Abstract
Using molecular techniques and microsensors for H2S and CH4, we studied the population structure of and the activity distribution in anaerobic aggregates. The aggregates originated from three different types of reactors: a methanogenic reactor, a methanogenic-sulfidogenic reactor, and a sulfidogenic reactor. Microsensor measurements in methanogenic-sulfidogenic aggregates revealed that the activity of sulfate-reducing bacteria (2 to 3 mmol of S2− m−3 s−1 or 2 × 10−9 mmol s−1 per aggregate) was located in a surface layer of 50 to 100 μm thick. The sulfidogenic aggregates contained a wider sulfate-reducing zone (the first 200 to 300 μm from the aggregate surface) with a higher activity (1 to 6 mmol of S2− m−3 s−1 or 7 × 10−9 mol s−1 per aggregate). The methanogenic aggregates did not show significant sulfate-reducing activity. Methanogenic activity in the methanogenic-sulfidogenic aggregates (1 to 2 mmol of CH4 m−3 s−1 or 10−9 mmol s−1 per aggregate) and the methanogenic aggregates (2 to 4 mmol of CH4 m−3 s−1 or 5 × 10−9 mmol s−1 per aggregate) was located more inward, starting at ca. 100 μm from the aggregate surface. The methanogenic activity was not affected by 10 mM sulfate during a 1-day incubation. The sulfidogenic and methanogenic activities were independent of the type of electron donor (acetate, propionate, ethanol, or H2), but the substrates were metabolized in different zones. The localization of the populations corresponded to the microsensor data. A distinct layered structure was found in the methanogenic-sulfidogenic aggregates, with sulfate-reducing bacteria in the outer 50 to 100 μm, methanogens in the inner part, and Eubacteria spp. (partly syntrophic bacteria) filling the gap between sulfate-reducing and methanogenic bacteria. In methanogenic aggregates, few sulfate-reducing bacteria were detected, while methanogens were found in the core. In the sulfidogenic aggregates, sulfate-reducing bacteria were present in the outer 300 μm, and methanogens were distributed over the inner part in clusters with syntrophic bacteria.
Methanogenic and sulfidogenic granular sludge consists of well-settling microbial aggregates that develop by the mutual attachment of bacterial cells in the absence of a carrier material (28). These aggregates contain a variety of bacterial species involved in the anaerobic degradation of organic matter, including hydrolytic, fermentative, acidogenic, acetogenic, homoacetogenic, sulfate-reducing, and methanogenic bacteria. These aggregates develop spontaneously in wastewater treatment systems of the upflow anaerobic sludge bed (UASB) reactor design under a variety of operation conditions (28).
The formation, composition, and functioning of UASB granules has been investigated by a variety of analytical techniques (60). To characterize the bacterial species present, various types of activity tests, in combination with molecular, microbial, and physiological assays, have been applied to granular sludge samples, as well as to individual aggregates (60). The spatial distribution of the bacterial species present within UASB aggregates has been studied by using light, electron, and laser-scanning microscopy on either intact or sectioned individual aggregates (31). Based on these observations, conceptual models have been postulated to describe the distribution of acidogens, syntrophic and methanogenic bacteria within UASB aggregates (12, 14, 31). These models proposed a multilayered structure with H2-consuming bacteria located at the outside of the aggregate, methanogens located in the inner part, and H2-producing bacteria located between the two layers.
By the same analytical techniques, however, a homogeneous distribution of the different populations present in UASB granules was determined as well (13). The use of molecular techniques has been particularly useful for the identification and localization of the different microbial populations present in methanogenic granular sludge. Slot-dot blot hybridization of the 16S rRNA extracted from granules allow to both identify and quantify the methanogenic, syntrophic, and sulfate-reducing populations (40, 42, 46). The application of fluorescent in situ hybridization (FISH) with specific probes for a bacterial species or population further allows their detection and localization within granular sludge (16, 52). By using the FISH technique, different layered population structures have been determined in different UASB aggregates cultivated on different substrates (15, 16, 52).
Although these investigations considerably improved our understanding of the granular sludge composition and anatomy, there is still a lack of knowledge about the distribution of microbial activities within granular sludge. Population distributions determined by molecular probes do not necessarily correspond to microbial activity distributions, since bacterial populations can have very low or unusual activities. Activity distributions in anaerobic granular sludge are poorly documented, since in situ activity measurements require specific analytical tools, e.g., microsensors. The activity distribution of fermentative and methanogenic populations in UASB aggregates has been measured with micrometer resolution by using microsensors for pH and glucose (10, 26, 27). One study indicated an inhomogeneous activity distribution, with acetogenic and methanogenic activity being predominantly located in, respectively, the outer layer (150 to 200 μm) and the center of the aggregates (27).
The population dynamics between sulfate-reducing bacteria (SRB) and methanogenic bacteria (MB) are crucial in governing the metabolic properties of granular sludge. Their dynamics were studied in detail by using molecular techniques (40, 42, 46), but their in situ activity distributions have not yet been reported. Recently, two novel microsensors, i.e., a CH4 biosensor (8) and a H2S microsensor (19), have been developed to study the microbial ecology of sediments. In the present study, both microsensors were used to determine the in situ methanogenic and sulfate-reducing activity in anaerobic aggregates. These localized activity measurements were combined with molecular techniques to analyze on a microscale the structure, population (sulfate-reducing, methanogenic, and syntrophic bacteria) and activity distribution of three different types of aggregates with different degrees of sulfate-reducing activity.
MATERIALS AND METHODS
Aggregates.
Aggregates were retrieved from three different UASB reactors. (i) Methanogenic-sulfidogenic aggregates originate from a UASB reactor treating wastewater from a potato-processing plant (Leusden, Belgium). These aggregates were subcultured for several months at 30°C by batchwise feeding twice a week. The feed (pH 6 to 7) contained a volatile-fatty-acids (VFA) mixture (13.8 mM acetate, 4.7 mM propionate, and 2.3 mM butyrate), supplemented with 20 mM NaSO4, 4 mM NH4Cl, 0.32 mM NaHPO4, 0.2 mM MgCl2, and trace elements. (ii) Methanogenic aggregates were obtained from a UASB reactor treating paper mill wastewater (Eerbeek, The Netherlands). These aggregates were subcultured for 3 months at 30°C by batchwise feeding three times a week. The feed (pH 7) contained a VFA mixture (6.25 mM acetate, 7.15 mM propionate, and 5.00 mM butyrate), supplemented with 5.2 mM (NH4)Cl, 10.2 mM NaH2PO4, 6.8 mM K2HPO4, 0.45 mM MgSO4 · 7H2O, 0.04 mM CaCl2, and trace elements. (iii) Sulfidogenic aggregates were sampled directly from a full-scale UASB reactor treating ethanol (12 mM) and sulfate (7 mM) containing wastewater (pH 7 to 7.5, 30 to 35°C) at Emmen (The Netherlands). All types of aggregates had diameters that were between 1 and 2 mm. The methanogenic and methanogenic-sulfidogenic aggregates were smooth and almost spherical, whereas the sulfidogenic aggregates had a rather loose structure and an irregular surface.
Microsensor measurements.
For microsensor measurements, aggregates were attached to insect needles (150 μm, Minutie; Entomologie Vermandel b.v., Hulst, The Netherlands) in an incubation cell at 30°C. The medium in the cell was kept anaerobic by continuous bubbling with N2 or argon, which resulted in circulation and mixing of the medium. The flow rate, judged from movement of suspended particles, was 1 to 3 mm/s. Anoxicity was checked with an oxygen microsensor (47). Before measurements, the aggregates were preincubated 1 day in the measurement medium. The latter consisted of 5.2 mM (NH4)Cl, 10.2 mM NaH2PO4, 6.8 mM K2HPO4, 0.45 mM MgCl2, and 0.04 mM CaCl2, plus micronutrients, at pH 7.0 for the methanogenic-sulfidogenic and methanogenic aggregates. The measurement medium for the sulfidogenic aggregates contained 0.19 mM (NH4)Cl and 10 mM KH2PO4 at pH 7.2. During the microsensor measurements, the media were supplemented with different concentrations of acetate, propionate, ethanol, H2, and sulfate. The phosphate buffer in the media ensured a constant pH in the aggregates, which was confirmed with pH microelectrodes (48).
Microsensors, mounted on micromanipulators, were positioned at the aggregate surface with the aid of a dissection microscope. Microprofiles were recorded by penetrating the aggregate with microsensors in steps of 50 or 100 μm.
