Abstract
Objective:
We aimed to characterize the phenotypic spectrum and functional consequences associated with variants in the gene GABRB2, coding for the GABAA receptor subunit β2.
Methods:
We recruited and systematically evaluated 25 individuals with variants in GABRB2, 17 of whom are newly described and 8 previously reported with additional clinical data. Functional analysis was performed using a Xenopus laevis oocyte model system.
Results:
Our cohort of 25 individuals from 22 families with variants in GABRB2 demonstrated a range of epilepsy phenotypes from genetic generalized epilepsy to developmental and epileptic encephalopathy. Fifty-eight percent of individuals had pharmaco-resistant epilepsy; response to medications targeting the GABAergic pathway was inconsistent. Developmental disability (present in 84%) ranged from mild intellectual disability to severe global disability; movement disorders (present in 44%) included choreoathetosis, dystonia and ataxia. Disease-associated variants cluster in the extracellular N-terminus and transmembrane domains 1–3, with more severe phenotypes seen in association with variants in transmembrane domains 1 and 2 and the allosteric binding site between transmembrane domains 2 and 3. Functional analysis of 4 variants in transmembrane domains 1 or 2 (p.Ile246Thr, p.Pro252Leu, p.Ile288Ser, p.Val282Ala) revealed strongly reduced amplitudes of GABA-evoked anionic currents.
Interpretation:
GABRB2-related epilepsy ranges broadly in severity from genetic generalized epilepsy to developmental and epileptic encephalopathies. Developmental disability and movement disorder are key features. The phenotypic spectrum is comparable to other GABAA receptor-encoding genes. Phenotypic severity varied by protein domain. Experimental evidence supports loss of GABA-ergic inhibition as the mechanism underlying GABRB2-associated neurodevelopmental disorders.
Introduction
Variants in γ–aminobutyric acid type A (GABAA) receptor subunits (including GABRA1, GABRB1, GABRB3, and GABRG2) are well-established causes of epilepsy and neurodevelopmental disorders.(1–10) The associated neurologic phenotype ranges from genetic generalized epilepsies, including absence and myoclonic epilepsies and genetic generalized epilepsy with febrile seizures plus (GEFS+), to severe developmental and epileptic encephalopathies (DEE), such as Ohtahara syndrome (OS), infantile spasms (IS), Dravet syndrome and Lennox-Gastaut syndrome (LGS).(1, 2, 5, 7, 8, 11–21)
GABA receptors are comprised of a pentamer of subunits that form an anionic ligand-gated channel, including a combination of 2 α-subunits, 2 β-subunits, and 1 γ- or δ-subunit.(22) The subtype composition varies by brain region, cell type, timing of development, and neurotrophic factors, but the most common GABAA receptor composition is 2α1/2β2/1γ2.(23) Rodent data suggest that both β2- and β3-subunits are expressed during development, that the expression of β3 declines postnatally, and that β2 increases during development and is highly expressed in adult brain.(4, 24, 25) Given the established role of GABRB3 in disease,(7, 8, 19) we hypothesized that GABRB2 variants cause a similar range of neurodevelopmental disease with a prominent epilepsy phenotype. In so doing, we present evidence for a strong association between GABRB2 and neurodevelopmental disease.
Prior literature describes individuals or families with variants in GABRB2, presented as single reports or individuals within studies focusing on gene discovery.(4, 26–35) A recent clinical series of 15 patients with GABRB2 variants summarizes a broad phenotypic spectrum and highlights more severe phenotypes associated with variants in the transmembrane domains.(34) The associated phenotypes range from febrile seizures and genetic generalized epilepsy to early onset DEE with severe neurodevelopmental handicaps and variable associated symptoms.(4, 26–30, 34–36) Functional analysis of one GABRB2 variant associated with Early Myoclonic Encephalopathy showed abnormal trafficking of β2- and γ2-subunits.(31) We sought to characterize the phenotypic spectrum of individuals with variants in the gene GABRB2 by evaluating cases in an international collaboration and to evaluate functional characteristics for a subset of variants in a cell-based model.
Materials and Methods
Clinical ascertainment.
