Abstract
An automated method of ribosomal intergenic spacer analysis (ARISA) was developed for the rapid estimation of microbial diversity and community composition in freshwater environments. Following isolation of total community DNA, PCR amplification of the 16S-23S intergenic spacer region in the rRNA operon was performed with a fluorescence-labeled forward primer. ARISA-PCR fragments ranging in size from 400 to 1,200 bp were next discriminated and measured by using an automated electrophoresis system. Database information on the 16S-23S intergenic spacer was also examined, to understand the potential biases in diversity estimates provided by ARISA. In the analysis of three natural freshwater bacterial communities, ARISA was rapid and sensitive and provided highly reproducible community-specific profiles at all levels of replication tested. The ARISA profiles of the freshwater communities were quantitatively compared in terms of both their relative diversity and similarity level. The three communities had distinctly different profiles but were similar in their total number of fragments (range, 34 to 41). In addition, the pattern of major amplification products in representative profiles was not significantly altered when the PCR cycle number was reduced from 30 to 15, but the number of minor products (near the limit of detection) was sensitive to changes in cycling parameters. Overall, the results suggest that ARISA is a rapid and effective community analysis technique that can be used in conjunction with more accurate but labor-intensive methods (e.g., 16S rRNA gene cloning and sequencing) when fine-scale spatial and temporal resolution is needed.
rRNA intergenic spacer analysis (RISA) is a method of microbial community analysis which provides estimates of microbial diversity and community composition without the bias imposed by culture-based approaches or the labor involved with small-subunit rRNA gene clone library construction. RISA was used originally to contrast diversity in soils (6) and more recently to examine microbial diversity in the rhizosphere and marine environments (1, 22). The method involves PCR amplification from total bacterial community DNA of the intergenic region between the small (16S) and large (23S) subunit rRNA genes in the rRNA operon, with oligonucleotide primers targeted to conserved regions in the 16S and 23S genes. The 16S-23S intergenic region, which may encode tRNAs depending on the bacterial species, displays significant heterogeneity in both length and nucleotide sequence. Both types of variation have been extensively used to distinguish bacterial strains and closely related species (4, 11, 14, 20, 23). In RISA, the length heterogeneity of the intergenic spacer is exploited. The PCR product (a mixture of fragments contributed by community members) is electrophoresed in a polyacrylamide gel, and the DNA is visualized by silver staining. The result is a complex banding pattern that provides a community-specific profile, with each DNA band corresponding to at least one organism in the original assemblage.
Although RISA provided relatively rapid estimates of microbial community composition, in practice polyacrylamide gel electrophoresis tended to be time-consuming and cumbersome. In addition, because silver staining is a relatively insensitive method of DNA detection, large amounts of PCR product were necessary for analysis and resolution tended to be low. The limitations of the existing methodology led us to develop an improved version of RISA, which we refer to as automated RISA (ARISA). This method is similar to the recently published terminal restriction fragment length polymorphism and length heterogeneity analysis by PCR community analysis techniques (13, 24). In the automated approach, the initial steps of DNA extraction and PCR amplification are the same as in RISA, except that PCR is conducted with a fluorescence-tagged oligonucleotide primer. The electrophoretic step is subsequently performed with an automated system, which provides laser detection of fluorescent DNA fragments. ARISA-PCR may generate DNA fragments up to 1,400 bp in length (6). Discrimination of these larger size fragments represented a new application for the capillary electrophoresis system employed.
In this work, the sensitivity and reproducibility of ARISA were demonstrated through application of the technique to natural freshwater bacterial communities from three sites in northern Wisconsin. In addition, information on the length heterogeneity of the 16S-23S intergenic spacer among cultured organisms available through the GenBank database was evaluated, so as to gain a better understanding of the potential biases inherent in the estimates of bacterial diversity that ARISA provides. Overall, the results suggest that ARISA is a rapid and effective method for assessing microbial community diversity and composition that can be especially useful at the fine spatial and temporal scales necessary in ecological studies.
MATERIALS AND METHODS
Study sites and sample collection.
Samples were collected in June, August, and September 1998 from Crystal Bog Lake and Sparkling Lake in the Northern Highland Lake District in northern Wisconsin (Oneida County) and Lake Mendota, adjacent to the University of Wisconsin—Madison campus in southern Wisconsin (Dane County). All three lakes are part of the North Temperate Lakes Long-Term Ecological Research Site. Crystal Bog Lake is a shallow (maximum depth, 2.5 m), dystrophic lake; Sparkling Lake is an oligotrophic lake; and Lake Mendota is a deep, eutrophic lake. Sparkling Lake and Lake Mendota were stratified at the time of sampling. In Crystal Bog and Sparkling Lakes, whole water samples were collected above the point of maximum depth by using either a Van Dorn or integrated water column sampler. In Lake Mendota, surface grab samples were taken at the end of a pier in front of the Center for Limnology on the University of Wisconsin campus. Whole water samples were screened through a 10-μm-pore-size nylon mesh (Spectrum) in the field, transported to the laboratory on ice, and concentrated in aliquots of 400 ml to 1 liter onto 0.2-μm-pore-size filters (Supor-200; Gelman). The filtration apparatus was rinsed with filter-sterilized, deionized water and sample water in between samples. Filters were placed in cryovials, frozen immediately in liquid nitrogen, and stored at −80°C until further processing.