Hydrogen sulfide microsensors.
Sulfide concentration profiles were measured with H2S microsensors (19, 25) with a tip diameter of ca. 10 μm and a 90% response time of <0.5 s. The microsensors were calibrated at 30°C in a dilution series as described previously (24). The concentration of total dissolved sulfide (H2S + HS− + S2−) in the dilution series was determined by a spectrophotometric method (6). Since the sensor was calibrated in medium of the same pH as the measuring medium and the aggregate, no pH correction was necessary. The sensor had a linear response to H2S concentrations of up to 1,000 μM. The detection limit of the microsensors was 1 to 3 μM total sulfide.
Methane microsensors.
Methane microsensors were constructed and inoculated with the methane-oxidizing bacterium Methylosinus trichosporium (8). Microsensor tip diameters were 25 to 30 μm, and 90% response times were 30 to 100 s. Since all measurements were performed under anoxic conditions, an oxygen-scavenging guard capillary (9) was not applied. Calibrations were performed at 30°C, before and after measurements, as described previously (9). Interference from H2S, CO2, and H2 were tested by exposing the sensor tip to known concentrations or mixing ratios of these compounds. The response to H2S was 25% of the response to CH4. Corrections of the methane profile were made by subtracting 25% of the corresponding H2S concentration, measured with the H2S microsensor. CO2 did not interfere. Some methane biosensors exhibited similar responses to H2 and methane due to culture contamination. However, H2 interference was insignificant since microsensor measurements in aggregates showed H2 concentrations to be less than 5 μM. Methane profiles were only measured in methanogenic-sulfidogenic and methanogenic aggregates since no methane biosensor was available for measurements with sulfidogenic aggregates.
Diffusivity microsensors.
Microscale diffusivity sensors with a diameter of approximately 70 μm were used (49). Acetylene was used as the tracer substance instead of H2 (7) to avoid interference by the H2 metabolism in the aggregates. A two-point calibration in agar and glass beads (49) was performed before the measurements were made. The spatial resolution of this sensor was estimated to be ca. 300 μm, a value insufficient for detection of detailed spatial distribution of diffusivity. Therefore, the stable readings in the center of the aggregate were taken to represent the entire aggregate.
Flux and activity calculations.
Diffusive fluxes were calculated by using Fick’s first law:
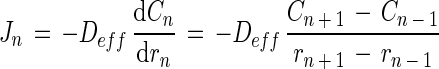 |
where Jn is the flux at point n (mmol m−2 s−1), Deff is the effective diffusion coefficient (m2 s−1), dCn/drn is the concentration gradient at point n (mmol m−3 m−1), C is the substrate concentration (mmol m−3), and r is the distance from the aggregate center. The molecular diffusion coefficient (Dw) for oxygen at 30°C is 2.75 × 10−9 m2 s−1 (4). The Dw for methane and sulfide was determined by multiplying the Dw of oxygen by 0.8495 and 0.7573, respectively (4), yielding values of 2.34 × 10−9 m2 s−1 for methane and 2.08 × 10−9 m2 s−1 for sulfide. The Deff of these compounds within the aggregates was found by correcting Dw with the ratio of the diffusivity in aggregates and in water, as determined with the diffusivity microsensor. The local activities of methanogenesis and sulfate reduction were calculated by assuming spherical geometry, i.e., considering the aggregate buildup from concentric layers. The rates (Rn [mmol m−3 s−1]) in each layer n were found by subtracting the fluxes into and out of the layer, divided by the volume of the layer:
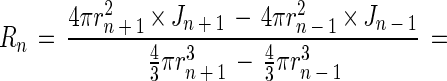 |
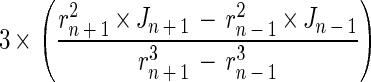 |
Aggregate fixation and slicing.
After microsensor analysis, the aggregates were fixed for in situ hybridization by overnight incubation in paraformaldehyde (4% [wt/vol] in phosphate-buffered saline [PBS]) at 4°C and subsequently washed in PBS. Then they were embedded for ca. 12 h in OCT compound (Sakura Finetek USA, Torrance, Calif.) and frozen at −20°C. The aggregates were sectioned with a cryomicrotome (Microm HM 505 E) at −18°C. The slices (10 μm thick) were collected on gelatin-coated microscopic slides, air dried, and dehydrated in an ethanol concentration series (50, 80, and 96% [vol/vol]).
Nucleic acid extraction and PCR amplification.
DNA and RNA was extracted from the aggregates by a combined bead beating (2 min at maximum speed with 0.75 to 1.0-mm glass beads), lysis (10 mg of lysosyme per ml for 1 h at 37°C, 1% [wt/vol] sodium dodecyl sulfate (SDS), and 0.25 mg of protinase K per ml for 30 min at 55°C), and hot phenol-chloroform-isoamyl alcohol treatment (57). The ribosomal DNA (rDNA) was enzymatically amplified as described by Muyzer et al. (35) by using the eubacterial primer GM5F with GC-clamp and the universal primer 907R (Table 1). The rRNA was amplified according to the method of Teske et al. (57). A hot-start, touch-down PCR program was used for all amplifications to minimize nonspecific amplification (35).
TABLE 1.
Oligonucleotide probes used for PCR amplification and in situ hybridization analyses
| Probe | Positiona | Sequence (5′→3′) | Target (reference) | Formamide (%)b | NaCl (mM)c |
|---|---|---|---|---|---|
| GM5Fd | 341–357 | CCTACGGGAGGCAGCAG | Bacteria (36) | –e | –e |
| 907R | 907–928 | CCGTCAATTCCTTTGAGTTT | All organisms (universal probe) (36) | – | – |
| ARC915 | 915–934 | GTGCTCCCCCGCCAATTCCT | Archaea (55) | 40 | |
| EUB338 | 338–355 | GCTGCCTCCCGTAGGAGT | Bacteria (1) | 20 | 225f |
| SRB385 | 338–355 | CGGCGTCGCTGCGTCAGG | SRB of the delta proteobacteria (2) plus several gram-positive bacteria (e.g., Clostridium) (45) | 35 | 80f |
| DSV698 | 698–717 | GTTCCTCCAGATATCTACGG | Desulfovibrio (33) | 35 | 88 |
| DSD131 | 131–148 | CCCGATCGTCTGGGCAGG | Desulfovibrio (33) | 20 | 250 |
| DSV407 | 407–424 | CCGAAGGCCTTCTTCCCT | Desulfovibrio (33) | 50 | 31.2 |
| DSV1292 | 1,292–1,309 | CAATCCGGACTGGGACGC | Desulfovibrio (33) | 35 | 88 |
| DSV214 | 214–230 | CATCCTCGGACGAATGC | Desulfomicrobium (33) | 10 | 500 |
| DSS658 | 658–678 | TCCACTTCCCTCTCCCAT | Desulfosarcina (33) | 60 | 15.6 |
| DSB985 | 985–1,004 | CACAGGATGTCAAACCCAG | Desulfobacter (33) | 20 | 250 |
| DSBO224 | 224–242 | GGGACGCGGACTCATCCTC | Desulfobotulus (33) | 60 | 15.6 |
| DSMA488 | 488–507 | GCCGGTGCTTCCTTTGGCGG | Desulfomonile (33) | 60 | 15.6 |
| DSR651 | 651–668 | CCCCCTCCAGTACTCAAG | Desulforhopalus (33) | 35 | 88 |
| 221 | 221–240 | TGCGCGGACTCATCTTCAAA | Desulfobacterium (11) | 35 | 88 |
| 660 | 660–679 | GAATTCCACTTTCCCCTCTG | Desulfobulbus (11) | 60 | 15.6 |
| 687 | 687–702 | TACGGATTTCACTCCT | Desulfovibrio (11) plus members of the genera Geobacter, Desulfomonas, Desulfuromonas, Desulfomicrobium, Bilophila, and Pelobacter spp. | – | – |
| 804 | 804–821 | CAACGTTTACTGCGTGGA | Desulfobacterium spp., Desulfobacter, Desulfococcus, Desulfosarcina, and Desulfobotulus spp. (11) | – | – |
| MPOB | 222–240 | ACGCAGGCCCATCCCCGAA | Syntrophic propionate-oxidizing strains MPOB (Syntrophobacter fumaroxidans) and KOPROP1 and Desulforhabdus amnigenus (although these last two strains have one mismatch with the probe) (16, 17) | 20 | 900 |
| NON338 | 338–355 | ACTCCTACGGGAGGCAGC | None (negative control) (32) | 0 | 900 |
Position in the 16S rRNA of E. coli (5).
