Abstract
Background
Prostate cancer (Pca) is a public health problem that affects men, usually of middle age or older. It is the second most common cancer diagnosed in men and the fifth leading cause of death. The RNASEL gene located in 1q25 and identified as a susceptibility gene to hereditary prostate cancer, has never been studied in relation to prostate cancer in Burkina Faso. The aim of this study was to analyze the carriage of RNASEL R462Q and D541E mutations and risks factors in patients with prostate cancer in the Burkina Faso.
Methods
This case–control study included of 38 histologically diagnosed prostate cancer cases and 53 controls (cases without prostate abnormalities). Real-time PCR genotyping of R462Q and D541E variants using the TaqMan® allelic discrimination technique was used. Correlations between different genotypes and combined genotypes were investigated.
Results
The R462Q variant was present in 5.3% of cases and 7.5% of controls. The D541E variant was present in 50.0% of cases and 35% of controls. There is no association between R462Q variants (OR = 0.60; 95%IC, 0.10–3.51; p = 0.686) and D541E variants (OR = 2.46; 95%IC, 0.78–7.80; p = 0.121) and genotypes combined with prostate cancer. However, there is a statistically significant difference in the distribution of cases according to the PSA rate at diagnosis (p ˂ 0.001). For the Gleason score distribution, only 13.2% of cases have a Gleason score greater than 7. There is a statistically significant difference in the Gleason score distribution of cases (p ˂ 0.001).
Conclusions
These variants, considered in isolation or in combination, are not associated with the risk of prostate cancer.
Keywords: RNASEL, R462Q, D541E, Prostate cancer, Burkina Faso
Background
Prostate cancer (Pca) is the second most frequently diagnosed malignant tumour in humans in the world. It is the fifth leading cause of cancer death in humans, with an estimated 1.4 million new cases and 375,304 deaths in 2020 in the world. In Burkina Faso, in the same year, the number of new cases of prostate cancer was 997 out of 4,305 new cases of cancer, with 608 deaths caused. It is the first cancer in terms of incidence in men (and the fifth cancer in both sexes), followed by liver cancer [1]. The etiology of prostate cancer has been the subject of numerous studies but remains largely unknown. The risk factors that remain well established are advanced age, ethnicity, family history [2–4]. Indeed, the incidence of prostate cancer is estimated to be 1 in 350 for men under 50 years [5]; 1 in 52 for 50- to 59-year-olds; then 60% in men over 65 years. Almost 30% of men over 50 years who died from causes other than prostate cancer have been shown to have histological evidence of prostate cancer at the time of the autopsy [6]. Populations of African descent, such as African Americans, Caribbean, and blacks in Europe had the highest incidences, early disease and more aggressive form compared to other racial and ethnic groups [7]. Men of African descent are estimated to have a relative risk of 9.7 versus 3.9 in Caucasians and 1.6 in Asians when two or more first-degree relatives have prostate cancer [8]. Regarding family history, more than 20% of patients with prostate cancer report a family history. This can be explained on the one hand by the common sharing of genes; but also on the other hand by a similar pattern [9] of exposure to certain environmental carcinogens and to common lifestyles [10]. The relative risk of prostate cancer for men with a first-degree relative with prostate cancer is estimated to be about 2.5. This risk increases to 5.3 when three or more first-degree relatives are affected. Serum prostate antigen assay and rectal touch are currently the primary screening methods for prostate cancer [11]. However, with the ultimate goal of developing new, more accurate and beneficial biomarkers in the detection, prevention and treatment of this disease, several studies have been conducted to elucidate the molecular mechanisms involved in the genesis and progression of prostate cancer [12]. The high incidence of prostate cancer in African men suggests a genetic predisposition. Initial quantitative genetic analyses of homo and dizygous twins estimated that germ mutations contributed to prostate cancer risk at approximately 40–58% [13–15]. Linkage analysis and positional cloning were used to successfully map inherited chromosomal regions containing prostate cancer susceptibility genes. The HPC1 (Hereditary Prostate Cancer 1) locus, located on chromosome region 1q24-25, was the first of these loci to be identified in 1996 [16, 17]. Since then, several other loci of predisposition to hereditary forms of prostate cancer have been identified: HPCX (Xq27-28), HPC20 (20q13), HPC2 (17p11), PG1 (prostate cancer susceptibility gene 1) (8p22-23), CAPB (1p36) [18]. Three genes for hereditary prostate cancer susceptibility have been identified in three of these loci. This is the RNASEL(2'-5' oligoadenylate synthetase-dependent ribonuclease) gene (HPC1); of ELAC2 (ElaC Ribonuclease Z 2) (HPC2) which encodes a metallo-dependent hydrolase potentially involved in the repair of the inter-strand cross-linking of DNA and the editing of mRNA and finally the MSR1(Macrophage Scavenger Receptor1) (PG1) gene which encodes subunits a macrophage scavenger receptor which is capable of binding to a variety of ligands. [19–23]. Mutations in these different genes have low or moderate penetrance. They influence the way the prostate works and are responsible for about 30% of prostate cancer [24]. Other high penetration mutations have been identified in the genes regulating: the critical stages of the development process, namely the G84E mutation of the HOXB13 gene [25, 26]; the Q356R, 185delAG, 5382insC and 6174delT mutations in the BRCA2 gene [27]. Studies of these different regions related to prostate cancer in different populations have provided inconsistent results. These observations show the genetic complexity and heterogeneity (environmental and genetic factors) of prostate cancer predisposition. The RNASEL gene, located at 1q24-25, with a size of about 15 kilos pair of bases, and comprising 8 exons; code for ribonuclease 2'-5'-oligoadenaylate (2-5A) -dependent. RNASEL regulates cell proliferation and apoptosis through the interferon-induced 2’-5’A pathway through its antiviral and antiproliferative activity [28]. There are many nucleotide variants identified in the RNASEL gene. Seven of them cause changes in the protein sequence. Six variants cause false sense alterations and a rare variant creates a nonsense mutation [29]. The most commonly studied synonymous variants in association with prostate cancer in different types of populations are R462Q and D541E. The different expression studies did not prove that the two polymorphisms of RNASEL can influence the expression of the gene; but the functional studies were able to show that the R462Q reduces the ability of the cell to cause apoptosis in response to 2’-5’A activation and also has three times less enzymatic activity than normal, while D541E does not affect the function of the Rnase L protein [18, 19]. The results of these studies remain contradictory. The AA genotype in R462Q has been associated with both an increased risk of prostate cancer in the United States and in some Caucasian population groups [30, 31] and a decreased risk in Caucasian and Japanese sample groups. Previous studies on the RNASEL variant D541E indicated that the GG and TT genotypes were associated with an increased risk of prostate cancer in some Japanese [32] and European-American [33] populations, respectively. On the other hand, a negative association of the TT genotype with prostate cancer in Swedish Caucasian samples was reported by Wiklund et al. in 2004 [34]. In summary, several studies provide strong support, both functional and epidemiological, that RNASEL plays a role in prostate cancer, but other studies have suggested a lack of role based on the ethno-geographic origins of study populations. In West Africa, several studies of prostate cancer in different populations have focused on the epidemic and morphological aspects of prostate cancer [35–37]. Very few studies have examined the genetic background of African populations and its contribution to prostate cancer susceptibility. This limits the use of genetic data at all levels of prostate cancer management such as screening, diagnosis, treatment and follow-up in the African context. The study described here was undertaken to determine the involvement of R462Q and D541E variants of the RNASEL gene in prostate cancer in the Burkinabe population. This could provide additional information that could potentially be exploited to improve early detection and diagnosis of high-risk individuals for early therapeutic intervention or ease of management.