Individuals with variants in the gene GABRB2 and neurologic disease were recruited for clinical and functional characterization. Recruitment mechanisms included the following: 1) patients seen through our Epilepsy Genetics Program at Boston Children’s Hospital, 2) families who contacted the Epilepsy Genetics Research Program at Boston Children’s Hospital with interest in enrollment, 3) contact with clinicians of patients with variants in the gene GABRB2 identified by the referring clinical laboratory GeneDx or through GeneMatcher,(37) and 4) individuals recruited through a direct collaboration between centers in the US, Europe and South America. Our cohort included 17 novel individuals and 8 individuals from 6 families for whom additional clinical information was available.(26–29, 33) This study was approved by the Boston Children’s Hospital Institutional Review Board (IRB), and data shared from other centers were collected with approval from local IRBs.
Literature search.
To identify cases in the literature, a PubMed search was conducted, using the terms “GABR*”, “GABRB2” and “GABAA”, last updated October 1st, 2020. All cases including case reports, case series, and large genetic series with variants in GABRB2 and seizures or developmental delay were reviewed.
Phenotypic analysis.
Data were collected through a combination of direct patient evaluation, detailed review of medical records, and a standardized phone interview with the subset of families enrolled through the Epilepsy Genetics Program at Boston Children’s Hospital (N = 9). Thorough clinical and neurodevelopmental history, epilepsy phenotype, imaging, and metabolic and genetic testing were reviewed and collected using a standardized data collection form across centers (N=25). Primary MRI and EEG data were reviewed by the primary study team for a subset of individuals (N=7). Imaging was systematically reviewed for 6 individuals by a neuroradiologist (EY), and EEGs were reviewed for 6 individuals by 2 epileptologists (HEO, CMA) (Table S5). We defined individuals as having DEE when there was a combination of infantile/childhood onset epilepsy with an encephalopathy pattern on EEG and developmental delay or intellectual disability (ID) with both epileptic and non-epileptic factors thought to contribute to the developmental disorder.(38) Included encephalopathy patterns comprised burst suppression, hypsarrhythmia, electrical status epilepticus of sleep (ESES), and generalized slowing with focal or generalized epileptiform activity. Medication-refractory epilepsy was defined as not responsive to 2 or more tolerated and appropriately chosen anti-seizure medications at therapeutic doses.(39) Epilepsy syndromes were as defined by the International League Against Epilepsy (www.epilepsydiagnosis.org).(38)
Genotypic analysis.
The genetic findings were obtained through either targeted epilepsy panels (N=3), whole exome sequencing (N=19) or Sanger sequencing (N=3, family 2, individuals 21a-c). Variant classification was performed using the most recent American College of Medical Genetics and Genomics criteria (ACMG).(40) All variants are reported based on transcript NM_021911.2. We include scores from in silico prediction models SIFT, PolyPhen2, and CADD. We excluded variants of uncertain significance.
Functional analysis.
Functional analysis was performed as described previously.(29)
Mutagenesis and RNA preparation.
The Quick Change kit (Stratagene) was used for mutagenesis of the GABRB2 cDNA (pcDNA3vector). cRNA was prepared using the T7 RNA polymerase kit (Roche). Primers are available upon request.
Oocyte preparation and injection.
Oocytes were obtained from the Institute of Physiology I, Tuebingen, or purchased from EcoCyte Bioscience (Castrop-Rauxel). Experiments were approved by local authorities (Regierungspraesidium Tuebingen, Tuebingen, Germany). The preparation of oocytes for 2-microelectrode voltage clamp recordings included treatment with collagenase (1mg/ml of type CLS II collagenase, Biochrom KG) in OR-2 solution (mM: 82.5 NaCl, 2.5 KCl, 1 MgCl2 and 5 Hepes, pH 7.6), followed by thorough washing and storing at 16 °C in Barth solution (mM: 88 NaCl, 2.4 NaHCO3, 1 KCl, 0.41 CaCl2, 0.82 MgSO4 and 5 Tris/HCl, pH 7.4 with NaOH) supplemented with 50μg/ml gentamicin (Biochrom KG) as described previously.(29) Seventy nanoliter encoding the WT or mutant cDNA (2 μg/μl) were injected into oocytes using Robooinject® (Multi Channel Systems). Oocytes were stored for 1–3 days (at 17 °C) before the experiment. The combination of the different subunits used was α1β2γ2s in a 1:1:2 ratio.
