Significance Statement
Proinflammatory macrophages that infiltrate the kidney after ischemia-reperfusion injury later transition to a proreparative state characterized by expression of multiple proteins including arginase-1 (Arg1). By comparing the kidney repair response after ischemia-reperfusion injury in mice that lack macrophage Arg1 expression with littermate controls, we show that macrophage Arg1 plays a critical role in renal recovery in part by promoting renal epithelial cell proliferative repair. Thus, therapeutic interventions that enhance Arg1 expression may improve renal recovery after kidney injury.
Keywords: kidney repair, macrophage, arginase-1, kidney tubules, regeneration
Abstract
Background
After kidney injury, macrophages transition from initial proinflammatory activation to a proreparative phenotype characterized by expression of arginase-1 (Arg1), mannose receptor 1 (Mrc1), and macrophage scavenger receptor 1 (Msr1). The mechanism by which these alternatively activated macrophages promote repair is unknown.
Methods
We characterized the macrophage and renal responses after ischemia-reperfusion injury with contralateral nephrectomy in LysM-Cre;Arg1fl/fl mice and littermate controls and used in vitro coculture of macrophages and tubular cells to determine how macrophage-expressed arginase-1 promotes kidney repair.
Results
After ischemia-reperfusion injury with contralateral nephrectomy, Arg1-expressing macrophages were almost exclusively located in the outer stripe of the medulla adjacent to injured S3 tubule segments containing luminal debris or casts. Macrophage Arg1 expression was reduced by more than 90% in injured LysM-Cre;Arg1fl/fl mice, resulting in decreased mouse survival, decreased renal tubular cell proliferation and decreased renal repair compared with littermate controls. In vitro studies demonstrate that tubular cells exposed apically to dead cell debris secrete high levels of GM-CSF and induce reparative macrophage activation, with those macrophages in turn secreting Arg1-dependent factor(s) that directly stimulate tubular cell proliferation.
Conclusions
GM-CSF–induced, proreparative macrophages express arginase-1, which is required for the S3 tubular cell proliferative response that promotes renal repair after ischemia-reperfusion injury.
Multiple studies have shown that macrophages are important regulators of the renal response to injury.1–4 Injury-induced signals from endothelial and epithelial cells, including chemokines and danger-associated molecular patterns, stimulate homing and proinflammatory activation of macrophages that can initially worsen tubule dysfunction. This is followed by downregulation of proinflammatory gene expression and a locally regulated switch to a macrophage phenotype that promotes renal repair.3 We have shown that a critical inducer of the proreparative phenotype is the expression of GM-CSF by surviving tubular cells, and that GM-CSF induces a unique macrophage gene expression profile including Arg1, Msr1, and Mrc1 that is distinct from the canonical M2 activation induced by IL-4.5
Our group and others have shown that depletion of macrophages as they switch to proreparative activation reduces tubular cell proliferation and repair.3,6 Arginase-1, the protein product of the Arg1 gene, converts arginine to ornithine, which in turn is converted to a series of polyamines that have been implicated in cell proliferative responses.7,8 To understand the mechanism by which macrophages mediate repair, we generated LysM-Cre;Arg1fl/fl mice to selectively deplete Arg1 from myelomonocytic cells. Mice lacking macrophage Arg1 expression exhibited reduced survival and reduced tubular cell proliferation after ischemia-reperfusion injury (IRI) with contralateral nephrectomy (CL-NX) (termed IRI/CL-NX). In vitro coculture studies demonstrate that primary renal cells exposed to urinary casts or dead cell debris secreted high levels of GM-CSF, inducing Arg1 expression in wild-type macrophages that in turn secreted factor(s) that directly stimulated tubular cell proliferation. This response was lost in coculture with Arg1 knockout macrophages.
Methods
Animal Model
All animal protocols were approved by the Yale University Institutional Animal Care and Use Committee. Mice were kept under a 12-hour day/night cycle with food and water provided ad libitum. All mice were purchased from The Jackson Laboratory: arginase-1 flox/flox (Stock #: 008817), LysM-Cre/+ (Stock #: 004781), and mTmG (Stock #: 007576). Mice were crossed to generate Arg1con (Arg1 control: Arg1+/+;LysMCre/+) and Arg1mko (Arg1 macrophage knockout: Arg1fl/fl;LysMCre/+). Arg1mko mice were further crossed with reporter mTmG mice to identify Cre expression and confirm arginase-1 knockout in Cre-expressing cultured macrophages. All mice were backcrossed at least nine times and maintained on a C57b6/NJ genetic background (The Jackson Laboratory, Stock #: 005304). Male mice, 8–10 weeks old, were anesthetized and subjected to IRI/CL-NX as previously described.5 Male mice were used for IRI due to substantial variability in susceptibility to IRI between male and female mice.9 Briefly, the left renal pedicle was clamped for 27 minutes using a nontraumatic vasculature clamp (Cat. #: 00398-02; Fine Science Tools), whereas the right renal pedicle and ureter were ligated with two sutures and the right kidney was nephrectomized. Reperfusion after release was considered successful if the kidney rapidly recovered its original color. Mice were kept at 37°C using a warming pad during the whole procedure and closely monitored during their recovery phase. Mice were given 0.5 mL of normal saline intraperitoneally to prevent dehydration. Blood for serum creatinine and BUN was obtained before and at the indicated times after reperfusion. Creatinine and BUN assays were performed at the George O’Brien Kidney Center at Yale University. All experiments were repeated on at least two separate occasions.
