Abstract
Egg-laying mammals (monotremes) are a sister clade of therians (placental mammals and marsupials) and a key clade to understand mammalian evolution. They are classified into platypus and echidna, which exhibit distinct ecological features such as habitats and diet. Chemosensory genes, which encode sensory receptors for taste and smell, are believed to adapt to the individual habitats and diet of each mammal. In this study, we focused on the molecular evolution of bitter taste receptors (TAS2Rs) in monotremes. The sense of bitter taste is important to detect potentially harmful substances. We comprehensively surveyed agonists of all TAS2Rs in platypus (Ornithorhynchus anatinus) and short-beaked echidna (Tachyglossus aculeatus) and compared their functions with orthologous TAS2Rs of marsupial and placental mammals (i.e., therians). As results, the agonist screening revealed that the deorphanized monotreme receptors were functionally diversified. Platypus TAS2Rs had broader receptive ranges of agonists than those of echidna TAS2Rs. While platypus consumes a variety of aquatic invertebrates, echidna mainly consumes subterranean social insects (ants and termites) as well as other invertebrates. This result indicates that receptive ranges of TAS2Rs could be associated with feeding habits in monotremes. Furthermore, some orthologous receptors in monotremes and therians responded to β-glucosides, which are feeding deterrents in plants and insects. These results suggest that the ability to detect β-glucosides and other substances might be shared and ancestral among mammals.
Keywords: TAS2R, β-glucoside, molecular evolution, platypus, echidna
Introduction
Mammals are classified into three clades: placental mammals (eutherians), pouched mammals (marsupials), and egg-laying mammals (monotremes). The monotreme lineage is the sister clade of therians (eutherians and marsupials). All mammals do breast-feed, but the egg-laying trait in monotremes is only shared with the reptiles and birds (sauropsids), the amniote sister of mammals. There are only five species of monotremes at present, platypus (Ornithorhynchus anatinus), short-beaked echidna (Tachyglossus aculeatus), and three -species of long-beaked echidna (Zaglossus spp.). They are only distributed in Australia, Papua New Guinea, and Indonesia. The estimated divergence time of platypus (the family Ornithorhynchidae) and echidna (the family Tachyglossidae) is 54.6 million years ago (Mya), ∼10 million years after the Cretaceous–Paleogene (K–Pg) mass extinction event (Zhou et al. 2021). Platypus and echidna exhibit distinct ecological features such as habitats and diet. Platypus is semi-aquatic feeding on aquatic invertebrates, while echidna is terrestrial with an invertebrate diet, especially ants and termites. To hunt prey, platypus mainly uses electroreception and mechanoreception (Pettigrew et al. 1998), whereas echidnas may rely on olfaction. Behavioral and neuroanatomical studies demonstrated the outstanding abilities of electroreception and mechanoreception (Ashwell 2013). Molecular and neuroanatomical studies also demonstrated the chemosensory differences between platypus and echidna (Zhou et al. 2021). For example, platypus has more vomeronasal type-1 receptor (V1Rs) genes with a larger accessory olfactory bulb than echidna. By contrast, echidna has more olfactory receptor (ORs) genes with a larger main olfactory bulb than platypus (Zhou et al. 2021). Eco-evolutionary comparison between these two egg-laying mammals is fundamental for understanding chemosensory and dietary adaptation as well as knowing the mammal origin of the chemical senses.
Another chemical sense to olfaction, gustation (taste perception) is an important sense for foraging behavior in monotremes because they masticate prey using horny pads (Griffiths 1965; Grant 2007). Of the five basic taste qualities (sweet, umami, bitter, salty, and sour), bitterness is perceived via bitter taste receptors (TAS2Rs) and associated with the detection of potentially harmful substances in diet. As well as ORs and V1Rs, TAS2Rs are G protein-coupled receptors (GPCRs) and construct a multi-gene family (Adler et al. 2000; Chandrashekar et al. 2000). The repertoire size of the TAS2R gene family in the genome is very diversified among mammals. Humans (Homo sapiens) and mice (Mus musculus) potentially have ∼26 and ∼40 intact TAS2R genes (Go et al. 2005; Hayakawa et al. 2014), whereas platypus and short-beaked echidna have only 7 and 3, respectively (Zhou et al. 2021). TAS2R gene repertoire and each receptor function are believed to be related to animals’ feeding habits and foraging behavior (Hayakawa et al. 2014; Li and Zhang 2014; Liu et al. 2016; Purba et al. 2020; Itoigawa et al. 2021). However, studies focusing on marsupials and monotremes are limited (Johnson et al. 2018; Zhou et al. 2021); therefore, exploring monotreme TAS2R functions will provide insight into not only their molecular adaptation in each monotreme species but may also inform diversification of bitter taste receptors in early mammalian evolution.