DNA extraction and purification.
DNA was extracted and purified by using a modification of the FastPrep bead beater method (Bio 101), following Borneman et al. (5). In the homogenization step, one half of each filter was placed into separate extraction tubes after first cutting the filter half into several pieces. The mixture was shaken with a single large bead on the FastPrep instrument (Bio 101) for 30 s at speed 5. By comparing the extracted DNA to known quantities of standard DNA in an agarose gel, this homogenization procedure was found to result in the highest yields of DNA without excessive shearing. In the final steps of purification, DNA extracted from the two filter halves for each sample was pooled.
ARISA.
ARISA-PCR was performed following the method of Borneman and Triplett (6) with modifications. Reaction mixtures contained 1× PCR buffer (Promega), 2.5 mM MgCl2, 500 μg of bovine serum albumin per ml, a 200 μM concentration of each dNTP, a 400 μM concentration of each primer, 2.5 U of Taq polymerase, and approximately 350 ng of template DNA in a final volume of 50 μl. The primers were 1406f, 5′ TGYACACACCGCCCGT 3′ (universal, 16S rRNA gene), and 23Sr, 5′ GGGTTBCCCCATTCRG 3′ (bacterial-specific, 23S rRNA gene), and primer 1406f was 5′ end labeled with the phosphoramidite dye 5-FAM. Reaction mixtures were held at 94°C for 2 min, followed by 30 cycles of amplification at 94°C for 15 s, 55°C for 15 s, and 72°C for 45 s and a final extension of 72°C for 2 min. To investigate the effect of the PCR cycle number on ARISA profiles, PCR was also performed with 15, 20, and 25 rounds of amplification by using samples from Crystal Bog and Sparkling Lakes. Reaction volumes were 10, 20, and 50 μl, with reagent concentrations as described above, except for the template DNA, which was increased by 1.5 to 3 times the usual amount.
The concentration of labeled PCR product was estimated by comparing it to known quantities of standard DNA, as described above. Based on these estimates, a standardized amount (1 to 2 μl) of PCR product, along with 1 μl of an internal size standard, was added to 20 μl of deionized formamide, and the mixture was denatured at 95°C for 5 min, followed by 2 min on ice. Sample fragments were then discriminated by using the ABI 310 genetic analyzer (Perkin-Elmer), in which DNA is electrophoresed in a capillary tube filled with electrophoresis polymer rather than in a polyacrylamide gel. The samples were run under standard ABI 310 denaturing electrophoresis conditions for 1 h each, with the POP-4 polymer, and the data were analyzed by using the GeneScan 3.1 software program (Perkin-Elmer). The program output is a series of peaks (an electropherogram), the sizes of which are estimated by comparison to fragments in the internal size standard. The performances of two Rhodamine X-labeled internal size standards, the GeneScan-2500 size standard (Perkin-Elmer) and a custom 200- to 2,000-bp standard (Bioventures, Inc.), were compared for the sizing of large fragments (up to 1,200 bp). In addition, the GeneScan software calculates the fluorescence contained in each peak, which is proportional to the quantity of DNA in the fragment. The relative amount of each fragment in the PCR product was estimated as the ratio between the fluorescence (peak area) of the fragment of interest and the total fluorescence of all fragments in the profile.
ARISA profiles for samples collected in June and September 1998 from Crystal Bog Lake were compared by using Sorenson’s index, Cs = 2j/(a + b), a pairwise similarity coefficient (15), where j is the number of fragments common to both samples and a and b are the total number of fragments in samples A and B, respectively. A Cs value of 0 indicates that the two samples are completely different, whereas a Cs value of 1 indicates that they are identical.
Cloning and sequencing of ARISA-PCR products.
An unlabeled ARISA-PCR product from an integrated water column sample collected in the Sparkling Lake hypolimnion (SH) was chosen for cloning and sequencing because of the complexity of its profile. Products from two replicate PCRs were pooled and purified by using the Wizard DNA purification kit (Promega). Purified DNA was then cloned into vector pSTBlue-1 by using the Novagen Perfectly Blunt cloning kit and the clones screened for α-complementation with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and IPTG (isopropyl-β-d-thiogalactopyranoside). Plasmid DNA was isolated from positive colonies by using the Qiaprep Spin Miniprep kit (Qiagen) and was digested with EcoRI to verify the presence of inserts.
Plasmid DNA was sequenced with the ABI PRISM Big Dye Terminator Cycle sequencing kit and 20 to 30 pmol of sequencing primers T7 and Sp6 and/or PCR primer 1406f by using standard cycle sequencing parameters. Extra dye terminators were removed from the reactions with AutoSeq Sephadex G-50 columns (Amersham Pharmacia), and the reactions were electrophoresed on an ABI 377 sequencer (Perkin-Elmer). Sequences were edited and aligned in Sequencer 3.1 (Gene Codes) and submitted to BLAST (2) for an initial phylogenetic placement.
