron phase, we explored how different exposure histories could confer different levels of protection against infection with homologous and heterologous variants. Accordingly, in our model, protection varied both with the variant that conferred immunity (either through infection or vaccine) and the hypothetical variant circulating in the post-Omicron future. Protection was expressed as relative risk compared to naïve individuals (where indicates no protection while indicates perfect protection). For simplicity, we assumed that protection against transmission remained constant at 60% ( =0.4 ( 26 )). We assessed the risk of future recurrences of different SARS-CoV-2 variants after the initial Omicron wave had subsided, assuming possible changes in contacts to account for erosion in adherence to SARS-CoV-2 interventions over time (models detailed in Materials and Methods Section 5).
Even at the contact rate estimated during the Delta wave, the extent of population immunity would not be able to prevent a recuring Omicron epidemic unless past Omicron infection conferred high and durable protection against itself (fig. S10A). If contact rates increased 100% relative to current rates, a return of the Omicron variant would likely cause outbreaks irrespective of the protection afforded by prior Omicron infections (where outbreak conditions are defined as growth rate larger than zero, fig. S10B). A 100% increase in contacts may be plausible given the estimated transmission reduction in South Africa at the end of 2021, which reflects the combined effect of population behavior and seasonality (Materials and Methods Section 4.5 and table S5). In contrast, if contact rates were to remain the same as those observed during the Delta wave, the Delta variant would be unlikely to return and cause outbreaks across all ranges of Omicron-specific immunity assumptions against Delta (fig. S10C). With 100% higher contact rates, some scenarios favored a re-emergence of Delta if immunity induced by Omicron did not protect well against Delta (fig. S10D). We also explored scenarios involving a hypothetical new variant X with the same basic reproduction number and generation time as the Delta variant, and at equal antigenic distance from Omicron and pre-Omicron variants. Accordingly, the relative risk of reinfection with variant X was assumed to be the same irrespective of whether an individual was primed with pre-Omicron or Omicron antigens. With an increased contact rate compared to Delta (fig. S10D), we found more opportunities for variant X to cause recuring epidemic waves in the explored ranges of parameters, primarily by escaping immunity conferred by pre-Omicron variants (fig. S10E). Emerging variants with such antigenic features need to be closely monitored in the future. On the other hand, if heterologous prime and boost infections (accounting for 45.7% of the population, Fig. 4F) were found to elicit broadly protective antibodies and confer high cross-protection against variant X, only a small parameter space would be favorable to variant X epidemics (fig. S10E). Given that a large fraction of the world’s population has been primed by vaccination or infection with pre-Omicron antigens, it is important to understand how heterologous boosting by Omicron may broaden the immune repertoire, and how this could translate into clinical protection against different antigenic variants.
DISCUSSION
For a period of 13 months, the PHIRST-C study carefully monitored SARS-CoV-2 infections in 222 households at a rural and an urban site in South Africa. These data provide a unique opportunity to characterize variant-specific shedding kinetics, transmission dynamics within the household, and the degree of immune protection conferred by prior infection before the Omicron surge. Longitudinal rRT-PCR data available for each infection episode at roughly 3-day resolution allowed for reconstruction of the intensity of SARS-CoV-2 exposures exerted on each household member based on Ct values. We found that individuals were more infectious in the RNA proliferation than clearance stage. Prior to the emergence of Omicron, substantial population immunity had accumulated through prior infection, with high and durable protection against symptomatic and asymptomatic reinfection, in line with prior findings ( 31 , 32 ). These detailed cohort data allowed us to project the full scope of the Omicron epidemic and assess possible futures. Overall, even with a high degree of immunity post the Omicron wave (with over 90% of the population previously exposed to SARS-CoV-2 antigens), recurrence of past or antigenically novel variants is plausible, especially if post-Omicron behavioral changes increase contacts.
The disease burden of future SARS-CoV-2 epidemic waves depends on the intrinsic severity of the variants themselves as well as the degree of protection conferred by pre-existing immunity ( 33 ). Current evidence suggests that although Omicron is able to evade immunity against infection to a significant degree, protection against severe outcomes remains high ( 17 , 34 , 35 ). Our model’s projections also suggest a low clinical burden of Omicron, with a greater than 10-fold reduction in infection case ratio relative to prior waves. If existing immunity can sustain protection against severe outcomes over long timescales, the disease burden of future epidemic waves would be attenuated even if infections were widespread. However, if protection against severe outcomes waned over time, vaccine boosting would likely be needed to compensate for loss of protection.
As the PHIRST-C sampling scheme was not symptom-driven, it allowed us to capture shedding kinetics in both symptomatic and asymptomatic individuals. We observed that most infections 86.8% (581/669) were asymptomatic, but asymptomatically infected individuals transmitted the virus within their households. For context, in the South African winter seasons of 2017-2018, approximately half of the influenza infections within the PHIRST-C cohorts were asymptomatic ( 36 ). The transmission potential of asymptomatic or pre-symptomatic SARS-CoV-2 infections identified in our study is in sharp contrast with SARS-CoV-1, where most transmission occurs after symptom onset ( 37 ). It is also worth noting in our cohort data, prior infections, whether symptomatic or not, conferred durable protection against reinfection. We found that the shedding kinetics of SARS-CoV-2 were characterized by a rapid RNA proliferation stage until peak viral load, followed by a more gradual RNA clearance stage. The median duration of rRT-PCR positivity lasted 10.5 days (IQR 6.3 – 14.0 days) with median peak Ct = 23.1 (IQR 20.0 – 27.2), in agreement with other high-frequency sampling studies ( 21 , 22 ). Reinfections had shorter durations of rRT-PCR positivity and lower shedding peaks compared to primary infections, which would be expected to decrease the probability of onward transmission. Our findings align with reports of reduction in viral shedding among vaccine breakthroughs relative to primary infections, prior to the occurrence of the Omicron variant ( 38 – 41 ). We observed variation in infectiousness through the course of infection, after adjusting for Ct values, whereby an individual in the proliferation stage tends to be more infectious than one in the clearance stage. The post-peak decline in infectiousness coincides with the onset of adaptive immune responses that work to suppress the on-going infection ( 42 ). The observed decline in infectiousness in the RNA clearance stage also could be due to neutralization of some viral particles by antibodies, precluding productive transmission.