Isolation of Primary Cultured Renal Cells and Generation of Dead Cell Debris As a Cast-Mimetic
Primary cultured renal cells (PCRCs) were isolated using a modified protocol described by Schafer et al.10,11 Briefly, kidneys were harvested and minced in Liberase Research Grade (0.5 mg/ml in 1× PBS, Cat. #: 05401127001; Roche) supplemented with 0.1% MgCl2 and DNase I (0.1 mg/ml, Cat. #: 10104159001; Roche) and incubated in CO2 incubator for 30 minutes with gentle mixing every 10 minutes. The digested mixture was pipetted up and down until no obvious tissue clamps were visible then passed through a 40-μm cell strainer. The flow-through was centrifuged at 300 × g for 5 minutes at 4°C. Pellets were resuspended in DMEM with 10% FBS with antibiotics and seeded on 10-cm cell culture dishes (six plates for each kidney) in 5% CO2 incubator at 37°C. Cells were expanded for 5 days (culture media was changed after 48 hours). On day 5, P0 PCRCs were harvested (approximately 12 million per kidney) and cryopreserved. P1 PCRCs were used for coculture throughout this study. Single-cell RNA sequencing (scRNA-seq) was performed on P1 PCRCs, identifying them as 71.8% proximal tubule, 22.6% other epithelial cells, and 5.6% fibroblasts (Supplemental Figure 1).
To generate PCRC debris as a cast-mimetic, PCRCs (1 × 106) were seeded onto a 10-cm tissue culture plate and expanded for 3 days (Corning DMEM/Hams F-12 50/50 Mix with 10% FBS). Cells were then mechanically scraped off the plate and frozen at –80°C for a minimum of 24 hours. Freeze-fractured cell debris was generated from approximately 1 × 106 PCRCs and was plated per well ±2.5 × 105 PCRCs on a 6-well Transwell plate. Methodology for single cell isolation and RNA sequencing of both cultured PCRC and whole kidney is provided in the Supplemental Material.
Immune Cell Isolation from Kidneys
Kidneys were harvested at the indicated time points, minced, and digested for 45 minutes at 37°C using the Liberase enzyme cocktail (500 μg/ml, Cat. #: 05401127001; Roche) in the presence of DNase-1 (100 μg/ml, Cat. #: D45131-1VL; Sigma) and MgCl2 (10 μg/ml; Sigma) in PBS. The cell homogenate was sieved at 70 μm and 40 μm to remove undigested tubules, washed, and resuspended at 4°C in 0.5% BSA, 2 mM EDTA, and 0.1% pluronic acid in 1× PBS. After a 15-minute blocking step using CD16/CD34 antibodies (Fc receptor block), cells were labeled for 30 minutes on ice using specific antibody cocktails supplemented with DAPI 100 ng/ml, washed, and flow sorted on a FACSAria flow cytometer (BD Biosciences). Antibodies used for flow are provided in Supplemental Table 1.
Immunostaining
Kidneys were perfusion fixed with 4% paraformaldehyde and processed for histology (hematoxylin and eosin, paraffin blocks) or immunostaining (4-μm cryosections). The sections were blocked at room temperature using saline containing 0.1% BSA and 10% goat serum.
The following primary antibodies were used: anti-BrdU (Sigma), anti-F4/80 (clone BM8; eBioscience), and anti-Ki67 (clone SP6; NeoMarkers). DNA strand breaks were identified using terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining (Roche In Situ Cell Death Detection Kit). Quantification of cells expressing the specified marker/stain was performed in a blinded fashion by counting the number of positive cells/total cells (identified as DAPI+ nuclei) in 10 randomly chosen 400× fields from the outer medulla. At least two sections per kidney from at least four kidneys were counted for each experiment. Images were taken at 200×, 400×, or 1000× using a Nikon microscopy system. Image analysis was performed with ImageJ software (National Institutes of Health, Bethesda, MD).
Analysis of mRNA
RNA was extracted with an RNeasy Mini kit (Qiagen) and reverse transcribed using the iScript cDNA Synthesis Kit (Bio-Rad). Gene expression analysis was determined by quantitative real-time PCR (rtPCR) (iTaq Universal SybrGreen Supermix; Bio-Rad) using a CFX96 machine (Bio-Rad) and normalized to hypoxanthine guanine phosphoribosyl transferase (Hprt). Primer sequences are provided in Supplemental Table 2.
In Vitro Coculture Assays
Bone marrow macrophages (BMMs) were isolated and cultured as per the protocol of Cui et al.12 with the following minor modifications: bone marrow cells were flushed from the femurs of 8- to 12-week-old Arg1con and Arg1mko mice and red blood cells were lysed in ACK buffer (0.15 M NH4Cl, 10 mM KHCO3, 0.1 mM EDTA). Remaining cells were incubated in α-MEM medium (Life Technologies) containing 10% FBS (Sigma), 10% L929 fibroblast cell supernatant, 1% glutamine (Life Technologies), 1% MEM vitamin (Life Technologies), and 1% penicillin/streptomycin (Life Technologies). After 24 hours, nonadherent cells (containing the majority of BMMs) were transferred to a new dish and incubated in 30% L929 supernatant for an additional 7 days to generate naïve BMMs. Immortalized mouse proximal tubule cells (MPTs) from Boston University were provided by Dr. John Schwartz,13 and PCRCs were cultured as described above.