Database examination.
The GenBank database (National Center for Biotechnology Information) was searched for information on the length heterogeneity of the 16S-23S rRNA intergenic region among described microbial taxa. The lengths of 307 16S-23S intergenic spacer sequences, which were accessed by searching with the Entrez browser, were determined. Included in the analysis were data for species of both Bacteria and Archaea, for multiple strains within single species, and for multiple rRNA operons within the genomes of the same organisms. When multiple sequences were recorded for the same species or strain, if the sequences were of different length or were clearly labeled as different operon versions (e.g., rrnA and rrnB) but were of the same length, they were included in the analysis. However, multiple sequences reported for the same strain were excluded if they did not differ in length and were labeled only as different clones.
Nucleotide sequence accession numbers.
The nucleotide sequences of the cloned ARISA-PCR fragments that were sequenced in both directions and whose lengths are reported in Table 1 were deposited in the GenBank database and given accession no. AF164144 to AF164150.
TABLE 1.
ARISA-PCR clones obtained from the SHa
| Clone | Closest database match (cultured taxa)b | % Similarity | Putative phylum | Product length (bp) |
|---|---|---|---|---|
| SH1c | Shewanella alga (X81622) | 97 | γ-Proteobacteria | 657 |
| SH3c | Azoarcus evansii (X77679) | 94 | β-Proteobacteria | |
| SH4c | Sphaerotilus sp. (AF072915) | 97 | β-Proteobacteria | |
| SH5c | Mycobacterium bovis (M20940) | 93 | Gram-positive | 581 |
| SH6 | Trichodesmium sp. (X70767) | 93 | Cyanobacteria | |
| SH7 | Ferrimonas balearica (X93021) | 96 | γ-Proteobacteria | |
| SH8 | Mycobacterium kansasii (L42263) | 95 | Gram-positive | |
| SH9cd | Env. clone, activated sludge (Z94005) | 86 | 406 | |
| SH11c | Mycobacterium smegmatis (Y08453) | 91 | Gram-positive | 548 |
| SH12c | Sphaerotilus sp. (AF072914) | 98 | β-Proteobacteria | |
| SH13 | Bordetella bronchiseptica (X57026) | 97 | β-Proteobacteria | |
| SH14 | Mycobacterium bovis (M20940) | 97 | Gram-positive | |
| SH15 | Mycobacterium gordonae (L42260) | 96 | Gram-positive | |
| SH16 | Trichodesmium sp. (X70767) | 93 | Cyanobacteria | |
| SH17c | Mycobacterium bovis (M20940) | 92 | Gram-positive | 554 |
| SH18c | Pelobacter propionicus (X70954) | 96 | δ-Protobacteria | 745 |
| SH19 | Chlorobium vibrioforme (M62791) | 97 | Green sulfur | |
| SH20c | Chlorobium vibrioforme (M62791) | 91 | Green sulfur | 720 |
Tentative phylogenetic placement and percent similarity values were determined by using BLAST and are based on 99 to 138 bp of the 16S rRNA gene sequence for each clone.
Accession numbers for closest database matches are given in parentheses. Env., environmental.
Sequenced from both ends of the PCR product.
This clone did not show high sequence similarity with any sequences from cultured organisms available through the database.
RESULTS
Database examination.
We examined 307 16S-23S intergenic spacer sequences from the GenBank database contributed by approximately 60 genera and 165 species of prokaryotes belonging to the Archaea domain and 8 major lineages of the Bacteria domain. However, the majority of currently available sequences are from taxa belonging to either the gram-positive phyla or the γ-proteobacteria. Sequences from these two groups comprised roughly 60 and 20%, respectively, of the total number examined. The reason for the dominance of these groups in the current database is likely due to the large number of medically important organisms within these lineages. Because of the utility of intergenic spacer heterogeneity for distinguishing closely related strains and species, analysis of this region is increasingly used to identify clinical isolates which are often difficult to distinguish phenotypically (9). Thus, because we wanted to include as many sequences as possible in our examination of the database, overrepresentation of these taxa in our analyses was unavoidable.
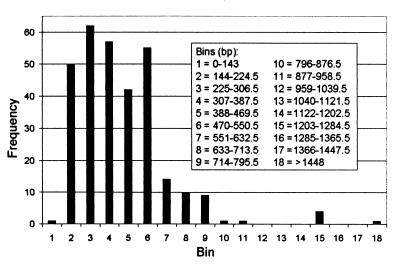
Within the compiled data set from GenBank, the intergenic region ranged in length from 143 to 1,529 bp, with 85 to 90% of the spacer lengths falling within 150 to 600 bp (Fig. 1). This particular size distribution (skewed toward smaller sizes) might in part be explained by the dominance of gram-positive sequences in the data set. Many gram-positive organisms examined to date have no tRNAs in the spacer region (9) and thus might be expected to have shorter spacer lengths on average. In the compiled data set, the mean spacer size was 533 bp (standard deviation [SD] = 233) and 327 bp (SD = 111) for the gram-negative (n = 106) and gram-positive (n = 191) organisms, respectively, and these means were significantly different (t test, P < 0.001). In addition, only 12% of the spacer lengths above 500 bp (n = 66) were contributed by gram-positive species.