The peak of COVID-19 hospitalizations during the Omicron wave was lower than that during the Delta wave in South Africa ( 43 ), despite our model projecting a much higher infection rate for Omicron than Delta. This is compatible, however, with some lines of evidence suggesting a lower severity of the Omicron variant in naïve individuals, combined with robust infection and vaccine-induced immunity against severe Omicron disease ( 17 , 44 ). However, it is important to stress that, in South Africa, immunity accumulated prior to the Omicron wave was mostly through prior infections due to a delayed start and slow rollout of the vaccine campaign ( 45 ). As a result, the proportion of the population infected by pre-Omicron variants was substantially higher in South Africa than in countries that experienced faster vaccine rollout or effective mitigation strategies.
Our study has several limitations. First, our findings about the persistence of infection-induced immunity are based on a 13-month study. The duration and quality of protective immunity over longer timescales remain open questions. Recent studies have found that antibody responses improve over time through affinity maturation ( 46 , 47 ) and that long-lived plasma cells can be identified in the bone marrow at least one year after infection, suggesting that immunity conferred by infection or vaccination could be potent and durable against non-immune evasive variants ( 48 ). Persistent germinal center responses and durable T cell memory have also been observed among vaccine recipients ( 49 – 51 ). However, how the protection holds up against immune-evasive variant such as Omicron remains an outstanding question. Unfortunately, the PHIRST-C cohorts did not cover the Omicron wave, thus we could not directly measure immune protection at the individual level and relied on modelling of population-level dynamics. Post-Omicron serologic surveys following up the cohort population could provide deeper insight into the full impact of the Omicron wave. In our projections of SARS-CoV-2 resurgences, we did not consider waning explicitly because our cohort data did not support pronounced waning of infection-induced immunity. Accordingly, our projections are most relevant to short time scales, in the order of a few months. We however found that resurgences are likely even over short time horizons. A second limitation relates to missed infection episodes, despite frequent rRT-PCR testing. In total, 21% (303407 person-days/1472400 person days) of the total person-days of observation were excluded from the regression during the entire study period due to missing nasal swab visits, missing serologic status, or experiencing an active infection episode. For example, 14% (90/639) of individuals who seroconverted during the study lacked rRT-PCR confirmation of active infections (that is, their first serologic test was negative, but they seroconverted later). This could possibly be due to delayed seroconversion from infection episodes occurring prior to the first nasal specimen, infection episodes that occurred during missed routine household visits, a shorter duration of rRT-PCR positivity than the interval between consecutive nasal swabbing (3 days), false positives in serology test results, or false negatives of the rRT-PCR assay specimen (due to specimen quality issues or detection limit of the rRT-PCR). A third limitation relates to the simplicity of the contact structure in our transmission models. Projections of the trajectories of Delta, Omicron, and hypothetical variant X did not address heterogeneity due to age-specific susceptibility, transmissibility, and contact patterns. Nor did we consider individual variation in infection and vaccine-derived protection. Heterogeneity in mixing patterns and immune protection could lead to a lower infection rate when compared to homogeneous models with the same basic reproduction number ( 52 , 53 ), thus the size of the Omicron epidemic could be inflated in our projections. In addition, our size projections could be inflated if the Omicron’s serial interval was shorter (corresponding to a lower basic reproduction number) than that the range of values explored. Population surveys on active infections during the Omicron wave as well as paired-sera surveys before and after the Omicron wave will be necessary to confirm the true scale of the Omicron epidemic.
In conclusion, our study provides an in-depth analysis of the kinetics of viral shedding, transmission dynamics, and persistence of immunity conferred by sequential exposures to different SARS-CoV-2 variants, and how these factors contribute to shaping the Omicron and post-Omicron phases. We found durable cross-protective immunity conferred by prior infection against pre-Omicron variants. However, Omicron successfully breached population immunity due to a combination immune escape and increased transmissibility, reinfecting a large fraction of the population and leaving a complex immune landscape in its aftermath. With increasing contacts as the Omicron wave subsides, several possible scenarios for SARS-CoV-2 recurrences are possible, involving both old and hypothetical new variants. Further work on how immunity may strengthen and broaden upon sequential exposures with different variants and vaccination episodes will be important to clarify the next phase of the pandemic.
MATERIALS AND METHODS
Study design: We conducted a prospective household cohort study of SARS-CoV-2 transmission at an urban and a rural site in South Africa from July 2020 to August 2021 ( 4 ). The rural site was in Agincourt, a rural community in Mpumalanga Province, which has been a longstanding health and socio-demographic surveillance system. The urban site was in Klerksdorp, an urban community located in the North West Province. This study was built upon the larger multi-year Prospective Household cohort study of Influenza, Respiratory Syncytial virus and other respiratory pathogens community burden and Transmission dynamics (PHIRST), which was conducted from 2016 to 2018 to monitor transmission of respiratory pathogens ( 54 ). The study was repurposed for SARS-CoV-2 during the pandemic. To study infection and reinfection with SARS-CoV-2, a total of 222 households (114 in the rural site and 108 in the urban site) with at least 3 household members were enrolled between July 2020 and August 2021, consisting of 638 and 557 participants in the rural and urban site, respectively. In the rural site, we first approached households from the 2017 and 2018 cohorts, and in the urban site, those from the 2016, 2017, and 2018 cohorts. To supplement the sample size of the cohorts, we enrolled additional households at each site using the same methods as for the initial PHIRST study. Baseline demographic factors and information on underlying medical conditions were collected at enrollment (Table 1). Throughout the study period, household members were visited twice a week by study nurses and trained lay field workers for collection of biological and clinical data. At each visit, upper respiratory tract specimens were collected using mid-turbinate nasal swabs, irrespective of symptom presentation. Data on symptoms, healthcare seeking behavior, hospitalization, and death were captured at each follow up visit on a REDCap tablet-based real-time database. Respiratory specimens were tested by rRT-PCR for SARS-CoV-2. The lineage of positive rRT-PCR specimens was determined by variant-specific rRT-PCR assay. Sera were collected at enrollment and approximately every 2 months during the 11-month follow-up period from all participants (see Fig. 1 for timeline) and tested for the presence of SARS-CoV-2 antibodies.