Coculture and Generation of Conditioned Medium
MPTs or PCRCs were plated on 0.4-μm polyester membrane Transwell inserts for a 12-well plate (Corning Incorporated) and grown to a confluent monolayer, whereas 1 × 106 naïve BMMs were plated in individual wells of a 12-well plate for 24 hours. The inserts containing either tubular MPTs or PCRCs were then cocultured ± BMMs in the bottom well for 24 hours. The MPTs, PCRCs, and macrophages were then harvested for RNA isolation and rtPCR analysis of the indicated genes. Conditioned medium was generated by 24-hour PCRC-BMM Transwell coculture in RPMI 1640 media (Thermo Fisher Scientific) + 1% FBS.
Isolation of Arg1 Null Macrophages from Arg1fl/fl;LysMCre/+; Rosa26mTmG/+ Mice
Arg1mko mice were crossed with Rosa26mTmG/+ mice, which constitutively express the tdTomato transgene until Cre-mediated activation of green fluorescent protein (GFP) expression. BMMs from Arg1fl/fl;LysMCre/+;Rosa26mTmG/+ bone marrow were cultured for 7 days as described above, revealing a mix of GFP+ and tdTomato+ macrophages (Supplemental Figure 2A). Cre-expressing GFP+ cells were then flow sorted via FACSAria (BD Biosciences) and loss of arginase-1 expression confirmed by Western blot (Supplemental Figure 2B) and rtPCR (Supplemental Figure 2C).
PCRC Proliferation Quantification
Isolated PCRCs were plated on a 96-well plate at 10,000 cells per well. Cell numbers were quantified by nuclear counting at time 0 and after 24 hours of culture ± conditioned medium or FBS stimulation. The nuclei of PCRC were stained with DAPI, imaged at 10× using a Nikon microscopy system, and quantified in a blinded fashion by counting DAPI-stained nuclei from five randomly selected fields per well using ImageJ software (National Institutes of Health, Bethesda, MD).
Tubular Injury Scoring
All scoring was performed in a blinded fashion as per the methodology of Shih et al.14 Briefly, hematoxylin and eosin–stained sections were scored for the percentage of tubules in the outer medulla showing tubular atrophy, tubular dilation, epithelial desquamation, apical blebbing, or brush border loss in 10 randomly chosen, nonoverlapping fields per section. Lotus tetragonolobus lectin (LTL)-stained sections were quantified by the percentage of tubules in the outer medulla in five randomly chosen, nonoverlapping fields per section. The degree of tubular injury was designated as follows: 0 if <5% of tubules in the field were designated as damaged; 1 for 6%–25%; 2 for 26%–50%; 3 for 51%–75%; and 4 if damage involved >75% of the observed tubules.
Statistical Analyses
All data are presented as mean±SD. Multigroup comparisons were performed using one-way ANOVA with Tukey multiple comparison test for subgroup comparisons; two-group comparisons were analyzed by t tests using Prism 8.0 (GraphPad Software, Inc.). P values <0.05 were considered statistically significant.
Results
Arginase-1 Knockout in Intrarenal Macrophages
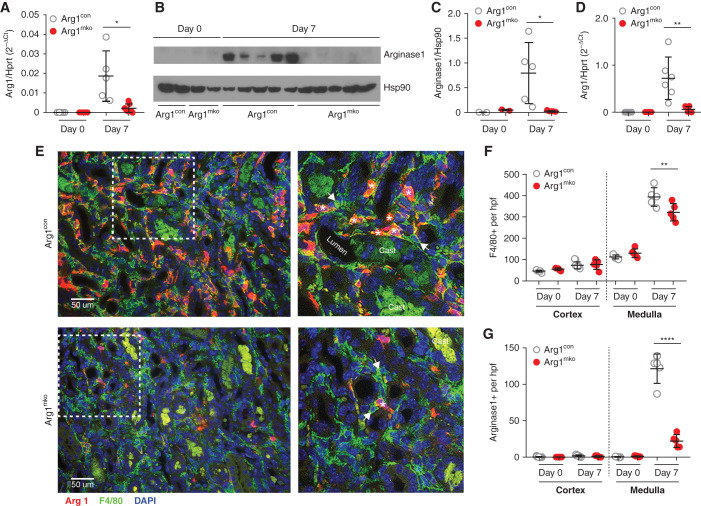
Analysis of kidneys from Arg1mko mice at baseline and 7 days after IRI/CL-NX demonstrated a 90% reduction in whole kidney Arg1 mRNA and protein expression on day 7 compared with Arg1con mice (Figure 1, A–C). mRNA analysis of FACS-sorted CD45+, F4/80+ intrarenal macrophages from Arg1mko mice confirmed a 91.4% reduction of Arg1 mRNA on day 7 (Figure 1D). Immunofluorescence staining for F4/80 and arginase-1 demonstrated the expected increase in F4/80+ macrophages in the medulla on day 7 after IRI/CL-NX in both Arg1con and Arg1mko mice, albeit with 20% less increase in the Arg1mko mice (Figure 1E, quantified in Figure 1F). The number of outer medullary macrophages expressing arginase-1 was reduced by 84% in the Arg1mko mice (Figure 1G), with quantitation of the few remaining arginase-1-positive cells demonstrating that 91% coexpress F4/80 and thus likely reflect incomplete knockout in the macrophages.
Figure 1.
