SUMMARY
In tumors, nutrient availability and metabolism are known to be important modulators of growth signaling. However, it remains elusive whether cancer cells that are growing out in the metastatic niche rely on the same nutrients and metabolic pathways to activate growth signaling as cancer cells within the primary tumor. We discovered that breast-cancer-derived lung metastases, but not the corresponding primary breast tumors, use the serine biosynthesis pathway to support mTORC1 growth signaling. Mechanistically, pyruvate uptake through Mct2 supported mTORC1 signaling by fueling serine biosynthesis-derived α-ketoglutarate production in breast-cancer-derived lung metastases. Consequently, expression of the serine biosynthesis enzyme PHGDH was required for sensitivity to the mTORC1 inhibitor rapamycin in breast-cancer-derived lung tumors, but not in primary breast tumors. In summary, we provide in vivo evidence that the metabolic and nutrient requirements to activate growth signaling differ between the lung metastatic niche and the primary breast cancer site.
INTRODUCTION
Metastasis formation is the leading cause of death in cancer patients (Elia et al., 2016). In breast cancer, metastatic dissemination is an early event that can occur even before primary tumor detection (Harper et al., 2016; Hosseini et al., 2016). To transition from dissemination to metastasis formation, cancer cells need to activate growth signaling (Lambert et al., 2017). Although growth signaling has been extensively studied in primary tumors (Sever and Brugge, 2015), less is known about the growth signaling requirements and mechanisms in cancer cells that undergo metastatic outgrowth.
An important growth signal is mediated through mammalian target of rapamycin (mTOR) (Condon and Sabatini, 2019), which is a serine/threonine-specific protein kinase that exists in two complexes, namely, mTORC1 and mTORC2. Aberrant activity of mTORC1 has been found in multiple cancers and contributes to uncontrolled proliferation (Cargnello et al., 2015). Early metastatic steps that allow cancer cells to disseminate from the primary tumor have been linked to the regulation of mTORC1 signaling. In this respect, it has been found that epithelial-mesenchymal transition, migration, and invasion of nasopharyngeal carcinoma (Wang et al., 2018), non-small cell lung cancer (Yu et al., 2015, 2017), and breast cancer (Yang et al., 2016) cells is promoted by mTORC1 activity. Yet it remains largely unknow whether and how mTORC1 signaling is activated in cancer cells that have successfully disseminated to a distant organ.
mTORC1 is regulated by growth-factor-mediated activity of the tuberous sclerosis complex (Condon and Sabatini, 2019) and by nutrients and metabolites such as amino acids, α-ketoglutarate, and nucleotides (Ben-Sahra and Manning, 2017; Lorendeau et al., 2015; Mossmann et al., 2018). Interestingly, it has recently emerged that nutrient availability and metabolism differ between primary cancers and metastases (Elia et al., 2018; Schild et al., 2018). Thus, this raises the question of whether cancer cells in a primary breast cancer and a corresponding metastasis activate mTORC1 signaling through different nutrients and metabolic activities.
Here, we discovered that both primary breast tumors and their corresponding lung metastases use mTORC1 signaling for proliferation. However, unlike in primary breast tumors, pyruvate-driven activity of serine biosynthesis in lung metastases potentiated the activation of mTORC1 signaling.
RESULTS
Pyruvate Supports mTORC1 Signaling in Lung Metastases
Growth signaling is relevant for primary tumor proliferation and outgrowth of metastases. We therefore asked whether primary breast cancers and their corresponding lung metastases rely on the same growth signaling pathway, namely, mTORC1 signaling, for proliferation and outgrowth. Specifically, we used 4T1 mouse breast cancer cells to initiate primary tumors in the mammary fat pad, which resulted in spontaneous lung metastasis formation in mice. Subsequently, we resected matched pairs of primary 4T1 breast tumors and lung metastases and evaluated mTORC1 signaling based on the phosphorylation of S6 and eukaryotic translation initiation factor 4E (eIF4E) binding protein 1 (4EBP1). We observed that both the proliferating primary breast tumors and the outgrowing metastases showed substantial S6 and 4EBP1 phosphorylation compared with adjacent healthy lung tissue (Figure 1A; Figure S1A). Thus, we concluded that 4T1 primary breast tumors and their corresponding metastases use mTORC1-dependent growth signaling.
Figure 1. Pyruvate Supports mTORC1 Signaling in Lung Metastases.
(A) Phosphorylation of mTORC1 targets S6 and 4EBP1 in the lung (L), primary tumor (PT), and metastasis (M) of BALB/c mice injected with 4T1 cells in the mammary fat pad (m.f.). n = 4.
(B) Protein quantification of S6 (P-Ser240/244) and 4EBP1 (P-Ser65) shown in (A). Data are presented as fold changes compared with PT levels. Data are normalized on actin and the respective band for total protein. Tissues from 4 additional mice were run on a second western blot (Figure S1A), and the quantification of S6 (P-Ser240/244) was included in the graph. Two-tailed unpaired Student’s t test. S6, n = 8; 4EBP1, n = 4.
(C) mTORC1 activity based on a publicly available phosphoproteomics dataset in human M compared with matched PT from 5 patients.
(D and E) Correlation of GLUT1, MCT2, ASCT2, SCL7A2, and SLC7A1 gene expression with mTOR activity as measured by a sum score of the phosphotargets by reverse phase protein assay (RPPA) protein level. Samples were divided in quartiles, with the 1st quartile comprising all samples with the lowest mTOR activity and the 4th quartile comprising all samples with the highest mTOR activity. Data are normalized to the average gene expression calculated for the 1st quartile. Two-sided Mann-Whitney test. n = 859.
(F) Concentration of glucose in the interstitial fluid and in the blood plasma of BALB/c mice. Two-tailed unpaired Student’s t test. Lung interstitial fluid, n = 4; blood plasma, n = 10.
(G) Total protein synthesis in the presence or absence of 2 mM pyruvate and puromycin (10 μg/mL) in 4T1 cells based on the incorporation of puromycin into newly synthesized proteins. 4T1 cells were starved from serum for 16 h, starved in Hank’s balanced salt solution (HBSS) for 1 h, and subsequently reactivated (30 min) in serum containing culture medium with or without 2 mM sodium pyruvate. The protein synthesis inhibitor cycloheximide (CHX, 100 nM) was used as negative control (CTR). One representative image is shown. n = 3.
(H) Concentration of pyruvate in the interstitial fluid of lung samples collected from human patients and BALB/c mice. Two-tailed unpaired Student’s t test. Human, n = 7; mouse, n = 5.
(I and J) Phosphorylation of the mTORC1 targets S6 and 4EBP1 in 4T1 PT and lung M upon acute inhibition of pyruvate uptake using the MCT2 inhibitor α-cyano-4-hydroxycinnamic acid (60 mg/kg i.p.). 4EBP1 phosphorylation increases from the α to the γ (δ) band. Quantification of phospho-S6 is shown in the graph. Data are normalized on total S6 and actin and are shown as fold changes compared with the group of mice treated with the vehicle. Two-tailed unpaired Student’s t test. n = 6.
Error bars represent SD (B, F, and H–J) and SEM (D and E) from the mean of biologically independent samples.
Although both the primary breast tumors and their corresponding lung metastases showed activated mTORC1 growth signaling, we found that the metastases exhibited a higher ratio of phosphorylated to total S6 compared with primary tumors (Figure 1B; Figure S1A). Accordingly, we found that four of five matched metastases samples from human breast cancer patients exhibited an elevated mTOR activity signature compared with corresponding primary breast cancer samples based on publicly available phosphoproteomics data (Figure 1C; Table S1). This indicates increased activity of mTORC1 signaling specifically in the outgrowing metastases.
mTORC1 signaling is highly influenced by nutrient availability (Condon and Sabatini, 2019; Mossmann et al., 2018). Therefore, we correlated the gene expression of a selection of transporters for glucose, pyruvate, glutamine, and arginine with mTOR activity in breast cancer samples from patients (SLC2A1 and GLUT1 for glucose, SLC16A7 and MCT2 for pyruvate, SLC1A5 and ASCT2 for glutamine, and SLC7A1/SLC7A2 and CAT1/2 for arginine). Although SLC7A1 and SLC7A2 showed opposing results, we observed a positive correlation between GLUT1, as well as MCT2 gene expression, and mTOR activity in breast cancer samples from patients (Figures 1D and 1E). Because the link between glucose concentrations and mTOR activity is well characterized (Leprivier and Rotblat, 2020; Orozco et al., 2020) and glucose concentrations in lung interstitial fluid compared with blood were reduced in mice (Figure 1F), we decided to focus on pyruvate. Specifically, we hypothesized that pyruvate in the lung environment may stimulate growth signaling. Accordingly, pyruvate supplementation to 4T1 breast cancer cultured in vitro increased protein translation (Figure 1G) based on puromycin incorporation (Schmidt et al., 2009) and resulted in faster phosphorylation of some mTORC1 targets after starvation and reactivation in 4T1 and HCT116 cancer cells (Figures S1B and S1C). Thus, pyruvate may be important to activate growth signaling.
We had previously shown that the nutrient pyruvate is available in the lung environment (i.e., lung interstitial fluid) of mice (Christen et al., 2016) and that Mct2 inhibition selectively impairs formation of lung metastases, but not of primary breast tumors (Elia et al., 2019). To relate this prior finding to humans, we measured the absolute pyruvate concentration in the interstitial fluid of non-cancerous lung tissue of human individuals. Strikingly, we observed a similar pyruvate concentration in human and mouse lung interstitial fluid (Figure 1H; Table S2). These data show that pyruvate availability not only is a particularity of the mouse lung environment but is also reflected in the human lung environment. Thus, we hypothesized that pyruvate availability in the lung metastatic niche may support mTORC1 growth signaling. A prerequisite for this hypothesis is that pyruvate abundance is increased in metastases compared with primary tumors. In line with our previous work (Christen et al., 2016), we confirmed a 2.5-fold increase in pyruvate abundance in lung metastases compared with primary 4T1 breast tumors (Figure S2D). Next, we addressed the question of whether inhibition of pyruvate uptake modulates mTORC1 signaling, particularly in metastases. We initiated 4T1 breast-cancer-derived lung metastases by mammary fat pad injection and treated the mice acutely with two doses of the MCT2 inhibitor α-cyano-4-hydroxycinnamic acid (60 mg/kg/day intraperitoneally [i.p.], 24 and 2 h before sacrificing the mice), which we had previously shown to inhibit pyruvate uptake of lung metastases (Elia et al., 2019). Subsequently, we resected metastases and primary tumors from mice treated with the MCT2 inhibitor or vehicle and assessed mTORC1 signaling based on the phosphorylation of S6 and 4EBP1. In primary breast tumor tissue, phosphorylation of mTORC1 targets did not change (Figure 1I). In contrast, we observed a marked decrease in S6 phosphorylation and an increased fraction of lower-phosphorylated (α and β band) 4EBP1 in metastases (Figure 1J). Based on these data, we concluded that pyruvate uptake through Mct2 supports mTORC1 signaling in breast-cancer-derived lung metastases, but not in primary breast tumors.