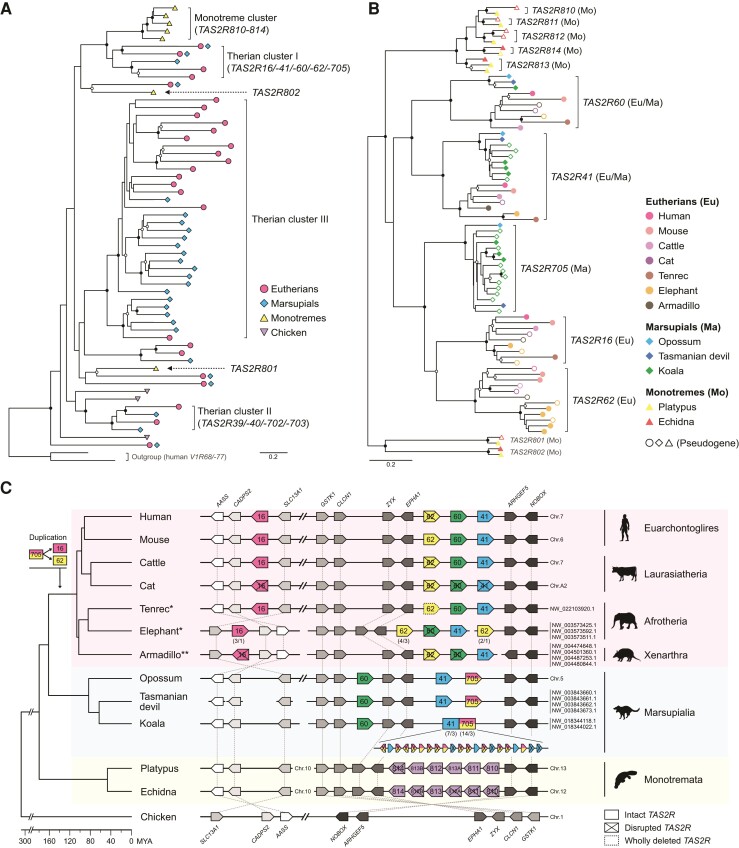
Monotreme TAS2R genes consist of TAS2R801, TAS2R802, and one phylogenetic cluster, “Monotreme cluster” (TAS2R810, TAS2R811, TAS2R812, TAS2R813, and TAS2R814) (fig. 1A). Monotreme cluster is the sister phylogenetic cluster of “Therian cluster I” (TAS2R16, TAS2R41, TAS2R60, TAS2R62, and TAS2R705) (Zhou et al. 2021) (fig. 1A and B). Of TAS2Rs in the Therian cluster I, TAS2R16 is known as a sensor for β-glucosides, which are feeding deterrents in plants and insects (Bufe et al. 2002). β-glucosides include cyanogenic glucosides (CNglcs), highly deleterious and widely distributed biomolecules for feeding deterrents (Beran et al. 2019). While TAS2R16 of primates, rodents, and bats have sensitivity to β-glucosides (Bufe et al. 2002; Imai et al. 2012; Jiao et al. 2018, 2021; Itoigawa et al. 2019; Yang et al. 2021), some primates which regularly feed on plants containing much CNglcs have TAS2R16 low sensitive to β-glucosides (Itoigawa et al. 2021). Therefore, β-glucoside detection would be important for survival in various mammals. Here we hypothesize that the Monotreme cluster (TAS2R810–814) is the orthologous gene group of Therian cluster I and responds to β-glucosides which are potentially harmful to mammals.
Fig. 1.
Phylogenetic and syntenic relationships between Monotreme and Therian clusters. (A) Phylogenetic positions of the Monotreme and Therian clusters in the mammalian TAS2R gene tree. Each tip indicates a single orthologous TAS2R gene group supported by ≥ 95% bootstrap values. Internal branches of each orthologous group are compressed for clarity (see supplementary fig. S1, Supplementary Material online for the full view of mammalian TAS2R gene tree). The nomenclature of TAS2R clusters followed Zhou et al. (2021). (B) The phylogenetic relationships among TAS2Rs of the Monotreme cluster and Therian cluster I. The nodes with ≥ 70% and ≥ 95% bootstrap values in (A) and (B) are marked with open and black circles, respectively. (C) Syntenic relationships between the Monotreme cluster and Therian cluster I. TAS2R and adjacent genes are drawn by the colored and grey-scaled boxes, respectively. Regions are not drawn to scale (see supplementary table S2, Supplementary Material online for the actual positions). The numbers below the boxes indicate “total copy number of genes/copy number of intact genes” for tandemly repeated TAS2R genes. The chromosome or scaffold numbers are shown at the right of synteny illustrations (The topmost number corresponds to the leftmost scaffold name). The nomenclature of mouse receptors is unified with that of human receptors for easily understanding (e.g., mouse Tas2r118 to TAS2R16). *All the scaffolds of Afrotherians are shown inverted for clarity. **The CADPS2 gene of the armadillo is located across two scaffolds (NW_004474648.1 and NW_004501360). The species phylogeny and divergence time are from TimeTree (http://www.timetree.org/; accessed on October 25, 2021) (Kumar et al. 2017). The animal silhouettes are from PhyloPic (http://phylopic.org/). The Silhouettes of platypus and Tasmanian devil drawn by S. Werning are reused under the CC BY 3.0 (https://creativecommons.org/licenses/by/3.0/).
To investigate this, we performed synteny analysis of mammalian genomes and the agonist screening of all intact TAS2Rs of platypus and short-beaked echidna using cell-based functional assays. We also screened agonists of the therian TAS2R16 orthologous groups (eutherian TAS2R16 and marsupial TAS2R705) to explore the functional conservation and the evolutionary history of β-glucoside sensing by TAS2Rs in mammals.
Results
Conserved Synteny of TAS2Rs in Monotreme and Therian Mammals
First, to clarify the orthologous relationship between the Monotreme cluster and Therian cluster I, we surveyed whole-genome assemblies of two monotremes, three marsupials, seven eutherians covering the four main clades (Euarchontoglires, Laurasiatheria, Afrotheria, and Xenarthra), and chicken as an outgroup. We found well-conserved chromosomal synteny surrounding the TAS2R genes in the Monotreme cluster and Therian cluster I across mammals. Monotreme TAS2R810-814 were located in tandem between EPHA1 and ARHGEF5 gene loci where almost all the TAS2Rs in Therian cluster I (TAS2R41, TAS2R60, and TAS2R62) are located (fig. 1C). Eutherian TAS2R16 had been translocated from this region after a duplication event of TAS2R705. In chicken, this region did not include any TAS2R genes. Taking the phylogenetic closeness between the Monotreme cluster and Therian cluster I into consideration (fig. 1A and B) (Zhou et al. 2021), this result supports that the Monotreme cluster is the orthologous group of Therian cluster I.
Deorphanization of Monotreme TAS2Rs
To characterize the response profiles of monotreme TAS2Rs and therian TAS2R16 orthologs, we performed cell-based calcium assays for seven platypus and three short-beaked echidna TAS2Rs, three eutherian TAS2R16, and five marsupial TAS2R705 using 24 commercially available bitter substances reported as agonists for human TAS2Rs (supplementary table S3, Supplementary Material online). We deorphanized seven of ten monotreme TAS2Rs, all eutherian TAS2R16, and three of five marsupial TAS2R705 receptors (table 1 and supplementary fig. S2, Supplementary Material online). The agonist screenings revealed that tandemly repeated TAS2Rs in monotremes (TAS2R810-814) showed divergent response profiles. TAS2R813 paralogs in platypus even showed distinct response profiles. This finding is an example of functional divergence among lineage-specific duplicated TAS2Rs as seen in mice, bats, and hummingbirds (Lossow et al. 2016; Jiao et al. 2018; Wang et al. 2019). Interestingly, TAS2R811 and TAS2R813A in platypus responded to twelve out of twenty-four and seven out of 24 tested compounds, respectively. The two platypus receptors can cover 14 of 24 (58.3%) tested substances, while platypus has only seven TAS2Rs. In contrast, Echidna does not have intact TAS2R811, and echidna TAS2R813 had fewer agonists than platypus ortholog TAS2R813A whose agonists overlapped. The total number of agonists identified in echidna is also less compared to platypus. These results suggest that platypus has relatively broadly tuned receptors and that the receptive ranges of bitter substances are also broader in platypus than in echidna.