Formamide concentration in hybridization buffer.
Sodium chloride concentration in washing buffer.
This primer has the following GC-clamp at the 5′ end: 5′-CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCG-3′.
–, Probe not used for FISH.
Plus 5 mM EDTA in washing buffer.
DGGE analysis of 16S rDNA fragments.
Denaturing gradient gel electrophoresis (DGGE) was performed by using the D-Gene system (Bio-Rad) and the following specifications: 1× TAE (40 mM Tris, 20 mM acetic acid, and 1 mM EDTA at pH 8.3), 1-mm thick gels, a denaturant gradient from 35 to 65% urea-formamide, a temperature of 60°C, and a constant voltage of 100 V for 17 h (35, 36). DGGE gels were photographed on a UV transillumination table (302 nm) with a Polaroid camera. Photos were scanned and inversed.
Blotting and hybridization analysis of DGGE gels.
DGGE gels were blotted and hybridized with group-specific probes for SRB as described previously (50). The probes used (Table 1) were probe 660 (specific for Desulfobulbus species), 687 (targeting Desulfovibrio species, as well as some members of the Geobacter, Desulfomonas, Desulfuromonas, Desulfomicrobium, Bilophila, and Pelobacter genera), and probe 804 (targeting Desulfobacter, Desulfobacterium, Desulfosarcina, Desulfococcus, and Desulfobotulus species) developed by Devereux et al. (11).
Excision and amplification of DGGE bands.
DGGE bands were carefully excised on a UV transillumination table and transferred to a 1.5-ml tube with 500 μl of water and approximately 500 μl of glass beads 0.75 to 1.0 mm in diameter. The acrylamide bands were disrupted by bead beating at maximum speed twice for 1 min. The samples were left overnight at 4°C, and the DNA was subsequently amplified by adding 1 to 10 μl of the samples’ supernatant to the PCR mixture. The PCR was then performed as described above. A second DGGE was run to confirm that the amplified bands had the same position in the gel as the excised bands. Prior to sequencing, the PCR products were purified by using the QIAquick PCR purification kit (Qiagen, Inc.).
Sequencing and phylogenetic analysis.
Amplified DGGE bands were sequenced by using the Applied Biosystems PRISM Dye Terminator Cycle Sequencing Ready reaction kit supplied with AmpliTaq DNA polymerase. The sequencing products were analyzed with the Applied Biosystems 377 DNA sequencer. The partial sequences, which were 536 to 581 nucleotides long, were added to the 16S rRNA parsimony tree of the Technical University of Munich by using the program package ARB (56).
FISH.
The protocol described by Manz et al. (32) was used for FISH of the aggregate slices with probe ARC915 for Archaea bacteria (55); probe SRB385 for the detection of general sulfate reducers of the delta subdivision; probes 221 and 660 for group-specific SRB (Devereux et al. [11]); probes DSV698, DSD131, DSV407, DSV1292, DSV214, DSS658, DSB985, DSBO224, DSMA488, and DSR651 developed by Manz et al. (33); and probe NON338 as a negative control (Table 1). The probe MPOB described by Harmsen et al. (16) was used for detection of syntrophic bacteria (Table 1).
The probes were synthesized and labeled with a hydrophilic sulfoindocyanine dye CY3 or CY5 by Interactiva GmbH (Ulm, Germany). The hybridization buffer contained 0.9 M NaCl, a percentage (vol/vol) of formamide as shown in Table 1, 20 mM Tris-HCl (pH 7.4), and 0.01% (wt/vol) SDS. The probe concentrations were 5 ng/μl. Hybridization was performed for 1 to 2 h at 46°C. The aggregate slices were washed at 48°C for 15 min in a washing buffer containing 20 mM Tris-HCl (pH 7.4), 0.01% (wt/vol) SDS, and a concentration of NaCl as mentioned in Table 1. The specimens were microscopically examined with a Zeiss LSM 510 confocal laser scanning microscope (Carl Zeiss, Jena, Germany), equipped with two HeNe lasers (543 and 633 nm). The hybridizations shown in the figures are representative for several independent hybridizations on several aggregates.
Sulfur analysis.
For sulfur (S0 and sulfanes in polysulfides) determination, three to five aggregates were put into a reaction tube and immediately fixed with 20 μl of ZnCl2 (2%). During this treatment, polysulfides are converted to S0 and ZnS. S0 was extracted by shaking the samples with pure methanol (high-pressure liquid chromatography [HPLC] grade) for 6 h. Identification, and quantification of zerovalent sulfur was performed by HPLC by using a Sykam S1100 pump (Gilching, Germany), a Zorbax ODS column (125 by 4 mm, 5 μm; Knauer, Germany), and an Sykam S3000 UV detector (265 nm). A mixture of 0.25% acetic acid (pH 4) and 100% methanol (10/90 [vol/vol]) was used as eluent; the flow rate was 1.2 ml/min. Under these conditions S0 eluted as cyclo-octasulfur (S8) at 5.4 min. The precision for injection of a 100 μM S0-standard was 0.5% s.d. (n = 8), the detection limit was about 1 μM. A second method was used to confirm the identity of S0, based on the reaction of S0 with SO32−: S0 + SO32− → S2O32− (22). A few aggregates were incubated with 1 ml of 5% Na2SO3 solution for 2 h at 90°C after fixation with ZnCl2, followed by extraction and analysis as described above. The presence or absence (after the sulfite treatment) of S0 in the methanol extracts was also confirmed by UV spectroscopy.
RESULTS
Diffusivity measurements.
Diffusivity microsensor measurements showed an approximately constant Dapp in the methanogenic-sulfidogenic aggregates of 50% of that in water. Therefore, the Dapp of methane and sulfide were assumed to be 50% of their Dw, i.e., 1.17 × 10−9 m2 s−1 for methane and 1.04 × 10−9 m2 s−1 for sulfide. These values were used to calculate the fluxes and activities in the aggregates from the microprofiles.
Endogeneous sulfide and methane microprofiles.
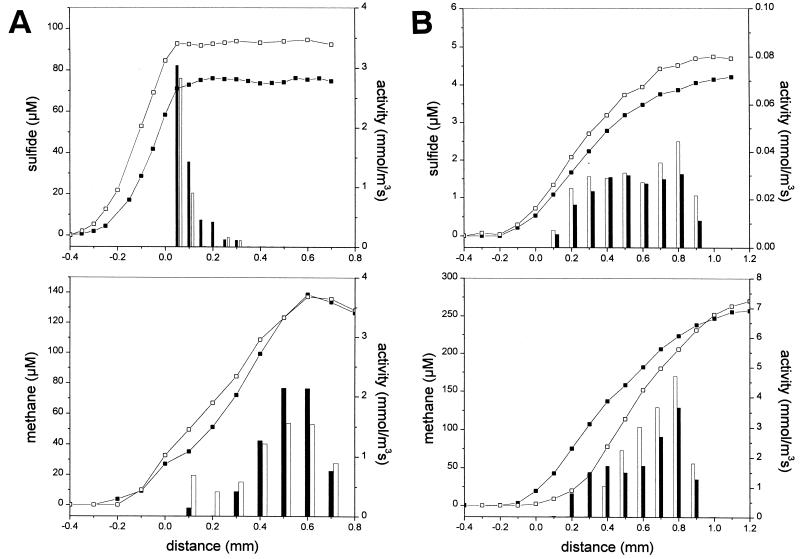
In all methanogenic-sulfidogenic (Fig. 1A), methanogenic (Fig. 1B), and sulfidogenic (Fig. 2A) aggregates, endogenous sulfide production was measured in the absence of sulfate, even after one night’s incubation in a sulfate- and electron donor-free medium. In addition, endogeneous methane production was observed in the methanogenic-sulfidogenic and methanogenic aggregates (Fig. 1). The addition of 10 mM sulfate significantly increased the sulfide production, without affecting the methane production in the methanogenic-sulfidogenic aggregates (Fig. 1A). In the methanogenic aggregates, only a negligible amount of sulfide (<6 μM) was measured which, like the methanogenic activity, was not affected by the presence or absence of sulfate (Fig. 1B). All three types of aggregates contained large amounts (up to 59 mol m−3) of S0 (Table 2).
FIG. 1.