Pyruvate Activates the Serine Synthesis Pathway
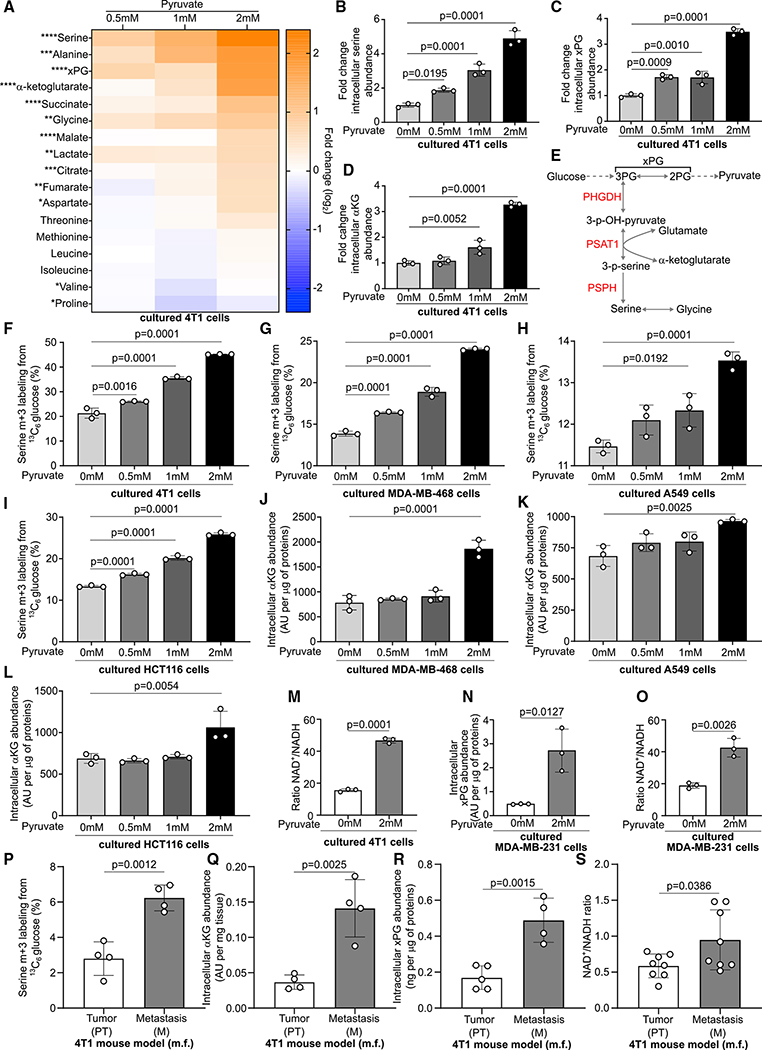
Next, we investigated the mechanism by which pyruvate supports mTORC1 growth signaling. We hypothesized that the metabolites that changed the most in abundance upon pyruvate supplementation could mechanistically link pyruvate to mTORC1 signaling. Thus, we cultured 4T1 breast cancer cells in the presence of increasing amounts of pyruvate and performed a metabolomics analysis. We defined the highest pyruvate concentration so that we achieved in vitro an increase in intracellular pyruvate abundance similar to the one observed in vivo (metastases versus primary tumors) (Figures S1D–S1F). We found that upon pyruvate supplementation, several metabolites changed in abundance. Serine, alanine, x-phosphoglycerate (the sum of 2- and 3-phosphoglycerate), and α-ketoglutarate abundance showed the most pronounced increase (Figures 2A–2D). However, dihydroxyacetone phosphate (DHAP), which was recently discovered to signal glucose availability to mTOR (Orozco et al., 2020), was not altered in abundance (Figure S2A). Serine and α-ketoglutarate can be directly linked via the serine biosynthesis pathway, which produces serine and equimolar amounts of α-ketoglutarate, because the enzyme phosphoserine aminotransferase 1 (PSAT1) converts 3-phosphohydroxypyruvate and glutamate to 3-phospho-L-serine and α-ketoglutarate (Figure 2E). Thus, we hypothesized that pyruvate stimulates serine biosynthesis. Therefore, we measured de novo serine biosynthesis using 13C tracer analysis (Buescher et al., 2015) in 4T1, MDA-MB-468, A549, and HCT116 cells. We observed that pyruvate supplementation increased the fraction of serine produced from glucose (Figures 2F–2I), which is indicative of elevated de novo serine biosynthesis in the presence of pyruvate. Moreover, MDA-MB-468, A549, and HCT116 cells showed, similar to 4T1 cells, elevated α-ketoglutarate abundance upon pyruvate supplementation (Figures 2J–2L). Based on these data, we concluded that pyruvate stimulates the serine biosynthesis pathway in vitro.
Figure 2. Pyruvate Activates De Novo Serine Biosynthesis.
(A) Heatmap representing metabolite abundance changes upon supplementation of different concentrations of sodium pyruvate to the culture medium of 4T1 cells after 24 h of incubation. Data represent the fold changes compared with the no-pyruvate condition. One-way ANOVA. n = 3. Asterisks represent statistical significance as follows: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
(B–D) Fold changes of serine, α-ketoglutarate, and x-phosphoglycerate (xPG) (3-phosphoglycerate [3PG] and 2-phosphoglycerate [2PG]) extracted from the heatmap in (A). One-way ANOVA with Dunnett’s multiple comparison test (comparison between 0 mM conditions and all others). n = 3.
(E) Schematic representation of the de novo serine biosynthesis pathway branching from glycolysis at the level of 3PG. Black, metabolites; red. enzymes of serine biosynthesis.
(F–I) Activation of de novo serine biosynthesis assessed through measurement of serine m+3 labeling enrichment after incubation of 4T1, MDA-MB-468, A549, and HCT116 cells for 24 h in culture medium containing 13C6 glucose and increasing concentrations of sodium pyruvate. One-way ANOVA with Dunnett’s multiple comparison test (comparison between the 0 mM conditions and all others). n = 3.
(J–L) Intracellular levels of α-ketoglutarate in MDA-MB-468, A549, and HCT116 cells incubated for 24 h in culture medium containing increasing concentrations of sodium pyruvate. One-way ANOVA with Dunnett’s multiple comparison test (comparison between 0 mM conditions and all others). n = 3.
(M–O) Intracellular xPG abundance and NAD+/NADH ratio in 4T1 and MDA-MB-231 cells incubated for 24 h in medium without or with 2 mM sodium pyruvate. Two-tailed unpaired Student’s t test. n = 3.
(P–S) Activation of de novo serine biosynthesis, α-ketoglutarate, and xPG abundance, NAD+/NADH ratio in 4T1 PT and lung M. De novo serine synthesis was assessed through measurement of serine m+3 labeling enrichment after a 6 h infusion of 13C6 glucose to tumor harboring BALB/c mice. Two-tailed unpaired Student’s t test. Serine synthesis and α-ketoglutarate (α KG) levels, n = 4; xPG PT, n = 5; xPG M, n = 4; NAD+/NADH, n = 8.
All error bars represent SD from the mean of biologically independent samples or mice.
Serine biosynthesis is metabolically regulated by 3-phosphoglycerate (Oslund et al., 2017) and constrained by the NAD+ to NADH ratio (Diehl et al., 2019). Accordingly, we found that pyruvate supplementation increased x-phosphoglycerate abundance and the NAD+ to NADH ratio in 4T1 and MDA-MB-231 cells (Figures 2C and 2M–2O). To investigate the relevance of this finding, we used the electron acceptor α-ketobutyrate (α-KB) (Diehl et al., 2019) and lactate (which decreases the NAD+ to NADH ratio) and measured pyruvate, x-phosphoglycerate, and α-ketoglutarate, as well as 13C glucose contribution to serine in 4T1 cells. We found that α-KB elevated pyruvate, x-phosphoglycerate and α-ketoglutarate abundance, as well as 13C glucose contribution to serine (Figures S2B–S2E), whereas lactate supplementation increased pyruvate and x-phosphoglycerate abundance but only elevated to a minor extent α-ketoglutarate abundance and 13C glucose contribution to serine (Figures S2F–S2I). These data suggest that both 3-phosphoglycerate and a favorable NAD+ to NADH ratio are needed to activate serine biosynthesis in vitro.
To relate these results to the in vivo situation, we performed an in vivo 13C tracer infusion (Fernández-García et al., 2020; Altea-Manzano et al., 2020) and metabolomics analysis in mice harboring primary breast tumors and lung metastases. We found that 4T1 breast-cancer-derived lung metastases exhibited a higher fraction of serine biosynthesis from glucose (Figure 2P), elevated α-ketoglutarate, and x-phosphoglycerate abundance (Figures 2Q and 2R) and an increased NAD+ to NADH ratio (Figure 2S) compared with the corresponding primary breast tumors. Thus, these data are consistent with the notion that pyruvate activates serine biosynthesis in lung metastases by elevating 3-phosphoglycerate abundance and the NAD+ to NADH ratio.
Serine Biosynthesis Promotes mTORC1 Signaling in Lung Metastases
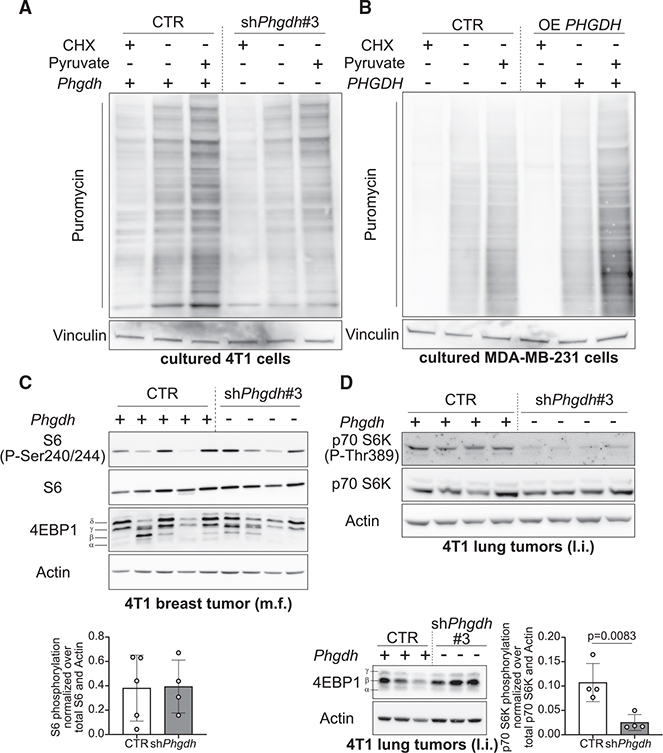
Next, we hypothesized that the serine biosynthesis pathway potentiates mTORC1 signaling in lung metastases. To address this hypothesis, we silenced and overexpressed phosphoglycerate dehydrogenase (PHGDH) (Figures S3A–S3C), which is an essential enzyme of the serine biosynthesis pathway, in 4T1 and MDA-MB-231 breast cancer cells, respectively. As expected, silencing Phgdh decreased the fraction of serine produced from glucose, intracellular α-ketoglutarate abundance, and protein translation (Figure 3A; Figures S3D–S3I), whereas overexpression resulted in the opposite effect (Figure 3B; Figures S3J and S3K). Moreover, treatment of PHGDH-overexpressing (OE PHGDH) MDA-MB-231 breast cancer cells with the PHGDH inhibitor PH-755 decreased phosphorylation of some mTORC1 targets (Figure S3L). Next, we injected 4T1 control, Phgdh-silenced, and OE PHGDH breast cancer cells into the lung or mammary fat pad of mice and assessed mTORC1 growth signaling in the arising tumors. In primary breast tumors arising from mammary fat pad injections, Phgdh silencing did not alter phosphorylation of mTORC1 targets (Figure 3C). However, in Phgdh-silenced breast-cancer-derived lung tumors, p70 S6K phosphorylation was greatly decreased compared with control tumors (Figure 3D). We observed no statistically significant increase in p70 S6K phosphorylation in breast-cancer-derived OE PHGDH lung tumors (Figure S3M), likely because of the larger variation across mice with overexpression tumors. Thus, we concluded that inhibition of the serine biosynthesis pathway decreased mTORC1 signaling in cancer cells in the lung environment, but not in the primary tumor site.