Arg1fl/fl;LysMCre/+ mice have reduced arginase-1 in the kidney after ischemic injury. Eight-week-old male Arg1mko (Arg1fl/fl;LysMCre/+) mice underwent IRI, and kidney lysates were collected for RNA and protein on day 0 and day 7 after surgery. (A) Arg1 mRNA expression level (relative to Hprt1) was significantly decreased in Arg1mko mice at day 7 within the whole kidney (*P<0.05, t test). (B and C) Western blot analysis (B) demonstrates significantly decreased arginase-1 protein levels on day 7 after IRI in Arg1mko mice, quantified in (C). Each lane represents one mouse kidney (*P<0.05, t test). (D) CD45+F4/80+ macrophages were isolated from kidneys at the indicated time by FACS and Arg1 mRNA expression quantified by quantitative rtPCR. Arg1 mRNA is significantly lower in day 7 macrophages from Arg1mko compared with control mice (**P<0.01, t test). (E) Immunofluorescence staining of injured kidneys from Arg1con mice (top panels) and Arg1mko mice (bottom panels) on day 7 after IRI (arginase-1 [red], F4/80+ [green], DAPI [blue]) shows significantly reduced arginase-1 staining in F4/80+ macrophages from the Arg1mko mouse. The boxed area in the left hand panels is magnified in the right panels. Asterisks indicate arginase-1+ and F4/80+ macrophage; arrows indicate arginase-1− and F4/80+ macrophage. (F) Quantification of total F4/80+ cells demonstrates increased medullary macrophages on day 7 after IRI Arg1con mice with a modest reduction seen in Arg1mko mice. (**P<0.01, ANOVA). (G) Quantification of arginase-1+ cells shows an 84% reduction in the Arg1mko mice. (****P<0.0001, ANOVA). n=5 mice per group. Casts exhibit green autofluorescence.
Arginase-1 Is Required for Recovery of Renal Function
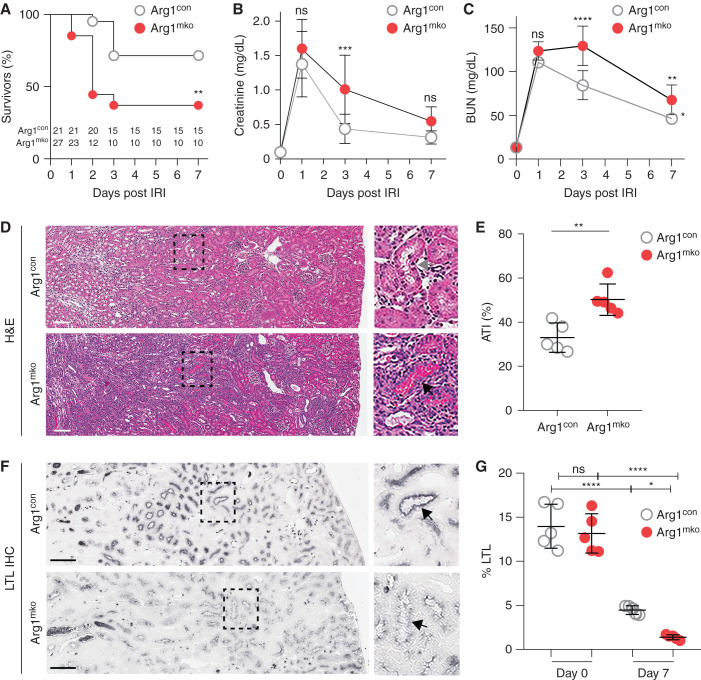
Arg1mko and Arg1con mice were subjected to IRI/CL-NX at time 0 and the injury and repair responses were investigated over 7 days. Survival of the Arg1mko mice was significantly reduced starting at 2 days after IRI (Figure 2A), consistent with upregulated expression of Arg1 at this early time point (Supplemental Figure 3). Initial injury, as judged by day 1 creatinine and BUN, was equivalent in the two groups (Figure 2, B and C). Creatinine and BUN values improved less after day 1 in the surviving Arg1mko mice, accompanied by a significantly higher day 7 tubular injury score (Figure 2, D and E) and less recovery of proximal tubule brush border LTL expression (Figure 2, F and G). These data show that macrophage expression of arginase-1 is required for proximal tubule repair after IRI/CL-NX.
Figure 2.
Macrophage expression of Arg1 is required for renal recovery. Arg1con (n=21) and Arg1mko mice (n=27) were subjected to 27 minutes of IRI/CL-NX. (A) Survival was significantly decreased in Arg1mko mice compared with control mice (**P<0.01, Log-rank Mantel–Haenszel test). (B) Serum creatinine and (C) BUN are equally increased on day 1, but reflect reduced recovery of GFR in the surviving Arg1mko mice on day 3 (****P<0.0001, ***P<0.001, ANOVA). (D) Histology demonstrates persistent injury in the Arg1mko mice on day 7 after IRI compared with control. Boxed area in the left panel is magnified in the right panel (gray arrow indicates clear tubule; black arrow indicates cast-filled tubule). (E) Acute tubular injury (ATI) score of kidneys from five mice on day 7 after IRI (**P<0.01, t test). (F) LTL staining of brush border at 7 days post IRI. (G) Quantification of LTL staining on days 0 and 7 shows reduced recovery of proximal tubule brush border in Arg1mko mice (****P<0.0001, *P<0.05, ANOVA). Scale bar, 100 μm.