Sulfide and methane microsensor profiles (lines) and activity values (bars) in methanogenic-sulfidogenic (A) and methanogenic (B) aggregates in the presence (□, open bars) or absence (■, closed bars) of sulfate. No external electron donor was supplied during the measurements. The aggregate surface is at distance of 0 mm, the center of the aggregates is at a distance of ca. 0.9 mm.
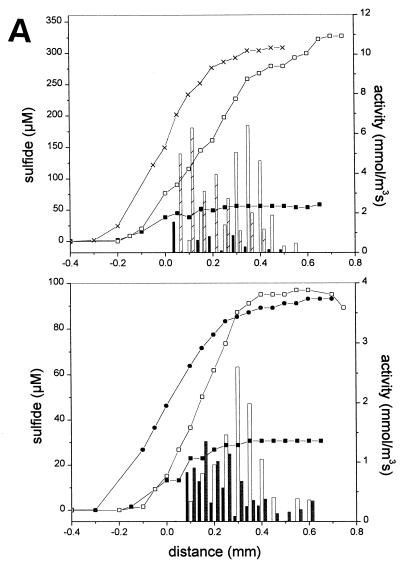
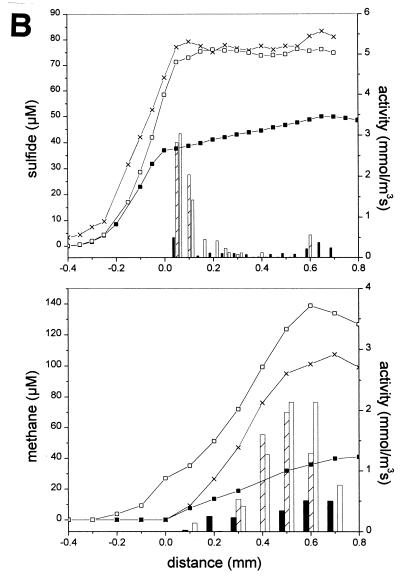
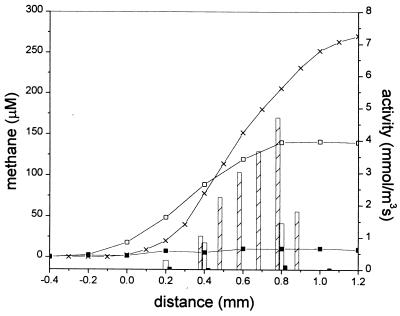
FIG. 2.
Steady-state microsensor profiles (lines) and activity values (bars) of sulfide and methane in aggregates in the presence of 10 mM SO42−. (A) Sulfidogenic aggregate after one night without electron donor (■, closed bars), with the addition of 7 mM ethanol (□, open bars), with H2 saturation (×, crossed bars), and with the addition of 7 mM acetate (●, shaded bars). (B) Methanogenic-sulfidogenic aggregate after one night without electron donor (■, closed bars), with addition of 1 mM acetate (×, crossed bars), and with 1 mM propionate (□, open bars). (C) Methanogenic aggregate after one night of incubation without electron donor (■, closed bars) and with the addition of a VFA mixture (6 mM acetate, 7 mM propionate, and 5 mM butyrate; □, open bars). The aggregate surface is at a distance of 0 mm, the center of the aggregate is approximately at a distance of 0.6 mm for the sulfidogenic aggregates (A) and at a distance of 0.9 mm for both the methanogenic-sulfidogenic (B) and the methanogenic (C) aggregates. The profiles in the upper and lower parts of graph A are obtained from two different sulfidogenic aggregates and were measured at different times.
TABLE 2.
Characteristics and activities of the different aggregates used in this study
| Bacterial type | Growth conditions
|
Activity valuesa
|
Sulfur concn inside aggregates (mM)b | |||||
|---|---|---|---|---|---|---|---|---|
| Original substrate | Subculture medium | Sulfate concn during subculturing (mM) | Methane
|
Sulfide
|
||||
| Production (mmol m−3 s−1) | Conversion/ aggregate (mmol/s) | Production (mmol m−3 s−1) | Conversion/ aggregate (mmol/s) | |||||
| Sulfidogenic | Ethanol | NAc | 7 | ND | ND | 1–6 | 7 × 10−9 | ∼28 |
| Methanogenic-sulfidogenic | Potato starch wastewater | VFA mixtured | 20 | 1–2 | 1 × 10−9 | 2–3 | 2 × 10−9 | ∼21 |
| Methanogenic | Papermill wastewater | VFA mixture | 0.45 | 2–4 | 4.5 × 10−9 | 0.02–0.06 | 8 × 10−11 | ∼59 |
Calculated from the microprofile measurements (conversion per aggregate values were calculated from integrated local values). ND, not determined.
Elemental sulfur and polysulfide.
NA, not applicable.
Low-chain VFA were the main electron donors present in the reactor liquid.
Microprofiles after addition of substrates.
The production of methane and sulfide were stimulated by volatile fatty acids (acetate, propionate, and/or butyrate), ethanol, or H2 (Fig. 2). The sulfidogenic sludge, fed only ethanol for more than 1 year, was also able to metabolize both H2 and acetate, since upon their addition sulfide developed instantly (Fig. 2A). The sulfide production rates by H2 were comparable to that by ethanol. Figure 2A shows that these substrates were consumed in distinctly different zones within the aggregate: sulfate reduction was stimulated by H2 in the outer 200 μm depth and by ethanol at a depth of between 200 to 400 μm. Sulfidogenesis with acetate as the substrate was mainly located in the outer 200 μm of the aggregate. Acetate was metabolized with a specific sulfidogenic activity half of that with ethanol as the substrate (Fig. 2A).
Both acetate and propionate induced sulfidogenic activity in the outer 100 to 150 μm of the methanogenic-sulfidogenic aggregates. These substrates also induced methanogenic activity, which only started from 300 μm onwards inside the aggregate (Fig. 2B). Sulfate reduction was not affected when methanogenic-sulfidogenic aggregates were supplied with 1 mM nitrate (data not shown).
The addition of the VFA mixture to the methanogenic aggregates did not affect the sulfide microprofile but induced a substantial methane production predominantly in the core of the aggregates (Fig. 2C). Addition of 50 mM BES completely inhibited the methane production after 3.5 h of incubation (Fig. 3).
FIG. 3.
Microsensor profiles (lines) and activity values (bars) in methanogenic aggregates of methane after the addition of 50 mM BES at time zero (×, crossed bars), after 1 h and 15 min (□, open bars), and after 3.5 h (■, closed bars). The aggregate surface is at a distance of 0 mm, the center of the aggregate is approximately at a distance of 0.9 mm from the surface.
Activity profiles.
Sulfide production was in all aggregates restricted to the outer layer, while methane was produced deeper in the aggregates. In the sulfidogenic aggregates, sulfate reduction is restricted to the outer 200 to 300 μm (Fig. 2A). In the methanogenic-sulfidogenic aggregates, sulfide production was localized in the outer 50 to 100 μm, while methane production was exclusively detected below a depth of 100 μm (Fig. 2B). Also in the methanogenic aggregates, methane is produced only in the core, starting at 200 μm from the surface (Fig. 2C). Table 2 summarizes the average activity values for the three types of aggregates, as derived from the profiles presented in Fig. 1 to 3.
Population analysis by DGGE.