Ethics statement: The PHIRST-C protocol was approved by the University of Witwatersrand Human Research Ethics Committee (Reference 150808) and the U.S. Centers for Disease Control and Prevention’s Institutional Review Board relied on the local review (#6840). The protocol was registered on clinicaltrials.gov on 6 August 2015 and updated on 30 December 2020 (https://clinicaltrials.gov/ct2/show/NCT02519803). Participants received grocery store vouchers of ZAR50 (USD 3) per visit to compensate for time required for specimen collection and interview.
Statistical analysis: For regression analyses of SARS-CoV-2 viral RNA peak shedding, we fitted a generalized linear model with gamma distributed error and identity link function, with two-tailed z-test used to determine statistical significance. For regression analyses of SARS-CoV-2 viral RNA shedding duration, we fitted a linear regression model to the data. The fitted model passed both homoscedasticity and normality tests, with two-tailed t test used to determine statistical significance. For regression analyses of risk factors for SARS-CoV-2 infection and re-infection, we used mixed-effects Poisson regression (Materials and Methods Section 2.5), and two-tailed z-tests to determine statistical significance. We also present a sensitivity analysis of risk factors associated with SARS-CoV-2 infection using mixed-effects logistic regression (Materials and Methods Section 2.5), with two-tailed z-tests to determine statistical significance. P<0.05 was considered significant. All regression analysis were performed using R package lme4 version 1.1-27.1 under R version 4.1.2.
Details on the SARS-CoV-2 transmission models and calibration procedures to project the trajectory of the Omicron and post-Omicron waves can be found in Materials and Methods Section 3-5.
Acknowledgments
Funding: This work was supported by the National Institute for Communicable Diseases of the National Health Laboratory Service and the U.S. Centers for Disease Control and Prevention [cooperative agreement number: 6 U01IP001048] and Wellcome Trust (grant number 221003/Z/20/Z) in collaboration with the Foreign, Commonwealth and Development Office, United Kingdom. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the NIH or the U.S. Centers for Disease Control and Prevention.
Author contributions: KS, ST, JK, AvG, MLM, NW, JM, NAM, KK, STo, LL, CV, CC designed the experiments. CC, JK, and ST accessed and verified the underlying data. ST, JK, AvG, MLM, NW, JNB, JM, MdP, MC, AB, NAM, KK, STo, LL, FW, JdT, FXG, FSD, TMK, CC collected the data and performed laboratory experiments. KS, ST, JK, AvG, MLM, NW, JNB, JM, MdP, MC, AB, NAM, KK, STo, LL, FW, JdT, FXG, FSD, TMK, CV, and CC analyzed the data and interpreted the results. KS, ST, JK, AvG, CV, and CC drafted the manuscript. All authors critically reviewed the article and had access to all the data reported in the study.
Competing interests: CC has received grant support from Sanofi Pasteur, Advanced Vaccine Initiative, and payment of travel costs from Parexel. AvG has received grant support from Sanofi Pasteur, Pfizer related to pneumococcal vaccine, CDC and the Bill & Melinda Gates Foundation. NW reports grants from Sanofi Pasteur and the Bill & Melinda Gates Foundation. NAM has received a grant to his institution from Pfizer to conduct research in patients with pneumonia and from Roche to collect specimens to assess a TB assay. JM has received grant support from Sanofi Pasteur.
Data and materials availability: All data associated with this study are present in the paper or supplementary materials. Code and aggregate data to reproduce the figures, regression analyses, and transmission model for the Omicron wave are available at 10.5281/zenodo.6544507. To access data, including individual participant data and a data dictionary defining each field in the data set, please submit a proposal to Professor Cheryl Cohen. These data can be made available through a data access agreement or materials transfer agreement.
This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using this material.
Supplementary Materials
This PDF file includes:
Other Supplementary Material for this manuscript includes the following:
MDAR Reproducibility Checklist
References and Notes
- 1. Buss L. F., Prete C. A. Jr., Abrahim C. M. M., Mendrone A. Jr., Salomon T., de Almeida-Neto C., França R. F. O., Belotti M. C., Carvalho M. P. S. S., Costa A. G., Crispim M. A. E., Ferreira S. C., Fraiji N. A., Gurzenda S., Whittaker C., Kamaura L. T., Takecian P. L., da Silva Peixoto P., Oikawa M. K., Nishiya A. S., Rocha V., Salles N. A., de Souza Santos A. A., da Silva M. A., Custer B., Parag K. V., Barral-Netto M., Kraemer M. U. G., Pereira R. H. M., Pybus O. G., Busch M. P., Castro M. C., Dye C., Nascimento V. H., Faria N. R., Sabino E. C., Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science 371, 288–292 (2021). 10.1126/science.abe9728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bhuiyan T. R., Hulse J. D., Hegde S. T., Akhtar M., Islam T., Khan Z. H., Khan I. I., Ahmed S., Rashid M., Rashid R., Gurley E. S., Shirin T., Khan A. I., Azman A. S., Qadri F., SARS-CoV-2 Seroprevalence before Delta Variant Surge, Chattogram, Bangladesh, March-June 2021. Emerg. Infect. Dis. 28, 429–431 (2022). 