Figure 3. Serine Biosynthesis Potentiates mTORC1 Signaling in Lung M.
(A and B) Total protein synthesis in the presence or absence of 2 mM pyruvate in 4T1 CTR or Phgdh-silenced (shPhgdh) or MDA-MB-231 CTR or OE PHGDH cells based on the incorporation of puromycin (10 μg/mL) into newly synthesized proteins. Cells were starved from serum for 16 h, starved in HBSS for 1 h, and subsequently reactivated (30 min) in serum containing culture medium with or without 2 mM sodium pyruvate. The protein synthesis inhibitor CHX (100 nM) was used as negative CTR. One representative image is shown. n = 3.
(C) Phosphorylation of mTORC1 targets S6 and 4EBP1 in 4T1 PT. Quantification of phospho-S6 is shown in the graph. Data are normalized on total p70 S6K and actin and are shown as fold changes compared with the CTR group. Two-tailed unpaired Student’s t test. 4T1 CTR n = 5, 4T1 shPhgdh n= 4.
(D) Phosphorylation of the mTORC1 targets p70 S6K and 4EBP1 in 4T1 CTR or shPhgdh breast-cancer-derived lung tumors. l.i. refers to lung injection into NMRI nu/nu mice. Quantification of phospho-p70 S6K is shown in the graph. Data are normalized on total p70 S6K and actin. Two-tailed unpaired Student’s t test. n = 4.
Error bars represent SD from the mean of biologically independent mice.
α-Ketoglutarate Produced by the Serine Biosynthesis Pathway Supports mTORC1 Signaling
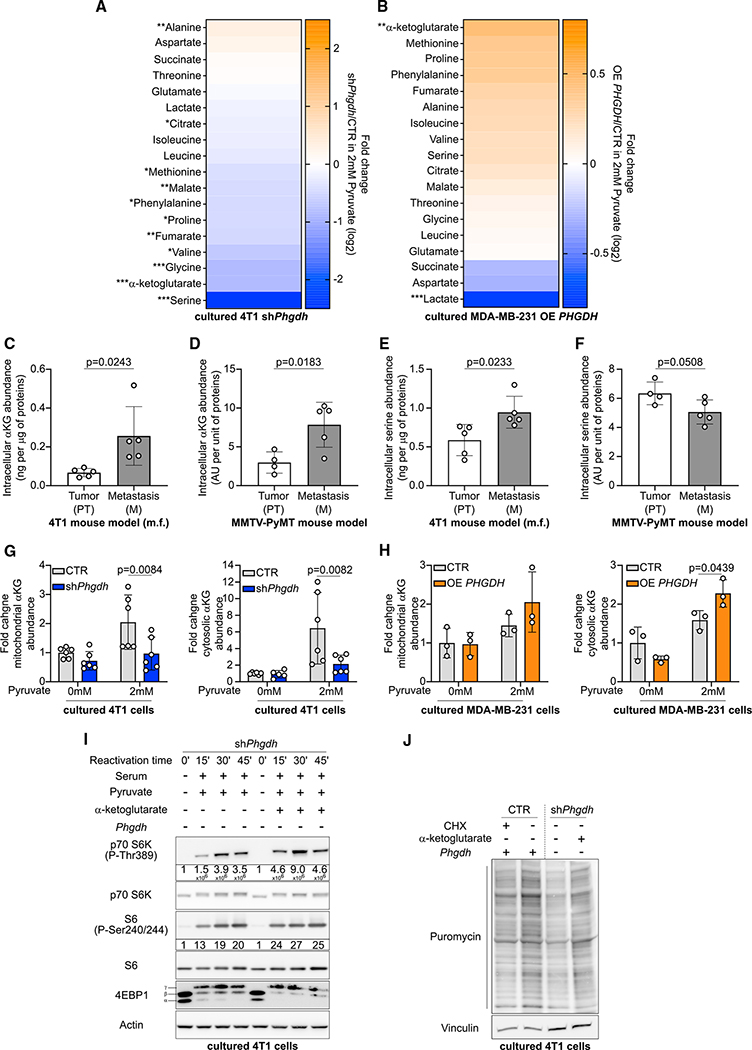
Next, we asked which aspect of serine biosynthesis is required for the activation of mTORC1 growth signaling. Thus, we measured which metabolite products of the serine biosynthesis pathway changed in a PHGDH-dependent manner using mass spectrometry. We observed that predominantly α-ketoglutarate, serine, and glycine decreased in abundance upon Phgdh silencing in 4T1 cells in the presence of pyruvate, whereas α-ketoglutarate showed the most pronounced increase in MDA-MB-231 cells upon OE PHGDH in the presence of pyruvate (Figures 4A and 4B). Notably, nucleotides, which are downstream metabolites of serine biosynthesis, did not consistently change in abundance upon Phgdh silencing or OE PHGDH in the presence of pyruvate (Figure S4). Based on the literature, both serine and α-ketoglutarate could support mTORC1 signaling (Bernfeld et al., 2018; Durán et al., 2012, 2013; Ye et al., 2012). However, serine was abundantly available in our culture conditions (286 μM in RPMI and 400 μM in DMEM) and in the lung environment (199 ± 58.6 μM in mouse and 94 ± 24.1 μM in human) (Figure S1G), which suggests that this metabolite is not limiting in availability. Moreover, α-ketoglutarate, but not serine, was consistently increased in abundance in 4T1 and MMTV-PyMT-derived lung metastases compared with the corresponding primary breast tumors (Figures 4C–4F). Thus, we focused on α-ketoglutarate.
Figure 4. α-Ketoglutarate Produced by the Serine Biosynthesis Pathway Supports mTORC1 Signaling.
(A and B) Heatmap representing metabolite abundance changes in 4T1 shPhgdh cells or MDA-MB-231 OE PHGDH cells compared with the respective CTR cells in culture media containing 2 mM of sodium pyruvate. Two-tailed unpaired Student’s t test. n = 3. Asterisks represent statistical significance as follows: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
(C–F) Intracellular α-ketoglutarate and serine abundance in 4T1 or PyMT PT and lung M. Two-tailed unpaired Student’s t test. n = 5 for the 4T1 model, n = 4 for the PT group of the PyMT model, n = 5 for the M group of the MMTV-PyMT model.
(G and H) Mitochondrial and cytosolic α-ketoglutarate abundance in 4T1 CTR or shPhgdh cells, as well as MDA-MB-231 CTR or OE PHGDH cells after 24 h of incubation with culture medium with or without 2 mM sodium pyruvate. 40 μg/mL (4T1 cells) or 25 μg/mL (MDA-MB-231) digitonin treatment of 2 min was used to permeabilize the cells and separate the cytosolic fraction from the mitochondrial fraction. Two-way ANOVA with Sidak’s multiple comparison test (comparison between CTR and shPhgdh or OE PHGDH). 4T1, n = 6; MDA-MB-231, n = 3.
(I) Phosphorylation of mTORC1 targets p70 S6K, S6, and 4EBP1 in the presence or absence of 1 mM α-ketoglutarate in 4T1 shPhgdh cells. 4T1 cells were starved for 16 h of serum, starved in HBSS for 1 h, and subsequently reactivated (from 15 to 45 min) in serum and 2 mM sodium pyruvate containing culture medium with and without 1 mM α-ketoglutarate. Protein quantification of phosho-p70 S6K and phospho-S6 is shown below the immunoblot. Data are normalized on total p70 S6K or S6 protein and actin and are shown as fold changes compared with time 0. One representative image is shown. n = 3.
(J) Total protein synthesis in the presence or absence of 1 mM α-ketoglutarate and puromycin in 4T1 CTR or shPhgdh cells based on the incorporation of puromycin into newly synthesized proteins. 4T1 cells were starved from serum for 16 h, starved in HBSS for 1 h, and subsequently reactivated (30 min) in serum containing culture medium with or without 1 mM α-ketoglutarate. The protein synthesis inhibitor CHX (100 nM) was used as negative CTR. One representative image is shown. n = 3.
Error bars represent SD from the mean of biologically independent samples.
To activate mTORC1 signaling, cytosolic abundance, but not mitochondrial abundance, of α-ketoglutarate is relevant. Therefore, we measured cytosolic versus mitochondrial α-ketoglutarate abundance in 4T1 control and Phgdh-silenced cells, as well as in MDA-MB-231 control and OE PHGDH cells, upon pyruvate supplementation using a digitonin-based cell permeabilization method (Nonnenmacher et al., 2019). In line with the possibility that α-ketoglutarate produced by the serine biosynthesis pathway influences mTORC1 signaling, we found increased α-ketoglutarate abundance, in the cytosolic fraction of 4T1 cells and MDA-MB-231 OE PHGDH cells compared with 4T1 cells silenced for Phgdh and MDA-MB-231 control cells upon pyruvate supplementation (Figures 4G and 4H). This shows that cytosolic α-ketoglutarate abundance is consistently altered upon silencing or overexpression of PHGDH, which indicates that this metabolite could be involved in serine biosynthesis-driven regulation of mTORC1 signaling.
Next, we reasoned that α-ketoglutarate supplementation rescues the activation of mTORC1 signaling upon Phgdh silencing in 4T1 cells. Indeed, we observed that α-ketoglutarate supplementation elevated the phosphorylation dynamics of p70 S6K, S6, and 4EBP1 (decreased α and β band) in Phgdh-silenced 4T1 cells after starvation and reactivation with serum and pyruvate (Figure 4I). Accordingly, α-ketoglutarate supplementation increased protein translation in PHDGH-silenced 4T1 cells (Figure 4J). These data show that α-ketoglutarate can rescue mTORC1 signaling in vitro upon Phgdh silencing.
Serine Biosynthesis Defines Rapamycin Sensitivity of Lung Metastases
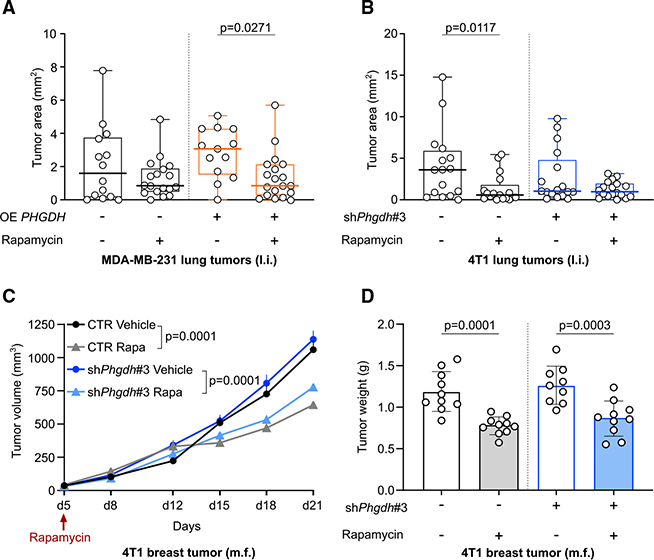
Next, we asked whether serine biosynthesis defines the sensitivity of breast-cancer-derived lung metastases to mTORC1 inhibition. To address this question, we injected MDA-MB-231 control and OE PHGDH cells into the lung of nude mice and treated them with rapamycin (1 mg/kg i.p. every 2nd day) once tumors established. Subsequently, we assessed growth of the breast-cancer-derived lung tumors based on H&E staining. We observed that only OE PHGDH MDA-MB-231 lung tumors, but not control tumors, were sensitive to rapamycin treatment (3.5-fold decrease in area upon rapamycin treatment in OE PHGDH lung tumors compared with vehicle treatment) (Figure 5A). This shows that active serine biosynthesis results in rapamycin sensitivity in breast-cancer-derived lung tumors.