Arg1-Expressing Macrophages Promote Renal Epithelial Cell Proliferation
scRNA-seq analysis of kidneys from wild-type mice subjected to IRI/CL-NX demonstrated increased proliferating proximal tubule cells compared with wild-type control kidneys (Supplemental Figure 4). There was a corresponding increase in Ccr2+ trafficking macrophages, with a lesser increase in resident macrophages and no change in Cx3cr1+ trafficking macrophages. Ccr2+ macrophages were found to express all three GM-CSF–induced alternative activation genes (Arg1, Mrc1, Msr1, Supplemental Figure 5A), with Arg1 expression almost entirely confined to the Ccr2+ macrophage population (Supplemental Figure 5B). Ccr2+Arg1+ macrophages are predicted to induce the greatest number of ligand-receptor interactions and target gene responses to promote proximal tubule cell proliferation (Supplemental Figure 5, C and D).
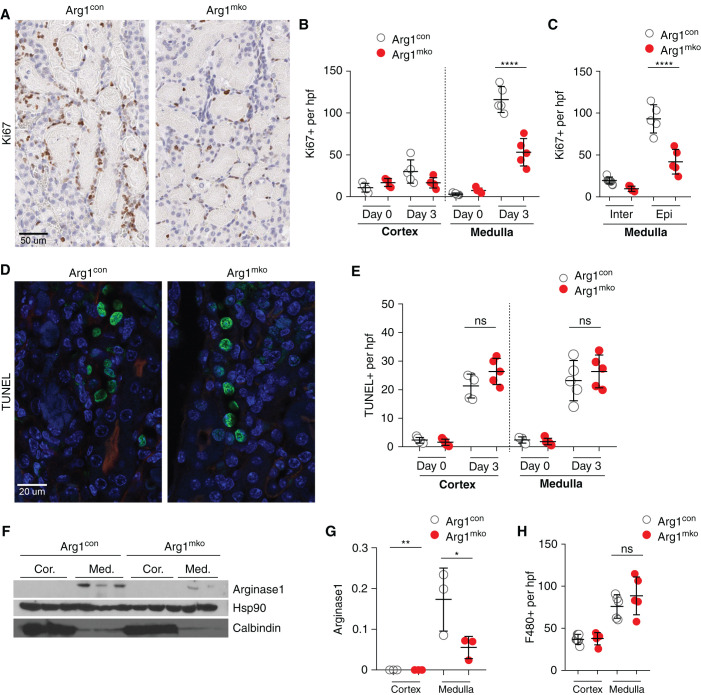
Kidneys from Arg1mko mice and Arg1con mice on day 3 after IRI/CL-NX, the time of peak tubule cell proliferation, were immunostained for Ki67 as a marker of proliferating cells (Figure 3A, quantified in Figure 3B (total cells) and Figure 3C (interstitial and epithelial cells)). Ki67+ epithelial cells were localized primarily to the outer medulla, and were significantly decreased in the Arg1mko mice compared with Arg1con mice. In contrast, quantification of TUNEL-positive cells on day 3 after IRI/CL-NX did not demonstrate a difference in programmed cell death in the absence of macrophage-expressed arginase-1 (Figure 3, D and E). Arginase-1 protein was detected only in the medulla on day 3 after IRI/CL-NX in Arg1con mice and was significantly reduced in Arg1mko mice (Figure 3, F and G), even though the number of F4/80+ macrophages was identical between the two groups at this time point (Figure 3H). Profiling of FACS-sorted renal macrophages on day 3 after IRI confirmed that Arg1 was predominantly expressed in trafficking macrophages with a significant decrease in Arg1mko cells as compared with Arg1con cells (Supplemental Figure 6A), but did not demonstrate a change in the expression of the proinflammatory cytokines IL1β or Il6, or the alternative activation genes (Mrc1 or Msr1, Supplemental Figure 6B). Of note, the loss of macrophage Arg1 expression did not significantly alter renal Arg2, Csf2, or Ccl2 expression at baseline or 7 days after IRI (Supplemental Figure 6C). These data demonstrate macrophage arginase-1 expression promotes proliferation of tubular cells during renal repair.
Figure 3.
Arginase-1+ macrophages promote epithelial cell proliferation. (A–C) Ki67 staining of Arg1con and Arg1mko kidneys on day 3 after IRI/CL-NX is shown in (A) and quantified in (B) (total cells) and (C) (interstitial and epithelial cells). Outer medullary Ki67+-proliferating epithelial cells are significantly reduced in Arg1mko kidneys (****P<0.0001, ANOVA). (D and E) TUNEL staining of Arg1con and Arg1mko kidneys on day 3 after IRI/CL-NX is shown in (D) and quantified in (E) (ns, not significant; t test). (F and G) Western blot analysis of the cortex and medulla of Arg1con and Arg1mko kidneys 3 days after IRI/CL-NX reveals upregulation of arginase-1 selectively in the medulla (F), with a significant reduction in the Arg1mko kidneys (G) (**P<0.01, *P<0.05, ANOVA). (H) Quantification of F4/80+ cells 3 days after IRI/CL-NX demonstrates no difference in macrophage numbers in the two groups. n=5 mice per group.