10.3201/eid2802.211689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huete-Pérez J. A., Ernst K. C., Cabezas-Robelo C., Páiz-Medina L., Silva S., Huete A., Prevalence and risk factors for SARS-CoV-2 infection in children with and without symptoms seeking care in Managua, Nicaragua: Results of a cross-sectional survey. BMJ Open 11, e051836 (2021). 10.1136/bmjopen-2021-051836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cohen C., Kleynhans J., von Gottberg A., McMorrow M. L., Wolter N., Bhiman J. N., Moyes J., du Plessis M., Carrim M., Buys A., Martinson N. A., Kahn K., Tollman S., Lebina L., Wafawanaka F., du Toit J. D., Gómez-Olivé F. X., Dawood F. S., Mkhencele T., Sun K., Viboud C., Tempia S.; PHIRST-C Group , SARS-CoV-2 incidence, transmission, and reinfection in a rural and an urban setting: Results of the PHIRST-C cohort study, South Africa, 2020-21. Lancet Infect. Dis. 22, 821–834 (2022). 10.1016/S1473-3099(22)00069-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tegally H., Wilkinson E., Lessells R. J., Giandhari J., Pillay S., Msomi N., Mlisana K., Bhiman J. N., von Gottberg A., Walaza S., Fonseca V., Allam M., Ismail A., Glass A. J., Engelbrecht S., Van Zyl G., Preiser W., Williamson C., Petruccione F., Sigal A., Gazy I., Hardie D., Hsiao N.-Y., Martin D., York D., Goedhals D., San E. J., Giovanetti M., Lourenço J., Alcantara L. C. J., de Oliveira T., Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat. Med. 27, 440–446 (2021). 10.1038/s41591-021-01255-3 [DOI] [PubMed] [Google Scholar]
- 6. Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E. J., Msomi N., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A. J., Engelbrecht S., Van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao M., Korsman S., Davies M.-A., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkordi A., Godzik A., Wibmer C. K., Sewell B. T., Lourenço J., Alcantara L. C. J., Kosakovsky Pond S. L., Weaver S., Martin D., Lessells R. J., Bhiman J. N., Williamson C., de Oliveira T., Emergence of a SARS-CoV-2 variant of concern with mutations in spike glycoprotein. Nature (2021). 10.1038/s41586-021-03402-9 [DOI] [PubMed] [Google Scholar]
- 7. Viana R., Moyo S., Amoako D. G., Tegally H., Scheepers C., Althaus C. L., Anyaneji U. J., Bester P. A., Boni M. F., Chand M., Choga W. T., Colquhoun R., Davids M., Deforche K., Doolabh D., du Plessis L., Engelbrecht S., Everatt J., Giandhari J., Giovanetti M., Hardie D., Hill V., Hsiao N.-Y., Iranzadeh A., Ismail A., Joseph C., Joseph R., Koopile L., Kosakovsky Pond S. L., Kraemer M. U. G., Kuate-Lere L., Laguda-Akingba O., Lesetedi-Mafoko O., Lessells R. J., Lockman S., Lucaci A. G., Maharaj A., Mahlangu B., Maponga T., Mahlakwane K., Makatini Z., Marais G., Maruapula D., Masupu K., Matshaba M., Mayaphi S., Mbhele N., Mbulawa M. B., Mendes A., Mlisana K., Mnguni A., Mohale T., Moir M., Moruisi K., Mosepele M., Motsatsi G., Motswaledi M. S., Mphoyakgosi T., Msomi N., Mwangi P. N., Naidoo Y., Ntuli N., Nyaga M., Olubayo L., Pillay S., Radibe B., Ramphal Y., Ramphal U., San J. E., Scott L., Shapiro R., Singh L., Smith-Lawrence P., Stevens W., Strydom A., Subramoney K., Tebeila N., Tshiabuila D., Tsui J., van Wyk S., Weaver S., Wibmer C. K., Wilkinson E., Wolter N., Zarebski A. E., Zuze B., Goedhals D., Preiser W., Treurnicht F., Venter M., Williamson C., Pybus O. G., Bhiman J., Glass A., Martin D. P., Rambaut A., Gaseitsiwe S., von Gottberg A., de Oliveira T., Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 603, 679–686 (2022). 10.1038/s41586-022-04411-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pearson C. A. B., Silal S. P., Li M. W. Z., Dushoff J., Bolker B. M., Abbott S., van Schalkwyk C., Davies N. G., Barnard R. C., Edmunds W. J., Bingham J., Meyer-Rath G., Jamieson L., Glass A., Wolter N., Govender N., Stevens W. S., Scott L., Mlisana K., Moultrie H., Pulliam J. R. C., Bounding the levels of transmissibility and immune evasion of the Omicron variant in South Africa. bioRxiv (2021), doi:. 10.1101/2021.12.19.21268038 [DOI]
- 9.Alaa Abdel Latif, Julia L. Mullen, Manar Alkuzweny, Ginger Tsueng, Marco Cano, Emily Haag, Jerry Zhou, Mark Zeller, Emory Hufbauer, Nate Matteson, Chunlei Wu, Kristian G. Andersen, Andrew I. Su, Karthik Gangavarapu, Laura D. Hughes, and the Center for Viral Systems Biology, Omicron Variant Report. outbreak.info (2022) (available at https://outbreak.info/situation-reports/omicron?loc=ZAF&loc=GBR&loc=USA&selected=ZAF).
- 10.the Nextstrain team, Genomic epidemiology of novel coronavirus - Global subsampling. Nextstrain (2022) (available at https://nextstrain.org/ncov/gisaid/global).
- 11. Pulliam J. R. C., van Schalkwyk C., Govender N., von Gottberg A., Cohen C., Groome M. J., Dushoff J., Mlisana K., Moultrie H., Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science 376, eabn4947 (2022). 10.1126/science.abn4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.N. Ferguson, A. Ghani, A. Cori, A. Hogan, W. Hinsley, Erik Volz on behalf of the Imperial College COVID-19 response team, Report 49 - Growth, population distribution and immune escape of Omicron in England (MRC Centre for Global Infectious Disease Analysis, Imperial College London, 2021; https://www.imperial.ac.uk/mrc-global-infectious-disease-analysis/covid-19/report-49-omicron/).