Materials and methods
Design of study
The study was conducted between October 2019 and April 2021. The study population (Burkinabe) consisted of 38 patients, histologically diagnosed with prostate cancer (cases) and 53 males at least 45 years of age with either a total PSA levels less than 4 ng/ml or normal PSA derivatives (free PSA, free / total ratio, velocity and density of PSA) or a negative prostate biopsy (controls). They are all monitored at the Saint Camille hospital in Ouagadougou (HOSCO) or at the NINA clinic in Ouagadougou. Biomolecular analyzes were carried out at the Molecular and Genetic Biology Laboratory (LABIOGENE) and at the Pietro Annigoni Biomolecular Research Center (CERBA).
Sample collection
After obtaining consent from patients and controls, a questionnaire was administered to collect sociodemographic, anthropometric, and clinical data from participants. Venous blood from consenting participants was collected on Ethylene- Diamine-Tetra-Acetic (EDTA) filled tubes. After centrifugation, at 3,500 revolutions per minute for 15 min, the plasma and pellets were separated and stored at − 20 °C.
PSA assay
PSA levels were assayed at the HOSCO laboratory on the Cobas 6000 automated system using the "Elecsys Total PSA" reagent. This test is an “ECLIA” electro chemiluminescence immunoassay. It is based on the “sandwich” method.
DNA extraction and genotyping
The DNA was isolated from the total blood of the participants by the ‘salting out’ technique as described by Miller et al. (1988). TaqMan® allelic discrimination was used to genotype nucleotide variants R462Q (rs486907) and D541E (rs627928) of the RNASEL gene. The primers and probes for R462Q were as follows [38]: forward primer 5’-GGAAGATGTGGAAAATGAGGAAGA-3’, reverse primer 5’-TGCA GATCCTGGTGGGTGTA-3’, and probes 5’-VIC-CAGGACATTTCGGG CAA-MGB and 5’-FAM-CAGGACATTTTGGGCAA-MGB. Primers and probes for D541E were as follows: forward primer 5'-TCTATGTGGTAAAGAAGGGAAGCA-3’, reverse primer 5’-TTGAAC CACCTCTTCATTACTTTGAG-3’ and probes 5’-VIC-TTTCAGATCCT CAAAT-MGB and 5’-FAM-TTTCAGCTCCTCAAAT-MGB.
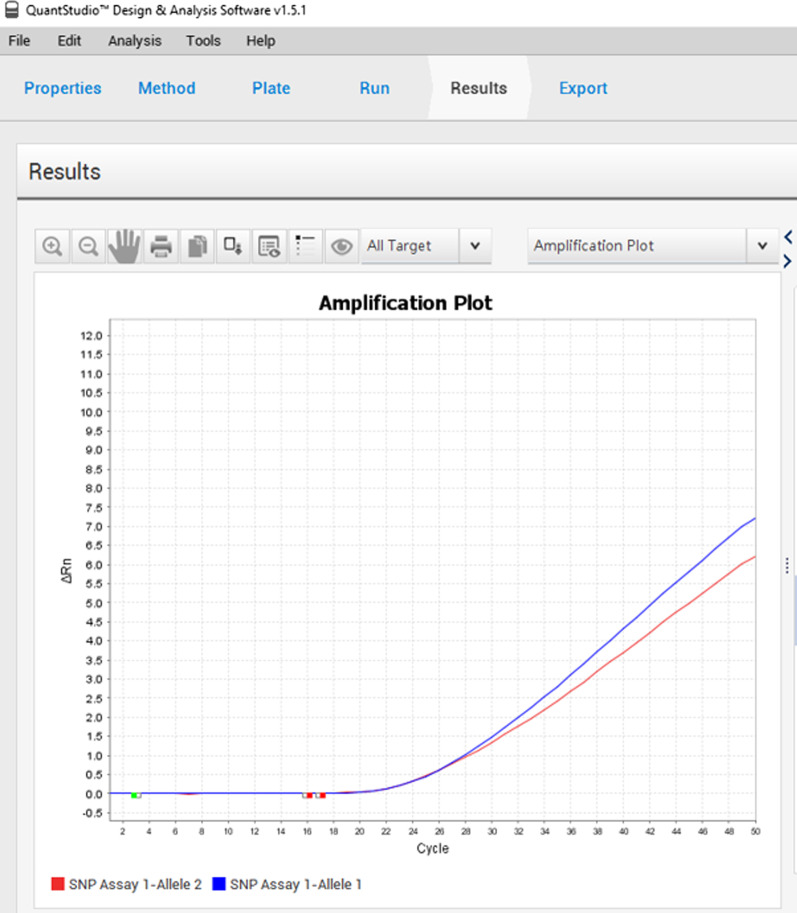
The target sequences were amplified by Real-time PCR in a 25 µL reaction mixture consisting of 5 µL of DNA, 1 µL of each primer at 200 nmol/L and 0.2 µL of each probe at 900 nmol/L, 8 µL of TaqMan® Universal PCR Master Mix II 2X (Applied Biosystems), and the remainder is completed with sterile water. PCR were run on a 95 °C program for 10 min followed by 50 cycles of denaturing at 95 °C for 15 s and hybridization/extension at 60 °C for 1 min on QuantStudio 5 (Applied Biosystems) detection system. TaqMan® Genotyper 1.6.0 software (Applied Biosystems) was used to determine genotypes (Fig. 1).