Dead Cell Debris Induces Expression of GM-CSF by Cultured Renal Cells and Promotes Arg1 Expression in BMMs, with Reciprocal Increased Tubular Cell Proliferation
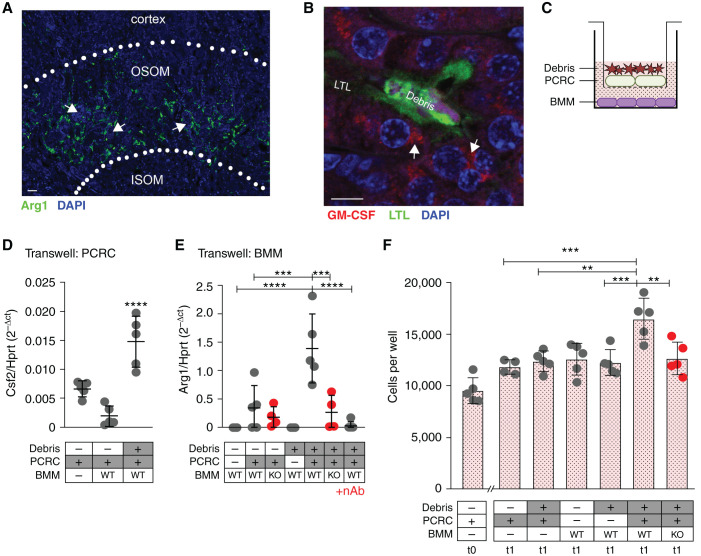
Immunofluorescent staining confirmed that arginase-1 expressing macrophages were predominantly located in the outer stripe of the renal medulla (Figure 4A). Staining for the Arg1 inducer GM-CSF revealed that surviving tubular cells adjacent to intraluminal debris were the predominant source of detectable GM-CSF (Figure 4B). Transwell coculture of PCRCs (top well) ± dead cell debris (on the apical surface of the PCRCs) ± naïve BMMs (bottom well) to mimic this outer medullary milieu revealed a significant increase in Csf2 expression by the debris-induced PCRCs, with a concomitant increase in Arg1 in cocultured wild-type BMMs (Figure 4, C–E). This effect was also seen when immortalized MPTs were exposed to urinary casts collected 1–3 days after IRI (Supplemental Figure 7). The induction of Arg1 was significantly reduced in Arg1 null macrophages and in the presence of a GM-CSF neutralizing antibody (Figure 4E).
Figure 4.
Luminal debris induces bidirectional tubular cell:macrophage cross-talk to promote tubule proliferative repair. (A) Immunofluorescent staining of mouse kidney for F4/80 after IRI/CL-NX reveals that arginase-1 (green) is limited to the outer stripe of the outer medulla near sites of intraluminal casts. (OSOM, outer stripe of the outer medulla; ISOM, inner stripe of the outer medulla; arrows indicate casts). Scale bar, 20 μm. (B) Immunofluorescent staining of mouse kidney for GM-CSF shows expression in epithelial cells adjacent to intraluminal cell debris (casts) after IRI/CL-NX (arrows indicate GM-CSF staining, LTL indicates brush border staining). Scale bar, 10 μm. (C) Schematic of Transwell experiments showing PCRCs and cultured naïve BMMs. (D) Csf2 mRNA expression is significantly higher in PCRCs cultured with cell debris on the apical surface (****P<0.0001, ANOVA). (E) Arg1 mRNA expression is significantly higher in wild-type (WT) BMMs cocultured with PCRCs exposed to debris versus PCRCs alone. Arg1 expression is significantly reduced in Arg1mko macrophages (KO) (****P<0.0001, ***P<0.001, ANOVA). (F) PCRCs were seeded at 10,000 cells per well (t0) and treated with conditioned media from Transwell cultures containing the components indicated under each bar, and cell numbers were counted at 24 hours (t1) (***P<0.001, **P<0.01, ANOVA).
Conditioned medium was generated by 24-hour culture of PCRCs ± apical cell debris ± Arg1con or Arg1mko BMMs in RPMI 1640 + 1% FBS. Conditioned medium from cultures in which Arg1 is not induced (PCRCs cultured in the absence of BMMs or BMMs cultured in the absence of PCRCs) failed to induce proliferation of PCRCs (Figure 4F). In contrast, conditioned medium from culture conditions in which Arg1 is maximally induced (debris-stimulated PCRCs cocultured with WT BMMs) stimulated a significant increase in PCRC numbers that was totally abrogated when the culture was performed with conditioned medium from Arg1mko BMMs (Figure 4F) or conditioned medium generated in the presence of a GM-CSF neutralizing antibody (Supplemental Figure 8).
Discussion
Our current experiments demonstrate that arginase-1, one of the proteins that macrophages express during the transition to alternative activation beginning on day 2 after IRI, is specifically required for the tubular cell proliferation that underlies kidney repair after injury. The loss of macrophage expression of this protein not only recapitulates the finding of slowed repair seen after nonselective macrophage depletion,3 but also leads to significantly decreased mouse survival. These experiments also provide several additional insights into how reparative activation is regulated after kidney injury and highlight the extensive cross-talk between trafficking macrophages and endogenous cell populations in the kidney.