Table 1.
Response Profiles of Monotreme TAS2Rs, Eutherian TAS2R16, and Marsupial TAS2R705.
| Oan | Tac | Hsa | Bta | Laf | Mdo | Dvi | Sha | Meu | Pci | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound/TAS2R | 801 | 802 | 810 | 811 | 812 | 813A | 813B | 802 | 813 | 814 | 16 | 16 | 16C | 705 | 705 | 705 | 705B | 705C |
| Acesulfame K | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Arbutin | — | — | — | ● | — | — | — | — | — | — | ● | ● | ● | — | ● | — | ● | ● |
| Caffeine | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Camphor | — | — | ● | ● | — | — | ● | — | — | — | — | — | — | — | — | — | — | — |
| Chloramphenicol | — | — | — | ● | — | ● | — | — | — | — | — | — | ● | — | — | — | — | — |
| Colchicine | — | — | — | — | ● | ● | ● | — | ● | — | — | — | — | — | — | — | — | — |
| Coumarin | — | — | — | — | — | — | — | — | — | — | — | ● | — | — | — | — | — | — |
| Denatonium benzoate | — | — | ● | ● | ● | ● | — | — | ● | — | — | ● | — | — | — | — | — | — |
| Diphenidol | — | — | — | — | ● | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Flufenamic acid | — | — | — | — | — | — | — | — | — | ● | — | — | — | — | — | — | — | — |
| Helicin | — | — | — | ● | — | ● | — | — | — | — | ● | ● | ● | — | ● | — | ● | ● |
| Linamarin | — | — | — | ● | — | — | — | — | — | — | ● | ● | ● | — | ● | — | ● | ● |
| Noscapine | — | — | — | ● | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Phenylthiocarbamide | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Picrotoxin | — | — | — | ● | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Quinine | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Saccharin | — | — | — | — | — | — | ● | — | — | — | — | — | — | — | — | — | — | — |
| Salicin | — | — | — | ● | — | ● | — | — | ● | — | ● | ● | ● | — | ● | — | ● | ● |
| Sinigrin | — | — | — | ● | — | ● | — | — | — | — | — | — | ● | — | — | — | — | — |
| Sodium benzoate | — | — | — | ● | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Sodium thiocyanate | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Strychnine | — | — | — | ● | ● | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Thiamine | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Yohimbine | — | — | — | — | — | ● | — | — | ● | — | — | — | — | — | — | — | — | — |
| Total | 0 | 0 | 2 | 12 | 4 | 7 | 3 | 0 | 4 | 1 | 4 | 6 | 6 | 0 | 4 | 0 | 4 | 4 |
Identified agonists are presented as dots in each TAS2R column. Agonists are detected by the statistical comparisons between fluorescence values (ΔF/F) of TAS2R-expressing and mock-transfected cells (n = 3–5) using Dunnett’s test (p < 0.01). β-glucoside analogs are presented in boldface. The number of compounds that activated each TAS2R is presented in the bottom row. Species abbreviations are as follows; Oan, platypus; Tac, echidna; Hsa, human; Bta, cattle; Laf, elephant; Mdo, opossum; Dvi, quoll; Sha, Tasmanian devil; Meu, wallaby; Pci, koala.
Focusing on the responses to β-glucosides, TAS2R811 and TAS2R813A in platypus and TAS2R813 in echidna responded to several β-glucosides. These receptors did not exclusively respond to them but various types of substances. On the other hand, eutherian TAS2R16 and marsupial TAS2R705 were highly specific for β-glucosides, while cattle and elephant TAS2R16 responded to a few non-β-glucoside substances (table 1). These results indicate that all the three mammalian lineages can recognize β-glucosides and that functional features of β-glucoside sensitive TAS2Rs are divided between monotremes and therians.
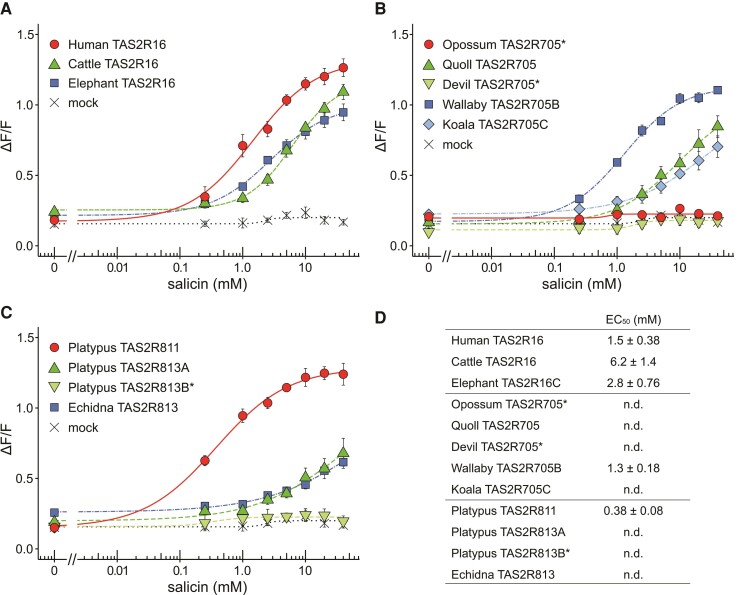
Next, to evaluate the inter-species or inter-paralog differences in β-glucoside sensitivity, dose-response curves were obtained for a subset of TAS2R receptors using salicin, a plant-derived and well-studied β-glucoside (Bufe et al. 2002). Salicin dose-dependently activated almost all TAS2R16, TAS2R705, TAS2R811, and TAS2R813 orthologs except for opossum and Tasmanian-devil TAS2R705, and platypus TAS2R813B but the sensitivities were varied among receptors. Eutherian TAS2R16 exhibited salicin sensitivity like primate TAS2R16 previously investigated (EC50 = 0.48–7.5 mM) (Imai et al. 2012) (fig. 2A and D). In marsupial TAS2R705, wallabies exhibited salicin sensitivity like human TAS2R16, but the other marsupial species showed lower than the wallaby receptor (fig. 2B and D). In monotremes, platypus TAS2R813A exhibited responses to salicin like the TAS2R813 ortholog in echidna (fig. 2C). TAS2R811 in platypus exhibited markedly high sensitivity to salicin compared with TAS2R813 orthologs, suggesting that platypus would be more sensitive to salicin than echidna.