- 13. van der Straten K., Guerra D., van Gils M. J., Bontjer I., Caniels T. G., van Willigen H. D. G., Wynberg E., Poniman M., Burger J. A., Bouhuijs J. H., van Rijswijk J., Lavell A. H. A., Appelman B., Sikkens J. J., Bomers M. K., Han A. X., Nichols B. E., Prins M., Vennema H., Reusken C., de Jong M. D., de Bree G. J., Russell C. A., Eggink D., Sanders R. W., Mapping the antigenic diversification of SARS-CoV-2. bioRxiv (2022), doi:. 10.1101/2022.01.03.21268582 [DOI]
- 14. Cele S., Jackson L., Khoury D. S., Khan K., Moyo-Gwete T., Tegally H., San J. E., Cromer D., Scheepers C., Amoako D. G., Karim F., Bernstein M., Lustig G., Archary D., Smith M., Ganga Y., Jule Z., Reedoy K., Hwa S.-H., Giandhari J., Blackburn J. M., Gosnell B. I., Abdool Karim S. S., Hanekom W., von Gottberg A., Bhiman J. N., Lessells R. J., Moosa M. S., Davenport M. P., de Oliveira T., Moore P. L., Sigal A.; NGS-SA; COMMIT-KZN Team , Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature 602, 654–656 (2022). 10.1038/s41586-021-04387-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carreño J. M., Alshammary H., Tcheou J., Singh G., Raskin A. J., Kawabata H., Sominsky L. A., Clark J. J., Adelsberg D. C., Bielak D. A., Gonzalez-Reiche A. S., Dambrauskas N., Vigdorovich V., Srivastava K., Sather D. N., Sordillo E. M., Bajic G., van Bakel H., Simon V., Krammer F.; PSP-PARIS Study Group , Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature 602, 682–688 (2022). 10.1038/s41586-022-04399-5 [DOI] [PubMed] [Google Scholar]
- 16. Cameroni E., Bowen J. E., Rosen L. E., Saliba C., Zepeda S. K., Culap K., Pinto D., VanBlargan L. A., De Marco A., di Iulio J., Zatta F., Kaiser H., Noack J., Farhat N., Czudnochowski N., Havenar-Daughton C., Sprouse K. R., Dillen J. R., Powell A. E., Chen A., Maher C., Yin L., Sun D., Soriaga L., Bassi J., Silacci-Fregni C., Gustafsson C., Franko N. M., Logue J., Iqbal N. T., Mazzitelli I., Geffner J., Grifantini R., Chu H., Gori A., Riva A., Giannini O., Ceschi A., Ferrari P., Cippà P. E., Franzetti-Pellanda A., Garzoni C., Halfmann P. J., Kawaoka Y., Hebner C., Purcell L. A., Piccoli L., Pizzuto M. S., Walls A. C., Diamond M. S., Telenti A., Virgin H. W., Lanzavecchia A., Snell G., Veesler D., Corti D., Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature 602, 664–670 (2022). 10.1038/s41586-021-04386-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.UKHSA Genomics Cell UKHSA Outbreak Surveillance Team UKHSA Epidemiology Cell UKHSA Immunisations Team UKHSA Contact Tracing Data Team UKHSA Environmental Monitoring for Health Protection Team UKHSA SIREN Study Team UKHSA Public Health Incident Directors Contributions from the Variant Technical Group Members, SARS-CoV-2 variants of concern and variants under investigation in England Technical briefing 34 (UK Health Security Agency, 2022; https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1046853/technical-briefing-34-14-january-2022.pdf).
- 18. Lyngse F. P., Mortensen L. H., Denwood M. J., Christiansen L. E., Møller C. H., Skov R. L., Spiess K., Fomsgaard A., Lassaunière M. M., Rasmussen M., Stegger M., Nielsen C., Sieber R. N., Cohen A. S., Møller F. T., Overvad M., Mølbak K., Krause T. G., Kirkeby C. T., SARS-CoV-2 Omicron VOC transmission in Danish households. bioRxiv (2021), doi:. 10.1101/2021.12.27.21268278 [DOI]
- 19. Ulloa A. C., Buchan S. A., Daneman N., Brown K. A., Early estimates of SARS-CoV-2 Omicron variant severity based on a matched cohort study, Ontario, Canada. bioRxiv (2021), doi:. 10.1101/2021.12.24.21268382 [DOI]
- 20. WEEKLY EPIDEMIOLOGICAL BRIEF - NICD , (2020) (available at https://www.nicd.ac.za/diseases-a-z-index/disease-index-covid-19/surveillance-reports/weekly-epidemiological-brief/).
- 21. Kissler S. M., Fauver J. R., Mack C., Olesen S. W., Tai C., Shiue K. Y., Kalinich C. C., Jednak S., Ott I. M., Vogels C. B. F., Wohlgemuth J., Weisberger J., DiFiori J., Anderson D. J., Mancell J., Ho D. D., Grubaugh N. D., Grad Y. H., Viral dynamics of acute SARS-CoV-2 infection and applications to diagnostic and public health strategies. PLOS Biol. 19, e3001333 (2021). 10.1371/journal.pbio.3001333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones T. C., Biele G., Mühlemann B., Veith T., Schneider J., Beheim-Schwarzbach J., Bleicker T., Tesch J., Schmidt M. L., Sander L. E., Kurth F., Menzel P., Schwarzer R., Zuchowski M., Hofmann J., Krumbholz A., Stein A., Edelmann A., Corman V. M., Drosten C., Estimating infectiousness throughout SARS-CoV-2 infection course. Science 373, eabi5273 (2021). 10.1126/science.abi5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Backer J. A., Eggink D., Andeweg S. P., Veldhuijzen I. K., van Maarseveen N., Vermaas K., Vlaemynck B., Schepers R., van den Hof S., Reusken C. B., Wallinga J., Shorter serial intervals in SARS-CoV-2 cases with Omicron BA.1 variant compared with Delta variant, the Netherlands, 13 to 26 December 2021. Euro Surveill. 27, (2022). 10.2807/1560-7917.ES.2022.27.6.2200042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Institute of Infectious Diseases Disease Control and Prevention Center, National Center for Global Health and Medicine, Active epidemiological investigation on SARS-CoV-2 infection caused by Omicron variant (Pango lineage B.1.1.529) in Japan: preliminary report on infectious period国立感染症研究所 (2022) (available at https://www.niid.go.jp/niid/en/2019-ncov-e/10884-covid19-66-en.html).