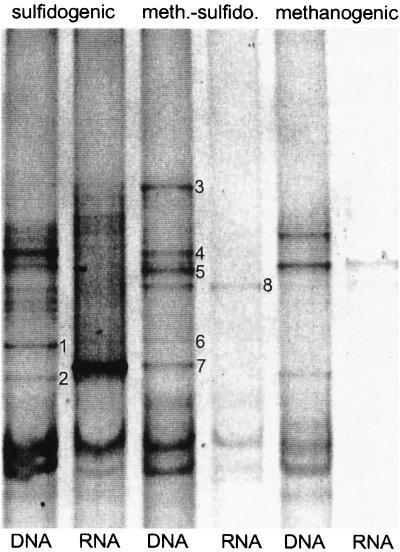
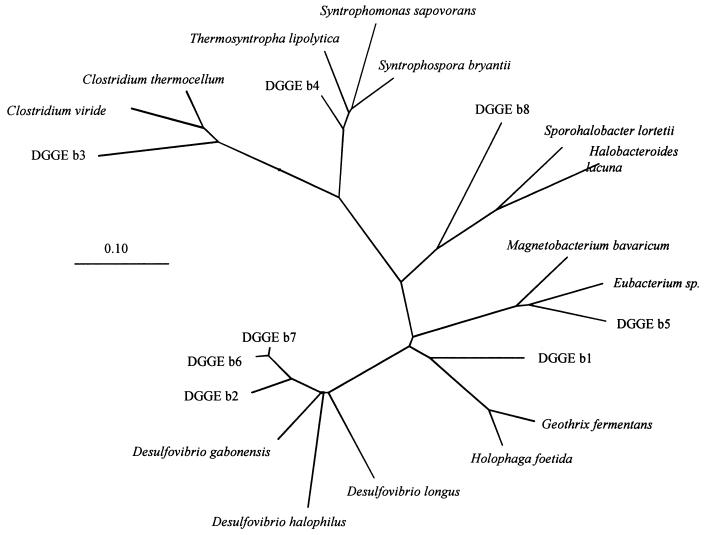
The DGGE analyses revealed that the three aggregates contained a different community composition on the DNA level as well as on the RNA level (Fig. 4). In all aggregates less cDNA bands (reverse transcriptase PCR [RT-PCR]-amplified 16S rRNA fragments) were seen than rDNA bands (PCR-amplified 16S rDNA fragments). Blotting of DGGE gels and hybridization of these blots with group-specific probes of SRB (660, 687, and 804) resulted for all samples in a positive hybridization signal with probe 687 (two to three bands) and probe 660 (two to four bands), but no hybridization was obtained with probe 804 (data not shown), indicating the presence of Desulfovibrio and Desulfobulbus species. Eleven bands were excised from the DGGE gel, from which eight were successfully reamplified and sequenced (numbers are indicated in Fig. 4). The sequences were phylogenetically analyzed and depicted in a phylogenetic tree (Fig. 5). Three of the DNA fragments resembled sequences of Desulfovibrio species (DGGE bands 2, 6, and 7), and one partial sequence (DGGE band 4) resembled the syntrophic bacteria Syntrophobacter wolinii and Syntrophomonas wolfei. The other four DNA fragments (DGGE bands 1, 3, 5, and 8) were found in diverse clusters, resembling sequences of Holophaga, Clostridium, Eubacterium, and Halobacteroides species (Fig. 5).
FIG. 4.
DGGE of 16S rDNA and 16S rRNA PCR fragments from the sulfidogenic, methanogenic-sulfidogenic, and methanogenic aggregates. The numbers refer to the numbers of the excised and sequenced bands. The curved bands in the lower part of the DGGE gel are single-stranded DNA and should be disregarded.
FIG. 5.
Phylogenetic analysis of the partial sequences derived from excised DGGE bands depicted in Fig. 4. The phylogenetic parsimony tree was calculated with the ARB program. The bar indicates 0.1 estimated change per nucleotide.
Population analysis by FISH.
Phase-contrast light microscopy of the aggregate sections showed dense bacterial clusters, some void space, and a regular surface of the aggregates. If excited with blue light (488 nm), the cells and extracellular material exhibited strong green autofluorescence. Therefore, we exclusively applied CY3- or CY5-labelled probes. Comparison of 5 to 10 aggregates showed little variation between individual aggregates from each reactor, but the three types of aggregates had a different structure and population distribution.
(i) Methanogenic-sulfidogenic aggregates.
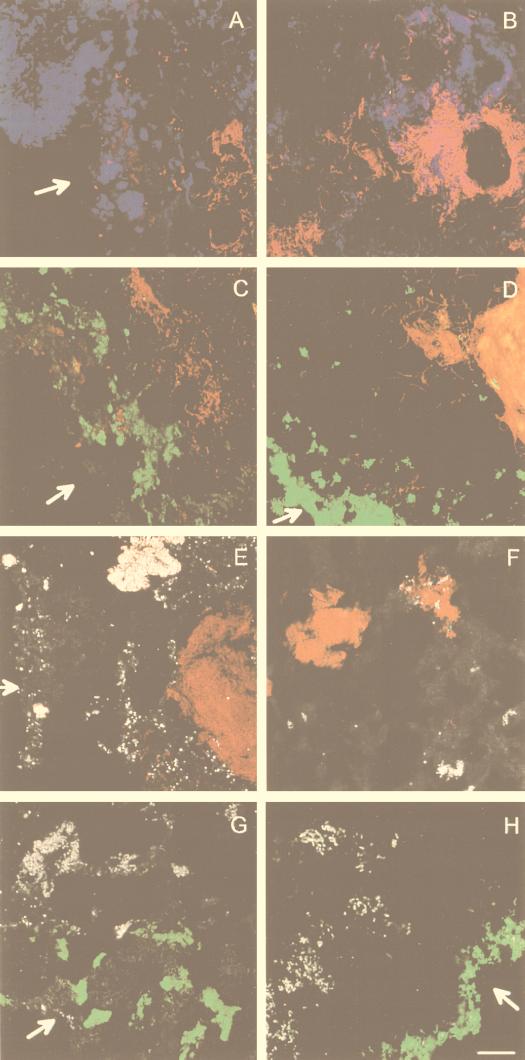
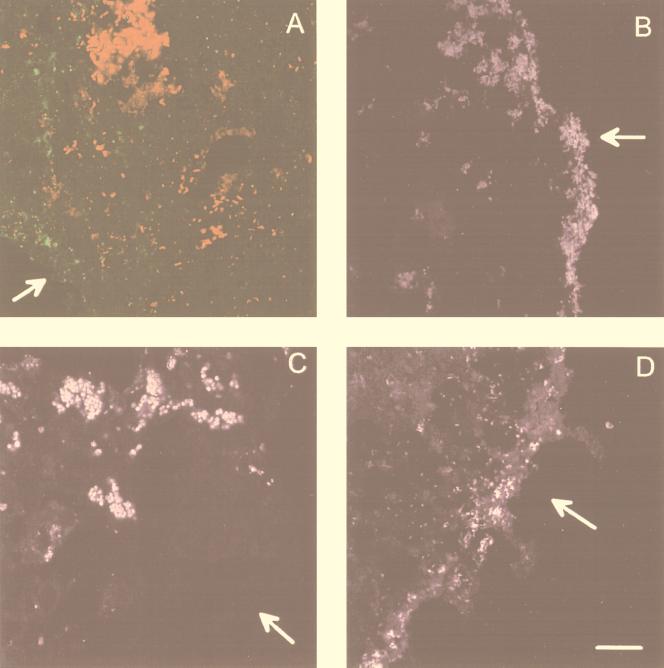
The methanogenic-sulfidogenic aggregates contained an inner core of Archaea (probe ARC915), below ca. 100 μm from the surface (Fig. 6B and D). Blue autofluorescence (F430) in the center of unfixed aggregates confirmed the presence of methanogenic Archaea. The outer shell (30 to 50 μm thick) contained dense populations of SRB (Fig. 6D). Between these two zones a low number of Eubacteria was found (Fig. 6B), of which some hybridized with probe MPOB (clusters with big coccoid cells, Fig. 6F and H). The use of group-specific probes for SRB resulted in hybridizations with probe DSV698 and 660 of cells in the outer layer (Fig. 7B and D). No hybridization was observed with the other group-specific SRB probes (Table 1).
FIG. 6.
FISH analysis to study the population distribution within sulfidogenic and methanogenic-sulfidogenic aggregates by using various probes labeled with CY3 and CY5. The photographs are overlays of two confocal microscopic images. Panels: A and B, EUB338 (artificial color blue) and ARC915 (artificial color red) hybridizations of sulfidogenic (A) and methanogenic-sulfidogenic (B) aggregates; C and D, SRB385 (artificial color green) and ARC915 (artificial color red) hybridization of sulfidogenic (C) and methanogenic-sulfidogenic (D) aggregates; E and F, ARC915 (artificial color red) and MPOB (artificial color white) hybridization of sulfidogenic (E) and methanogenic-sulfidogenic (F) aggregates; G and H, SRB385 (artificial color green) and MPOB (artificial color white) hybridization of sulfidogenic (G) and methanogenic-sulfidogenic (H) aggregates. The scale bar is 20 μm, and the arrows indicate the aggregate surface.
FIG. 7.
FISH analysis to localize specific SRB populations within the aggregates. (A) Probe SRB385 (artificial color green) and ARC915 (artificial color red) hybridization of methanogenic aggregates. This photograph is an overlay of two confocal microscopic images. (B and D) Hybridization with probe 660 (artificial color purple) (B) and probe 698 (artificial color purple) (D) in a methanogenic-sulfidogenic aggregate. (C) Hybridization with probe 658 (artificial color purple) in a sulfidogenic aggregate. The scale bar is 20 μm, and the arrows indicate the aggregate surface.