FIG. 1.
Frequency histogram of 307 16S-23S intergenic spacer lengths (in base pairs) contributed by approximately 60 genera and 165 species of prokaryotes. The data were compiled from the GenBank database.
The range in intergenic spacer length of 143 to 1,529 bp corresponds to an ARISA-PCR product range of approximately 400 to 1,775 bases, because roughly 125 to 140 bp of both the 16S and 23S genes are amplified in addition to the spacer region with our PCR primers. The lower end of this range corresponded well with the ARISA profiles for natural systems; generally, only a very small number of fragments shorter than 390 bp were observed (3 to 5 per profile; data not shown). These were considered to be artifacts and were not analyzed further. At the higher end of the range, ARISA-PCR products above 1,200 bp in size were not observed for the natural systems investigated in this study. The largest fragments in the ARISA profiles for the three freshwater sites, Crystal Bog Lake, Lake Mendota, and Sparkling Lake, were approximately 1,040, 1,140 and 1,150 bp, respectively, although fragments as large as 1,400 bp have been detected with RISA in soils (6, 22).
ARISA may underestimate diversity because unrelated microorganisms may possess spacer regions of identical length and thus be represented in the ARISA profile by a single peak. The database information was used to assess how frequently this might be expected to occur. Among the 307 spacer sequences examined, there were 200 size classes, and 59 spacer lengths among the 200 (29.5%) were shared by more than one organism. More specifically, there were 20 instances where spacer lengths were identical among 2 or more strains within a species, with up to 10 strains having the same spacer size. Other authors have shown that strains within species often have identical spacer lengths (4, 7, 11, 14). In 16 instances, spacer lengths were identical among two or more species within the same genus, and in 40 cases (20% of the size classes), different genera shared the same spacer size. Genera with intergenic regions of identical length were in general not closely related, often belonging to different phyla. The number of organisms (other than related strains) possessing identical spacer sizes of any particular length never exceeded four, and for the spacer lengths shared among different genera, most (90%) were common to only two genera for any particular size.
In bacterial genomes, the rRNA operon may be present in numbers varying from one to several copies, depending on the species (3, 8). These operon copies may exhibit length heterogeneity in the 16S-23S intergenic region so that in ARISA a single organism may contribute more than one peak to the community profile. Although 16S-23S intergenic spacer sequences are often only available for a subset of the total number of rRNA operon copies per genome (9), the database information was used to evaluate how often multiple operons within a single genome differed in the length of the spacer. In the compiled data set, there were 43 instances where multiple operon versions (range, 2 to 4) of the 16S-23S spacer were reported for the same organism, and in 40 of these, the spacer size differed among copies. The difference in spacer length between operons ranged from 2 to 301 bp, with a mean difference of 166 bp (SD = 89 bp).
Cloning and sequencing of the ARISA-PCR product.
Eighteen clones of an unlabeled ARISA-PCR product from a sample collected in the SH were sequenced, to confirm that the 16S-23S intergenic spacer was being amplified in the freshwater environmental samples. The SH sample was chosen because of its relatively high complexity; there were approximately 50 fragments in the ARISA profile from this site. All of the clones were sequenced from the 16S end of the PCR product, and 10 of the 18 were also sequenced from the 23S end. The partial 16S and/or 23S rRNA gene sequences (∼100 to 140 bp of each) were then submitted separately to BLAST. The names and accession numbers of cultured organisms that most closely matched each of the clones in 16S rRNA gene sequence (calculated by BLAST as percent similarity), as well as their tentative phylogenetic placements, are given in Table 1.
Based on the BLAST search, all 18 of the SH ARISA-PCR clones represented the 16S-23S intergenic spacer region, and for 7 of the 10 clones that were sequenced from both ends of the PCR product, the 16S- and 23S-end fragments were shown to overlap. For these latter clones, the length of the PCR product was determined, and none were of identical length (Table 1). In addition, none of the clones were identical in nucleotide sequence, although several were similar in 16S rRNA gene sequence, as reflected by their similar placements with BLAST (Table 1). For example, sequence types SH4 and SH12 were 96% similar over 138 bp of the 16S sequence, and clones SH5, SH14, and SH17 were 97% similar over 139 bp. However, in both of these cases, sequences in the intergenic region among the related clones diverged greatly, so as to be unalignable. BLAST further indicated that the clones grouped into several major lineages of Bacteria: the β-, δ-, and γ-proteobacteria, the cyanobacteria, the green sulfur bacteria, and the gram-positive bacteria. For the clones sequenced from both ends of the product, phylogenetic placement with BLAST was the same with either the 16S or 23S gene fragment.
Applications of ARISA to natural freshwater bacterial communities.