Fig. 1.
Curve of wild (blue) and mutated (red) genotypes
Statistical analyses
Data was entered using Excel 2016 software. For each polymorphism, allelic frequencies were determined and compared between cases and controls using the Chi2 and Fisher exact tests. Hardy Weinberg’s equilibrium was checked for each polymorphism. In order to verify the association between each polymorphism of the RNASEL gene and the risk of prostate cancer, the Odds ratios (OR) and 95% confidence intervals (95% CI) were determined by considering the age at the time of the cancer diagnosis in the cases. The analyses were carried out using R 4.1.1 software. Analyses were considered statistically significant at p ≤ 0.05.
Results
Socio-demographic characteristics of the study population
The characteristics of our study population are presented here in (Table 1). The mean age in years of the cases at the time of study was 69.81 ± 8.05 and 65.49 ± 8.90 that of the controls. The distribution by age at diagnosis shows that 60.5% of cases were diagnosed between 51 and 70 years old and 39.5% at over 70 years old. The average age at diagnosis is 67.13 ± 8.17 years. There is no statistically significant difference between the mean age at the diagnosis and that of the controls (p = 0.365). With regard to the family history of prostate cancer, 36.8% of cases and 32.1% of controls have a family history.
Table 1.
Socio-demographic characteristics
| Subgroup | Cases (n = 38) n (%) | Controls (n = 53) n (%) | P-value |
|---|---|---|---|
| Age during the study (years) | |||
| ≤ 50 | 0 (0) | 5 (9.4) | |
| 51–70 | 17 (44.7) | 35 (66.0) | |
| > 70 | 21(55.3) | 13 (24.5) | |
| Mean (SD) | 69.81 (8.05) | 65.49 (8.90) | 0.017 |
| Age at diagnostic (years) | |||
| ≤ 50 | 0 (0) | ||
| 51–70 | 23 (60.5) | ||
| > 70 | 15 (39.5) | ||
| Mean (SD) | 67.13 (8.17) | ||
| Family history | |||
| Yes | 14 (36.8) | 17 (32.1) | |
| No | 18 (47.4) | 25 (47.2) | |
| Unknown | 6 (15.8) | 11 (20.8) | |
statistically significant, p ≤ 0.05 is shown in bold
SD standard deviation
Biological characteristics of the study population
The distribution according to the PSA levels at diagnosis shows that the majority of cases, i.e. 81.6%, have a PSA level at diagnosis greater than 20 ng/ml. There is a statistically significant difference in the distribution of cases according to the PSA rate at diagnosis (p ˂ 0.001). For the Gleason score distribution, only 13.2% of cases have a Gleason score greater than 7. There is a statistically significant difference in the Gleason score distribution of cases (p ˂ 0.001) (Table 2).
Table 2.
Biological characteristics
| Subgroup | Cases (n = 38) n (%) | Controls (n = 53) n (%) | P-value |
|---|---|---|---|
| PSA during this study (ng/ml) | |||
| ≤ 4.0 | 37 (70) | ||
| 4.1–10.0 | 11 (21) | ||
| 10.1–20.0 | 3 (6) | ||
| > 20 | 2 (3) | ||
| Mean (SD) | 4.16 (4.70) | ||
| PSA at diagnosis (ng/ml) | |||
| ≤ 4.0 | 0 (0) | ||
| 4.1–10.0 | 3 (7.9) | ||
| 10.1–20.0 | 4 (10.5) | < 0.001 | |
| > 20 | 31 (81.6) | ||
| Mean (SD) | 627.85 (1153.42) | ||
| Gleason score | |||
| < 7 | 10 (26.3) | ||
| 7 | 23 (60.5) | < 0.001 | |
| > 7 | 5 (13.2) | ||
statistically significant, p ≤ 0.05 is shown in bold
PSA prostate specific antigen, SD standard deviation
Prostate cancer and lifestyle
No association between risk of prostate cancer and lifestyle such as physical inactivity (p = 0.31), alcohol intake (p = 0.80), smoking (p = 0.62), and the consumption of fatty meat (p = 0.67) (Table 3).
Table 3.
ORs for lifestyle and prostate cancer risk
| Cases (%) | Controls (%) | OR | IC 95% | P-value | |
|---|---|---|---|---|---|
| Physical activity | |||||
| Yes | 22 (73,3) | 43 (82,7) | 1 | Reference | |
| No | 8 (26,7) | 9 (17,3) | 1,74 | 0,59 – 5,13 | 0,314 |
| Alcohol | |||||
| No | 13 (43,3) | 24 (46,2) | 1 | Reference | |
| Yes | 17 (56,7) | 28 (53,8) | 1,12 | 0,45–2,77 | 0,805 |
| Smoking | |||||
| No | 25 (83,3) | 41 (78,8) | 1 | Reference | |
| Yes | 5 (16,7) | 11 (21,2) | 0,75 | 0,23–2,40 | 0,621 |
| Fatty meat | |||||
| No | 12 (33,3) | 20 (37,7) | 1 | Reference | |
| Yes | 24 (66,7) | 33 (62,3) | 1,21 | 0,50–2,95 | 0,671 |
OR Odd Ratio, CI confidence interval
Allelic frequencies
For the R462Q mutation, the [G] allele was the most frequent allele in both the case population 0.868 and the control population 0.802. There was no statistically significant difference between allele frequencies in cases and controls (p = 0.959). Conversely, the [G] allele for the D541E variant was more prevalent among cases 0.671 and controls 0.538. No statistically significant difference was also observed between allele frequencies in cases and controls (p = 0.881) (Table 4). The two polymorphisms studied were in Hardy–Weinberg equilibrium in the control population (p = 0.193 and p = 0.203).
Table 4.
Alleles frequencies
| SNP | Allèle | Cases (n = 38) | Controls (n = 53) | P-value |
|---|---|---|---|---|
| RNASEL R462Q | G | 0.868 | 0.802 | 0.959 |
| A | 0.132 | 0.198 | ||
| RNASEL D541E | T | 0.329 | 0.462 | 0.881 |
| G | 0.671 | 0.538 |
Associations of SNPs RNASEL R462Q and D541E with prostate cancer risk
No statistically significant association between the R462Q mutation and the risk of prostate cancer was found in our study population (OR, 0.60; 95% CI, 0.10–3.51; p = 0.686) (Table 5).