Fig. 2.
Dose-dependent responses to salicin in various mammalian TAS2Rs. HEK293T cells expressing (A) TAS2R16, (B) TAS2R705, (C) TAS2R811, TAS2R813A, and TAS2R813B in platypus and TAS2R813 in echidna with Gα16/gust44 were stimulated with increasing concentrations of salicin. Cells transfected with the empty pEAK10 vector served as negative controls (mock). Changes in fluorescence (ΔF/F) are plotted (mean ± SEM, n = 3–5). * indicate no response within tested concentrations (Dunnett’s test, P < 0.01). EC50 values of TAS2Rs are presented in (D). n.d. indicates that EC50 values were not determined due to not well saturation of responses within tested concentrations.
Discussion
In this study, we deorphanized five of the seven platypus TAS2Rs and two of the three short-beaked echidna TAS2Rs. TAS2R16/TAS2R705 of several therians (cattle, elephant, koala, wallaby, and quoll) were also deorphanized. Monotremes have considerably smaller TAS2R gene repertoires than therians (Zhou et al. 2021), but our screening assays found that the tandemly repeated TAS2R receptors in monotremes (TAS2R810–814) are functionally diversified (table 1). Particularly, a wide range of bitter substances is covered by platypus TAS2R811 and TAS2R813A as “broadly tuned receptors.” These results indicate that platypus can detect a wider range of bitter substances despite its small TAS2R gene repertoire. The broad range of bitter perception in platypus is presumably involved in the diversified prey selection (McLachlan-Troup et al. 2010) as a result of hunting by electroreception and mechanoreception. This kind of discrepancy between the repertoire size and receptive range was previously reported in birds and fishes. Both lineages generally have small TAS2R gene repertoires (Wang and Zhao 2015; Shiriagin and Korsching 2019), but chicken, turkey, and zebrafish have broadly tuned receptors (Behrens et al. 2014, 2021). The strategy of broadly tuned receptors to adapt to a wide range of environmental bitterants, even the small TAS2R repertoire size, might be shared across vertebrates.
The broadly tuned receptors are found in the Monotreme cluster but not in Therian cluster I (Meyerhof et al. 2010; Lossow et al. 2016). Then, how did such broadly tuned receptors appear in monotremes? There is a similar case in a different TAS2R cluster of humans. Human TAS2R46 is the only broadly tuned receptor in the paralogous TAS2Rs specifically duplicated in hominoids (TAS2R30/-31/-43/-45/-46) (Hayakawa et al. 2014) and likely acquired the broaden receptive ranges after duplication (Meyerhof et al. 2010; Lossow et al. 2016). Likewise, the Monotreme cluster may have acquired broadly tuned receptors as a part of functional differentiation after the duplication event. On the other hand, the structural requirements of broadly tuned TAS2Rs are still unclear. Previous studies demonstrated that several positions (e.g., positions 7.39 and 7.42; BW numberings) are involved in ligand selectivity of human broadly tuned receptors (TAS2R10, TAS2R14, and TAS2R46) (Brockhoff et al. 2010; Born et al. 2013; Nowak et al. 2018). However, the binding mode of a common ligand, strychnine, is different between TAS2R10 and TAS2R46, and the effects of substitutions at positions 7.39 and 7.42 are also different (Born et al. 2013; Xue et al. 2018). Furthermore, TAS2R10 has a complete loss of function due to the substitution at position 7.39, whereas another broadly tuned receptor TAS2R14 retains responses to ligands. This indicates that there are additional key residues for broad tuning (Nowak et al. 2018). Thus, the structural requirements for broad tuning may differ for each receptor. To understand the structural basis of broad tuning in monotreme TAS2Rs, further computational and functional analyses are required.
The receptive ranges of TAS2R811 and TAS2R813 in short-beaked echidna were different from platypus. These are broadly tuned receptors in platypus, but echidna TAS2R811 is pseudogenized and echidna TAS2R813 had only half the tuning breadth of the platypus ortholog (table 1). These results indicate that bitter taste space is relatively smaller in echidna than in platypus. Echidna mainly consumes subterranean social insects (ants and termites) for diet (Abensperg-Traun and De Boer 1992) and has only three intact TAS2Rs. Such reduction of TAS2R repertoire size is also observed in Chinese pangolin (Manis pentadactyla), which is a eutherian ant-and-termite specialist and has only two TAS2R genes (Liu et al. 2016) even though pangolin TAS2Rs are still orphan. Our results suggest that specialized insectivory drives not only the reduction of TAS2R gene repertoire but also narrowed receptive ranges of bitter substances in TAS2Rs. Notedly, as our ligand library is biased toward compounds that are bitter for humans, the receptive ranges of echidna TAS2Rs are possibly underestimated at present. To approach this concern, further functional assays are required using secretion or extracts from food items of echidna.