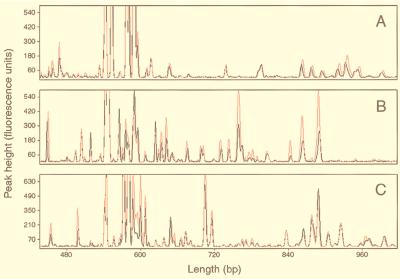
ARISA was used to assess the composition and diversity of three freshwater bacterial communities in eplimnetic samples from Crystal Bog Lake (CB) and Lake Mendota (LM) and in a metalimnetic sample from Sparkling Lake (SM), which provided a range of community complexity. For a single sample from each site, three to four PCR replicates were performed. The ARISA profiles (electropherograms) for two representative PCRs from each lake are shown in Fig. 2. In general, PCR reproducibility was very high, as evidenced by the fact that the electropherograms representing the duplicate PCRs for each site are almost entirely superimposed. The only major variation observed was for the CB site, in which two strong products were generated in one of the PCR replicates that were absent or nearly so from the other two (data not shown). Subsequently, a fourth replicate was performed that gave the profile in Fig. 2A.
FIG. 2.
Partial ARISA profiles of the bacterial communities in Crystal Bog Lake (A), Lake Mendota (B), and Sparkling Lake (C) during the summer of 1998. In each panel, the red and black electropherograms represent duplicate PCRs that were performed on a single sample from each site.
The reproducibility of ARISA was further assessed by quantifying the variability in size estimation for a series of fragments (three to five per profile) from CB, LM, and SM that represented a wide range in length. Triplicate PCR products were analyzed over a series of six to eight independent separations on the ABI 310 to give the results shown in Table 2. In general, sizing precision was very high, with standard deviations of <1 bp for fragments up to 800 bp and generally below 2 bp for fragments above this size. The largest SD observed, 5.21 bp, was for the longest fragment (1,147 bp) from the LM profile. However the coefficient of variation (CV) for this value was still only 0.45% (Table 2).
TABLE 2.
Variability in size estimation and the estimated relative abundance (percentage of total fluorescence) of selected fragments that represented a range in both parametersa
| Sample and peak | Fragment size (bp)
|
Fragment relative abundanceb
|
||
|---|---|---|---|---|
| Mean | SD/CV | Mean | SD/CV | |
| CB | ||||
| Peak 1 | 542.97 | 0.19/0.03 | 25.8 | 1.4/5.5 |
| Peak 2 | 589.08 | 0.28/0.05 | 22.8 | 0.7/3.2 |
| Peak 3 | 863.57 | 1.98/0.23 | 0.6 | 0.07/12.2 |
| LM | ||||
| Peak 1 | 546.35 | 0.52/0.10 | 22.8 | 1.4/6.1 |
| Peak 2 | 758.98 | 0.28/0.04 | 4.1 | 1.0/25.0 |
| Peak 3 | 889.69 | 1.61/0.18 | 3.4 | 1.1/31.1 |
| Peak 4 | 1,147.37 | 5.21/0.45 | 3.0 | 1.2/40.4 |
| SM | ||||
| Peak 1 | 544.32 | 0.17/0.03 | 6.1 | 0.7/11.8 |
| Peak 2 | 705.31 | 0.69/0.10 | 4.1 | 0.5/12.6 |
| Peak 3 | 879.17 | 1.67/0.19 | 2.1 | 0.4/18.4 |
| Peak 4 | 889.66 | 1.80/0.20 | 2.8 | 0.9/32.4 |
| Peak 5 | 1,136.43 | 0.75/0.07 | 3.3 | 0.8/25.3 |
The mean and SD values were determined from three PCR replicates for each sample analyzed over six to eight independent separations on the ABI 310 (total replicates = 9 to 10 for each).
Calculated as the ratio between the fluorescence of each fragment (peak area) and the total fluorescence of all fragments in the profile multiplied by 100.
Fragment size estimation with two internal size standards, the GS-2500 (Perkin-Elmer), originally designed to be used under nondenaturing conditions and a custom denaturing standard (Bioventures, Inc.), was compared by using the freshwater samples. Overall, the performances of the two standards were very similar. For ARISA fragments up to ∼1 kb, the difference in sizing of sample fragments between them was only 1 to 2 bp. Above 1 kb, sizing with the two standards increasingly diverged, with the GS-2500 standard consistently undersizing peaks, compared to the Bioventures standard. However, the difference was still not large (3 to 5 bp) for the size range considered (up to 1,150 bp). The largest sizing discrepancy noted was for a fragment with a known length of 1,200 bp. The Bioventures standard consistently sized this fragment at 1,193 bp, whereas the GS-2500 sized it at 1,180 bp (difference = 13 bp).
Variability in the relative abundance (percentage of total fluorescence) of individual ARISA fragments was also determined for the same series of peaks used to assess sizing variability (Table 2). Fragment relative abundance appeared to be most reproducible for those peaks that contributed the greatest amount to total fluorescence. For example, peak 1 in the CB and LM profiles made up 25.8 and 22.8% of the total fluorescence for each profile, with CVs of 5.5 and 6.1%, respectively. For the other fragments in the series, each of which contributed <10% to the total fluorescence of its respective profile and was usually larger in size than the more robust peaks, variability tended to be higher (CV range, ∼12 to 40%).