Automated oocyte 2-microelectrode voltage clamp.
GABA-evoked ionic currents in oocytes were recorded at a holding potential of −70 mV at room temperature (20–22°C) using GABA concentrations from 1 to 1000 μM and the Roboocyte2® (Multi Channel Systems). Prepulled and prepositioned intracellular glass microelectrodes had a resistance of 0.3–1 MΩ when filled with 1 M KCl/1.5 M KAc. The extracellular solution was ND96 (im mM: 93.5 NaCl, 2 KCl, 1.8 CaCl2, 2 MgCl2, 5 Hepes, pH 7.5). Currents were sampled at 1 kHz.
Electrophysiological data analysis.
Amplitudes of GABA-induced currents were analyzed using Clampfit (pClamp 8.2, Axon Instruments), Microsoft Excel (Microsoft) and Graphpad Prism (GraphPad Software) softwares. The current response of each GABA concentration was normalized to the maximum response evoked by the highest GABA concentration (1 mM). Current amplitudes in response to 1 mM GABA application for WT and mutant channels were recorded on the same day using the same batch of oocytes, and normalized to the mean value of WT response on the respective day. Normalized data were then pooled from different days.
A 4-parameter logistic equation was fit to the normalized GABA responses of each cell:
with max and min being the maximum and minimum evoked responses and X the corresponding GABA concentration. The EC50 value is the concentration of the agonist at which half of the maximum response is achieved while nH is the Hill coefficient. EC50 values were determined for each oocyte, and averaged values are presented as mean ± SEM.
Statistical analysis.
For statistical evaluation of functional data, GraphPad Prism 6 was used. Normalized current amplitudes were compared between different groups (WT and different variants) using one-way ANOVA on ranks (Kruskal Wallis rank sum test) with Dunńs post-hoc test. For descriptive statistical analysis of clinical data, SAS version 9.4 was used.
Results
Clinical features
We report a cohort of 25 individuals (15 male, 10 female), from 22 unrelated families, with variants in the gene GABRB2 identified in children and adults referred for evaluation of epilepsy, movement disorder, or other neurodevelopmental disorders (Table 1). The median age at last evaluation was 8 years, range 2 to 62 years. Three individuals were deceased, 1 of respiratory failure in setting of pneumonia (#6), 1 of probable SUDEP (#9), and one of respiratory failure in setting of status epilepticus (#15). We describe the clinical findings in 17 newly reported individuals and additional data from 8 individuals in 6 families who were reported previously as part of genetic studies and on whom additional information became available.(26–29, 33)
Table 1:
Summary of key genetic and phenotypic characteristics of 25 individuals with variants in GABRB2. Protein domains are delineated by alternating shading in order: extracellular 1 (#1–4), helical 1 (#5–10), helical 2 (#11–13), loop M2-M3 + allosteric binding (#14a-20), helical 3 (21a-22).