Figure 5. Serine Biosynthesis Defines Rapamycin Sensitivity of Lung M.
(A) MDA-MB-231 lung tumor area in NMRI nu/nu mice treated with rapamycin (1 mg/kg i.p. every 2 days, starting 9 days after tumor initiation) based on H&E staining. The thick black or orange line represents the median. The box extends from the 25th to 75th percentile, and the whisker extends from the minimum to the maximum value. Two-way ANOVA with Sidak’s multiple comparison test. MDA-MB-231 CTR vehicle, n = 14; MDA-MB-231 CTR rapamycin, n = 18;, MDA-MB-231 OE PHGDH vehicle, n = 13; MDA-MB-231 OE PHGDH rapamycin, n = 19.
(B) 4T1 lung tumor area in NMRI nu/nu mice treated with rapamycin (1 mg/kg i.p. every two days, starting the day after tumor initiation) based on H&E staining. The thick black or blue line represents the median. The box extends from the 25th to 75th percentile, and the whisker extends from the minimum to the maximum value. Two-way ANOVA with Sidak’s multiple comparison test. 4T1 CTR vehicle, n = 16; 4T1 CTR rapamycin, n = 17; 4T1 shPHGDH vehicle, n = 17; 4T1 shPHGDH rapamycin, n = 18.
(C) 4T1 PT volume in NMRI nu/nu mice treated with rapamycin (1 mg/kg i.p. every 2 days, starting 5 days after tumor initiation). Two-way ANOVA with Sidak’s multiple comparison test on the tumor volumes measured on the last day (day 21). n = 10.
(D) 4T1 PT weight in NMRI nu/nu mice treated with rapamycin (1 mg/kg i.p. every 2 days, starting 5 days after tumor initiation). Two-way ANOVA with Sidak’s multiple comparison test. n = 9 for the shPhgdh group treated with vehicle, n = 10 for the other groups. Error bars represent SEM (C) and SD (D) from the mean of biologically independent samples.
Subsequently, we injected 4T1 control and Phgdh-silenced breast cancer cells into the lung or mammary fat pad of nude mice and treated them with rapamycin (1 mg/kg i.p. every 2nd day) once tumors established. In line with the overexpression results, we observed that only control 4T1-derived lung tumors with Phgdh expression were sensitivity to rapamycin (6.3-fold decrease in area upon rapamycin treatment in control lung tumors that express Phgdh compared with vehicle treatment) (Figure 5B). Although mammary tumors were sensitive to rapamycin, their sensitivity was the same in the control condition and upon Phgdh silencing (Figures 5C and 5D). Based on these data, we concluded that PHGDH activity (i.e., serine biosynthesis) defined the sensitivity of breast cancer cells in the lung environment, but not in the mammary fat pad, to mTORC1 inhibition.
DISCUSSION
We identified an organ microenvironment-specific mechanism of mTORC1 activation contributing to metastatic outgrowth. In particular, we found that lung metastases, which are in a pyruvate-enriched environment (Christen et al., 2016) that potentiates serine biosynthesis, exhibited an elevated mTORC1 growth signaling. Conversely, pyruvate uptake and serine biosynthesis were not required for mTORC1 growth signaling in breast cancer cells growing in the mammary fat pad. Thus, our work provides evidence that mTORC1 growth signaling is differently regulated between breast cancer metastases and primary tumor environments.
The serine biosynthesis enzyme PHGDH is often amplified in triple-negative breast cancers (Locasale et al., 2011; Possemato et al., 2011), and a serine-enriched diet has been found to support primary breast cancer growth (Sullivan et al., 2019). Moreover, activity of the serine biosynthesis pathway can support redox homeostasis in breast cancer cells (Diehl et al., 2019; Samanta et al., 2016). In addition, it has been found that more than 70% of breast-cancer-derived lung and brain metastases are PHGDH positive (Kim et al., 2014) and that breast cancer cells that evolved to metastasize to bone showed increased PHGDH expression (Pollari et al., 2011). Moreover, a recent study concluded that brain metastases rely on PHGDH to fulfill their biosynthetic needs for nucleotides because of the limited serine (28 μM) and glycine (5 μM) availability in this particular environment (Ngo et al., 2020). In addition to these findings, we identify serine biosynthesis as a determinant for sensitivity to mTORC1 inhibition in breast cancer cells growing in the lung environment.
mTORC1 signaling has been found to activate serine biosynthesis by increasing the expression of PHGDH and PSAT (Ben-Sahra et al., 2016). Moreover, a feedback loop has been established, suggesting that pyruvate kinase M2 in its less active dimeric form promotes the serine production needed for mTORC1 activity (Ye et al., 2012). In turn, it was observed that serine is an allosteric activator of pyruvate kinase M2 (Chaneton et al., 2012). Thus, a balance between mTORC1 activity and serine levels can be established. Beyond this intracellular serine and pyruvate kinase-driven mechanism, we discovered that in vivo pyruvate uptake through Mct2 potentiates mTORC1 growth signaling in the lung metastatic niche. Strikingly, this identifies pyruvate as a nutrient that supports multiple cellular functions essential for metastasis formation in lung. Previously, we have identified that pyruvate is essential for the ability of breast cancer cells to shape the extracellular matrix environment of the lung metastatic niche (Elia et al., 2019). Here, we identify pyruvate as an important nutrient to potentiate mTORC1 signaling during metastatic outgrowth. We further found that pyruvate supports the biosynthetic needs of proliferating breast-cancer-derived lung metastases by stimulating pyruvate carboxylase-dependent anaplerosis (Christen et al., 2016). Thus, it is tempting to speculate that inhibiting pyruvate metabolism can be exploited to prevent and treat breast-cancer-derived lung metastases and perhaps other cancers growing in this environment.
In conclusion, we discovered that the in vivo environment defines which nutrients and metabolic pathways cancer cells exploit to drive mTORC1 signaling. Consequently, lung metastases evolved to depend on different metabolic pathways than primary breast tumors to activate the same growth signaling pathway.
Limitations of Study
Here, we discovered that lung metastases of mice and their corresponding primary breast tumors rely on different nutrients and metabolic pathways to active growth signaling. Specifically, we provided evidence that in the in vivo environment of the lung pyruvate uptake and PHGDH activity potentiate mTORC1 signaling and that consequent loss of PHGDH dampens rapamycin sensitivity. Although we provided the physiological relevance of this finding through our in vivo data, the effects of pyruvate supplementation and PHGDH activity on mTORC1 target phosphorylation in vitro were less evident. This may be explained because in vitro, we cannot fully recapitulate the in vivo environment. Moreover, although we provided some in vitro evidence of the applicability of this research finding beyond breast cancer cells, further in vivo studies with cancer cells of different origin need to be conducted to provide conclusions on this aspect. Finally, it will be relevant to determine how mTORC1 signaling is supported in other metastatic sites beyond the lung.
STAR⋆METHODS
RESOURCE AVAILABILITY
Lead Contact
Further requests for resources should be directed to the lead contact, Sarah-Maria Fendt (sarah-maria.fendt@kuleuven.vib.be).
Materials Availability
This study did not generate new unique reagents, except of genetically manipulated cell lines based on commercially available constructs. Reagents generated in this study will be made available on request through the lead author or the collaboration partner that generated the resource, but we may require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.
Data and Code Availability
The published article includes all analyzed data. The mass spectrometry files supporting the current study have not been deposited in a public repository due to the lack of a commonly accepted and feasible metabolomics file repository but are available upon reasonable request from the lead contact. Original western blot images are on Mendeley at http://dx.doi.org/10.17632/hph5nb2rjy.1.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Human HEK293T epithelial cells, MDA-MB-231, and MDA-MB-468 breast adenocarcinoma cells, murine 4T1 mammary gland cancer cells, A549 human epithelial lung carcinoma cells and HCT 116 colorectal carcinoma cells were obtained from ATCC. HEK293 T, MDA-MB-231, MDA-MB-468, A549 and HCT 116 cells were cultured in high glucose (4.5 g/L) Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (Life Technologies), 1% penicillin (final concentration of 50 U/mL) (Life Technologies) and 1% streptomycin (final concentration of 50 μ/mL) (Life technology). 4T1 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 Medium (Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (Life Technologies), 1% penicillin (final concentration of 50 U/mL) (Life technologies) and 1% streptomycin (final concentration of 50 μg/mL) (Life Technologies). The heat-inactivation of the fetal bovine serum was performed at 55°C for 45 minutes. Hygromycin B and puromycin dihydrochloride (Life Technologies) were added to the growth medium for selection of overexpression and knockdown cell lines, respectively. Blasticidin S HCl (Life Technologies, R21001) was added to the growth medium for selection of Luciferase-expressing cells. All cell lines were confirmed to be mycoplasma-free by Mycoalert detection kit (Lonza). HEK293T, MDA-MB-231, and MDA-MB-468 cells were validated by DNA fingerprinting.
Mouse models
All animal experiments were approved by the local authorities in compliance with all relevant ethical regulations. For injection models, mice were randomized before injection of cancer cells. All samples were analyzed blinded. Sample size was determined using power calculations with B = 0.8 and p < 0.05 based on preliminary data and in compliance with the 3R system: Replacement, Reduction, Refinement.
Mammary fat pad model
Six-week-old female BALB/c (Envigo) or NMRI nu/nu (Taconic) mice were inoculated with 4T1, 4T1 CTR or 4T1 shPHGDH cells in the mammary fat pad (m.f.; 1×106 cells, in 5μL PBS using a 30G syringe). Rapamycin (Selleckchem, S1039) treatment was started 5 days after cell injection. Rapamycin (1 mg per kg) was intraperitoneally injected every 48h. Rapamycin was dissolved in a solution of 2% DMSO (Merck Sigma, D4540), 30% polyethylene glycol 300 (Merck Sigma, 90878), 5% Tween80 (Merck Sigma, P4780) and ddH20 and used promptly after mixing as indicated by the manufacturer. The drug was dissolved at a concentration of 0.0002 mg per μL. α-Cyano-4-hydroxycinnamic acid (Merck Sigma, C2020) acute treatment consisted of two injections in the last two days of the experiment to only investigate short-term effects of the molecular mechanisms triggered by pyruvate uptake inhibition on the phosphorylation of mTORC1 targets. α-Cyano-4-hydroxycinnamic acid (60 mg per kg) was intraperitoneally injected. α-Cyano-4-hydroxycinnamic acid was dissolved in a mix containing 1.5% DMSO, 60% β-cyclodextrin (Santa Cruz, SC-203461A) (from an initial 30% aqueous solution), 35% polyethylene glycol 300 and 5% ethanol and pH neutralized with NaOH. The drug was dissolved at a concentration of 0.03 mg per μL. The same mix without drug was injected as a vehicle to control animals. Treated and control mice were randomly chosen. Mice were euthanized 21 days after cell injection. The last dose of drug was given on the last day, two hours before euthanasia. To follow the effect on the primary tumor of Rapamycin treatment, tumor volumes were measured during the experiment using a caliper. Additionally, at the end of the experiment, primary tumors were dissected and weighted. Humane end points were determined as follows: tumor size of 1.8 cm3, loss of ability to ambulate, labored respiration, surgical infection or weight loss over 10% of initial body weight. Mice were monitored and upon detection of one of the previous mentioned symptoms, the animal was euthanized. BALB/c mice and NMRI nu/nu mice were housed respectively in filter top cages and IVC cages. Housing and experimental animal procedures were approved by the Institutional Animal Care and Research Advisory Committee of KU Leuven, Belgium. The animal study complies with ethical regulations and was approved by the KU Leuven ethics committee.