We first hypothesized that the mammalian TAS2R16 orthologous group (Therian cluster I including TAS2R16, and Monotreme cluster) responds to harmful β-glucosides. As supporting this hypothesis, we found that the β-glucoside sensitivity of the TAS2R16 orthologous group is common in all three mammalian lineages (table 1). Eutherian TAS2R16 (human, cattle, and elephant) and marsupial TAS2R705 (koala, wallaby, and quoll) were functionally similar and specifically activated by β-glucosides, indicating that the common ancestor of therians had a TAS2R705 ortholog responding to β-glucosides. On the other hand, platypus TAS2R811 and TAS2R813A and echidna TAS2R813 are not specialized but undoubtedly sensitive to β-glucosides. Duplicated TAS2Rs are often functionally diverged but sometimes share some ligands (Lossow et al. 2016; Jiao et al. 2018; Wang et al. 2019). Accordingly, a common ancestral receptor of TAS2R810-814 may have had β-glucoside sensitivity. Moreover, these results parsimoniously suggest that the last common ancestor of mammals (∼188 Mya) can perceive β-glucosides as bitter via an ancestral TAS2R of Monotreme cluster and Therian cluster I and that the function has been conserved for a long time. Interestingly, there are so far no identified TAS2Rs in non-mammalian vertebrates responding to β-glucosides except for TAS2R201 in common carp (Cyprinus carpio) that weakly responds to salicin (Behrens et al. 2014, 2021; Shimizu et al. 2021). This viewpoint provides an important insight into mammalian diversification in terms of coevolution with dietary organisms. Some of β-glucosides are harmful compounds in dietary plants and invertebrates for mammals. In particular, CNglcs are highly deleterious for mammals. For example, koala exclusively relies on dietary eucalypt leaves that contain harmful substances such as CNglcs. The fact that koala TAS2R705C responds to β-glucosides suggests that koala senses the quality of eucalypt leaves in terms of CNglc concentrations. CNglcs are ancient biomolecules and widely distributed in plants (angiosperm, gymnosperm, and fern) and invertebrates (e.g., butterfly and moth) (Beran et al. 2019). The CYP79s, key enzymes for the biosynthesis of CNglcs, is estimated to have evolved in the common ancestor of angiosperm and gymnosperm (at least 330 Mya) (Luck et al. 2017; Thodberg et al. 2020), presuming that the ancestor of extant mammals (synapsids) may have been exposed to CNglcs from food items. Therefore, the ability to detect β-glucosides might be important for ancestral mammalian lineages to avoid feeding on such toxic plants and invertebrates.
We also found inter-species differences in β-glucoside sensitivity (table 1, fig. 2). These differences are possibly caused by substitutions at positions involved in ligand binding or shaping the binding cavity (Sakurai et al. 2010; Thomas et al. 2017; Fierro et al. 2019) (supplementary table S4, Supplementary Material online). For example, opossum TAS2R705 had two species-specific substitutions in the key positions involved in the activation of human TAS2R16 (positions 3.91 and 3.93). There were three amino acid differences in the key positions among three TAS2R813 orthologs (positions 3.90, 3.91, and 3.94). On the other hand, there were 13 amino acid differences between Tasmanian devil and quoll TAS2R705, but all of them were not included in the key positions. Accordingly, this functional difference could be provided by indirect ways such as the effects on G protein coupling or ligand access to the binding cavity. To clarify the effects of these substitutions in receptor functions, further computational and functional analyses are required. The general tendency in salicin sensitivity associated with feeding habits was not found across mammals. As reported in the primate studies (Imai et al. 2012; Itoigawa et al. 2019; Purba et al. 2020; Itoigawa et al. 2021; Yang et al. 2021), functional changes of TAS2Rs in agonist sensitivity had occurred many times independently within the order, family, or genus level by each ecological and evolutionary background. To understand the differences of agonist sensitivity in each species tested in this study, comparative analyses within more closely related species are required. At present, we only tested closely related Tasmanian devil and quoll (subfamily Dasyurinae). Quoll TAS2R705 responded to β-glucosides but the Tasmanian devil receptor did not (fig. 2B). While the diet of quoll is highly variable, including insects, vertebrates, and plants, the diet of Tasmanian devil is mostly occupied by mammals and birds (Godsell 1983; Jones and Barmuta 1998; Andersen et al. 2017). Since β-glucosides are mainly contained in insects and plants, the quoll receptor may retain sensitivity to such substances. To address this speculation, we need to investigate the agonist profiles of the remained TAS2Rs in Therian cluster I (TAS2R41 and TAS2R60) and the behavioral responses to β-glucosides because they possibly respond to such substances as well as in mouse orthologs (Lossow et al. 2016).
In conclusion, our results characterized the response profiles for almost all monotreme TAS2Rs. While our screening assays used a relatively small number of tested substances, this profile information will be useful to study monotreme TAS2R receptors in gustation and extra-oral functions. We also uncovered a part of molecular evolution in the detection of natural bitter compounds, β-glucosides, revealing the potential ecological importance of this function in mammals. Such studies on the functional evolution of taste receptors for natural bitter substances will contribute to understanding the history of adaptation to the feeding deterrents produced by prey from the past to the present.
Materials and Methods
Synteny Analysis
TAS2R gene sequences in the Therian cluster I (TAS2R16, TAS2R41, TAS2R60, TAS2R62, and TAS2R705) and in the Monotreme cluster (TAS2R810-814) were obtained from previous studies (Hayakawa et al. 2014; Liu et al. 2016; Johnson et al. 2018; Zhou et al. 2021). We then identified these sequences in the latest available whole-genome assemblies of each species using BLASTN search. TAS2R genes were newly identified in whole-genome assemblies of lesser hedgehog tenrec (Echinops telrairi), African elephant (Loxodonta Africana), and nine-banded armadillo (Dasypus novemcinctus) using BLAST search as described previously (Hayakawa et al. 2014) (supplementary table S1, Supplementary Material online). The genes located in the upstream and downstream regions of these TAS2Rs were identified in NCBI Database according to the NCBI Annotation (supplementary table S2, Supplementary Material online). The positions of these adjacent genes were also identified in chicken (Gallus gallus) as an outgroup.
Gene Tree Reconstruction
All intact and pseudogenized TAS2R gene sequences of eutherians (human, mouse, cattle, and domestic cat) (Hayakawa et al. 2014; Liu et al. 2016), marsupials (grey short-tailed opossum, Tasmanian devil, and koala) (Johnson et al. 2018), monotremes (platypus and short-beaked echidna) (Zhou et al. 2021), and chicken (Dong et al. 2009) were obtained from previous studies. Human vomeronasal type-1 receptors (V1Rs) were used as outgroups (Moriya-Ito et al. 2018). A maximum likelihood gene tree was reconstructed using RAxML under the GTR + Γ model with 1000 bootstrap replicates (Stamatakis 2014) based on the multiple alignment of nucleotide sequences constructed by MAFFT version 7 with E-INS-i option (Katoh and Standley 2013). A subtree of TAS2Rs in the Monotreme cluster and Therian cluster I was reconstructed using the nucleotide sequences obtained from previous studies and TAS2R16/-41/-60/-62 sequences identified in this study (tenrec, elephant, and armadillo).