| ID | Sex/Age | Variant, classification, domain | Age at seizure onset | Epilepsy type | Encephalopathy on EEG | MD | DD | Microcephaly |
|---|---|---|---|---|---|---|---|---|
| 1 | F,19y | Ala159Ser,LP | 12m | Fo+G | N | N | Y, mild | N |
| 2 | F,8y | Met161Leu,LP | 9m | Fo | N | N | Y, mild | N |
| 3 | M, 8y | Tyr181Phe, LP | 8m | Fo + G | N | N | Y, mild | N |
| 4(28) | M, 5y | Tyr183His,LP | 6m | Fo+G | N | Y | Y, mild | N |
| 5 | F,10y | Phe245Leu,LP | 3m | Fo+G | Y | N | Y, moderate | N |
| 6 | M,2yD | Ile246Thr, P | 5m | Fo+G | Y | N | Y, severe | Y(A) |
| 7 | M,3y | Ile246Thr, P | 2m | Fo+G | Y | Y | Y, severe | N |
| 8(26) | M,17y | Pro252Ala,LP | 16y | Fo+G | Y | N | Y, moderate | N |
| 9 | M,6yD | Pro252Leu, LP | 2m | G | Y | Y | Y, moderate | Y(A) |
| 10 | M,3y | Leu255Val,LP | 5m | G | Y | Y | Y, moderate | N |
| 11(33) | F,16y | Val282Ala, LP | 3y | G | Y | Y | Y, moderate | Y(A) |
| 12 (33) | M,14y | Ile288Ser, LP | 15m | Fo+G | Y | Y | Y, severe | Y(C) |
| 13 | F,3y | Ile288Thr,LP* | 4m | N/A | N | Y | Y, moderate | Y(U) |
| 14a | F,21y | Arg293Trp,P# | 13m | U | N | N | Y, mild | N |
| 14b | M, 31y | Arg293Trp,P# | 18y | Fo | N | N | N | U |
| 15 | F,4yD | Ile299Leu,LP | 19m | Fo+G | Y | Y | Y, severe | Y(U) |
| 16 | M,3y | Ile299Ser,LP | 3m | Fo+G | Y | Y | Y, severe | Y(A) |
| 17 | F,4y | Pro300Leu,LP | 5m | Fo+G | N | N | Y, moderate | N |
| 18 | M,10y | Val302Met,P | 8m | Fo+G | Y | Y | Y, moderate | Macro |
| 19 | M,5y | Lys303Arg,P | 3d | Fo+G | Y | N | Y, severe | Y(A) |
| 20 | F,14y | Ala304Thr,LP | 3y | Fo+G | Y | Y | Y, moderate | N |
| 21a(29) | M,62y | Val316Ile,P## | 11y | G | N | N | N | N |
| 21b(29) | M,37y | Val316Ile,P## | 8y | G | N | N | N | N |
| 21c(29) | M,21y | Val316Ile,P## | 9y | G | N | N | N | N |
| 22 (27) | F, 4y | Val316Ile,P | 12m | Fo+G | N | N | Y, mild | N |
M: male; F: female; LP: likely pathogenic; P: pathogenic; VUS: variant of unknown significance; y: year; m: month; d: day; G: generalized; Fo: focal; Y: yes; N: no; U: unknown; MD: movement disorder, DD: developmental disorder, A: acquired; C: congenital
variant is mosaic in the patient
Family 1
Family 2
indicates age of death, from respiratory failure in setting of pneumonia, reported SUDEP, and status epilepticus with respiratory failure in individuals 6, 9, and 15 respectively
All 25 individuals reported here harbored GABRB2 missense variants, classified as pathogenic (N=5 in 10 individuals) or likely pathogenic (N=15 in 15 individuals) (Tables 1, 2, Fig 1). All variants were absent from population databases (gnomAD, http://gnomad.broadinstitute.org/ search updated after January 2020).(41) One variant was present with apparent mosaicism (allele fraction was not specific by the clinical laboratory). All variants either occurred de novo or segregated with phenotype in two families (Table 2). One variant was inherited in a family with affected father and daughter (p.Arg293Trp). The p.Val316Ile variant was present in one individual as a de novo variant and was also present in an affected family.
Table 2:
Genetic details for variants in GABRB2 identified in 25 individuals from 22 families. Genetic variants all use reference transcript NM_021911.2.