Table 5.
ORs for RNASEL 462 SNP and prostate cancer risk
| Genotypes | Cases (n = 38) n (%) | Controls (n = 5 3) n (%) | OR (95% CI) | P-value |
|---|---|---|---|---|
| GG | 30 (78.9) | 36 (67.9) | 1.0 (Reference) | |
| AG | 6 (15.8) | 13 (24.5) | 0.55 (0.19–1.63) | 0.281 |
| AA | 2 (5.3) | 4 (7.5) | 0.60 (0.10–3.51) | 0.686 |
| AA vs AG/GG (Rec A) | 0.68 (0.12, 3.92) | 1.000 | ||
| AA/AG vs GG (Dom A) | 0.56 (0.21, 1.49) | 0.245 |
Rec recessive, Dom dominant, OR odds ratios, CI confidence interval
The result found no statistically significant association between the D541E mutation and the risk of prostate cancer in our study population (OR, 2.46; 95% CI 0.78–7.80; p = 0.121) (Table 6).
Table 6.
ORs for RNASEL 541 SNP and prostate cancer risk
| Genotypes | Cases (n = 38) n (%) | Controls (n = 53) n (%) | OR (95% CI) | P-value |
|---|---|---|---|---|
| TT | 6 (15.8) | 14 (26.4) | 1.0 (Reference) | |
| TG | 13 (34.2) | 21(39.6) | 1.44 (0.44–4.70) | 0.541 |
| GG | 19 (50.0) | 18 (34.0) | 2.46 (0.78–7.80) | 0.121 |
| GG vs GT/TT (Rec G) | 1.94 (0.83, 4.56) | 0.125 | ||
| GG/GT vs TT (Dom G) | 1.91 (0.66, 5.55) | 0.227 |
Rec recessive, Dom dominant, OR odds ratios, CI confidence interval
Combined genotypes of RNASEL R462Q and D541E linked to prostate cancer
This study found no statistically significant association between the risk of prostate cancer and the different combinations of genotypes of the mutations of the R462Q and D541E polymorphisms (Table 7).
Table 7.
ORs for RNASEL 462/541 combined genotypes and prostate cancer risk
| Combined genotypes 462/541 | Cases (n = 38) | Controls (n = 51) | OR (95% CI) | P-value |
|---|---|---|---|---|
| GG/TT | 6 | 13 | 1.0 (Reference) | |
| GG/GT | 12 | 14 | 1.85 (0.54–6.40) | 0.497 |
| GG/GG | 12 | 9 | 2.89 (0.79–10.57) | 0.192 |
| AG/GG | 5 | 7 | 1.52 (0.34–6.94) | 0.852 |
| AG/GT | 1 | 6 | 0.36 (0.00–4.33) | 0.628 |
| AA/GG | 2 | 2 | 2.16 (0.12–35.61) | 0.589 |
Associations of SNPs RNASEL R462Q and D541E with Gleason score
The R462Q and D541E mutations were compared between patients according to the Gleason score (≤ 7 and ˃ 7).
For the R462Q mutation, 33.3% of carriers of the AG genotype and 10.0% of carriers of the GG genotype have a Gleason score greater than seven (7) while 100% of carriers of AA genotypes have a score of seven at more (Fig. 2). We found a statistically significant association between the R462Q mutation and the Gleason score (p ˂ 0.001).
Fig. 2.
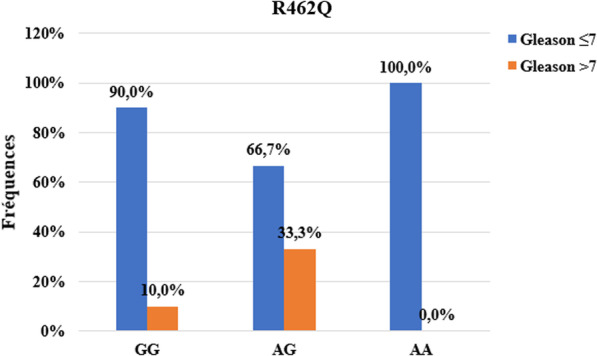
Association between Gleason score and R462Q mutation
For the D541E mutation, 21.1% of carriers of the GG genotype and 7.7% of carriers of the GT genotype had a Gleason score greater than 7 while 100% of carriers of TT genotypes had a score of 7 at more (Fig. 3). We found a statistically significant association between the D541E mutation and the Gleason score (p ˂ 0.001).
Fig. 3.
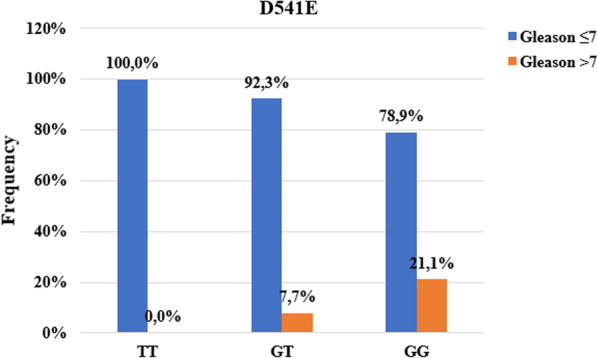
Association between Gleason score and D541E mutation
Associations of SNPs RNASEL R462Q and D541E with PSA at diagnosis
The PSA levels at diagnosis according to the different genotypes of the R462Q mutation indicate that 100% of carriers of the AA genotypes have PSA greater than 20 ng / ml. Only carriers of the GG genotype (13.3%) present PSA levels between 10.1 and 20 ng / ml (Fig. 4). No association was found between this mutation and the PSA level at diagnosis greater than 20 ng / ml (p = 0.773).
Fig. 4.
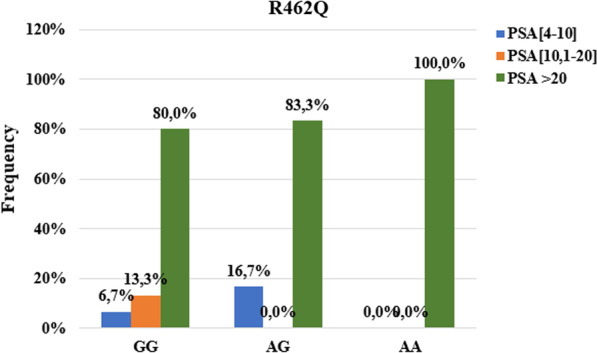
Association between PSA level at diagnostic and R462Q mutation
For the D541E mutation, 89.4% of the GG genotype have a PSA level greater than 20 ng / ml. 23.1% of carriers of the heterozygous GT genotype and 5.3% of carriers of the mutated GG genotype had a PSA level at diagnosis between 10.1 and 20 ng / ml (Fig. 5). No association was found between this mutation and PSA levels at diagnosis greater than 20 ng / ml (p = 0.346).