- 25.B. J. Willett, J. Grove, O. A. MacLean, C. Wilkie, N. Logan, G. D. Lorenzo, W. Furnon, S. Scott, M. Manali, A. Szemiel, S. Ashraf, E. Vink, W. Harvey, C. Davis, R. Orton, J. Hughes, P. Holland, V. Silva, D. Pascall, K. Puxty, A. da Silva Filipe, G. Yebra, S. Shaaban, M. T. G. Holden, R. M. Pinto, R. Gunson, K. Templeton, P. Murcia, A. H. Patel, J. Haughney, D. L. Robertson, M. Palmarini, S. Ray, E. C. Thomson, The COVID-19 DeplOyed VaccinE (DOVE) Cohort Study investigators, The COVID-19 Genomics UK (COG-UK) Consortium, The G2P-UK National Virology Consortium, The Evaluation of Variants Affecting Deployed COVID-19 Vaccines (EVADE) investigators, The hyper-transmissible SARS-CoV-2 Omicron variant exhibits significant antigenic change, vaccine escape and a switch in cell entry mechanism. bioRxiv (2022), doi:. 10.1101/2022.01.03.21268111 [DOI]
- 26. Braeye T., Cornelissen L., Catteau L., Haarhuis F., Proesmans K., De Ridder K., Djiena A., Mahieu R., De Leeuw F., Dreuw A., Hammami N., Quoilin S., Van Oyen H., Wyndham-Thomas C., Van Cauteren D., Vaccine effectiveness against infection and onwards transmission of COVID-19: Analysis of Belgian contact tracing data, January-June 2021. Vaccine 39, 5456–5460 (2021). 10.1016/j.vaccine.2021.08.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kleynhans J., Tempia S., Wolter N., von Gottberg A., Bhiman J. N., Buys A., Moyes J., McMorrow M. L., Kahn K., Gómez-Olivé F. X., Tollman S., Martinson N. A., Wafawanaka F., Lebina L., du Toit J., Jassat W., Neti M., Brauer M., Cohen C.; PHIRST-C Group , SARS-CoV-2 Seroprevalence in a Rural and Urban Household Cohort during First and Second Waves of Infections, South Africa, July 2020-March 2021. Emerg. Infect. Dis. 27, 3020–3029 (2021). 10.3201/eid2712.211465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rössler A., Riepler L., Bante D., von Laer D., Kimpel J., SARS-CoV-2 Omicron Variant Neutralization in Serum from Vaccinated and Convalescent Persons. N. Engl. J. Med. 386, 698–700 (2022). 10.1056/NEJMc2119236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.K. Khan, F. Karim, S. Cele, J. E. San, G. Lustig, H. Tegally, M. Bernstein, Y. Ganga, Z. Jule, K. Reedoy, N. Ngcobo, M. Mazibuko, N. Mthabela, Z. Mhlane, N. Mbatha, J. Giandhari, Y. Ramphal, T. Naidoo, N. Manickchund, N. Magula, S. S. Abdool Karim, G. Gray, W. Hanekom, A. von Gottberg, COMMIT-KZN Team, B. I. Gosnell, R. J. Lessells, P. L. Moore, T. de Oliveira, M.-Y. S. Moosa, A. Sigal, Omicron infection enhances neutralizing immunity against the Delta variant. medRxiv (2021), doi:. 10.1101/2021.12.27.21268439 [DOI]
- 30. Altarawneh H., Chemaitelly H., Tang P., Hasan M. R., Qassim S., Ayoub H. H., AlMukdad S., Yassine H. M., Benslimane F. M., Khatib H. A. A., Coyle P., Kanaani Z. A., Kuwari E. A., Jeremijenko A., Kaleeckal A. H., Latif A. N., Shaik R. M., Abdul Rahim H. F., Nasrallah G. K., Al Kuwari M. G., Butt A. A., Al Romaihi H. E., Al-Thani M. H., Khal A. A., Bertollini R., Abu-Raddad L. J., Protection afforded by prior infection against SARS-CoV-2 reinfection with the Omicron variant. bioRxiv (2022), doi:. 10.1101/2022.01.05.22268782 [DOI]
- 31.V. J. Hall, S. Foulkes, F. Insalata, A. Saei, P. Kirwan, A. Atti, E. Wellington, J. Khawam, K. Munro, M. Cole, C. Tranquillini, A. Taylor-Kerr, N. Hettiarachchi, D. Calbraith, N. Sajedi, I. Milligan, Y. Themistocleous, D. Corrigan, L. Cromey, L. Price, S. Stewart, E. de Lacy, C. Norman, E. Linley, A. D. Otter, A. Semper, J. Hewson, The SIREN Study Group, M. A. Chand, C. S. Brown, T. Brooks, J. Islam, A. Charlett, S. Hopkins, Effectiveness and durability of protection against future SARS-CoV-2 infection conferred by COVID-19 vaccination and previous infection; findings from the UK SIREN prospective cohort study of healthcare workers March 2020 to September 2021. medRxiv, 2021.11.29.21267006 (2021). 10.1101/2021.11.29.21267006 [DOI]
- 32. Hansen C. H., Michlmayr D., Gubbels S. M., Mølbak K., Ethelberg S., Assessment of protection against reinfection with SARS-CoV-2 among 4 million PCR-tested individuals in Denmark in 2020: A population-level observational study. Lancet 397, 1204–1212 (2021). 10.1016/S0140-6736(21)00575-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sigal A., Milder disease with Omicron: Is it the virus or the pre-existing immunity? Nat. Rev. Immunol. 22, 69–71 (2022). 10.1038/s41577-022-00678-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thompson M. G., Natarajan K., Irving S. A., Rowley E. A., Griggs E. P., Gaglani M., Klein N. P., Grannis S. J., DeSilva M. B., Stenehjem E., Reese S. E., Dickerson M., Naleway A. L., Han J., Konatham D., McEvoy C., Rao S., Dixon B. E., Dascomb K., Lewis N., Levy M. E., Patel P., Liao I.-C., Kharbanda A. B., Barron M. A., Fadel W. F., Grisel N., Goddard K., Yang D.-H., Wondimu M. H., Murthy K., Valvi N. R., Arndorfer J., Fireman B., Dunne M. M., Embi P., Azziz-Baumgartner E., Zerbo O., Bozio C. H., Reynolds S., Ferdinands J., Williams J., Link-Gelles R., Schrag S. J., Verani J. R., Ball S., Ong T. C., Effectiveness of a Third Dose of mRNA Vaccines Against COVID-19-Associated Emergency Department and Urgent Care Encounters and Hospitalizations Among Adults During Periods of Delta and Omicron Variant Predominance - VISION Network, 10 States, August 2021-January 2022. MMWR Morb. Mortal. Wkly. Rep. 71, 139–145 (2022). 10.15585/mmwr.mm7104e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lewnard J. A., Hong V. X., Patel M. M., Kahn R., Lipsitch M., Tartof S. Y., Clinical outcomes among patients infected with Omicron (B.1.1.529) SARS-CoV-2 variant in southern California. bioRxiv (2022), doi:. 10.1101/2022.01.11.22269045 [DOI] [PMC free article] [PubMed]