Lung injection model
Six-week-old female NMRI nu/nu (Taconic) mice were inoculated with 4T1 CTR, shPHGDH (l.i.; 4×104 cells) or MDA-MB-231 CTR, overexpressing PHGDH (OE PHGDH) (l.i.; 9×105 cells) in the lung. Mice were first anesthetized using a ketamine/xylazine solution (37.5% Ketamine, 12.5% xylazine, 50% saline solution at 0.9%). Ketamine initial concentration was 100 mg per mL. Xylazin initial concentration was 20mg per mL. Subsequently, 40μL of cell suspension (50% PBS, 50% Matrigel) were injected into the right side of the mouse, under the armpit between 5th and 6th costal rib, 5mm deep using a 29G syringe (Becton Dickinson Medical, BDAM324892). Immediately after injection, the mice were turned on their right side and monitored until they woke up.
When 4T1 cells were injected, Rapamycin (1mg per kg) treatment was started the day after cell injection. Rapamycin was injected intraperitoneally every 48h until the end of the experiment. Mice were euthanized after 10 days from cells injection. When MDA-MB-231 cells were injected, Rapamycin (1mg per kg) treatment was started 9 days after cell injection. Rapamycin was injected intraperitoneally every 48h until the end of the experiment. Mice were euthanized 21 days after cell injection. Treated and control mice were randomly chosen. The last dose of Rapamycin was given on the last day, two hours before euthanasia. Humane end points were determined as follows: loss of ability to ambulate, labored respiration or weight loss over 10% of initial body weight. Mice were monitored and upon detection of one of the previous mentioned symptoms, the animal was euthanized. NMRI nu/nu mice were housed in IVC cages. Housing and experimental animal procedures were approved by the Institutional Animal Care and Research Advisory Committee of KU Leuven, Belgium. The animal study complies with ethical regulations and was approved by the KU Leuven ethics committee.
MMTV-PyMT tumor generation
Mouse experiments were performed under the Animal Study Protocol LPG-002, approved by the National Cancer Institute (NCI) Animal Use and Care Committee. BL10 (MMTV-PyMT/FVB × C57BL/10J), BL6 (MMTV-PyMT/FVB × C57BL/6J), CAST (MMTV-PyMT/FVB × CAST/EiJ) or MOLF (MMTV-PyMT/FVB × MOLF/EiJ) mice were generated as previously reported (Lifsted et al., 1998). Mice were euthanized by cervical dislocation following anesthesia by Avertin at humane end point as determined by the institute veterinarian. Tumor tissue was excised and immediately flash-frozen in liquid nitrogen, then placed at −80°C for long term storage.
METHOD DETAILS
Knockdown and overexpression strategies
The plasmids pLKO expressing the shRNA sequences for Phgdh #1 and #2 were obtained from BCCM (Belgian co-ordinated collections of micro-organisms). Phgdh knock down #3 in 4T1 cells was generated using the plasmid pLKO-shRNA2 vector expressing the shRNA against Phgdh obtained from the Peter Carmeliet’s lab (VIB/KU Leuven) (Vandekeere et al., 2018, PHGDH KD1). The same plasmid expressing a non-targeting shRNA sequence was used as control. Lentiviral particles were produced in HEK293T cells. Transduction of 4T1 cells was performed overnight and the medium was replaced the next day. Cells were selected with puromycin at a concentration of 2 μg/mL. The overexpression of PHGDH in MDA-MB-231 cells was generated using the plasmid pLHCX expressing the PHGDH cDNA obtained from the lab of Prof. Matthew Vander Heiden (MIT). The empty plasmid pLHCX was used as a control. Retroviral particles were produced in HEK293T cells. Transduction of MDA-MB-231 cells was performed overnight with freshly prepared virus and the medium was replaced the next day. Cells were selected with hygromycin in a concentration of 400 μg/mL. Knock down or overexpression of PHGDH was validated by western blot analysis.
Starvation and reactivation
Cells were seeded in a 10 cm dish to be 70%–80% confluent at the end of the experiment. The day after seeding, cells were starved from serum over night for 16 h. Subsequently, cells were washed with Dulbecco’s phosphate-buffered saline (dPBS) and starved 1 additional hour in Hanks’ Balanced Salt Solution (HBSS) (Thermo scientific, 14025050). Next, cells were reactivated with medium containing dialyzed FBS in presence or absence of 2mM Sodium Pyruvate (Merck Sigma, P8574), in presence or absence of 1mM Dimethyl α-ketoglutarate (Merck Sigma, 349631) and collected after the indicated time points. Time 0’ conditions were collected immediately after the starvation with HBSS.
Protein extraction and western blot analysis
Cells were collected in dPBS and lysed in RIPA lysis and extraction buffer (Thermo Scientific, 89901) supplemented with protease (Merck Sigma, 5892970001) and phosphatase inhibitors (Merck Sigma, 4906845001). Frozen tissue samples were placed in 1.5 mL tubes with sufficient amount of RIPA buffer and grinded with a tissue lyser to help the lysis of the tissue.
Extracted proteins were quantified using a Pierce BCA Protein Assay Kit (Thermo Scientific, 23225). Subsequently, 15–40 μg of proteins were loaded on a precast gel NuPAGE Novex 4%–12% Bis-Tris (Thermo Scientific, NP0336BOX) or a freshly casted 16% Acrylamide/Bis-acrylamide 29:1 (Biorad, 1610156) gel for 4EBP1 separation. Proteins were then transferred on a nitrocellulose membrane using an iBlot dry blotting system with iBlot transfer stacks (Thermo Scientific, IB301031). Membranes were incubated for 1h at room temperature in a blocking solution of 5% milk TRIS Buffer Saline 0.05% Tween (TBS-T). Subsequently, membranes were incubated over night at 4°C with primary antibodies against either p70 S6 kinase (Cell Signaling Technology, 9202S), Phospho-p70 S6 Kinase (Thr389) (Cell Signaling Technology, 9205), S6 Ribosomal Protein (5G10) (Cell Signaling Technology, 2217S), Phospho-S6 Ribosomal Protein (Ser240/244) (D68F8) (Cell Signaling Technology, 5364S), β-Actin (Merck Sigma, A5441), PHGDH (Merck Sigma, HPA021241), PSAT1 (Bio-techne, H00029968-A01), PSPH (Merck Sigma, HPA020376), 4EBP1 (Cell Signaling Technology, 9452S), Phospho-4EBP1 (Ser65)(Cell Signaling Technology, 9451S), Vinculin (Cell Signaling Technology, 4650S). All primary antibodies were used in a 1:1000 dilution in 5% bovine serum albumin in TBS-T. The following day, membranes were incubated with HRP-linked secondary antibodies anti rabbit (Cell Signaling Technologies, 7074S) or mouse (Cell Signaling Technologies, 7076S) used 1:2000 in 5% milk in TBS-T. Bound antibodies were visualized using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific, 34095) or SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Scientific, 34580). Images were acquired using an ImageQuant LAS 4000.
To detect both the total and phosphorylated form of p70 S6 kinase, S6 Ribosomal protein and 4EBP1, samples were loaded twice on two different gels. β-Actin or Vinculin were used as loading control. In the figures showing the phosphorylation status of the mTOR target, one representative β-Actin blot is shown. The β-Actin shown is always coming from the very same gel were the phosphorylated proteins are blotted. Images were quantified using the software Image Studio Lite. The signals of the phosphorylated proteins were normalized on the signal of the total protein and on the loading control β-Actin.
Protein synthesis (SUnSET)
Puromycin incorporation into newly synthesized protein was used to quantify protein synthesis as previously described (Schmidt et al., 2009; Vendramin et al., 2018). Cells were seeded in a 6cm dish to be 70%–80% confluent at the end of the experiment. The day after seeding, cells were starved from serum over night for 16h in medium in presence or absence of 2mM Sodium Pyruvate, in presence or absence of Dimethyl α-ketoglutarate. Subsequently, cells were washed with Dulbecco’s phosphate-buffered saline (dPBS) and starved 1 additional hour in Hanks’ Balanced Salt Solution (HBSS) in presence or absence of 2mM Sodium Pyruvate, in presence or absence of 1mM Dimethyl α-ketoglutarate. Next, cells were reactivated with medium containing dialyzed FBS in presence or absence of 2mM Sodium Pyruvate, in presence or absence of 1mM Dimethyl α-ketoglutarate and 100nM cycloheximide or 0.001% DMSO for a total of 30 minutes. During the last 10 minutes of reactivation, puromycin was added to the medium at a concentration of 10μg per mL. After cell collection and protein extraction, puromycin incorporation was evaluated by western blot analysis using an anti-puromycin antibody (Merck, MABE343).
Surgery and 13C6-glucose infusion
The mammary fat pad model in BALB/c mice was used for the 13C6-glucose infusion experiment (Broekaert and Fendt, 2019). Mice were implanted with a jugular vein catheter and, after one week of recovery time, infused with 13C6-glucose as previously described. Briefly, after mice anesthetization with isoflurane, a catheter was inserted in the jugular vein and connected to an antenna protruding from the dorsal part of the mice. Mice were left recovering single housed for 1 week after surgery. Next, mice were infused for 6h with a solution of 500 mg per mL of 13C6-glucose, at a rate of 30 mg per kg per min. At the end of the infusion protocol, mice were sacrificed and all tissues of interest were collected and stored.
Tissue collection and preparation for protein and metabolite extraction
At the end of every in vivo experiment, mice were sacrificed by injecting approximately 50 μL of a 60 mg per mL Dolethal (pentobarbital sodium) solution (Vetoquinol). Blood was quickly collected via cardiac puncture, stored in heparin-coated tubes and centrifuged to separate the plasma. Primary tumors, lung metastases, lung tumors and lung healthy tissue were dissected and washed in ice-cold saline, placed into pre-labeled bags and frozen using a liquid-nitrogen-cooled Biosqueezer (Biospec Products). The bags were then placed in liquid nitrogen until all collections were finished and finally stored at −80°C until further processing.
For tissue preparation for metabolite extraction, the tissue was weighed (10–15 mg) and pulverized (Cryomill, Retsch) under liquid-nitrogen conditions. The pulverized tissue was next used for metabolite extraction as described below.
Hematoxylin and eosin staining
Hematoxylin and eosin staining were used to measure the area of cancerous lesions in the lung of the different mouse models. During dissection, the lungs were gently infused via the trachea with 10% neutral buffered formalin. Next, samples were embedded in paraffin and sliced in 5 μm thick sections that were stained with hematoxylin and eosin. Metastatic area was quantified by ZEN Blue software (Zeiss).
Lung interstitial fluid extraction
The lung interstitial fluid extraction method has been adapted and modified from Christen et al. (2016), Nagahashi et al. (2016), and Wiig et al. (2003). For collection of human samples, the study was approved by the local ethics committee (Medical Ethics Committee UZ/KU Leuven) under the protocol S57123. Human ‘normal’ lung tissue was collected from patients who underwent lung surgery for emphysematous lung volume reduction or tumorectomy. In the latter case, lung tissue from the resection specimen was taken as far away as possible from the tumor front. Following surgical resection, lung samples were taken and transported to our research facility. For the collection of lung samples from mice, five 9-weeks old healthy BALB/c female mice have been euthanized with 50mL of 60mg/ml of Dolethal (pentobarbital sodium), lungs were harvested by surgical resection, washed with a blood bank saline solution and dried from liquid excess by carefully tapping in a gauze. Subsequently, the organs were placed in a filtered centrifuge tube created by using a column-centrifuge tube in which the pre-existent filter has been substituted with a nylon mesh filter with 20 μm opening pores (Spectrum Labs, Product code: 145811). For the human samples, the tissue was cut in pieces of around 100 mg that were placed in distinct filtered centrifuge tubes. The interstitial fluids (around 2–5 μL for mouse samples, around 3–10 μL) was collected in the column-centrifuge tubes by centrifugation at 400 g, 4°C for 10 minutes. The lung interstitial fluid was stored in dry ice immediately after extraction.