Fig. 5.
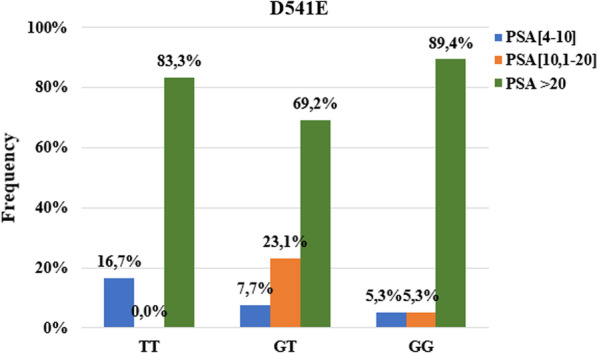
Association between PSA level at diagnostic and D541E mutation
Discussion
Sociodemographic characteristics of the study participants show that the mean age at diagnosis of cases was high, at 67.13 ± 8.17 years. This result is not different from that of Kaboré et al. [39] who report an average age of 71.5 years in Burkina Faso. These results indicate that the age at diagnosis in Burkina Faso is high as observed elsewhere in West Africa [40]. But our results are contrary to those obtained in various studies reporting when black men have an age at early diagnosis [41–43]. PSA levels at diagnosis were very high in our study with a mean of 627.85 ± 1153.42 ng / ml. Our results are in agreement with those obtained by Kaboré et al. in Burkina Faso with an average PSA of 537 ng / ml [35]. Our results corroborate those of Niang et al. in Senegal and Ofoha and Magnus in Nigeria [37, 44]. Tengue et al. in Togo also found PSA levels at diagnosis greater than 100 ng / ml [36]. Among the cases with their Gleason score, 82.14% have a score less than or equal to 7. This shows that the majority of these cases presented with a moderately differentiated tumor at diagnosis. These different results suggests that the diagnosis of prostate cancer is made at advanced stages of the disease and, the fact that there is absence of prostate cancer screening programs in this setting. Regarding the family history of prostate cancer, of the 32 cases with a family history, 43.8% have a family history while 56.2% did not. These results could show that the majority of prostate cancer cases in our study population are not familial. But this trend could be due to the fact that the information was collected through verbal testimonies and not from medical records. Indeed, patients could confuse other prostate conditions (example benign hypertrophy) and prostate cancer.
Regarding alcohol consumption, our results are similar to those obtained by Dennis et al. [45] who found only a strong association between alcohol consumption and prostate cancer mortality. Our results do not support those obtained by Middleton et al. in 2009 [46] and Rota et al. in 2012 [47] in their meta-analyzes. Concerning cigarette smoking, our results are different from those obtained by Jones et al. in England; Cerhan et al. in the United States and Giovannucci et al. also in the United States [48–50]. All these different studies have only shown a slight increase in the risk of developing prostate cancer while a strong association was found with mortality. No association was found between physical activity and prostate cancer in our study. Our results do not corroborate those of Guéritat in France. This study demonstrated that physical exercise prevents the progression of prostate cancer either by regulating redox status and redox-dependent signaling pathways, or via the modulation of cholesterolemia or even of the expression profile of miRNAs [51]. Considering the consumption of fatty meat, our results corroborate those of Park et al. in their study of a population of Hawaii and Los Angeles and those of Dennis et al. in their meta-analysis of 4 cohort studies [45, 52].
Linkage analyzes of families at high risk for prostate cancer have provided convincing evidence that the HPC1 locus is likely to harbor a prostate cancer susceptibility gene [53]. The RNASEL gene has been proposed as a putative tumor suppressor gene located in this region by the positional cloning technique and by the candidate gene approach [54]. Association analyzes of the R462Q and D541E variants within the RNASEL gene with the Prostate cancer have achieved controversial results. Analysis of the different genotypes of the R462Q variant in our study population showed no association of this variant with prostate cancer. Our results support the conclusions of Wei et al.; Noonan et al.; and Alvarez et al. [21, 29, 30] as well as those of Fredrik et al. [34]. These studies found no association between the R462Q variant and prostate cancer. However, Casey et al. and Xiang et al. [30, 31] show that the AA genotype of the R462Q variant is significantly associated with prostate cancer. Regarding the D541E variant, our study found no association with prostate cancer. This goes hand in hand with the studies of Wei et al.; Ignacio et al.; Shook et al. [38, 55, 56] as well as several other authors [29, 30, 57, 58]. Contrary to our results, Noonan-Wheeler et al. and Wiklund et al. [33, 34] in a Swedish population observed an association between the GG genotype and an increased risk of prostate cancer.
Our results showed an association between the R462Q mutation and the degree of tumor differentiation (p ˃ 0.001). Indeed, carriers of heterozygous AG genotype (33.3%) and normal GG genotype (10.0%) presented undifferentiated tumors (Gleason ˃7) unlike carriers of mutated genotype. Our results are identical to those obtained by Alvarez-Cubero et al. in Spain [59]. On the other hand, San Francisco et al. found no association between the R462Q mutation and Gleason score [56]. For the D541E mutation, we also found an association with the degree of tumor differentiation. It can be seen that 21.1% of the undifferentiated tumors were carriers of the mutated GG genotype against 7.7% and 0% for carriers of the heterozygous and homozygous TT genotype respectively (p ˃ 0.001). The same result was obtained by San Francisco et al. in Chile [56]. In contrast, Alvarez et al. found no association between this mutation and the Gleason score [59]. We found no association between R462Q and D541E mutations with PSA levels at diagnosis. This shows that these two mutations in the RNASEL gene are not associated with the level of risk of the tumor (PSA level at diagnosis). Indeed, the PSA level at diagnosis makes it possible to measure the level of risk of tumor progression. For PSA values at diagnosis greater than 20 ng / ml, the tumor is considered to be associated with a high risk of progression [60].