Sequence Determination of TAS2Rs
Genomic DNA of cattle (Bos taurus), eastern quoll (Dasyurus viverrinus), Tasmanian devil (Sarcophilus harrisii), koala (Phascolarctos cinereus) were isolated from the tissues using the DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany). Genomic DNA of platypus (Ornithorhynchus anatinus) and short-beaked echidna (Tachyglossus aculeatus) were obtained from wild animals of New South Wales and South Australia, respectively. Cattle TAS2R16, Tasmanian-devil and quoll TAS2R705, koala TAS2R705C (an intact TAS2R705 paralog), and all intact TAS2Rs of platypus and short-beaked echidna were amplified from the genomic DNA by polymerase chain reaction (PCR) using ExTaq or Tks Gflex DNA polymerase (TaKaRa Bio Inc., Kusatsu, Japan) with specific pairs of primers which were designed based on available whole-genome assemblies. All sequences of PCR products were then determined using the BigDye Terminator v. 3.1 Cycle Sequencing Kit and the ABI 3130xl Genetic Analyzer (Applied Biosystems, Foster City, CA). Subsequently, haplotypes of unphased heterozygous TAS2Rs were determined by sub-cloning the PCR amplicons with the TOPO TA Cloning Kit for Sequencing (Invitrogen, Carlsbad, CA). To identify the corresponding amino acid residues among tested receptors, a multiple alignment based on amino acid sequences was constructed using MAFFT version 7 (Katoh and Standley 2013). We then represented superscript numbers in the amino acid residues located in transmembrane domains following the Ballesteros–Weinstein (BW) numbering method (Ballesteros and Weinstein 1995). The BW numberings were based on the human TAS2R16 numbering reported in the GPCRdb (https://gpcrdb.org/) (Kooistra et al. 2021).
Construction of Expression Vectors
TAS2R16C from African elephant (Loxodonta africana), only one intact elephant TAS2R16 paralog, TAS2R705 from grey short-tailed opossum (Monodelphis domestica), and TAS2R705B from Tammar wallaby (Macropus eugenii), an intact wallaby TAS2R705 paralog, were obtained from previous studies (Johnson et al. 2018; Itoigawa et al. 2019) and synthesized by gBlocks DNA fragment services (Integrated DNA Technologies Inc., Coralville, IA). Synthesized DNA fragments and PCR products of TAS2R receptors were tagged at N-terminus with the first 45 amino acids of rat somatostatin receptor type 3 to improve cell-surface targeting and at the C-terminus with the last 8 amino acids of bovine rhodopsin as an epitope tag. The tagged fragments were inserted into the mammalian expression vector pEAK10 (Edge Biosystems Inc., Gaithersburg, MD) using In-Fusion HD Cloning Kit (Clontech, Fremont, CA). The expression vector of human TAS2R16 was prepared in a previous study (Imai et al. 2012). For the heterozygous TAS2R genes determined in the present study, one of the haplotypes was used for functional assays. The nucleotide sequences of all TAS2R genes used in functional assays were shown in the supplementary information (supplementary Data set S1, Supplementary Material online).
Calcium Assay
Functional characterization of each TAS2R receptor was performed by the cell-based calcium assay using the FlexStation 3 microplate reader with the Ca2+-sensitive fluorescence dyes, Calcium 4 and Calcium 5 (Molecular Devices, Sunnyvale, CA) as previously described (Itoigawa et al. 2019). Tested substances listed in supplementary table S3, Supplementary Material online were commercially purchased and dissolved in the assay buffer (130 mM NaCl, 10 mM glucose, 5 mM KCl, 2 mM CaCl2, 1.2 mM MgCl2, 10 mM HEPES, pH 7.4), or in dimethyl sulfoxide (DMSO) followed by dilution in the assay buffer not exceeding a final DMSO-concentration of 1% (v/v). The substance concentrations for screening were basically selected from the maximal concentrations in previous studies (Meyerhof et al. 2010; Lossow et al. 2016; Itoigawa et al. 2021). The calcium responses were expressed as the normalized peak response (F) relative to background fluorescence (F0): ΔF/F ( = [F − F0]/F0). Agonists of TAS2Rs were detected by the comparisons with the responses of mock-transfected cells using Dunnett’s test (p < 0.01). To estimate dose-response curves, cells expressing each TAS2R were stimulated by various concentrations of bitter substances. Subsequently, ΔF/F values were fitted to the nonlinear regression model f(x) = min + [(max − min)/(1 + x/EC50)h], where x is the test compound concentration and h is the Hill coefficient, using the drc package in R (Ritz et al. 2015).
Supplementary Material
Acknowledgments
We thank Dr Shuichi Matsumura in Gifu University for providing the tissue of cattle, Sapporo Maruyama Zoo for providing the tissue of Tasmanian devils, and Higashiyama Zoo and Botanical Gardens for providing the tissue of koalas, Jenny Newport of the Fenner School of Environment and Society for sampling and supplying the tissue of eastern quoll (from the Mulligans Flat—Goorooyarroo Woodland Experiment specimen collection). We also thank Dr Takashi Ueda in Nagoya City University, Dr Yoshiro Ishimaru, Dr Takumi Misaka, and Dr Keiko Abe in the University of Tokyo for providing Gα16/gust44 and pEAK10 vectors; Dr Hiroaki Matsunami in Duke University for providing HEK293T cells; Mihoko Umemura and members of Molecular Biology section in Primate Research Institute of Kyoto University for technical support in molecular and cellular experiments. This study was supported by KAKENHI from Japan Society for the Promotion of Science (JSPS) (Nos. 18J22288 to A.I., 16K18630, 19K16241, 21H04919, 21KK0106 to T.H., 18H04005, 19K21586, and 21KK0130 to H.I.), Sasakawa Scientific Research Grant from the Japan Science Society (29-534), Hokkaido University Sousei Tokutei Research, and JSPS Bilateral Joint Research Project “Earth-wide comparative genomics of endangered mammals in Japan and Australia” (JPJSBP 120219902) to T.H., and ARC FT160100267 to F.G. This work was partly supported by the Research Units for Exploring Future Horizons to H.I. Eastern quoll samples were collected with the support of the Mulligans Flat – Goorooyarroo Woodland Experiment, and the supporting Australian Research Council Linkage grant (LP140100209).