Metabolites measurements
All experiments where Sodium Pyruvate was supplemented in the culture medium, as well as labeling experiments, were performed in media with 10% dialyzed serum for the indicated time points. Metabolites for the subsequent mass spectrometry analysis were prepared by quenching the cells in liquid nitrogen followed by a cold two-phase methanol-water-chloroform extraction (van Gorsel et al., 2019). Phase separation was achieved by centrifugation at 4°C. The methanol-water phase containing polar metabolites was separated and dried using a vacuum concentrator. Dried metabolite samples were stored at −80°C.
The extraction of metabolites from the tissues of the MMTV-PyMT mouse model was conducted as following. Metabolites were extracted from tumors using a modification of the Bligh and Dyer method (Bligh and Dyer, 1959). Flash-frozen tumor chunks were immersed in a mixture of HPLC-grade methanol and water (5:3 ratio) kept at −20°C and homogenized on a Precellys Evolution tissue homogenizer (15 s − 1 cycle, 10,000 rpm). HPLC-grade chloroform was added (methanol:water:chloroform ratio to 5:3:5), the mixture vortexed for 10 min at 4°C, and centrifuged at maximum speed (16,000 × g) for 10 min at 4°C to achieve phase separation. The methanol-water phase containing polar metabolites was separated and dried under a stream of nitrogen gas. The dried metabolite samples were stored at −80°C until analysis. The protein interphase was also dried down and dissolved in 1 mL 0.2 mM potassium hydroxide, and the protein concentration was then quantified using the Pierce BCA Protein Assay Kit (Thermo Fisher). To normalize sample concentrations, the dried metabolite extracts were resuspended in HPLC-grade water at volumes corresponding to their protein quantification values.
Gas chromatography-mass spectrometry
Amino acids and organic acids of the central carbon metabolism were measured with gas chromatography-mass spectrometry. Polar metabolites were derivatized for 90 min at 37°C with 20 μL (13 μL in the case of the interstitial fluids) of 20 mg per ml methoxyamine (Merck Sigma, 226904) in pyridine (Merck Sigma, 270970). Subsequently, 15 μL of N-(tert-butyldimethylsilyl)-N-methyl-trifluoroacetamide, with 1% tert-butyldimethylchlorosilane were added to 7.5 μL of each derivative and incubated for 60 min at 60 °C (Merck Sigma, 375934) (Lorendeau et al., 2017). Isotopologue distributions and metabolite concentrations were measured with a 7890 A GC system (Agilent Technologies) combined with a 5975C Inert MS system (Agilent Technologies). 1 μL of sample was injected into a DB35MS column in split mode (ratio 1 to 3) using an inlet temperature of 270°C. The carrier gas was helium with a flow rate of 1 mL per min. Upon injection, the GC oven was set at 100 °C for 1 min and then increased to 105 °C at 2.5°C/min and with a gradient of 2.5 °C/ min finally to 320°C at 22°C/min. The measurement of metabolites has been performed under electron impact ionization at 70 eV using a selected-ion monitoring (SIM) mode. Isotopologue distributions were extracted from the raw ion chromatograms using a custom MATLAB M-file, which applies consistent integration bounds and baseline correction to each ion (Young et al., 2008). In addition, we corrected for naturally occurring isotopes using the method of Fernandez et al. (1996). All labeling fractions were transformed to a natural abundance-corrected mass distribution vector (MDV) (Buescher et al., 2015).
Liquid chromatography-mass spectrometry
Nucleotides, amino acids and α-ketoglutarate from the dynamic metabolomic experiment (Figure S4) and glucose in blood plasma versus lung interstitial fluid (Figure 1F) were measured on liquid chromatography-mass spectrometry system. The separation of metabolites was performed by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). Metabolite separation was performed at 28°C with an Ultra High-Performance Liquid Chromatography (UHPLC) from Thermo Scientific on a HILIC Fusion (P) column (150 × 2.1 mm, 5 μm). Polar metabolites were separated at a flow rate of 0.2 ml/min for 43 min as follows: 0–2 min: 8% B; 2–7 min: 8%–30% B, 7–20 min: 30%–47% B; 20–26 min: 90% B; 26–30 min: 90% B, 30–34 min: 90%–8% B and 34–43 min: 8% B (solvent A: acetonitrile; solvent B: ammonium acetate 10 mM pH = 9.3). The UHPLC autosampler was set at 4°C and 12 μL of sample were injected. A Thermo TSQ Quantiva triple quadrupole with an electrospray source was used for the analysis. The detection of metabolites was performed in Multiple reaction Monitoring mode (MRM) mode in negative mode. The ion transfer tube was set at 350°C and the vaporizer temperature at 400°C. Data were collected and integrated using Xcalibur software (Thermo Scientific).
The measurements of α-ketoglutarate and serine in the MMTV-PyMT mouse model was conducted as following. Liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis was performed with ion-pairing reverse phase chromatography using an Ascentis Express column (C18, 5 cm × 2.1 mm, 2.7 μm, Sigma-Aldrich) for separation and a Waters Xevo TQ-S triple quadrupole mass spectrometer operated in negative mode as mass analyzer. The LC parameters were as follows: autosampler temperature, 5°C; injection volume, 5 μl; column temperature, 50°C and flow rate, 400 μl/min. The LC solvents were solvent A: 10 mM tributylamine and 15 mM acetic acid in 97:3 water:methanol (pH 4.95) and solvent B: methanol. Elution from the column was performed over 12 min with the following gradient: t = 0, 0% solvent B, flow rate 0.4 ml/min; t = 1, 0% solvent B, flow rate 0.4 ml/min; t = 2, 20% solvent B, flow rate 0.3 ml/min; t = 3, 20% solvent B, flow rate 0.25 ml/min; t = 5, 55% solvent B, flow rate 0.15 ml/min; t = 8, 95% solvent B, flow rate 0.15 ml/min; t = 9.5, 95% solvent B, flow rate 0.15 ml/min; t = 10, 0% solvent B, flow rate 0.4 ml/min; t = 12, 0% solvent B, flow rate 0.4 ml/min. Mass spectra were acquired using negative-mode electrospray ionization operating in multiple reaction monitoring (MRM) mode. The capillary voltage was 3,000 V, and cone voltage was 50 V. Nitrogen was used as cone gas and desolvation gas, with flow rates of 150 and 600 l/h, respectively. The source temperature was 150°C, and desolvation temperature was 500°C. Argon was used as collision gas at a manifold pressure of 4.3 × 10−3 mbar. Precursor and product ion m/z, collision energies and source cone potentials were optimized using Waters QuanOptimize software. The MRM transition for alpha-ketoglutarate is as follows: parent m/z = 145, daughter m/z = 100.77, cone voltage = 22 V, collision energy = 5 V. Two replicate injections/runs of each sample was performed. Peaks were quantified using MAVEN (Clasquin et al., 2012).
Compartment specific-metabolic measurements
Selective permeabilization of the cytosolic membrane using digitonin was used to separate the cytosolic metabolic content from the mitochondrial metabolic content as previously described (Nonnenmacher et al., 2017, 2019). Briefly, cells were seeded in a 6-well plate to be 70%–80% confluent after 48h. After 24h from the seeding, the medium was replaced with one containing dialyzed FBS + or – 2mM Sodium Pyruvate. After 24h, the supernatant was removed and cells were washed with 2 mL of dPBS. Permeabilization of the cytosolic membrane was achieved by incubation with 500 μl digitonin solution (40 μg/ml in mitochondrial assay buffer for 4T1, 25 μg/ml in mitochondrial assay buffer for MDA-MB-231) for 2 min. Afterward, supernatants containing the cytosolic components were collected and stored at −80°C, while the 6-well plates containing the intact mitochondrial component were washed once with two volumes of saline, quenched in liquid nitrogen and stored at −80°C until metabolite were extracted as already described. The cytosolic supernatants were dried overnight at 4°C in a speedvac concentrator and next metabolite were extracted as already described.
Monitoring of tumor growth via optical imaging
In order to monitor tumor growth in the lung injection model, we generated MDA-MB-231 CTR and OE PHGDH expressing the firefly luciferase using the lentiviral particles pCH-EF1a-eGFP-T2A-Luc2-Ires-BsdR obtained from the Leuven Viral Vector Core. Transduction of 4T1 cells was performed overnight and the medium was replaced the next day. To minimize the possibility of multiple transductions of the same cells, we used a viral dilution that ensured only 20% of transduction efficacy. Transduction efficacy was evaluated by checking the percentage of GFP positive cells under a fluorescence microscope. Next, cells were selected with Blasticidin (Thermo Scientific, R21001) at a concentration of 5 μg per mL. Before each imaging session, the mice were injected intraperitoneally with 126 mg/kg D-luciferin (Promega, Madison, WI, USA) dissolved in PBS (15 mg/ml). Next, 5 animals per time, belonging to the same group, were positioned in the IVIS Spectrum (Perkin Elmer, Waltham, MA) and images were acquired after 10 min under 2% isoflurane inhalation. Images were acquired at day 4, 7, 9, 11, 14, 16, 18, 21 after cell injection. Bioluminescence images were analyzed using the LivingImage (Perkin Elmer, Waltham, MA) processing software. The same regions of interest (ROIs) were drawn around the lungs of every mouse, and measurements were generated as the total flux (photon per second) from the selected ROIs.
Analysis of mTOR pathway activity in human metastases and matched primary breast cancers
For comparison of mTOR pathway activity in human metastases versus matched primary breast cancers, publicly available phosphoproteomics RPPA data from The Cancer Genome Atlas (TCGA; RPPA profiles) were accessed and downloaded from the UCSC Xena browser (http://xena.ucsc.edu) (Vivian et al., 2017; Zhang et al., 2017), which contains five matched pairs. For each sample, an mTOR pathway signature could be scored based on their RPPA profiles, as defined previously (Zhang et al., 2017). This signature was defined as the sum of phosphoprotein levels of mTOR, 4EBP1 (S65, T37/T46, and T70 RPPA features), P70S6K, and S6 (S235/236 and S240/244 features) (Zhang et al., 2017). For each matched pair, we calculated the fold change of the sum score of the metastatic over the primary tumor sample. The TCGA sample identifier for the data presented are the following: #1 TCGA-BH-A1ES, #2 TCGA-BH-A1FE, #3 TCGA-E2-A15A, #4 TCGA-BH-A18V, #5 TCGA-E2-A15K.