The differences between our results and other studies may, on the one hand, be justified by the difference in sample sizes; the method of selection of controls and, on the other hand, by the ethno-geographic differences of the study populations. Indeed, the small sample size implies a lack of the statistical power to detect associations. Also, the genetic predisposition to prostate cancer is heterogeneous (contribution of environmental and genetic factors) in its hereditary form [61] and involves the predisposition genes in a variable way depending on ethno-geographic origins.
Conclusion
Our study is a first to explore the links that could exist between the Arg46Gln and D541E variants of the RNASEL gene and prostate cancer in Burkina Faso. Genetically, the [G] allele of the R462Q variant and the [G] allele of the D541E variant were the most common in our study population. There is no difference in allele frequencies between cases and controls. These variants, taken alone or in combination, are not associated with the risk of prostate cancer in Burkina Faso population.
Acknowledgements
We thank the Saint Camille hospital in Ouagadougou (HOSCO) and NINA clinic in Ouagadougou for hosting the study, the participants for agreeing to be part of the study, and CERBA/LABIOGENE for the technical platform for molecular testing.
Abbreviations
- D541E
Aspartic acid 541glutamic acid
- ECLIA
Electro chemiluminescence immunoassay
- HPC1
Hereditary Prostate Cancer 1
- MSR1
Macrophage Scavenger Receptor1 Pca: Prostate cancer
- PSA
Prostate specific antigen
- R462Q
Arginine 462 glutamine
- RNASEL
2'-5' Oligoadenylate synthetase-dependent ribonuclease
Author contributions
E.K. and J.S. designed this study. E.K., T.M.Z., B.D.K. and D.D.A.T. recruited patients and controls. E.K., A.A.Z., L.T., A.T.Y., B.V.J.T.E.B., H.K.S., P.A.S., S.F.A.T., K.T. and T.C.O. carried out the manipulations. E.K., A.A.Z. carried out statistical analyses and wrote the manuscript. F.W.D. and J.S. revised the manuscript. All authors have read and corrected the manuscript.
Funding
This study was funded by CERBA/LABIOGENE.
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Our study obtained the approval of the Institutional Ethics Committee (IEC) of CERBA / LABIOGENE. All participants gave their free and informed consent. The study scrupulously respected confidentiality and anonymity. All methods were performed in accordance with the relevant guidelines and regulations.
Consent for publication
Not applicable.
Competing interests
The authors state no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;4:1148. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Pienta KJ, Esper PS. Risk factors for prostate cancer. Ann Intern Med. 1993;118(10):793–803. doi: 10.7326/0003-4819-118-10-199305150-00007. [DOI] [PubMed] [Google Scholar]
- 3.Bostwick DG, Burke HB, Djakiew D, Euling S, Ho S, Landolph J, et al. Human prostate cancer risk factors. Cancer. 2004;101(S10):2371–2490. doi: 10.1002/cncr.20408. [DOI] [PubMed] [Google Scholar]
- 4.Dagnelie PC, Schuurman AG, Goldbohm RA, Van den Brandt PA. Diet, anthropometric measures and prostate cancer risk: a review of prospective cohort and intervention studies. BJU Int mai. 2004;93(8):1139–1150. doi: 10.1111/j.1464-410X.2004.04795.x. [DOI] [PubMed] [Google Scholar]
- 5.Perdana NR, Mochtar CA, Umbas R, Hamid ARA. The risk factors of prostate cancer and its prevention: a literature review. Acta Med Indones. 2016;48(3):228–238. [PubMed] [Google Scholar]
- 6.Scardino PT. Early detection of prostate cancer. Urol Clin North Am. 1989;16(4):635–655. [PubMed] [Google Scholar]
- 7.Kheirandish P, Chinegwundoh F. Ethnic differences in prostate cancer. Br J Cancer. 2011;105(4):481–5. doi: 10.1038/bjc.2011.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whittemore AS, Wu AH, Kolonel LN, John EM, Gallagher RP, Howe GR, et al. Family history and prostate cancer risk in black, white, and Asian men in the United States and Canada. Am J Epidemiol. 1995;141(8):732–40. doi: 10.1093/oxfordjournals.aje.a117495. [DOI] [PubMed] [Google Scholar]
- 9.Albright FS, Stephenson RA, Agarwal N, Cannon-Albright LA. Relative risks for lethal prostate cancer based on complete family history of prostate cancer death. Prostate. 2017;77(1):41–48. doi: 10.1002/pros.23247. [DOI] [PubMed] [Google Scholar]
- 10.Gallagher RP, Fleshner N. Prostate cancer: 3. Individual risk factors. CMAJ. 1998;159(7):807–13. [PMC free article] [PubMed] [Google Scholar]
- 11.Hernández J, Thompson IM. Prostate-specific antigen: a review of the validation of the most commonly used cancer biomarker. Cancer. 2004;101(5):894–904. doi: 10.1002/cncr.20480. [DOI] [PubMed] [Google Scholar]
- 12.Hernández J, Balic I, Johnson-Pais TL, Higgins BA, Torkko KC, Thompson IM, et al. Association between an estrogen receptor alpha gene polymorphism and the risk of prostate cancer in black men. J Urol. 2006;175(2):523–527. doi: 10.1016/S0022-5347(05)00240-5. [DOI] [PubMed] [Google Scholar]
- 13.Ahlbom A, Lichtenstein P, Malmström H, Feychting M, Hemminki K, Pedersen NL. Cancer in twins: genetic and nongenetic familial risk factors. J Natl Cancer Inst. 1997;89(4):287–93. doi: 10.1093/jnci/89.4.287. [DOI] [PubMed] [Google Scholar]
- 14.Page WF, Braun MM, Partin AW, Caporaso N, Walsh P. Heredity and prostate cancer: a study of World War II veteran twins. Prostate. 1997;33(4):240–5. doi: 10.1002/(sici)1097-0045(19971201)33:4<240::aid-pros3>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 15.Mucci LA, Hjelmborg JB, Harris JR, Czene K, Havelick DJ, Scheike T, et al. Familial risk and heritability of cancer among twins in Nordic countries. JAMA. 2016;315(1):68–76. doi: 10.1001/jama.2015.17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cussenot O, Cancel-Tassin G. Facteurs de risque génétiques pour le cancer de la prostate. MS. 2004;20(5):562–8. doi: 10.1051/medsci/2004205562. [DOI] [PubMed] [Google Scholar]