- 36. Cohen C., Kleynhans J., Moyes J., McMorrow M. L., Treurnicht F. K., Hellferscee O., Mathunjwa A., von Gottberg A., Wolter N., Martinson N. A., Kahn K., Lebina L., Mothlaoleng K., Wafawanaka F., Gómez-Olivé F. X., Mkhencele T., Mathee A., Piketh S., Language B., Tempia S.; PHIRST group , Asymptomatic transmission and high community burden of seasonal influenza in an urban and a rural community in South Africa, 2017-18 (PHIRST): A population cohort study. Lancet Glob. Health 9, e863–e874 (2021). 10.1016/S2214-109X(21)00141-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fraser C., Riley S., Anderson R. M., Ferguson N. M., Factors that make an infectious disease outbreak controllable. Proc. Natl. Acad. Sci. U.S.A. 101, 6146–6151 (2004). 10.1073/pnas.0307506101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pollett S. D., Richard S. A., Fries A. C., Simons M. P., Mende K., Lalani T., Lee T., Chi S., Mody R., Madar C., Ganesan A., Larson D. T., Colombo C. J., Colombo R., Samuels E. C., Broder C. C., Laing E. D., Smith D. R., Tribble D., Agan B. K., Burgess T. H., The SARS-CoV-2 mRNA vaccine breakthrough infection phenotype includes significant symptoms, live virus shedding, and viral genetic diversity. Clin. Infect. Dis. 74, 897–900 (2022). 10.1093/cid/ciab543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thompson M. G., Burgess J. L., Naleway A. L., Tyner H., Yoon S. K., Meece J., Olsho L. E. W., Caban-Martinez A. J., Fowlkes A. L., Lutrick K., Groom H. C., Dunnigan K., Odean M. J., Hegmann K., Stefanski E., Edwards L. J., Schaefer-Solle N., Grant L., Ellingson K., Kuntz J. L., Zunie T., Thiese M. S., Ivacic L., Wesley M. G., Mayo Lamberte J., Sun X., Smith M. E., Phillips A. L., Groover K. D., Yoo Y. M., Gerald J., Brown R. T., Herring M. K., Joseph G., Beitel S., Morrill T. C., Mak J., Rivers P., Poe B. P., Lynch B., Zhou Y., Zhang J., Kelleher A., Li Y., Dickerson M., Hanson E., Guenther K., Tong S., Bateman A., Reisdorf E., Barnes J., Azziz-Baumgartner E., Hunt D. R., Arvay M. L., Kutty P., Fry A. M., Gaglani M., Prevention and Attenuation of Covid-19 with the BNT162b2 and mRNA-1273 Vaccines. N. Engl. J. Med. 385, 320–329 (2021). 10.1056/NEJMoa2107058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Levine-Tiefenbrun M., Yelin I., Katz R., Herzel E., Golan Z., Schreiber L., Wolf T., Nadler V., Ben-Tov A., Kuint J., Gazit S., Patalon T., Chodick G., Kishony R., Initial report of decreased SARS-CoV-2 viral load after inoculation with the BNT162b2 vaccine. Nat. Med. 27, 790–792 (2021). 10.1038/s41591-021-01316-7 [DOI] [PubMed] [Google Scholar]
- 41. Regev-Yochay G., Amit S., Bergwerk M., Lipsitch M., Leshem E., Kahn R., Lustig Y., Cohen C., Doolman R., Ziv A., Novikov I., Rubin C., Gimpelevich I., Huppert A., Rahav G., Afek A., Kreiss Y., Decreased infectivity following BNT162b2 vaccination: A prospective cohort study in Israel. The Lancet Regional Health - Europe 7, 100150 (2021). 10.1016/j.lanepe.2021.100150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Dela Cruz C. S., Wang Y., Wu C., Xiao Y., Zhang L., Han L., Dang S., Xu Y., Yang Q.-W., Xu S.-Y., Zhu H.-D., Xu Y.-C., Jin Q., Sharma L., Wang L., Wang J., Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19). Clin. Infect. Dis. 71, 778–785 (2020). 10.1093/cid/ciaa310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.The National Institute For Communicable Diseases Of South Africa, NICD National COVID-19 Hospital Surveillance (The National Institute For Communicable Diseases Of South Africa, 2022; https://www.nicd.ac.za/wp-content/uploads/2022/02/NICD-COVID-19-Daily-Sentinel-Hospital-Surveillance-report-National-20220203.pdf).
- 44. Wolter N., Jassat W., Walaza S., Welch R., Moultrie H., Groome M., Amoako D. G., Everatt J., Bhiman J. N., Scheepers C., Tebeila N., Chiwandire N., du Plessis M., Govender N., Ismail A., Glass A., Mlisana K., Stevens W., Treurnicht F. K., Makatini Z., Hsiao N.-Y., Parboosing R., Wadula J., Hussey H., Davies M.-A., Boulle A., von Gottberg A., Cohen C., Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: A data linkage study. Lancet 399, 437–446 (2022). 10.1016/S0140-6736(22)00017-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.South Africa National Department Of Health, Latest Vaccine Statistics (South Africa)SA Corona Virus Online Portal (2021) (available at https://sacoronavirus.co.za/latest-vaccine-statistics/).