Comparison of mTOR activity with nutrient transporters gene expression levels
The activity of the mTOR pathway in n = 859 primary breast cancer samples from The Cancer Genome Atlas (TCGA) was assessed as previously described (Zhang et al., 2017) by generating activity sum scores of the following parameters from proteomic RPPA (reverse phase protein assay) data, which are publicly available via the UCSC Xena browser (https://xena.ucsc.edu/): MTOR_pS2448, X4EBP1_pS65, X4EBP1_pT37T46, X4EBP1_pT70, P70S6K_pT389, S6_pS235S236, and S6_pS240S244. Matching nutrient transporter gene expression data for the same primary tumors was retrieved from the NIH Genomic Data Commons (GDC) data portal (https://portal.gdc.cancer.gov/). The mTOR activity sum scores were stratified in quartiles and compared with the nutrient transporters expression levels using a two-sided Mann-Whitney test. The data are presented as relative fold change of each sample to the average expression value calculated for each gene across the samples in the 1st quartile.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical data analysis was performed using GraphPad Prism version 7.0 (GraphPad Software) on n ≥ 3 biological replicates. Details on statistical tests and post-tests are presented in the figure legends. Sample size for all experiments was chosen empirically. Independent experiments were pooled and analyzed together whenever possible as detailed in figure legends. Quantification of the western blots was performed using the software Image Studio Lite. For the human MDA-MB-231 mouse model, mice with tumors in the chest cavity and mice with no initial tumor growth were removed from the analysis based on a correlation between H&E and bioluminescence (90% prediction bands).
Data are presented as mean ± s.d., or as mean ± s.e.m., as indicated in the figure legends.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| p70 S6 kinase | Cell Signaling Technology | Cat# 9202, RRID:AB_331676 |
| Phospho-p70 S6 Kinase (Thr389) | Cell Signaling Technology | Cat# 9205, RRID:AB_330944 |
| S6 Ribosomal Protein (5G10) | Cell Signaling Technology | Cat# 2217, RRID:AB_331355 |
| Phospho-S6 Ribosomal Protein (Ser240/244) (D68F8) | Cell Signaling Technology | Cat# 5364, RRID:AB_10694233 |
| β-Actin | Merck Sigma | Cat# A5441, RRID:AB_476744 |
| PHGDH | Merck Sigma | Cat# HPA021241, RRID:AB_1855299 |
| PSAT1 | Bio-techne | Cat# H00029968-A01, RRID:AB_463717 |
| PSPH | Merck Sigma | Cat# HPA020376, RRID:AB_1855867 |
| 4EBP1 | Cell Signaling Technology | Cat# 9452, RRID:AB_331692 |
| Phospho-4EBP1 (Ser65) | Cell Signaling Technology | Cat# 9451, RRID:AB_330947 |
| Vinculin | Cell Signaling Technology | Cat# 4650, RRID:AB_10559207 |
| Anti-Puromycin Antibody, clone 12D10 | Merck Sigma | Cat# MABE343, RRID:AB_2566826 |
| HRP-linked secondary antibody anti rabbit | Cell Signaling Technology | Cat# 7074, RRID:AB_2099233 |
| HRP-linked secondary antibody anti mouse | Cell Signaling Technology | Cat# 7076, RRID:AB_330924 |
|
| ||
| Bacterial and Virus Strains | ||
|
| ||
| pCH-EF1a-eGFP-T2A-Luc2-Ires-BsdR | Leuven Viral Vector Core | N/A |
|
| ||
| Biological Samples | ||
|
| ||
| Human normal lung tissue | UZ Leuven | N/A |
|
| ||
| Chemicals, Peptides, and Recombinant Proteins | ||
|
| ||
| Hanks’ Balanced Salt Solution (HBSS) | Thermo scientific | 14025050 |
| Sodium Pyruvate | Merck Sigma | P8574 |
| Dimethyl α-ketoglutarate | Merck Sigma | 349631 |
| RIPA lysis and extraction buffer | Thermo scientific | 89901 |
| Protease inhibitors | Merck Sigma | 5892970001 |
| phosphatase inhibitors | Merck Sigma | 4906845001 |
| Acrylamide/Bis-acrylamide 29:1 | Biorad | 1610156 |
| Puromycin Dihydrochloride | Thermo scientific | A1113803 |
| Cycloheximide | Merck Sigma | C7698 |
| Rapamycin | Selleckchem | S1039 |
| DMSO | Merck Sigma | D4540 |
| polyethylene glycol 300 | Merck Sigma | 90878 |
| Tween80 | Merck Sigma | P4780 |
| α-Cyano-4-hydroxycinnamic acid | Merck Sigma | C2020 |
| β-cyclodextrin | Santa Cruz | SC-203461A |
| D-GLUCOSE (U-13C6, 99%) | Eurisotop | CLM-1396-5 |
| Methoxyamine | Merck Sigma | 226904 |
| Pyridine | Merck Sigma | 270970 |
| N-(tert-butyldimethylsilyl)-N-methyl-trifluoroacetamide | Merck Sigma | 375934 |
| Digitonin | Merck Sigma | D141 |
| Blasticidin | Thermo Scientific | R21001 |
| PH-755 | Raze Therapeutics | Ngo et al., 2020 |
|
| ||
| Critical Commercial Assays | ||
|
| ||
| Pierce BCA Protein Assay Kit | Thermo Scientific | 23225 |
| Precast gel NuPAGE Novex 4–12% Bis-Tris | Thermo Scientific | NP0336BOX |
| iBlot transfer stacks | Thermo Scientific | IB301031 |
| SuperSignal West Femto Maximum Sensitivity Substrate | Thermo Scientific | 34095 |
| SuperSignal West Pico PLUS Chemiluminescent Substrate | Thermo Scientific | 34580 |
|
| ||
| Deposited Data | ||
|
| ||
| Original western blot images | This paper; Mendeley Data | http://dx.doi.org/10.17632/hph5nb2rjy.1 |
|
| ||
| Experimental Models: Cell Lines | ||
|
| ||
| 4T1 | ATCC | CRL-2539 |
| MDA-MB-468 | ATCC | HTB-132 |
| MDA-MB-231 | ATCC | HTB-26 |
| A549 | ATCC | CCL-185 |
| 293T | ATCC | CRL-3216 |
| HCT116 | ATCC | CCL-247 |
|
| ||
| Experimental Models: Organisms/Strains | ||
|
| ||
| Six-week-old female BALB/c | Envigo | N/A |
| Six-week-old female NMRI nu/nu | Taconic | N/A |
| BL10 (MMTV-PyMT/FVB × C57BL/10J) | Prof. Kent Hunter | Lifsted et al., 1998 |
| BL6 (MMTV-PyMT/FVB × C57BL/6J) | Prof. Kent Hunter | Lifsted et al., 1998 |
| CAST (MMTV-PyMT/FVB × CAST/EiJ) | Prof. Kent Hunter | Lifsted et al., 1998 |
| MOLF (MMTV-PyMT/FVB × MOLF/EiJ) | Prof. Kent Hunter | Lifsted et al., 1998 |
|
| ||
| Recombinant DNA | ||
|
| ||
| pLKO-shPHGDH #1 | Belgian co-ordinated collections of micro-organisms | TRCN0000041625 |
| pLKO-shPHGDH #2 | Belgian co-ordinated collections of micro-organisms | TRCN0000041627 |
| pLKO-shPHGDH #3 | Provided by Prof. Peter Carmeliet | Vandekeere et al., 2018 |
| pLHCX overexpressing PHGDH | Provided by Prof. Matthew Vander Heiden | N/A |
|
| ||
| Software and Algorithms | ||
|
| ||
| Prism v.7.0 | GraphPad | https://www.graphpad.com/ |
| Image Studio Lite v.5.2 | LI-COR Biosciences | https://www.licor.com/bio/image-studio-lite/ |
| MATLAB | N/A | https://www.mathworks.com/products/matlab.html |
| TCGA Database | Open source | https://xena.ucsc.edu/ |
| NIH Genomic Data Commons (GDC) | NIH | https://portal.gdc.cancer.gov/ |
| ZEN Blue software | Zeiss | https://www.zeiss.com/microscopy/int/products/microscope-software/zen-lite.html |
ACKNOWLEDGMENTS
We thank Peter Carmeliet (VIB-KU Leuven) for providing short hairpin RNA (shRNA) against Phgdh, David Nittner (VIB histology core facility) for performing H&E staining, and John Blenis (Cornell Medical) and Roberto Zoncu (Berkeley) for their advice on analyzing mTORC1 activity. We also thank the Leuven Viral Vector Core for the luciferase virus, Stefan Soenen (KU Leuven MOSAIC) for help and assistance with the bioluminescence imaging, Matt Vander Heiden (MIT) for providing the OE PHGDH plasmid, Juan Fernandez Garcia and Martin Orth for advice on statistical analysis, and Roberta Schmieder for the setup of the in vivo glucose infusion methodology in the lab. We thank Raze Therapeutics for providing us with the PHGDH inhibitor PH-755. We also acknowledge http://www.somersault1824.com for image elements used in the graphical abstract (Creative Commons license CC BY-NC-SA 4.0). G.R. has received consecutive PhD fellowships from Kom op tegen Kanker and FWO (1137117N and 1137119N). E.P. has received a PhD fellowship supported by Region of Tuscany FSE (Pegaso project). G.D. has received consecutive PhD fellowships from Kom op tegen Kanker and FWO. The laboratory of T.G.P.G. is supported by grants from German Cancer Aid (DKH-70112257), the Gert & Susanna Mayer Foundation, and the Barbara and Wilfried Mohr Foundation. M.R. has received consecutive postdoc fellowships from FWO and Stichting Tegen Kanker. S.-M.F. acknowledges funding from the European Research Council under ERC Consolidator Grant Agreement 771486-MetaRegulation, FWO research projects (G088318N), KU Leuven-Methusalem co-funding, and Fonds Baillet Latour.
Footnotes
DECLARATION OF INTERESTS
S.-M.F. has received funding from Bayer AG, Merck, and Black Belt Therapeutics; has consulted for Fund+; and serves on the advisory board of Molecular Cell. All other authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.molcel.2020.11.027.