The relative diversity of the communities in each study site was determined by counting the peaks in Fig. 2 that were consistently present in two to three PCR replicates and were above a cutoff of 50 fluorescence units in peak height for at least one of the replicates. Although the use of this cutoff may have reduced the apparent diversity of the communities, it became difficult to distinguish sample fragments from background fluorescence with thresholds set below this value. Using the above criteria, the total number of fragments in the SM and LM profiles was very similar, at 45 and 41, respectively. The profile for the dystrophic CB site was somewhat different from the other two in that it was dominated by five extremely strong fragments that ranged in length from 543 to 589 bp and collectively contributed 87% to the total fluorescence of the profile (Fig. 2A). The profile for this site was also slightly less complex, with 34 fragments.
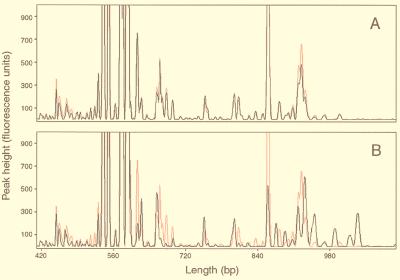
In addition to comparing the bacterial communities among the three lakes, ARISA was also used to assess bacterial diversity and community composition at finer scales within the CB site (Fig. 3). Figure 3A shows the electropherograms for two CB samples collected at 1 and 2 m depth in September 1998. The two profiles are almost identical, not only demonstrating the precision of the method at higher levels of replication but also suggesting that the bacterial community in this shallow (maximum depth, 2.5 m), well-mixed lake was vertically homogeneous on the sampling date. When one of the September samples was compared with the profile of the community for June, differences in the composition of the two communities were observed, although overall they were qualitatively very similar and readily comparable (Fig. 3B). The degree of similarity between the June and September communities was quantified by using Sorenson’s index (15), which has been used by others to assess the levels of similarity between denaturing gradient gel electrophoresis profiles of natural communities (12, 18). When all peaks greater than 50 fluorescence units in height were considered, the level of similarity between samples was 66%. However, when only major peaks were considered by raising the fluorescence cutoff to 100 U, the computed level of similarity between the communities increased to 74%.
FIG. 3.
Partial ARISA profiles of the bacterial communities in Crystal Bog Lake. (A) Red and black electropherograms representing samples collected from depths of 1 and 2 m, respectively, in June 1998. (B) Red and black profiles representing samples collected from a depth of 1 m in September and June 1998, respectively.
Influence of PCR cycle number on ARISA profiles.
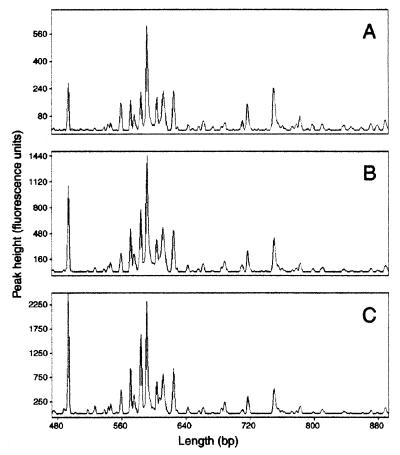
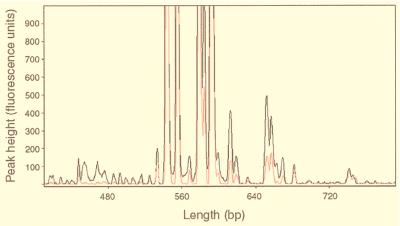
One of the greatest improvements of ARISA over the previous method is the increased sensitivity of DNA detection so that considerably less PCR product is needed for analysis. We took advantage of this feature to investigate how ARISA profiles might change as the PCR cycle number was reduced from 30, the number of amplification cycles normally used, to 15. In doing so, we were most interested in determining whether the pattern and number of detectable ARISA-PCR fragments would change with cycle number, thus influencing estimates of community composition and diversity. In this experiment, template DNA from CB, representing a relatively low complexity sample, and SH, which provided a more complex banding pattern, were used. In general, overall ARISA patterns were similar between PCRs performed with varying numbers of amplification cycles, especially for products that were present in higher concentrations. For example, the distribution of major peaks in the SH sample with 15, 20, and 25 rounds of amplification was highly reproducible (Fig. 4). As might be expected, however, the total number of fragments tended to be more sensitive to changes in cycle number because of the large number of fragments near the limit of detection (i.e., between 50 and 100 fluorescence units). For example, a series of eight and nine fragments ranging in size from ∼460 to 540 bp in the CB profile were consistently observed with 30 rounds of amplification but were undetectable at 25 (Fig. 5).
FIG. 4.
Results from an experiment in which the effect of the PCR cycle number on ARISA profiles was investigated by using a sample collected from the SH in September 1998. PCR was performed with 15, 20, and 25 rounds of amplification, giving the results displayed in panels A, B, and C, respectively.
FIG. 5.