- 46. Cho A., Muecksch F., Schaefer-Babajew D., Wang Z., Finkin S., Gaebler C., Ramos V., Cipolla M., Mendoza P., Agudelo M., Bednarski E., DaSilva J., Shimeliovich I., Dizon J., Daga M., Millard K. G., Turroja M., Schmidt F., Zhang F., Tanfous T. B., Jankovic M., Oliveria T. Y., Gazumyan A., Caskey M., Bieniasz P. D., Hatziioannou T., Nussenzweig M. C., Anti-SARS-CoV-2 receptor-binding domain antibody evolution after mRNA vaccination. Nature 600, 517–522 (2021). 10.1038/s41586-021-04060-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Muecksch F., Weisblum Y., Barnes C. O., Schmidt F., Schaefer-Babajew D., Wang Z., C Lorenzi J. C., Flyak A. I., DeLaitsch A. T., Huey-Tubman K. E., Hou S., Schiffer C. A., Gaebler C., Da Silva J., Poston D., Finkin S., Cho A., Cipolla M., Oliveira T. Y., Millard K. G., Ramos V., Gazumyan A., Rutkowska M., Caskey M., Nussenzweig M. C., Bjorkman P. J., Hatziioannou T., Bieniasz P. D., Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations. Immunity 54, 1853–1868.e7 (2021). 10.1016/j.immuni.2021.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Turner J. S., Kim W., Kalaidina E., Goss C. W., Rauseo A. M., Schmitz A. J., Hansen L., Haile A., Klebert M. K., Pusic I., O’Halloran J. A., Presti R. M., Ellebedy A. H., SARS-CoV-2 infection induces long-lived bone marrow plasma cells in humans. Nature 595, 421–425 (2021). 10.1038/s41586-021-03647-4 [DOI] [PubMed] [Google Scholar]
- 49. Turner J. S., O’Halloran J. A., Kalaidina E., Kim W., Schmitz A. J., Zhou J. Q., Lei T., Thapa M., Chen R. E., Case J. B., Amanat F., Rauseo A. M., Haile A., Xie X., Klebert M. K., Suessen T., Middleton W. D., Shi P.-Y., Krammer F., Teefey S. A., Diamond M. S., Presti R. M., Ellebedy A. H., SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature 596, 109–113 (2021). 10.1038/s41586-021-03738-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mateus J., Dan J. M., Zhang Z., Rydyznski Moderbacher C., Lammers M., Goodwin B., Sette A., Crotty S., Weiskopf D., Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science 374, eabj9853 (2021). 10.1126/science.abj9853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dan J. M., Mateus J., Kato Y., Hastie K. M., Yu E. D., Faliti C. E., Grifoni A., Ramirez S. I., Haupt S., Frazier A., Nakao C., Rayaprolu V., Rawlings S. A., Peters B., Krammer F., Simon V., Saphire E. O., Smith D. M., Weiskopf D., Sette A., Crotty S., Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 371, eabf4063 (2021). 10.1126/science.abf4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mistry D., Litvinova M., Pastore Y Piontti A., Chinazzi M., Fumanelli L., Gomes M. F. C., Haque S. A., Liu Q.-H., Mu K., Xiong X., Halloran M. E., Longini I. M. Jr., Merler S., Ajelli M., Vespignani A., Inferring high-resolution human mixing patterns for disease modeling. Nat. Commun. 12, 323 (2021). 10.1038/s41467-020-20544-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Britton T., Ball F., Trapman P., A mathematical model reveals the influence of population heterogeneity on herd immunity to SARS-CoV-2. Science 369, 846–849 (2020). 10.1126/science.abc6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cohen C., McMorrow M. L., Martinson N. A., Kahn K., Treurnicht F. K., Moyes J., Mkhencele T., Hellferscee O., Lebina L., Moroe M., Motlhaoleng K., Gómez-Olivé F. X., Wagner R., Tollman S., Wafawanaka F., Ngobeni S., Kleynhans J., Mathunjwa A., Buys A., Maake L., Wolter N., Carrim M., Piketh S., Language B., Mathee A., von Gottberg A., Tempia S.; PHIRST group , Cohort profile: A Prospective Household cohort study of Influenza, Respiratory syncytial virus and other respiratory pathogens community burden and Transmission dynamics in South Africa, 2016-2018. Influenza Other Respir. Viruses 15, 789–803 (2021). 10.1111/irv.12881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elecsys® Anti-SARS-CoV-2 (available at https://diagnostics.roche.com/us/en/products/params/elecsys-anti-sars-cov-2.html).
- 56. Peluso M. J., Takahashi S., Hakim J., Kelly J. D., Torres L., Iyer N. S., Turcios K., Janson O., Munter S. E., Thanh C., Donatelli J., Nixon C. C., Hoh R., Tai V., Fehrman E. A., Hernandez Y., Spinelli M. A., Gandhi M., Palafox M.-A., Vallari A., Rodgers M. A., Prostko J., Hackett J. Jr., Trinh L., Wrin T., Petropoulos C. J., Chiu C. Y., Norris P. J., DiGermanio C., Stone M., Busch M. P., Elledge S. K., Zhou X. X., Wells J. A., Shu A., Kurtz T. W., Pak J. E., Wu W., Burbelo P. D., Cohen J. I., Rutishauser R. L., Martin J. N., Deeks S. G., Henrich T. J., Rodriguez-Barraquer I., Greenhouse B., SARS-CoV-2 antibody magnitude and detectability are driven by disease severity, timing, and assay. Sci. Adv. 7, eabh3409 (2021). 10.1126/sciadv.abh3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.CDC, Frequently Asked Questions about Coronavirus (COVID-19) for Laboratories (2021) (available at https://www.cdc.gov/coronavirus/2019-ncov/lab/faqs.html).
- 58.Public Health England, Cycle threshold (Ct) in SARS-CoV-2 RT-PCR (2020) (available at https://www.gov.uk/government/publications/cycle-threshold-ct-in-sars-cov-2-rt-pcr).
- 59. Kang M., Xin H., Yuan J., Ali S. T., Liang Z., Zhang J., Hu T., Lau E. H. Y., Zhang Y., Zhang M., Cowling B. J., Li Y., Wu P., Transmission dynamics and epidemiological characteristics of Delta variant infections in China. bioRxiv (2021), doi:. 10.1101/2021.08.12.21261991 [DOI] [PMC free article] [PubMed]
- 60. Holford T. R., The analysis of rates and of survivorship using log-linear models. Biometrics 36, 299–305 (1980). 10.2307/2529982 [DOI] [PubMed] [Google Scholar]
- 61. Laird N., Olivier D., Covariance Analysis of Censored Survival Data Using Log-Linear Analysis Techniques. J. Am. Stat. Assoc. 76, 231–240 (1981). 10.1080/01621459.1981.10477634 [DOI] [Google Scholar]
- 62. Marc A., Kerioui M., Blanquart F., Bertrand J., Mitjà O., Corbacho-Monné M., Marks M., Guedj J., Quantifying the relationship between SARS-CoV-2 viral load and infectiousness. eLife 10, e69302 (2021). 10.7554/eLife.69302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cox D. R., Regression Models and Life-Tables. J. R. Stat. Soc. Series B Stat. Methodol. 34, 187–220 (1972). [Google Scholar]
- 64.Douglas Bates and Martin Machler and Ben Bolker and Steve Walker, Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 67, 1–48 (2015). [Google Scholar]
- 65. Neher R. A., Dyrdak R., Druelle V., Hodcroft E. B., Albert J., Potential impact of seasonal forcing on a SARS-CoV-2 pandemic. Swiss Med. Wkly. 150, w20224 (2020). 10.4414/smw.2020.20224 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MDAR Reproducibility Checklist