Contributor Information
Akihiro Itoigawa, Department of Cellular and Molecular Biology, Primate Research Institute, Kyoto University, Inuyama, Aichi, Japan; Department of Agricultural Chemistry, School of Agriculture, Meiji University, Kawasaki, Kanagawa, Japan.
Takashi Hayakawa, Faculty of Environmental Earth Science, Hokkaido University, Sapporo, Hokkaido, Japan; Japan Monkey Centre, Inuyama, Aichi, Japan.
Yang Zhou, BGI-Shenzhen, Shenzhen, China.
Adrian D. Manning, Fenner School of Environment and Society, The Australian National University, Canberra, ACT, Australia
Guojie Zhang, Department of Biology, University of Copenhagen, Kobenhavn, Denmark.
Frank Grutzner, School of Biological Sciences, The University of Adelaide, Adelaide, SA, Australia.
Hiroo Imai, Molecular Biology Section, Center for the Evolutionary Origins of Human Behavior, Kyoto University, Inuyama, Aichi, Japan.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author contributions
A.I., T.H., and H.I. conceived and designed the research. T.H., A.D.M., and F.G. collected samples. A.I. and T.H. performed DNA experiments. A.I. performed cellular assays. A.I., T.H., Y.Z., G.Z., and F.G. performed informatics analysis. All authors interpreted data. A.I. wrote the original draft. All authors edited and approved the final version of the manuscript.
Data availability
TAS2R sequences newly determined were deposited in DDBJ (LC672162-LC672182).
Conflict of interest
We declare no competing interests.
References
- Abensperg-Traun M, De Boer ES. 1992. The foraging ecology of a termite-and ant-eating specialist, the echidna Tachyglossus aculeatus (Monotremata: Tachyglossidae). J Zool. 226:243–257. [Google Scholar]
- Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJP, Zuker CS. 2000. A novel family of mammalian taste receptors. Cell 100:693–702. [DOI] [PubMed] [Google Scholar]
- Andersen GE, Johnson CN, Barmuta LA, Jones ME. 2017. Dietary partitioning of Australia’s two marsupial hypercarnivores, the Tasmanian devil and the spotted-tailed quoll, across their shared distributional range. PLoS One 12:e0188529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwell KWS. 2013. Neurobiology of monotremes: brain evolution in our distant mammalian cousins. Collingwood: (VIC: ): CSIRO Publishing. [Google Scholar]
- Ballesteros JA, Weinstein H. 1995. [19] Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Meth Neurosci. (Sealfon SC, editor) 25:366–428. [Google Scholar]
- Behrens M, Di Pizio A, Redel U, Meyerhof W, Korsching SI. 2021. At the root of T2R gene evolution: recognition profiles of coelacanth and zebrafish bitter receptors. Genome Biol Evol. 13:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens M, Korsching SI, Meyerhof W. 2014. Tuning properties of avian and frog bitter taste receptors dynamically fit gene repertoire sizes. Mol Biol Evol. 31:3216–3227. [DOI] [PubMed] [Google Scholar]
- Beran F, Köllner TG, Gershenzon J, Tholl D. 2019. Chemical convergence between plants and insects: biosynthetic origins and functions of common secondary metabolites. New Phytol. 223:52–67. [DOI] [PubMed] [Google Scholar]
- Born S, Levit A, Niv MY, Meyerhof W, Behrens M. 2013. The human bitter taste receptor TAS2R10 is tailored to accommodate numerous diverse ligands. J Neurosci. 33:201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhoff A, Behrens M, Niv MY, Meyerhof W. 2010. Structural requirements of bitter taste receptor activation. Proc Natl Acad Sci. 107:11110–11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bufe B, Hofmann T, Krautwurst D, Raguse J-D, Meyerhof W. 2002. The human TAS2R16 receptor mediates bitter taste in response to β-glucopyranosides. Nat Genet. 32:397–401. [DOI] [PubMed] [Google Scholar]
- Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJP. 2000. T2Rs function as bitter taste receptors. Cell 100:703–711. [DOI] [PubMed] [Google Scholar]
- Dong D, Jones G, Zhang S. 2009. Dynamic evolution of bitter taste receptor genes in vertebrates. BMC Evol Biol. 9:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierro F, Giorgetti A, Carloni P, Meyerhof W, Alfonso-Prieto M. 2019. Dual binding mode of “bitter sugars” to their human bitter taste receptor target. Sci Rep. 9:8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go Y, Satta Y, Takenaka O, Takahata N. 2005. Lineage-specific loss of function of bitter taste receptor genes in humans and nonhuman primates. Genetics 170:313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsell J. 1983. Ecology of the eastern quoll Dasyurus viverrinus (Dasyuridae: Marsupialia) [dissertation]. Canberra: The Australian National University.