Results from an experiment in which the effect of the PCR cycle number on ARISA profiles was investigated by using a sample collected from Crystal Bog Lake in September 1998. The red and black electropherograms represent PCRs that were performed with 25 and 30 cycles of amplification, respectively.
DISCUSSION
The need for a more rapid and reproducible method of microbial community analysis led us to develop an automated version of RISA, which previously relied on manual polyacrylamide gel electrophoresis and DNA detection by silver staining. ARISA has several advantages over the former method, thus increasing its utility as a technique for assessing the composition and diversity of naturally occurring bacterial communities. The new method was found to be very sensitive, thus requiring much less PCR product for analysis, and highly reproducible at all levels of replication tested. In addition, because of the technique’s ease, the effect of altered PCR conditions (e.g., amplification cycle number) on ARISA profiles could be rapidly assessed. In this study, a capillary electrophoresis system was employed to resolve large (up to 1,200 bp) ARISA-PCR size fragments from freshwater environmental samples, representing a new application for this automated system. Gel-based, automated DNA sequencing technology can also be used to discriminate large size fragments (16) and thus can also be applied to ARISA.
The results obtained with ARISA should be cautiously interpreted. As a molecular technique that relies upon total community DNA extraction and PCR amplification, ARISA is subject to the usual systematic biases introduced by these procedures (21, 25). In addition, specific biases associated with amplification of the 16S-23S intergenic region, such as possible preferential amplification of shorter templates and biases imposed by secondary structure or DNA flanking the template region (10), have not been well investigated. For these reasons, any conclusions regarding the relative abundance of bacterial populations represented in the ARISA profiles should be carefully made.
In addition to the above concerns, the relationship between the number of fragments in an ARISA profile and the absolute diversity of the community it represents requires further investigation. On the one hand, overlapping intergenic spacer size classes among unrelated organisms contributing to the profile may lead to underestimates of diversity. At the same time, interoperonic differences in spacer length frequently occur within the genomes of cultured organisms (9, 19), and this is probably true for uncultivated microbes in environmental samples as well. Thus, single organisms are likely to contribute more than one peak to an ARISA profile. The degree to which this occurs is currently unknown but perhaps may be inferred from data on spacer size variability among cultured isolates. For example, Jensen et al. (11) examined spacer length heterogeneity among 300 strains of bacteria belonging to eight genera and 28 species by using PCR amplification of the region. In addition to finding that most strains within species displayed the same pattern of amplification products, they observed that the majority of species (85%) exhibited between only one and three PCR products (i.e., one to three spacer lengths). In addition, in Escherichia coli K-12 there are only two types of intergenic spacer regions among the seven operons, and several clones of one of these regions obtained from K-12 and six other E. coli strains were recently shown to be identical in length (7). As more information of this type becomes available, we should gain a much clearer understanding of the biases associated with diversity estimates by ARISA. For now, assuming biases remain fairly constant between samples, we suggest that by counting the total number of reproducible fragments in a profile, ARISA can be used to estimate the relative diversity among sites, as we did for the three lakes in this study.
Despite the drawbacks outlined above, our results indicate that ARISA can be effectively used to estimate community composition in natural samples, especially for fine-scale comparative purposes, and to detect community shifts with experimental manipulation. Due to the high reproducibility and sensitivity of the method, we found that electropherograms could be readily compared among sites, and their similarity levels could be easily and precisely quantified by using the GeneScan analysis software. Indices such as Sorenson’s and Jaccard’s are increasingly used to quantitatively assess the similarity among communities (12, 13, 17, 18). These indices will obviously be influenced by the number of DNA fragments present in a community fingerprint, which in turn is dependent upon factors such as the detection level of the technique used, and characteristics of the particular gene fragment amplified. It would therefore be interesting to compare results from ARISA and other commonly used methods of community analysis, such as denaturing gradient gel electrophoresis and terminal restriction fragment length polymorphism analyses.
ARISA can be further developed by employing phylum-level (or below) oligonucleotide primers in order to investigate the dynamics of specific phylogenetic groups, as was done by Robleto et al. (22) for a specific group within the α-proteobacteria. Analysis of particular taxonomic groups rather than entire communities should result in much less complex fragment patterns, so that the biases discussed above could perhaps be more effectively investigated and reduced. In addition, several primers targeting different taxa could eventually be used on the same sample and the separate dynamics of each group could be evaluated simultaneously. Finally, although ARISA does not provide direct phylogenetic information on particular fragments in the profile, the precise sizing information afforded by the method could be used to identify the major bands of interest in a separate manual polyacrylamide gel. These fragments could then be further characterized through band excision and sequence analysis, as was performed by Robleto et al. (22). However, perhaps the most powerful and appropriate use of ARISA is as a rapid survey technique prior to, or in conjunction with, the application of more accurate but time-intensive methods for estimating community composition. Insight into ecological patterns gained from ARISA surveys at different spatial and temporal scales could allow for a more deliberate and effective application of techniques such as 16S rRNA gene cloning and sequencing and fluorescence in situ hybridization.