After ischemic injury, arginase-1 is almost exclusively produced by those macrophages located in the outer medulla even though macrophage numbers also increase significantly in the cortex. The outer medulla is the site of greatest tubular cell loss and intraluminal cast formation after IRI.15 Our ex vivo Transwell cultures and immunofluorescence staining of kidneys reveal that dead cell debris at the luminal (apical) surface of tubular cells can induce increased GM-CSF/Csf2 expression, leading to the expression of arginase-1/Arg1 selectively in the macrophages in this region of greatest tubular cell loss. These findings suggest that surviving tubular cells detect the presence of luminal debris from dying cells, possibly as danger-associated molecular pattern signaling via toll-like receptors, and respond by signaling in a paracrine manner to the adjacent macrophages that in turn help stimulate proliferation to replace the lost cells.
Although transient upregulation of GM-CSF by tubular cells appears to be beneficial by inducing macrophage Arg1 expression and reparative tubule cell proliferation in the model of IRI/CL-NX used in the current studies, it is worth noting that the sustained tubular cell GM-CSF expression that was observed after unilateral IRI with the contralateral kidney intact correlated with eventual downregulation of Arg1 and late profibrotic macrophage gene expression profile and kidney fibrosis.16 Even in the current studies, in which there was successful recovery of BUN and creatinine in control mice by day 7 after IRI, LTL staining demonstrated a 77% reduction in brush border density. These findings are consistent with those of other groups who have found that there is persistent structural damage after ischemic renal injury even when BUN and creatinine return to near baseline,15,17 leading to long-term loss of renal parenchyma and function.16,18
It is known that arginase-1 metabolizes arginine to produce ornithine and urea, with ornithine subsequently metabolized to produce multiple polyamines.19,20 Polyamines (spermidine, spermine, putrescine, and cadaverine) are found in all living cells and have been implicated in cellular functions ranging from cell growth and proliferation, regulation of ion channels, apoptosis, and cell migration.8,21,22 Exactly how polyamine production in macrophages induces proliferation by the nearby tubular cells remains unclear. The scRNA-seq analysis suggests that multiple ligand-receptor interactions may be involved, and the Transwell cultures confirm that a secreted factor or factors are responsible for the in vitro proliferative response. Of interest in this regard are the studies showing that spermidine is required for hypusination of the ribosomal protein ElF5a, which is needed for translation of mRNAs important for mitochondrial function and alternative activation in macrophages.23 Finally, to perform these studies we used LysMCre/+;Arg1fl/fl mice in which Cre recombinase is expressed in macrophages only when lysozyme M expression is induced. BMMs cultured from these mice were mosaic for Arg1 with only about 50% exhibiting Arg1 knockout (Supplemental Figure 1), and the small number of intrarenal macrophages present in healthy control kidneys did not have detectable Cre-mediated excision of the floxed Arg1 exon (data not shown). In contrast, Ccr2-positive macrophages that trafficked to the kidney after IRI injury in the LysMCre/+;Arg1fl/fl mice exhibited a 90% reduction in arginase-1 expression, suggesting that activated macrophages successfully induce LysM expression.
In summary, we demonstrate that injured outer medullary epithelial cells express GM-CSF that is increased in response to apical exposure to dead cell debris, leading to high levels of expression of Arg1 by adjacent macrophages, and this tubular cell-macrophage cross-talk promotes the secretion of an as yet unidentified factor or factors that can directly stimulate the proliferative response during kidney repair.
Disclosures
L. Cantley reports consultation fees from Drug Farm (<$5,000), Goldfinch Bio, and Johnson & Johnson (<$5,000), outside of the submitted work; and reports ownership interest with Arvinas, Inc. (spouse’s employer). D. Linberg reports employment and advisory or leadership role with eCLASS AB. All remaining authors have nothing to disclose.
Funding
This work was supported by National Institutes of Health grants R01 DK093771 (to L. Cantley) and T32 DK007276-41 (to N. Shin).
Supplementary Material
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “More than a Marker: Arginase-1 in Kidney Repair,” on pages 1051–1053.
Author Contributions
L. Cantley, J. Guo, A. Marlier, and N. Shin conceptualized the study; L. Cantley, N. Doilicho, J. Guo, A. Marlier, N. Shin, and L. Xu were responsible for investigation; L. Cantley, J. Guo, and A. Marlier supervised the study; L. Cantley, N. Doilicho, A. Marlier, and L. Xu validated the study; N. Doilicho, J. Guo, D. Linberg, A. Marlier, N. Shin, and L. Xu were responsible for data curation; J. Guo, D. Linberg, A. Marlier, N. Shin, and L. Xu were responsible for formal analysis; D. Linberg was responsible for methodology; and N. Shin and L. Xu wrote the original draft; and L. Cantley was responsible for funding acquisition and project administration and reviewed and edited the manuscript.
Data Sharing Statement
Original data reported in this paper of type “experimental data” have been deposited in the Gene Expression Omnibus under GSE188966.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2021121548/-/DCSupplemental.
Supplemental Figure 1. scRNA-seq analysis of PCRC composition.
Supplemental Figure 2. Arginase-1 in GFP+ macrophages from Arg1fl/fl;LysMCre/+;Rosa26mTmG+ mice.
Supplemental Figure 3. Time-course of arginase-1 expression after kidney injury.
Supplemental Figure 4. Integrated scRNA-seq analysis of differential cell populations between wild-type IRI and control kidneys.
Supplemental Figure 5. Ccr2+ macrophages express Arg1 and are projected to promote proximal tubular epithelial cell proliferation.
Supplemental Figure 6. Cytokine expression in renal macrophages and whole kidney.
Supplemental Figure 7. Cast-treated mouse proximal tubule cells (mPTs) express GM-CSF and induce arginase-1 expression in cultured bone marrow macrophages (BMMs).