| ID | cDNA | Protein | Inh. | Test | SIFT | P2 | CS | ACMG classification(40) |
|---|---|---|---|---|---|---|---|---|
| 1 | 475G>T | A159S | De novo | WES | 0.139 | 0.40 | 23.7 | LP: PM2,PM2,PP2,BP4 |
| 2 | 481A>C | M161L | De novo | Panel** | 0.009 | 0.76 | 26.1 | LP: PM2,PM6,PP2,PP3 |
| 3 | 542A>T | Y181F | De novo | Trio WES | 0.003 | 0.73 | 28.5 | LP: PM2,PM6,PP2,PP3 |
| 4 (28) | 547T>C | Y183H | De novo | Trio WES | 0.000 | 1.00 | 28.3 | LP: PS2,PM2,PP2,PP3 |
| 5 | 735T>A | F245L | De novo | Trio WES | 0.002 | 0.99 | 24.8 | LP: PS2,PM2,PP2,PP3 |
| 6 | 737T>C | I246T | De novo | Trio WES | 0.046 | 0.92 | 27.1 | P: PVS,PM2,PP2,PP3 |
| 7 | 737T>C | I246T | De novo | Trio WES | 0.046 | 0.92 | 27.1 | P: PVS,PM2,PP2,PP3 |
| 8 (26) | 754C>G | P252A | De novo | Trio panel** | 0.000 | 1.00 | 26.4 | LP: PS2,PM2,PP2,PP3 |
| 9 | 755C>T | P252L | De novo | Trio WES | 0.000 | 1.00 | 25.8 | LP: PS2,PM2,PM5,PP2 |
| 10 | 763C>G | L255V | De novo | Trio WES | 0.001 | 0.96 | 23.0 | LP: PS2,PM2,PP2,PP3 |
| 11 (33) | 845T>C | V282A | De novo | Trio WES | 0.000 | 1.00 | 27.4 | LP: PS[PS2+PM6],PM2,PP2,PP3 |
| 12 (33) | 863T>G | I288S | De novo | Trio WES | 0.000 | 1.00 | 28.1 | LP:PS2,PM2,PP2,PP3 |
| 13 | 863T>C | I288T | De novo * | Trio WES | 0.000 | 1.00 | 26.5 | LP: PS2,PM2,PM5,PP2 |