(ii) Sulfidogenic aggregates.
The different populations in sulfidogenic aggregates were more dispersed. Figure 6A illustrates their irregular surface, clearly showing eubacterial buddings at several locations. The SRB were mainly situated at the outer 200 to 300 μm of the aggregate (Fig. 6C) but did not form a compact shell as in methanogenic-sulfidogenic aggregates (Fig. 6D). Cells hybridizing with probe MPOB were scattered over the interior part (Fig. 6E and G). The methanogens were present in clusters in the inner part of the aggregate, often forming a very compact core (Fig. 6A, C, and E and Fig. 7A). DSS658 was the only specific SRB probe that hybridized with sulfidogenic aggregates, clearly showing clusters of coccoid cells at the surface and more inward (Fig. 7C). Strong autofluorescence, especially with higher formamide concentrations, hampered quantitative analysis of the FISH results.
(iii) Methanogenic aggregates.
The methanogenic aggregates contained hardly any SRB (Fig. 7A), and no cells hybridizing with the MPOB probe could be detected. The methanogens were present in clusters in the inner part of the aggregate.
DISCUSSION
Architecture of UASB aggregates.
The structural study with molecular techniques could not be performed on the same individual aggregates used for the functional analyses with microsensors; however, comparison of 5 to 10 aggregates from each reactor show little variation. Thus, the observations, obtained with different techniques on different individual aggregates, can be compared well. This study clearly showed that different spatial arrangements of the SRB and MB populations occur in anaerobic aggregates. SRB and MB were distributed in a layered structure in the methanogenic-sulfidogenic and sulfidogenic aggregates (Fig. 6 and 7), which resulted in a zonation where their activity is predominant (Fig. 1 to 3). The differences in structure of the different types of aggregates can be attributed to the wastewater compositions (Table 2). The development of different types of aggregates, depending on the substrate, has been reported previously (12, 13).
The core of all three types of aggregates was composed of MB, whereas SRB were mainly present in an outer shell, as was also found by Sekiguchi et al. (52). The low SRB population density in the center of the aggregates contrasts with the fact that SRB can outcompete methanogens because of their more favorable thermodynamics for acetate and H2 (23, 51, 62). Diffusional limitation of sulfate has been suggested as a reason for the maintenance of MB in the core of aggregates treating sulfate-rich wastewaters (43). From our in situ activity measurements, we calculated that this cannot be the case for the aggregates used in this study. We suggest that the MB core of the primary inoculum is conserved and that SRB colonize this core in a later development stage. A similar outgrowth of SRB after methanogenic granular sludge was fed a mixture of propionate and sulfate was observed by Harmsen et al. (16). Besides adaptation of the primary inoculum to a different substrate, also new granules are formed during reactor operation. The initial aggregation might be mediated by methanogens, which have better attachment characteristics than SRB (18, 38), while SRB attach later on during aggregate development.
The inner core of the methanogenic-sulfidogenic aggregates was one solid cluster of MB, surrounded by a layer of syntrophic bacteria (Fig. 6B, D, and E). The core of the sulfidogenic aggregates contained several smaller clusters of MB with dispersed syntrophs (Fig. 6A and C and Fig. 7A). Indeed, the formation of clusters of methanogens in juxtaposition with syntrophic bacteria offers both groups of bacteria a nutritional advantage.
Endogenous microprofiles.
Microsensor measurement of H2S and CH4 profiles indicated that both the SRB and MB populations were also active in the absence of externally supplied electron donor for at least 24 h (Fig. 1), as was also found by de Beer et al. (10). This indicates that both SRB and MB metabolize some pool of storage material, either bound to particles, polymers or cells. E-donors were, however, limited within the aggregates, and the activity of both the SRB and MB increased considerably upon addition of an electron donor (Fig. 2).
The S0 found might be the source of sulfide in sulfate-free medium (Fig. 1), either by sulfur reduction or sulfur disproportionation (58). However, it is unclear how the S0 was formed in the absence of an electron acceptor. We took care to avoid exposure to oxygen, so it is unlikely that this could cause the high S0 concentrations observed (Table 2). A reverse sulfur disproportionation might be an interesting possibility, but it has not yet been demonstrated and could not explain both the presence of sulfur and the formation of sulfide.
Characteristics of the SRB population.
The sulfate reduction rates in the aggregates are up to 1,000 times higher than those reported for sediments, i.e., 0.0009 mmol of S2− m−3 s−1 (25) and 0.002 to 0.01 mmol of S2− m−3 s−1 (3). However, a realistic comparison is only possible between systems with similar cell densities. Comparable activities have been measured in the anaerobic zones of biofilms, i.e., 0.2 mmol of S2− m−3 s−1 (24) or 8 mmol of S2− m−3 s−1 (37). From the H2S fluxes and the number of SRB (determined by FISH) in the methanogenic-sulfidogenic aggregates, the specific sulfate reduction rate was calculated to be 25 fmol SO42− cell−1 day−1. This value is in the high-end range of the specific sulfate reduction rates reported (21).
The methanogenic-sulfidogenic aggregates contained Desulfovibrio spp., as well as Desulfobulbus spp. These two SRB species were absent in the sulfidogenic granules, which contained predominantly the nutritionally versatile Desulfosarcina and/or Desulfococcus species. The H2S microprofiles measured in the sulfidogenic aggregates suggest that different, spatially separated, H2- and ethanol-consuming SRB populations are present in the outer 400 μm (Fig. 2A). These populations could, however, not be differentiated by the FISH probes applied. Few of the specific SRB probes (Table 1) yielded a positive signal in the aggregates (Fig. 7B to D). Probably, these aggregates contain SRB populations, for which specific probes still need to be developed. Several acetate-utilizing SRB species are known, such as Desulfobacter, Desulfobacterium, Desulfococcus, Desulfosarcina, and Desulfotomaculum species. Desulfobacter species were not detected by the DGGE and FISH analyses, confirming other studies that Desulfobacter is unimportant in methanogenic aggregates (40, 46). Desulfosarcina or Desulfococcus species are likely to be responsible for the degradation of acetate in the sulfidogenic aggregates. The presence of other acetotrophic SRB species cannot be excluded, as recently Desulforhabdus amnigenus (41) and Desulfobacca acetoxidans (39) have been isolated from granular sludge. More research with specific 16S rRNA probes for these newly described organisms is needed to examine their presence in the anaerobic aggregates.
The low SRB population density in the methanogenic aggregates (Fig. 2C and 7A) is somewhat surprising. SRB are generally assumed to be more robust than MB (62) and have the ability to use other metabolic pathways, such as fermentation (62) or syntrophic growth with methanogens (63). The medium on which the methanogenic aggregates were subcultured contained sulfate (0.45 mM), which could have supported the growth of SRB. Apparently, this small amount of sulfate could not support the development of a substantial SRB population.
Characteristics of the MB population.
The methanogenic rates in the aggregates were 10 times higher than the methanogenic activities measured in sediments (0.1 to 0.6 mmol of CH4 m−3 s−1 [34]) and during sewage sludge digestion (0.6 mmol of CH4 m−3 s−1 [54]). The rod-shaped morphology of the methanogens in the center of the aggregates resembles that of Methanosaeta species (64). This obligate acetoclastic methanogenic species was also found to be present in the core of other layered granules (14, 27, 31, 52). Methanosaeta species are the least sensitive among the methanogens for acetate diffusional limitations because of their higher affinity for acetate compared to other methanogens (13, 14, 20). This, together with their ability to form close frameworks and/or mats (44), explains their presence in the center of the aggregates.
Although MB were found in sulfidogenic granules (Fig. 6A, C, and E), no methane production was detected by activity tests (data not shown). Indeed, MB survive long starvation times (28), while maintaining their ribosomes (46).
The CH4 microprofiles showed that methanogenesis was not affected by the addition of sulfate (Fig. 1 and 2). The literature concerning this phenomenon is contradictory. Methanogenesis was inhibited by sulfate addition in sediments (30, 62) and anaerobic digestors (29, 46). Others reported no effect at all on methanogenesis under sulfate-rich conditions in the same environments (53, 59, 64). The differences between these observations might be explained by substrate availability or by the adaptation times of SRB populations.
Characteristics of the syntrophic population.