Supplemental Figure 8. Renal epithelial cell proliferation after exposure to various conditioned media.
Supplemental Table 1. Antibodies used to flow-sort cells.
Supplemental Table 2. Primers used to perform rtPCR.
References
- 1.Day YJ, Huang L, Ye H, Linden J, Okusa MD: Renal ischemia-reperfusion injury and adenosine 2A receptor-mediated tissue protection: Role of macrophages. Am J Physiol Renal Physiol 288: F722–F731, 2005 [DOI] [PubMed] [Google Scholar]
- 2.Li L, Okusa MD: Macrophages, dendritic cells, and kidney ischemia-reperfusion injury. Semin Nephrol 30: 268–277, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee S, Huen S, Nishio H, Nishio S, Lee HK, Choi BS, et al. : Distinct macrophage phenotypes contribute to kidney injury and repair. J Am Soc Nephrol 22: 317–326, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jo SK, Sung SA, Cho WY, Go KJ, Kim HK: Macrophages contribute to the initiation of ischaemic acute renal failure in rats. Nephrol Dial Transplant 21: 1231–1239, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Huen SC, Huynh L, Marlier A, Lee Y, Moeckel GW, Cantley LG: GM-CSF promotes macrophage alternative activation after renal ischemia/reperfusion injury. J Am Soc Nephrol 26: 1334–1345, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin SL, Li B, Rao S, Yeo EJ, Hudson TE, Nowlin BT, et al. : Macrophage Wnt7b is critical for kidney repair and regeneration. Proc Natl Acad Sci U S A 107: 4194–4199, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Odenlund M, Holmqvist B, Baldetorp B, Hellstrand P, Nilsson BO: Polyamine synthesis inhibition induces S phase cell cycle arrest in vascular smooth muscle cells. Amino Acids 36: 273–282, 2009 [DOI] [PubMed] [Google Scholar]
- 8.Miller-Fleming L, Olin-Sandoval V, Campbell K, Ralser M: Remaining mysteries of molecular biology: The role of polyamines in the cell. J Mol Biol 427: 3389–3406, 2015 [DOI] [PubMed] [Google Scholar]
- 9.Wei Q, Dong Z: Mouse model of ischemic acute kidney injury: Technical notes and tricks. Am J Physiol Renal Physiol 303: F1487–F1494, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schafer JA, Watkins ML, Li L, Herter P, Haxelmans S, Schlatter E: A simplified method for isolation of large numbers of defined nephron segments. Am J Physiol 273: F650–F657, 1997 [DOI] [PubMed] [Google Scholar]
- 11.Schmitt R, Marlier A, Cantley LG: Zag expression during aging suppresses proliferation after kidney injury. J Am Soc Nephrol 19: 2375–2383, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui W, Ke JZ, Zhang Q, Ke HZ, Chalouni C, Vignery A: The intracellular domain of CD44 promotes the fusion of macrophages. Blood 107: 796–805, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinha D, Wang Z, Price VR, Schwartz JH, Lieberthal W: Chemical anoxia of tubular cells induces activation of c-Src and its translocation to the zonula adherens. Am J Physiol Renal Physiol 284: F488–F497, 2003 [DOI] [PubMed] [Google Scholar]
- 14.Shih W, Hines WH, Neilson EG: Effects of cyclosporin A on the development of immune-mediated interstitial nephritis. Kidney Int 33: 1113–1118, 1988 [DOI] [PubMed] [Google Scholar]
- 15.Basile DP, Donohoe D, Roethe K, Osborn JL: Renal ischemic injury results in permanent damage to peritubular capillaries and influences long-term function. Am J Physiol Renal Physiol 281: F887–F899, 2001 [DOI] [PubMed] [Google Scholar]
- 16.Xu L, Sharkey D, Cantley LG: Tubular GM-CSF promotes late MCP-1/CCR2-mediated fibrosis and inflammation after ischemia/reperfusion injury. J Am Soc Nephrol 30: 1825–1840, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forbes JM, Hewitson TD, Becker GJ, Jones CL: Ischemic acute renal failure: Long-term histology of cell and matrix changes in the rat. Kidney Int 57: 2375–2385, 2000 [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Kumar S, Dolzhenko E, Alvarado GF, Guo J, Lu C, et al. : Molecular characterization of the transition from acute to chronic kidney injury following ischemia/reperfusion. JCI Insight 2: e94716, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM: M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol 164: 6166–6173, 2000. 10843666 [Google Scholar]
- 20.Munder M, Eichmann K, Morán JM, Centeno F, Soler G, Modolell M: Th1/Th2-regulated expression of arginase isoforms in murine macrophages and dendritic cells. J Immunol 163: 3771–3777, 1999 [PubMed] [Google Scholar]
- 21.Munder M: Arginase: An emerging key player in the mammalian immune system. Br J Pharmacol 158: 638–651, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang CI, Liao JC, Kuo L: Macrophage arginase promotes tumor cell growth and suppresses nitric oxide-mediated tumor cytotoxicity. Cancer Res 61: 1100–1106, 2001 [PubMed] [Google Scholar]
- 23.Puleston DJ, Buck MD, Klein Geltink RI, Kyle RL, Caputa G, O’Sullivan D, et al. : Polyamines and eIF5A hypusination modulate mitochondrial respiration and macrophage activation. Cell Metab 30: 352–363.e8, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.