- 17.Xu J. Combined analysis of hereditary prostate cancer linkage to 1q24-25: results from 772 hereditary prostate cancer families from the International Consortium for Prostate Cancer Genetics. Am J Hum Genet. 2000;66(3):945–957. doi: 10.1086/302807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simard J, Dumont M, Labuda D, Sinnett D, Meloche C, El-Alfy M, et al. Prostate cancer susceptibility genes: lessons learned and challenges posed. Endocr Relat Cancer. 2003;10(2):225–259. doi: 10.1677/erc.0.0100225. [DOI] [PubMed] [Google Scholar]
- 19.Agalliu I, Leanza SM, Smith L, Trent JM, Carpten JD, Bailey-Wilson JE, et al. Contribution of HPC1 (RNASEL) and HPCX variants to prostate cancer in a founder population. Prostate. 2010;70(15):1716–1727. doi: 10.1002/pros.21207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fesinmeyer MD, Kwon EM, Fu R, Ostrander EA, Stanford JL. Genetic variation in RNASEL and risk for prostate cancer in a population-based case-control study. Prostate. 2011;71(14):1538–47. doi: 10.1002/pros.21370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuo L, Ren K-W, Bai Y, Zhang L-F, Zou J-G, Qin X-H, et al. Association of a common genetic variant in RNASEL and prostate cancer susceptibility. Oncotarget. 2017;8(43):75141–50. doi: 10.18632/oncotarget.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rebbeck TR, Walker AH, Zeigler-Johnson C, Weisburg S, Martin AM, Nathanson KL, et al. Association of HPC2/ELAC2 genotypes and prostate cancer. Am J Hum Genet. 2000;67(4):1014–1019. doi: 10.1086/303096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alvarez-Cubero MJ, Saiz M, Martinez-Gonzalez LJ, Alvarez JC, Lorente JA, Cozar JM. Genetic analysis of the principal genes related to prostate cancer: a review. Urol Oncol. 2013;31(8):1419–1429. doi: 10.1016/j.urolonc.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 24.Tan S-H, Petrovics G, Srivastava S. Prostate cancer genomics: recent advances and the prevailing underrepresentation from racial and ethnic minorities. Int J Mol Sci. 2020;19(4):1102. doi: 10.3390/ijms19041255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, et al. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012;366(2):141–9. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pilie PG, Giri VN, Cooney KA. HOXB13 and other high penetrant genes for prostate cancer. Asian J Androl. 2016;18(4):530. doi: 10.4103/1008-682X.175785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, et al. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med. 2016;375(5):443–53. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou A, Hassel BA, Silverman RH. Expression cloning of 2–5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell. 1993;72(5):753–65. doi: 10.1016/0092-8674(93)90403-d. [DOI] [PubMed] [Google Scholar]
- 29.Rökman A, Ikonen T, Seppälä EH, Nupponen N, Autio V, Mononen N, et al. Germline alterations of the RNASEL gene, a candidate HPC1 gene at 1q25, in patients and families with prostate cancer. Am J Hum Genet. 2002;70(5):1299–1304. doi: 10.1086/340450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casey G, Neville PJ, Plummer SJ, Xiang Y, Krumroy LM, Klein EA, et al. RNASEL Arg462Gln variant is implicated in up to 13% of prostate cancer cases. Nat Genet. 2002;32(4):581–583. doi: 10.1038/ng1021. [DOI] [PubMed] [Google Scholar]
- 31.Xiang Y, Wang Z, Murakami J, Plummer S, Klein EA, Carpten JD, et al. Effects of RNase L mutations associated with prostate cancer on apoptosis induced by 2’,5’-oligoadenylates. Cancer Res. 2003;63(20):6795–801. [PubMed] [Google Scholar]
- 32.Nakazato H, Suzuki K, Matsui H, Ohtake N, Nakata S, Yamanaka H. Role of genetic polymorphisms of the RNASEL gene on familial prostate cancer risk in a Japanese population. Br J Cancer. 2003;89(4):691–6. doi: 10.1038/sj.bjc.6601075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noonan-Wheeler FC, Wu W, Roehl KA, Klim A, Haugen J, Suarez BK, et al. Association of hereditary prostate cancer gene polymorphic variants with sporadic aggressive prostate carcinoma. Prostate. 2006;66(1):49–56. doi: 10.1002/pros.20320. [DOI] [PubMed] [Google Scholar]
- 34.Wiklund F, Jonsson B-A, Brookes AJ, Strömqvist L, Adolfsson J, Emanuelsson M, et al. Genetic analysis of the RNASEL gene in hereditary, familial, and sporadic prostate cancer. Clin Cancer Res. 2004;10(21):7150–6. doi: 10.1158/1078-0432.CCR-04-0982. [DOI] [PubMed] [Google Scholar]
- 35.Kabore FA, Kambou T, Zango B, Ouédraogo A. Knowledge and awareness of prostate cancer among the general public in Burkina Faso. J Cancer Educ. 2014;29(1):69–73. doi: 10.1007/s13187-013-0545-2. [DOI] [PubMed] [Google Scholar]
- 36.Tengue K, Kpatcha TM, Botcho G, Leloua E, Amavi AK, Sikpa K, et al. Profil épidémiologique, diagnostique, thérapeutique et évolutif du cancer de la prostate au Togo. Afr J Urol. 2016;22(2):76–82. [Google Scholar]
- 37.Niang L, Ndoye M, Ouattara A, Jalloh M, Labou M, Thiam I, et al. Management of prostate cancer in Senegal: what is being done? Prog Urol. 2013;23(1):36–41. doi: 10.1016/j.purol.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Shook SJ, Beuten J, Torkko KC, Johnson-Pais TL, Troyer DA, Thompson IM, et al. Association of RNASEL variants with prostate cancer risk in hispanic caucasians and African Americans. Clin Cancer Res. 2007;13(19):5959–64. doi: 10.1158/1078-0432.CCR-07-0702. [DOI] [PubMed] [Google Scholar]
- 39.Kabore F, Zango B, Kambou T, Yameogo C, Kirakoya B. Cancer de la prostate au Burkina Faso: caractéristiques diagnostics et indications thérapeutiques initiales. Progrès en Urologie. 2013;23:1075. [Google Scholar]