- Grant T. 2007. Platypus. Forth edit (Illustrated by D. Fanning). Clayton South: CSIRO Publishing. [Google Scholar]
- Griffiths M. 1965. Digestion, growth and nitrogen balance in an egg-laying mammal, Tachyglossus aculeatus (Shaw). Comp Biochem Physiol. 14:357–375. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Suzuki-Hashido N, Matsui A, Go Y. 2014. Frequent expansions of the bitter taste receptor gene repertoire during evolution of mammals in the euarchontoglires clade. Mol Biol Evol. 31:2018–2031. [DOI] [PubMed] [Google Scholar]
- Imai H, Suzuki N, Ishimaru Y, Sakurai T, Yin L, Pan W, Abe K, Misaka T, Hirai H. 2012. Functional diversity of bitter taste receptor TAS2R16 in primates. Biol Lett. 8:652–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoigawa A, Fierro F, Chaney ME, Lauterbur ME, Hayakawa T, Tosi AJ, Niv MY, Imai H. 2021. Lowered sensitivity of bitter taste receptors to β-glucosides in bamboo lemurs: an instance of parallel and adaptive functional decline in TAS2R16? Proc R Soc B Biol Sci. 288:rspb.2021.0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoigawa A, Hayakawa T, Suzuki-Hashido N, Imai H. 2019. A natural point mutation in the bitter taste receptor TAS2R16 causes inverse agonism of arbutin in lemur gustation. Proc R Soc B Biol Sci. 286:20190884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao H, Wang Q, Wang B-J, Li K, Lövy M, Nevo E, Li Q, Su W, Jiang P, Zhao H. 2021. Local adaptation of bitter taste and ecological speciation in a wild mammal. Mol Biol Evol. (Zhang J, editor) 38:4562–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao H, Wang Y, Zhang L, Jiang P, Zhao H. 2018. Lineage-specific duplication and adaptive evolution of bitter taste receptor genes in bats. Mol Ecol. 27:4475–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RN, O’Meally D, Chen Z, Etherington GJ, Ho SYW, Nash WJ, Grueber CE, Cheng Y, Whittington CM, Dennison S, et al. 2018. Adaptation and conservation insights from the koala genome. Nat Genet. 50:1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ME, Barmuta LA. 1998. Diet overlap and relative abundance of sympatric dasyurid carnivores: a hypothesis of competition. J Anim Ecol. 67:410–421. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra AJ, Mordalski S, Pándy-Szekeres G, Esguerra M, Mamyrbekov A, Munk C, Keserű GM, Gloriam DE. 2021. GPCRdb in 2021: integrating GPCR sequence, structure and function. Nucleic Acids Res. 49:D335–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 34:1812–1819. [DOI] [PubMed] [Google Scholar]
- Li D, Zhang J. 2014. Diet shapes the evolution of the vertebrate bitter taste receptor gene repertoire. Mol Biol Evol. 31:303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Liu G, Hailer F, Orozco-terWengel P, Tan X, Tian J, Yan Z, Zhang B, Li M. 2016. Dietary specialization drives multiple independent losses and gains in the bitter taste gene repertoire of Laurasiatherian mammals. Front Zool. 13:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossow K, Hübner S, Roudnitzky N, Slack JP, Pollastro F, Behrens M, Meyerhof W. 2016. Comprehensive analysis of mouse bitter taste receptors reveals different molecular receptive ranges for orthologous receptors in mice and humans. J Biol Chem. 291:15358–15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck K, Jia Q, Huber M, Handrick V, Wong GK-S, Nelson DR, Chen F, Gershenzon J, Köllner TG. 2017. CYP79 P450 monooxygenases in gymnosperms: CYP79A118 is associated with the formation of taxiphyllin in Taxus baccata. Plant Mol Biol. 95:169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan-Troup TA, Dickman CR, Grant TR. 2010. Diet and dietary selectivity of the platypus in relation to season, sex and macroinvertebrate assemblages. J Zool. 280:237–246. [Google Scholar]
- Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, Appendino G, Behrens M. 2010. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem Senses 35:157–170. [DOI] [PubMed] [Google Scholar]
- Moriya-Ito K, Hayakawa T, Suzuki H, Hagino-Yamagishi K, Nikaido M. 2018. Evolution of vomeronasal receptor 1 (V1R) genes in the common marmoset (Callithrix jacchus). Gene 642:343–353. [DOI] [PubMed] [Google Scholar]
- Nowak S, Di Pizio A, Levit A, Niv MY, Meyerhof W, Behrens M. 2018. Reengineering the ligand sensitivity of the broadly tuned human bitter taste receptor TAS2R14. Biochim Biophys Acta - Gen Subj. 1862:2162–2173. [DOI] [PubMed] [Google Scholar]
- Pettigrew JD, Manger PR, Fine SLB. 1998. The sensory world of the platypus. Phil Trans R Soc Lond B Biol Sci. 353:1199–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purba LHPS, Widayati KA, Suzuki-Hashido N, Itoigawa A, Hayakawa T, Nila S, Juliandi B, Suryobroto B, Imai H. 2020. Evolution of the bitter taste receptor TAS2R38 in colobines. Primates 61:485–494. [DOI] [PubMed] [Google Scholar]
- Ritz C, Baty F, Streibig JC, Gerhard D. 2015. Dose-response analysis using R. PLoS One 10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Misaka T, Ishiguro M, Masuda K, Sugawara T, Ito K, Kobayashi T, Matsuo S, Ishimaru Y, Asakura T, et al. 2010. Characterization of the beta-D-glucopyranoside binding site of the human bitter taste receptor hTAS2R16. J Biol Chem. 285:28373–28378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Kubozono T, Asaoka R, Toda Y, Ishimaru Y. 2021. Expression profiles and functional characterization of common carp (Cyprinus carpio) T2Rs. Biochem Biophys Rep. 28:101123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiriagin V, Korsching SI. 2019. Massive expansion of bitter taste receptors in blind cavefish, Astyanax mexicanus. Chem Senses 44:23–32. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thodberg S, Sørensen M, Bellucci M, Crocoll C, Bendtsen AK, Nelson DR, Motawia MS, Møller BL, Neilson EHJ. 2020. A flavin-dependent monooxygenase catalyzes the initial step in cyanogenic glycoside synthesis in ferns. Commun Biol. 3:507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Sulli C, Davidson E, Berdougo E, Phillips M, Puffer BA, Paes C, Doranz BJ, Rucker JB. 2017. The bitter taste receptor TAS2R16 achieves high specificity and accommodates diverse glycoside ligands by using a two-faced binding pocket. Sci Rep. 7:7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiao H, Jiang P, Zhao H. 2019. Functional divergence of bitter taste receptors in a nectar-feeding bird. Biol Lett. 15:20190461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhao H. 2015. Birds generally carry a small repertoire of bitter taste receptor genes. Genome Biol Evol. 7:2705–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue AY, Di Pizio A, Levit A, Yarnitzky T, Penn O, Pupko T, Niv MY. 2018. Independent evolution of strychnine recognition by bitter taste receptor subtypes. Front Mol Biosci. 5:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Yang S, Fan F, Li Y, Dai S, Zhou X, Steiner CC, Coppedge B, Roos C, Cai X, et al. 2021. A new world monkey resembles human in bitter taste receptor evolution and function via a single parallel amino acid substitution. Mol Biol Evol. 38:5472–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Shearwin-Whyatt L, Li J, Song Z, Hayakawa T, Stevens D, Fenelon JC, Peel E, Cheng Y, Pajpach F, et al. 2021. Platypus and echidna genomes reveal mammalian biology and evolution. Nature 592:756–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TAS2R sequences newly determined were deposited in DDBJ (LC672162-LC672182).