REFERENCES
- Ben-Sahra I, and Manning BD (2017). mTORC1 signaling and the metabolic control of cell growth. Curr. Opin. Cell Biol. 45, 72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altea-Manzano P, Broekaert D, Duarte JAG, Ferná ndez-García J, Planque M, and Fendt S-M (2020). Analyzing the Metabolism of Metastases in Mice. Methods Mol. Biol. 2088, 93–118. [DOI] [PubMed] [Google Scholar]
- Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, and Manning BD (2016). mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351, 728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernfeld E, Menon D, Vaghela V, Zerin I, Faruque P, Frias MA, and Foster DA (2018). Phospholipase D-dependent mTOR complex 1 (mTORC1) activation by glutamine. J. Biol. Chem. 293, 16390–16401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, and Dyer WJ (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. [DOI] [PubMed] [Google Scholar]
- Broekaert D, and Fendt S-M (2019). Measuring In Vivo Tissue Metabolism Using 13C Glucose Infusions in Mice. In Metabolic Signaling: Methods and Protocols, Fendt S-M and Lunt SY, eds. (Springer; New York: ), pp. 67–82. [DOI] [PubMed] [Google Scholar]
- Buescher JM, Antoniewicz MR, Boros LG, Burgess SC, Brunengraber H, Clish CB, DeBerardinis RJ, Feron O, Frezza C, Ghesquiere B, et al. (2015). A roadmap for interpreting (13)C metabolite labeling patterns from cells. Curr. Opin. Biotechnol. 34, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargnello M, Tcherkezian J, and Roux PP (2015). The expanding role of mTOR in cancer cell growth and proliferation. Mutagenesis 30, 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaneton B, Hillmann P, Zheng L, Martin ACL, Maddocks ODK, Chokkathukalam A, Coyle JE, Jankevics A, Holding FP, Vousden KH, et al. (2012). Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature 491, 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen S, Lorendeau D, Schmieder R, Broekaert D, Metzger K, Veys K, Elia I, Buescher JM, Orth MF, Davidson SM, et al. (2016). Breast cancer-derived lung metastases show increased pyruvate carboxylase-dependent anaplerosis. Cell Rep. 17, 837–848. [DOI] [PubMed] [Google Scholar]
- Clasquin MF, Melamud E, and Rabinowitz JD (2012). LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr. Protoc. Bioinformatics 37, 14.11.1–14.11.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon KJ, and Sabatini DM (2019). Nutrient regulation of mTORC1 at a glance. J. Cell Sci. 132, jcs222570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl FF, Lewis CA, Fiske BP, and Vander Heiden MG (2019). Cellular redox state constrains serine synthesis and nucleotide production to impact cell proliferation. Nat. Metab. 1, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durán RV, Oppliger W, Robitaille AM, Heiserich L, Skendaj R, Gottlieb E, and Hall MN (2012). Glutaminolysis activates Rag-mTORC1 signaling. Mol. Cell 47, 349–358. [DOI] [PubMed] [Google Scholar]
- Durán RV, MacKenzie ED, Boulahbel H, Frezza C, Heiserich L, Tardito S, Bussolati O, Rocha S, Hall MN, and Gottlieb E (2013). HIF-independent role of prolyl hydroxylases in the cellular response to amino acids. Oncogene 32, 4549–4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia I, Schmieder R, Christen S, and Fendt S-M (2016). Organ-Specific Cancer Metabolism and Its Potential for Therapy. Handb. Exp. Pharmacol. 233, 321–353. [DOI] [PubMed] [Google Scholar]
- Elia I, Doglioni G, and Fendt S-M (2018). Metabolic Hallmarks of Metastasis Formation. Trends Cell Biol. 28, 673–684. [DOI] [PubMed] [Google Scholar]
- Elia I, Rossi M, Stegen S, Broekaert D, Doglioni G, van Gorsel M, Boon R, Escalona-Noguero C, Torrekens S, Verfaillie C, et al. (2019). Breast cancer cells rely on environmental pyruvate to shape the metastatic niche. Nature 568, 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez CA, Des Rosiers C, Previs SF, David F, and Brunengraber H (1996). Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J. Mass Spectrom. 31, 255–262. [DOI] [PubMed] [Google Scholar]
- Fernández-García J, Altea-Manzano P, Pranzini E, and Fendt S-M (2020). Stable Isotopes for Tracing Mammalian-Cell Metabolism In Vivo. Trends Biochem. Sci. 45, 185–201. [DOI] [PubMed] [Google Scholar]
- Harper KL, Sosa MS, Entenberg D, Hosseini H, Cheung JF, Nobre R, Avivar-Valderas A, Nagi C, Girnius N, Davis RJ, et al. (2016). Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature 540, 588–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini H, Obradović MMS, Hoffmann M, Harper KL, Sosa MS, Werner-Klein M, Nanduri LK, Werno C, Ehrl C, Maneck M, et al. (2016). Early dissemination seeds metastasis in breast cancer. Nature 540, 552–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HM, Jung WH, and Koo JS (2014). Site-specific metabolic phenotypes in metastatic breast cancer. J. Transl. Med. 12, 354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert AW, Pattabiraman DR, and Weinberg RA (2017). Emerging Biological Principles of Metastasis. Cell 168, 670–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprivier G, and Rotblat B (2020). How does mTOR sense glucose starvation? AMPK is the usual suspect. Cell Death Discov. 6, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifsted T, Le Voyer T, Williams M, Muller W, Klein-Szanto A, Buetow KH, and Hunter KW (1998). Identification of inbred mouse strains harboring genetic modifiers of mammary tumor age of onset and metastatic progression. Int. J. Cancer 77, 640–644. [DOI] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. (2011). Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat. Genet. 43, 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorendeau D, Christen S, Rinaldi G, and Fendt S-M (2015). Metabolic control of signalling pathways and metabolic auto-regulation. Biol. Cell 107, 251–272. [DOI] [PubMed] [Google Scholar]
- Lorendeau D, Rinaldi G, Boon R, Spincemaille P, Metzger K, Jäger C, Christen S, Dong X, Kuenen S, Voordeckers K, et al. (2017). Dual loss of succinate dehydrogenase (SDH) and complex I activity is necessary to recapitulate the metabolic phenotype of SDH mutant tumors. Metab. Eng. 43 (Pt B), 187–197. [DOI] [PubMed] [Google Scholar]
- Mossmann D, Park S, and Hall MN (2018). mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer 18, 744–757. [DOI] [PubMed] [Google Scholar]
- Nagahashi M, Yamada A, Miyazaki H, Allegood JC, Tsuchida J, Aoyagi T, Huang WC, Terracina KP, Adams BJ, Rashid OM, et al. (2016). Interstitial Fluid Sphingosine-1-Phosphate in Murine Mammary Gland and Cancer and Human Breast Tissue and Cancer Determined by Novel Methods. J. Mammary Gland Biol. Neoplasia 21, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo B, Kim E, Osorio-Vasquez V, Doll S, Bustraan S, Liang RJ, Luengo A, Davidson SM, Ali A, Ferraro GB, et al. (2020). Limited Environmental Serine and Glycine Confer Brain Metastasis Sensitivity to PHGDH Inhibition. Cancer Discov. 10, 1352–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonnenmacher Y, Palorini R, d’Herouël AF, Krämer L, Neumann-Schaal M, Chiaradonna F, Skupin A, Wegner A, and Hiller K (2017). Analysis of mitochondrial metabolism in situ: Combining stable isotope labeling with selective permeabilization. Metab. Eng. 43 (Pt B), 147–155. [DOI] [PubMed] [Google Scholar]
- Nonnenmacher Y, Palorini R, and Hiller K (2019). Determining Compartment-Specific Metabolic Fluxes. Methods Mol. Biol. 1862, 137–149. [DOI] [PubMed] [Google Scholar]
- Orozco JM, Krawczyk PA, Scaria SM, Cangelosi AL, Chan SH, Kunchok T, Lewis CA, and Sabatini DM (2020). Dihydroxyacetone phosphate signals glucose availability to mTORC1. Nat. Metab. 2, 893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oslund RC, Su X, Haugbro M, Kee J-M, Esposito M, David Y, Wang B, Ge E, Perlman DH, Kang Y, et al. (2017). Bisphosphoglycerate mutase controls serine pathway flux via 3-phosphoglycerate. Nat. Chem. Biol. 13, 1081–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollari S, Käkön en S-M, Edgren H, Wolf M, Kohonen P, Sara H, Guise T, Nees M, and Kallioniemi O (2011). Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res. Treat. 125, 421–430. [DOI] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo H-K, Jang HG, Jha AK, et al. (2011). Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476, 346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta D, Park Y, Andrabi SA, Shelton LM, Gilkes DM, and Semenza GL (2016). PHGDH Expression Is Required for Mitochondrial Redox Homeostasis, Breast Cancer Stem Cell Maintenance, and Lung Metastasis. Cancer Res. 76, 4430–4442. [DOI] [PubMed] [Google Scholar]
- Schild T, Low V, Blenis J, and Gomes AP (2018). Unique Metabolic Adaptations Dictate Distal Organ-Specific Metastatic Colonization. Cancer Cell 33, 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, and Pierre P (2009). SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277. [DOI] [PubMed] [Google Scholar]
- Sever R, and Brugge JS (2015). Signal transduction in cancer. Cold Spring Harb. Perspect. Med. 5, a006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MR, Mattaini KR, Dennstedt EA, Nguyen AA, Sivanand S, Reilly MF, Meeth K, Muir A, Darnell AM, Bosenberg MW, et al. (2019). Increased Serine Synthesis Provides an Advantage for Tumors Arising in Tissues Where Serine Levels Are Limiting. Cell Metab. 29, 1410–1421.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gorsel M, Elia I, and Fendt S-M (2019). 13C Tracer Analysis and Metabolomics in 3D Cultured Cancer Cells. In Metabolic Signaling: Methods and Protocols, Fendt S-M and Lunt SY, eds. (Springer; New York: ), pp. 53–66. [DOI] [PubMed] [Google Scholar]
- Vandekeere S, Dubois C, Kalucka J, Sullivan MR, García-Caballero M, Goveia J, Chen R, Diehl FF, Bar-Lev L, Souffreau J, et al. (2018). Serine Synthesis via PHGDH Is Essential for Heme Production in Endothelial Cells. Cell Metab. 28, 573–587. [DOI] [PubMed] [Google Scholar]
- Vendramin R, Verheyden Y, Ishikawa H, Goedert L, Nicolas E, Saraf K, Armaos A, Delli Ponti R, Izumikawa K, Mestdagh P, et al. (2018). SAMMSON fosters cancer cell fitness by concertedly enhancing mitochondrial and cytosolic translation. Nat. Struct. Mol. Biol. 25, 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivian J, Rao AA, Nothaft FA, Ketchum C, Armstrong J, Novak A, Pfeil J, Narkizian J, Deran AD, Musselman-Brown A, et al. (2017). Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 35, 314–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M-H, Sun R, Zhou X-M, Zhang M-Y, Lu J-B, Yang Y, Zeng L-S, Yang X-Z, Shi L, Xiao R-W, et al. (2018). Epithelial cell adhesion molecule overexpression regulates epithelial-mesenchymal transition, stemness and metastasis of nasopharyngeal carcinoma cells via the PTEN/AKT/mTOR pathway. Cell Death Dis. 9, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiig H, Aukland K, and Tenstad O (2003). Isolation of interstitial fluid from rat mammary tumors by a centrifugation method. Am. J. Physiol. Heart Circ. Physiol. 284, H416–H424. [DOI] [PubMed] [Google Scholar]
- Yang H, Guan L, Li S, Jiang Y, Xiong N, Li L, Wu C, Zeng H, and Liu Y (2016). Mechanosensitive caveolin-1 activation-induced PI3K/Akt/mTOR signaling pathway promotes breast cancer motility, invadopodia formation and metastasis in vivo. Oncotarget 7, 16227–16247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Mancuso A, Tong X, Ward PS, Fan J, Rabinowitz JD, and Thompson CB (2012). Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc. Natl. Acad. Sci. USA 109, 6904–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD, Walther JL, Antoniewicz MR, Yoo H, and Stephanopoulos G (2008). An elementary metabolite unit (EMU) based method of isotopically nonstationary flux analysis. Biotechnol. Bioeng. 99, 686–699. [DOI] [PubMed] [Google Scholar]
- Yu T, Li J, Yan M, Liu L, Lin H, Zhao F, Sun L, Zhang Y, Cui Y, Zhang F, et al. (2015). MicroRNA-193a-3p and −5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene 34, 413–423. [DOI] [PubMed] [Google Scholar]
- Yu T, Zhao Y, Hu Z, Li J, Chu D, Zhang J, Li Z, Chen B, Zhang X, Pan H, et al. (2017). MetaLnc9 Facilitates Lung Cancer Metastasis via a PGK1-Activated AKT/mTOR Pathway. Cancer Res. 77, 5782–5794. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kwok-Shing Ng P, Kucherlapati M, Chen F, Liu Y, Tsang YH, de Velasco G, Jeong KJ, Akbani R, Hadjipanayis A, et al. (2017). A Pan-Cancer Proteogenomic Atlas of PI3K/AKT/mTOR Pathway Alterations. Cancer Cell 31, 820–832.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all analyzed data. The mass spectrometry files supporting the current study have not been deposited in a public repository due to the lack of a commonly accepted and feasible metabolomics file repository but are available upon reasonable request from the lead contact. Original western blot images are on Mendeley at http://dx.doi.org/10.17632/hph5nb2rjy.1.