- 40.Ajape AA, Ibrahim KOO, Fakeye JA, Abiola OO. An overview of cancer of the prostate diagnosis and management in Nigeria: the experience in a Nigerian tertiary hospital. Ann Afr Med. 2010;9(3):113–117. doi: 10.4103/1596-3519.68353. [DOI] [PubMed] [Google Scholar]
- 41.Freedland SJ, Sutter ME, Naitoh J, Dorey F, Csathy GS, Aronson WJ. Clinical characteristics in black and white men with prostate cancer in an equal access medical center. Urology. 2000;55(3):387–390. doi: 10.1016/s0090-4295(99)00461-6. [DOI] [PubMed] [Google Scholar]
- 42.Barros MS, Silva VRS, Santos GB, Hughes A, Silveira MA. Prevalence of prostate adenocarcinoma according to race in an university hospital. Int Braz J Urol. 2003;29(4):306–312. doi: 10.1590/s1677-55382003000400004. [DOI] [PubMed] [Google Scholar]
- 43.Odedina FT, Ogunbiyi JO, Ukoli FAM. Roots of prostate cancer in African-American men. J Natl Med Assoc. 2006;98(4):539–543. [PMC free article] [PubMed] [Google Scholar]
- 44.Ofoha CG, Magnus FE. Presentation, characteristics and co-morbidities of men with prostate cancer in Nigeria. JAMMR. 2019;2:1–7. [Google Scholar]
- 45.Dennis LK. Meta-analysis for combining relative risks of alcohol consumption and prostate cancer. Prostate. 2000;42(1):56–66. doi: 10.1002/(sici)1097-0045(20000101)42:1<56::aid-pros7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 46.Middleton Fillmore K, Chikritzhs T, Stockwell T, Bostrom A, Pascal R. Alcohol use and prostate cancer: a meta-analysis. Mol Nutr Food Res. 2009;53(2):240–255. doi: 10.1002/mnfr.200800122. [DOI] [PubMed] [Google Scholar]
- 47.Rota M, Scotti L, Turati F, Tramacere I, Islami F, Bellocco R, et al. Alcohol consumption and prostate cancer risk: a meta-analysis of the dose-risk relation. Eur J Cancer Prev. 2012;21(4):350–359. doi: 10.1097/CEJ.0b013e32834dbc11. [DOI] [PubMed] [Google Scholar]
- 48.Jones MR, Joshu CE, Kanarek N, Navas-Acien A, Richardson KA, Platz EA. Cigarette smoking and prostate cancer mortality in four US States, 1999–2010. Prev Chronic Dis. 2021;14:13. doi: 10.5888/pcd13.150454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cerhan JR, Torner JC, Lynch CF, Rubenstein LM, Lemke JH, Cohen MB, et al. Association of smoking, body mass, and physical activity with risk of prostate cancer in the Iowa 65+ Rural Health Study (United States) Cancer Causes Control. 1997;8(2):229–238. doi: 10.1023/a:1018428531619. [DOI] [PubMed] [Google Scholar]
- 50.Giovannucci E, Rimm EB, Ascherio A, Colditz GA, Spiegelman D, Stampfer MJ, et al. Smoking and risk of total and fatal prostate cancer in United States health professionals. Cancer Epidemiol Biomarkers Prev. 1999;8(4):277–82. [PubMed] [Google Scholar]
- 51.Guéritat J. Exercice physique et progression du cancer de la prostate: effets combinés avec la prise d’antioxydants naturels ou la radiothérapie externe : identification de voies de signalisation redox-dépendantes [Internet] [phdthesis] Université Rennes. 2015;2:12258. [Google Scholar]
- 52.Park S-Y, Murphy SP, Wilkens LR, Henderson BE, Kolonel LN. Fat and meat intake and prostate cancer risk: the multiethnic cohort study. Int J Cancer. 2007;121(6):1339–1345. doi: 10.1002/ijc.22805. [DOI] [PubMed] [Google Scholar]
- 53.Smith JR, Freije D, Carpten JD, Grönberg H, Xu J, Isaacs SD, et al. Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science. 1996;274(5291):1371–4. doi: 10.1126/science.274.5291.1371. [DOI] [PubMed] [Google Scholar]
- 54.Carpten J, Nupponen N, Isaacs S, Sood R, Robbins C, Xu J, et al. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat Genet. 2002;30(2):181–184. doi: 10.1038/ng823. [DOI] [PubMed] [Google Scholar]
- 55.Wei B, Xu Z, Ruan J, Zhu M, Jin K, Zhou D, et al. RNASEL Asp541Glu and Arg462Gln polymorphisms in prostate cancer risk: evidences from a meta-analysis. Mol Biol Rep. 2012;39(3):2347–2353. doi: 10.1007/s11033-011-0985-x. [DOI] [PubMed] [Google Scholar]
- 56.San Francisco IF, Rojas PA, Torres-Estay V, Smalley S, Cerda-Infante J, Montecinos VP, et al. Association of RNASEL and 8q24 variants with the presence and aggressiveness of hereditary and sporadic prostate cancer in a Hispanic population. J Cell Mol Med. 2014;18(1):125–133. doi: 10.1111/jcmm.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L, McDonnell SK, Elkins DA, Slager SL, Christensen E, Marks AF, et al. Analysis of the RNASEL gene in familial and sporadic prostate cancer. Am J Hum Genet. 2002;71(1):116–123. doi: 10.1086/341281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maier C, Haeusler J, Herkommer K, Vesovic Z, Hoegel J, Vogel W, et al. Mutation screening and association study of RNASEL as a prostate cancer susceptibility gene. Br J Cancer. 2005;92(6):1159–64. doi: 10.1038/sj.bjc.6602401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alvarez-Cubero MJ, Pascual-Geler M, Martinez-Gonzalez LJ, Expósito Ruiz M, Saiz M, Cozar JM, et al. Association between RNASEL, MSR1, and ELAC2 single nucleotide polymorphisms and gene expression in prostate cancer risk. Urol Oncol. 2016;34(10):431.e1–8. doi: 10.1016/j.urolonc.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 60.Hull GW, Rabbani F, Abbas F, Wheeler TM, Kattan MW, Scardino PT. Cancer control with radical prostatectomy alone in 1,000 consecutive patients. J Urol. 2002;167(2 Pt 1):528–534. doi: 10.1016/S0022-5347(01)69079-7. [DOI] [PubMed] [Google Scholar]
- 61.Karayi MK, Neal DE, Markham AF. Current status of linkage studies in hereditary prostate cancer: linkage studies in hereditary prostate cancer. BJU Int. 2001;86(6):659–69. doi: 10.1046/j.1464-410x.2000.00837.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.