Methanogens cannot use propionate as an electron donor. The increase in methanogenic activity by propionate addition (Fig. 2B) illustrates the dependency of methanogens on propionate-oxidizing syntrophic bacteria. The DGGE analysis suggested the presence of a relative of Syntrophobacter wolinii and Syntrophomonas wolfei (Fig. 5), whereas the FISH analysis with probe MPOB (Fig. 6G and H) suggested the presence of strain MPOB, classified as Syntrophobacter fumaroxidans (17). According to recent studies, Syntrophobacter spp. were capable of oxidizing propionate by sulfate reduction (15, 62). The Desulfobulbus layer at the surface of the methanogenic-sulfidogenic aggregates (Fig. 7B) indicates that syntrophic bacteria cannot outcompete Desulfobulbus spp. in this granular sludge developed under sulfate-rich conditions.
The MPOB-like cells grew more inwards than did the SRB. Methanogens can grow syntrophically with acetate- and H2-producing bacteria either in juxtaposition in the core or as adjacent layers within the aggregate (16, 52, 63). The latter type of syntrophy was found in methanogenic-sulfidogenic aggregates (Fig. 6F). Syntrophic bacteria were found between layers of SRB and MB, providing both groups with H2 and acetate, as postulated by MacLeod et al. (31). In the sulfidogenic granules (Fig. 6E), MPOB-like cells were surrounded by MB cells, indicating that the MPOB-like cells indeed grow syntrophically with MB.
Concluding remarks.
Combining microsensors and molecular techniques provided direct information about sulfate reduction and methanogenesis in UASB aggregates. Data on the community structure could be related to the metabolic functions of the respective populations. SRB were mainly found in the outer layer (<200 μm) and MB were predominantly in the core of UASB aggregates. For further detailed investigation, microsensors for the substrate H2 and additional oligonucleotide probes for newly isolated syntrophic and SRB populations are under development.
ACKNOWLEDGMENTS
This work was financially supported by a personal grant to Bo Barker Jørgensen from the Körber Foundation (Hamburg, Germany), the Max Planck Society (Munich, Germany), and the European Union (project MICROMARE contract MASCT 950029).
We thank Helle Ploug for fruitful discussions; Rudi Amann for his support; Gaby Eickert, Anja Eggers, Vera Hübner, and Lars B. Pedersen for technical assistance with the microsensors; and Ramón Rosselló-Mora for his help with the phylogenetic analysis.
REFERENCES
- 1.Amann R, Krumholz I, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R, Stromley J, Devereux R, Key R, Stahl D A. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl Environ Microbiol. 1992;58:614–623. doi: 10.1128/aem.58.2.614-623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaabjerg V, Mouritsen K N, Finster K. Diel cycles of sulphate reduction rates in sediments of a Zostera marina bed (Denmark) Aquat Microb Ecol. 1998;15:97–102. [Google Scholar]
- 4.Broecker W S, Peng T-H. Gas exchange rates between air and sea. Tellus. 1974;26:21–35. [Google Scholar]
- 5.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 6.Cline J D. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr. 1969;14:454–458. [Google Scholar]
- 7.Damgaard, L. R., and N. P. Revsbech. 1999. Unpublished data.
- 8.Damgaard L R, Revsbech N P. A microscale biosensor for methane containing methanotrophic bacteria and an internal oxygen reservoir. Anal Chem. 1997;69:2262–2267. doi: 10.1021/ac9611576. [DOI] [PubMed] [Google Scholar]
- 9.Damgaard L R, Revsbech N P, Reichard W. Use of an oxygen-insensitive microscale biosensor for methane to measure methane concentration profiles in a rice paddy. Appl Environ Microbiol. 1998;64:864–870. doi: 10.1128/aem.64.3.864-870.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Beer D, Huisman J W, Van den Heuvel J C, Ottengraf S P P. The effect of pH profiles in methanogenic aggregates on the kinetics of acetate conversion. Water Res. 1992;26:1329–1336. [Google Scholar]
- 11.Devereux R, Kane M D, Wilfrey J, Stahl D A. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst Appl Microbiol. 1992;15:601–609. [Google Scholar]
- 12.Fang H H P, Chui H K, Li Y Y. Microbial structure and activity of UASB granules treating different wastewaters. Water Sci Technol. 1994;30:87–96. [Google Scholar]
- 13.Grotenhuis J T C, Smit M, Plugge C M, Yuansheng X, Van Lammeren A A M, Stams A J M, Zehnder A J B. Bacteriological composition and structure of granular sludge adapted to different substrates. Appl Environ Microbiol. 1991;57:1942–1949. doi: 10.1128/aem.57.7.1942-1949.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guiot S R, Pauss A, Costerton J W. A structured model of the anaerobic granule consortium. Water Sci Technol. 1992;25:1–10. [Google Scholar]
- 15.Harmsen H J M, Akkermans A D L, Stams A J M, De Vos W M. Population dynamics of propionate-oxidizing bacteria under methanogenic and sulfidogenic conditions in anaerobic granular sludge. Appl Environ Microbiol. 1996;62:2163–2168. doi: 10.1128/aem.62.6.2163-2168.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harmsen H J M, Kengen H M P, Akkermans A D L, Stams A J M, De Vos W M. Detection and localization of syntrophic propionate-oxidizing bacteria in granular sludge by in situ hybridization using 16S rRNA-based oligonucleotide probes. Appl Environ Microbiol. 1996;62:1656–1663. doi: 10.1128/aem.62.5.1656-1663.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harmsen H J M, van Kuijk B L M, Plugge C M, Akkermans A D L, de Vos W M, Stams A J M. Description of Syntrophobacter fumaroxidans sp. nov., a syntrophic propionate-degrading sulfate-reducing bacterium. Int J Syst Bacteriol. 1998;48:1383–1387. doi: 10.1099/00207713-48-4-1383. [DOI] [PubMed] [Google Scholar]
- 18.Isa Z, Grusenmeyer S, Verstraete W. Sulfate reduction relative to methane production in high-rate anaerobic digestion: microbial aspects. Appl Environ Microbiol. 1986;51:580–587. doi: 10.1128/aem.51.3.580-587.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeroschewski P, Steuckart C, Kühl M. An amperometric microsensor for the determination of H2S in aquatic environments. Anal Chem. 1996;68:4351–4357. [Google Scholar]
- 20.Jetten M S M, Stams A J M, Zehnder A J B. Acetate threshold values and acetate activating enzymes in methanogenic bacteria. FEMS Microbiol Ecol. 1990;73:339–344. [Google Scholar]
- 21.Jørgensen B B. A comparison of methods for the quantification of bacterial sulfate reduction in coastal marine sediments. Geomicrobiology. 1978;1:49–64. [Google Scholar]
- 22.Karchmer J H. The analytical chemistry of sulfur and its compounds. New York, N.Y: Wiley-Interscience; 1979. [Google Scholar]
- 23.Kristjansson J K, Schönheit P, Thauer R K. Different Ks values for hydrogen of methanogenic bacteria and sulfate reducing bacteria: an explanation for the apparent inhibition of methanogenesis by sulfate. Arch Microbiol. 1982;131:278–282. [Google Scholar]
- 24.Kühl M, Jørgensen B B. Microsensor measurements of sulfate reduction and sulfide oxidation in compact microbial communities of aerobic biofilms. Appl Environ Microbiol. 1992;58:1164–1174. doi: 10.1128/aem.58.4.1164-1174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kühl M, Steuckart C, Eickert G, Jeroschewski P. A H2S microsensor for profiling biofilms and sediments: application in an acidic lake sediment. Aquat Microb Ecol. 1998;15:201–209. [Google Scholar]
- 26.Lens P, De Beer D, Cronenberg C, Ottengraf S, Verstraete W. The use of microsensors to determine population distribution in UASB aggregates. Water Sci Technol. 1995;31:273–280. [Google Scholar]
- 27.Lens P N L, De Beer D, Cronenberg C C H, Houwen F P, Ottengraf S P P, Verstraete W H. Heterogeneous distribution of microbial activity in methanogenic aggregates: pH and glucose microprofiles. Appl Environ Microbiol. 1993;59:3803–3815. doi: 10.1128/aem.59.11.3803-3815.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lettinga G. Anaerobic digestion and wastewater treatment systems. Antonie Leeuwenhoek. 1995;67:3–28. doi: 10.1007/BF00872193. [DOI] [PubMed] [Google Scholar]