ACKNOWLEDGMENTS
This work was supported in part by an LTER grant from NSF (DEB 9632853) to the Center for Limnology at the University of Wisconsin—Madison. We thank the College of Agriculture and Life Sciences and the graduate school at the University of Wisconsin—Madison for their funding support of this project.
In addition, we thank Monica Zurawski of Perkin-Elmer for help with the 310 genetic analyzer and GeneScan software, Shannon Smith for performing the DNA sequencing, and Marisa Chelius for numerous helpful discussions and a review of the manuscript.
REFERENCES
- 1.Acinas S G, Anton J, Rodriguez-Valera F. Diversity of free-living and attached bacteria in offshore western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl Environ Microbiol. 1999;65:514–522. doi: 10.1128/aem.65.2.514-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amikam D, Glaser G, Razin S. Mycoplasmas (Mollicutes) have a low number of rRNA genes. J Bacteriol. 1984;158:376–378. doi: 10.1128/jb.158.1.376-378.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aubel D, Renaud F N R, Freney J. Genomic diversity of several Corynebacterium species identified by amplification of the 16S-23S rRNA gene spacer regions. Int J Syst Bacteriol. 1997;47:767–772. [Google Scholar]
- 5.Borneman J, Skroch P W, O’Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Neinhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borneman J, Triplett E W. Molecular microbial diversity in soils from Eastern Amazonia: evidence for unusual microorganisms and population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Martinez J, Martinez-Murcia A, Anton A I, Rodriguez-Valera F. Comparison of the small 16S to 23S intergenic spacer region (ISR) of the rRNA operons of some Escherichia coli strains of the ECOR collection and E. coli K-12. J Bacteriol. 1996;178:6374–6377. doi: 10.1128/jb.178.21.6374-6377.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garnier T, Canard B, Cole S T. Cloning, mapping, and molecular characterization of the rRNA operons of Clostridium perfringens. J Bacteriol. 1991;173:5431–5438. doi: 10.1128/jb.173.17.5431-5438.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurtler V, Stanisich V A. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 10.Hansen M C, Tolker-Nielsen T, Givskov M, Molin S. Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol Ecol. 1998;26:141–149. [Google Scholar]
- 11.Jensen M A, Webster J A, Straus N. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl Environ Microbiol. 1993;59:945–952. doi: 10.1128/aem.59.4.945-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindstrom E S. Bacterioplankton community composition in a boreal forest lake. FEMS Microbiol Ecol. 1998;27:163–174. [Google Scholar]
- 13.Liu W-T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maes N, Gheldre Y D, Ryck R D, Vaneechoutte M, Meugnier H, Etienne J, Struelens M J. Rapid and accurate identification of Staphylococcus species by tRNA intergenic spacer length polymorphism analysis. J Clin Microbiol. 1997;35:2477–2481. doi: 10.1128/jcm.35.10.2477-2481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maguerran A E. Ecological diversity and its measurement. Princeton, N.J: Princeton University Press; 1988. [Google Scholar]
- 16.McEvoy C R E, Seshadri R, Firgaira F A. Large DNA fragment sizing using native acrylamide gels on an automated DNA sequencer and GENESCANTM software. BioTechniques. 1998;25:464–470. [PubMed] [Google Scholar]
- 17.Murray A E, Hollibaugh J T, Orrego C. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denturing gradient gel electrophoresis of 16S rDNA fragments. Appl Environ Microbiol. 1996;62:2676–2680. doi: 10.1128/aem.62.7.2676-2680.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray A E, Preston C M, Massana R, Taylor L T, Wu B K, DeLong E F. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol. 1998;64:2585–2595. doi: 10.1128/aem.64.7.2585-2595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagpal M L, Fox K F, Fox A. Utility of 16S-23S rRNA spacer region methodology: how similar are interspace regions within a genome and between strains for closely related organisms? J Microbiol Methods. 1998;33:211–219. [Google Scholar]
- 20.Navarro E, Simonet P, Normand P, Bardin R. Characterization of natural populations of Nitrobacter spp. using PCR/RFLP analysis of the ribosomal intergenic spacer. Arch Microbiol. 1992;157:107–115. doi: 10.1007/BF00245277. [DOI] [PubMed] [Google Scholar]
- 21.Polz M F, Cavanaugh C M. Bias in template-to-product ratios in multitemplate PCR. Appl Environ Microbiol. 1998;64:3724–3730. doi: 10.1128/aem.64.10.3724-3730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robleto E A, Borneman J, Triplett E W. Effects of bacterial antibiotic production on rhizosphere microbial communities from a culture-independent perspective. Appl Environ Microbiol. 1998;64:5020–5022. doi: 10.1128/aem.64.12.5020-5022.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheinert P, Krausse R, Ullman U, Soller R, Krupp G. Molecular differentiation of bacteria by PCR amplification of the 16S-23S rRNA spacer. J Microbiol Methods. 1996;26:103–117. [Google Scholar]
- 24.Suzuki M, Rappe M S, Giovannoni S J. Kinetic bias estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl Environ Microbiol. 1998;64:4522–4529. doi: 10.1128/aem.64.11.4522-4529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wintzingerode F V, Gobel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]