- 29.Li Y-Y, Lam S, Fang H H P. Interactions between methanogenic, sulfate-reducing and syntrophic acetogenic bacteria in the anaerobic degradation of benzoate. Water Res. 1996;30:1555–1562. [Google Scholar]
- 30.Lovley D R, Dwyer D F, Klug M J. Kinetic analysis of competition between sulfate reducers and methanogens for hydrogen in sediments. Appl Environ Microbiol. 1982;43:1373–1379. doi: 10.1128/aem.43.6.1373-1379.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacLeod F A, Guiot S R, Costerton J W. Layered structure of bacterial aggregates produced in an upflow anaerobic sludge bed and filter reactor. Appl Environ Microbiol. 1990;56:1598–1607. doi: 10.1128/aem.56.6.1598-1607.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 33.Manz W, Eisenbrecher M, Neu T R, Szewzyk U. Abundance and spatial organization of gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol Ecol. 1998;25:43–61. [Google Scholar]
- 34.Mountfort D O, Asher R A. Role of sulfate reduction versus methanogenesis in terminal carbon flow in polluted intertidal sediment of Waimea Inlet, Nelson, New Zealand. Appl Environ Microbiol. 1981;42:252–258. doi: 10.1128/aem.42.2.252-258.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. 3.4.4. Dordrecht, The Netherlands: Kluwer; 1998. pp. 1–27. [Google Scholar]
- 36.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes encoding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okabe S, Matsuda T, Satoh H, Itoh T, Watanabe Y. Sulfate reduction and sulfide oxidation ion aerobic mixed population biofilms. Water Sci Technol. 1998;37:131–138. [Google Scholar]
- 38.Omil F, Oude Elferink S J W H, Lens P, Hulshoff-Pol L, Lettinga G. Effect of the inoculation with Desulforhabdus amnigenus and pH or O2 shocks on the competition between sulfate reducing and methanogenic bacteria in an acetate fed UASB reactor. Biores Technol. 1997;60:113–122. [Google Scholar]
- 39.Oude Elferink, S. J. W. H., W. M. Akkermans-van Vliet, J. J. Bogte, and A. J. M. Stams. Desulfobacca acetooxidans gen. nov. sp. nov., a novel acetate-degrading sulfate reducer isolated from sulfidogenic granular sludge. Int. J. Syst. Bacteriol., in press. [DOI] [PubMed]
- 40.Oude Elferink S J W H, Boschker H T S, Stams A J M. Identification of sulfate reducers and Syntrophobacter sp. in anaerobic granular sludge by fatty-acid biomarkers and 16S rRNA probing. Geomicrobiology. 1998;15:3–17. [Google Scholar]
- 41.Oude Elferink S J W H, Maas R N, Harmsen H J M, Stams A J M. Desulforhabdus amnigenus gen. nov. sp. nov., a sulfate reducer isolated from anaerobic granular sludge. Arch Microbiol. 1995;164:119–124. [PubMed] [Google Scholar]
- 42.Oude Elferink S J W H, Vorstman W J C, Sopjes A, Stams A J M. Characterization of the sulfate-reducing and syntrophic population in granular sludge from a full-scale anaerobic reactor treating papermill wastewater. FEMS Microbiol Ecol. 1998;27:185–194. [Google Scholar]
- 43.Overmeire A, Lens P, Verstraete W. Mass transfer limitation of sulfate in methanogenic aggregates. Biotechnol Bioeng. 1994;44:387–391. doi: 10.1002/bit.260440318. [DOI] [PubMed] [Google Scholar]
- 44.Patel G B. Characterization and nutritional properties of Methanotrix concilii sp. nov., a mesophilic, aceticlastic methanogen. Can J Microbiol. 1984;30:1383–1396. [Google Scholar]
- 45.Ramsing N B, Fossing H, Ferdelman T G, Andersen F, Thamdrup B. Distribution of bacterial populations in a stratified fjord (Mariager Fjord, Danmark) quantified by in situ hybridization and related to chemical gradients in the water column. Appl Environ Microbiol. 1996;62:1391–1404. doi: 10.1128/aem.62.4.1391-1404.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raskin L, Rittmann B E, Stahl D A. Competition and coexistence of sulfate-reducing and methanogenic populations in anaerobic biofilms. Appl Environ Microbiol. 1996;62:3847–3857. doi: 10.1128/aem.62.10.3847-3857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Revsbech N P. An oxygen microelectrode with a guard cathode. Limnol Oceanogr. 1989;55:1907–1910. [Google Scholar]
- 48.Revsbech N P, Jørgensen B B. Microelectrodes: their use in microbial ecology. Adv Microb Ecol. 1986;9:293–352. [Google Scholar]
- 49.Revsbech N P, Nielsen L P, Ramsing N B. A novel microsensor for determination of apparent diffusivity in sediments. Limnol Oceanogr. 1998;43:986–992. [Google Scholar]
- 50.Santegoeds C M, Ferdelman T G, Muyzer G, De Beer D. Structural and functional dynamics of sulfate-reducing populations in bacterial biofilms. Appl Environ Microbiol. 1998;64:3731–3739. doi: 10.1128/aem.64.10.3731-3739.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schönheit P, Kristjansson J K, Thauer R K. Kinetic mechanism for the ability of sulfate reducers to outcompete methanogens for acetate. Arch Microbiol. 1982;132:285–288. [Google Scholar]
- 52.Sekiguchi Y, Kamagata Y, Nakamura K, Ohashi A, Harada H. Fluorescence in situ hybridization using 16S rRN-targeted oligonucleotides reveals localization of methanogens and selected uncultured bacteria in mesophilic and thermophilic sludge granules. Appl Environ Microbiol. 1999;65:1280–1288. doi: 10.1128/aem.65.3.1280-1288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Senior E, Lindström E B, Banat I M, Nedwell D B. Sulfate reduction and methanogenesis in the sediment of saltmarsh on the East Coast of the United Kingdom. Appl Environ Microbiol. 1982;43:987–996. doi: 10.1128/aem.43.5.987-996.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith P H, Mah D T. Kinetics of acetate metabolism during sludge digestion. Appl Microbiol. 1966;14:368–371. doi: 10.1128/am.14.3.368-371.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stahl D A, Amann R. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Vol. 8. London, England: John Wiley & Sons, Ltd.; 1991. pp. 207–248. [Google Scholar]
- 56.Strunk O, Gross O, Reichel B, May M, Hermann S, Stuckmann N, Nonhoff B, Lenke M, Ginhart T, Vilbig A, Ludwig T, Bode A, Schleifer K-H, Ludwig W. ARB: a software environment for sequence data. Munich, Germany: Department of Microbiology, Technische Universität München; 1998. [Google Scholar]
- 57.Teske A, Wawer C, Muyzer G, Ramsing N B. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl Environ Microbiol. 1996;62:1405–1415. doi: 10.1128/aem.62.4.1405-1415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thamdrup B, Finster K, Hansen J W, Bak F. Bacterial disproportionation of elemental sulfur coupled to chemical reduction of iron or manganese. Appl Environ Microbiol. 1993;59:101–108. doi: 10.1128/aem.59.1.101-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ueki K, Ueki A, Takahashi K, Iwatsu M. The role of sulfate reduction in methanogenic digestion of municipal sewage sludge. J Gen Appl Microbiol. 1992;38:195–207. [Google Scholar]
- 60.Verstraete W, de Beer D, Pena M, Lettinga G, Lens P. Anaerobic bioprocessing of waste. World J Microbiol Biotechnol. 1996;12:221–238. doi: 10.1007/BF00360919. [DOI] [PubMed] [Google Scholar]
- 61.Wallrabenstein C, Hauschild E, Schink B. Syntrophobacter pfenningii sp. nov., new syntrophically propionate-oxidising anaerobe growing in pure culture with propionate and sulfate. Arch Microbiol. 1995;164:346–352. [Google Scholar]
- 62.Widdel F. Microbiology and ecology of sulfate- and sulfur-reducing bacteria. In: Zehnder A D J, editor. Biology of anaerobic microorganisms. New York, N.Y: John Wiley & Sons; 1988. pp. 469–585. [Google Scholar]
- 63.Winfrey M R, Zeikus J G. Effect of sulfate on carbon and electron flow during microbial methanogenesis in freshwater sediments. Appl Environ Microbiol. 1977;33:275–281. doi: 10.1128/aem.33.2.275-281.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu W-M, Jain M K, De Macario E C, Thiele J H, Zeikus J G. Microbial composition and characterization of prevalent methanogens and acetogens isolated from syntrophic methanogenic granules. Appl Microbiol Biotechnol. 1992;38:282–290. [Google Scholar]