| 14a | 877C>T | R293W | I# | Trio WES | 0.000 | 1.00 | 28.2 | P: PS1,PS3,PM2 |
| 14b | 877C>T | R293W | U# | Trio WES | 0.000 | 1.00 | 28.2 | P: PS1,PS3,PM2 |
| 15 | 895A>C | I299L | De novo | Trio WES | 0.000 | 0.76 | 26.2 | LP: PM2,PM6,PP2,PP3 |
| 16 | 896T>G | I299S | De novo | Panel* | 0.000 | 1.00 | 28.8 | LP: PM2,PM6,PP2,PP3 |
| 17 | 899C>T | P300L | De novo | Trio WES | 0.002 | 0.99 | 29.5 | LP: PM2,PM6,PP2,PP3 |
| 18 | 904G>A | V302M | De novo | Trio WES | 0.001 | 0.99 | 28.3 | P: PS1,PS2,PM2,PP2 |
| 19 | 908A>G | K303R | De novo | Trio WES | 0.000 | 1.00 | 27.5 | P: PS1,PS2,PM2,PP |
| 20 | 910G>A | A304T | De novo | Trio WES | 0.001 | 1.00 | 28.8 | LP: PS1,PM2,PM6,PP2 |
| 21a(29) | 946G>A | V316I | U## | SS | 0.032 | 1.00 | 24.7 | P: PS1,PS3,PM2,PP1 |
| 21b (29) |
946G>A | V316I | I## | SS | 0.032 | 1.00 | 24.7 | P: PS1,PS3,PM2,PP1 |
| 21c (29) |
946G>A | V316I | I## | SS | 0.032 | 1.00 | 24.7 | P: PS1,PS3,PM2,PP1 |
| 22 (27) | 946G>A | Va316I | De novo | WES | 0.032 | 1.00 | 24.7 | P: PS1,PS3,PM2,PP1 |
Inh.=Inheritance, U=Unknown, P2=polyPhen-2, CS= CADD Score, SS=Sanger Sequencing, I=Inherited
Family 1,
Family 2
mosaic,
NGS panel,
Singleton test
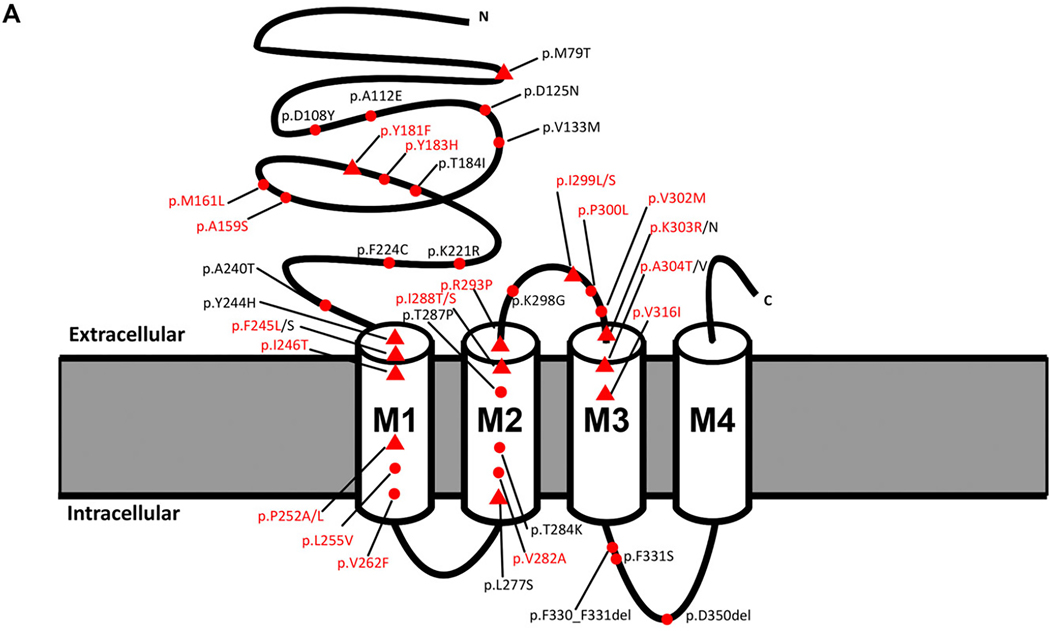
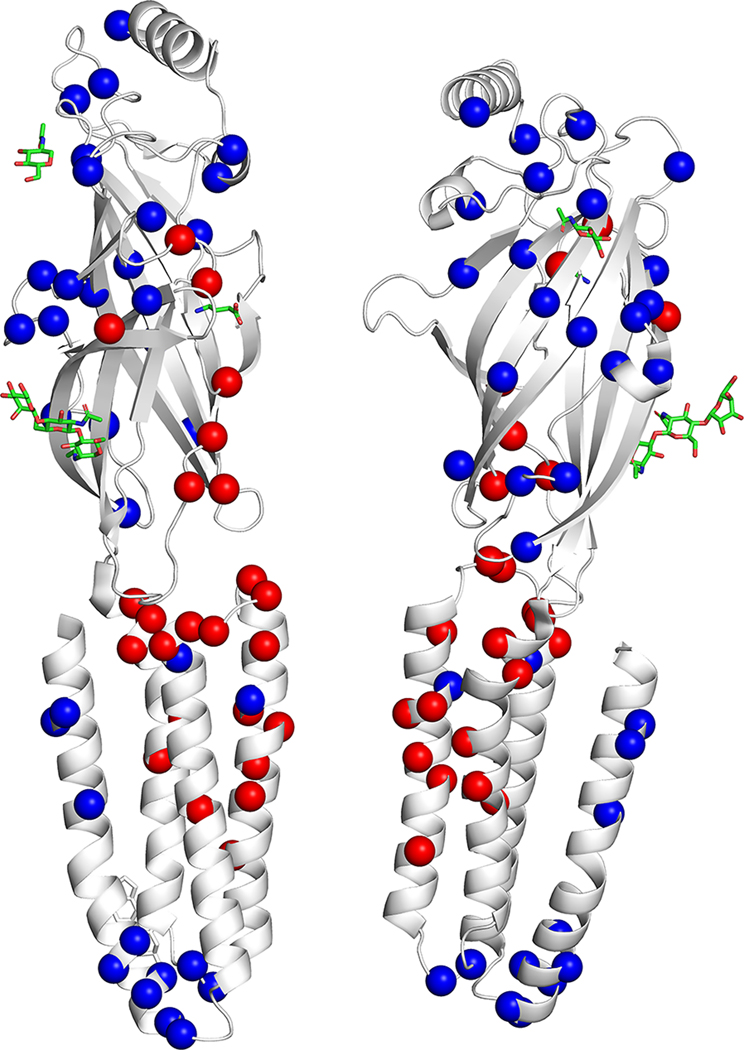
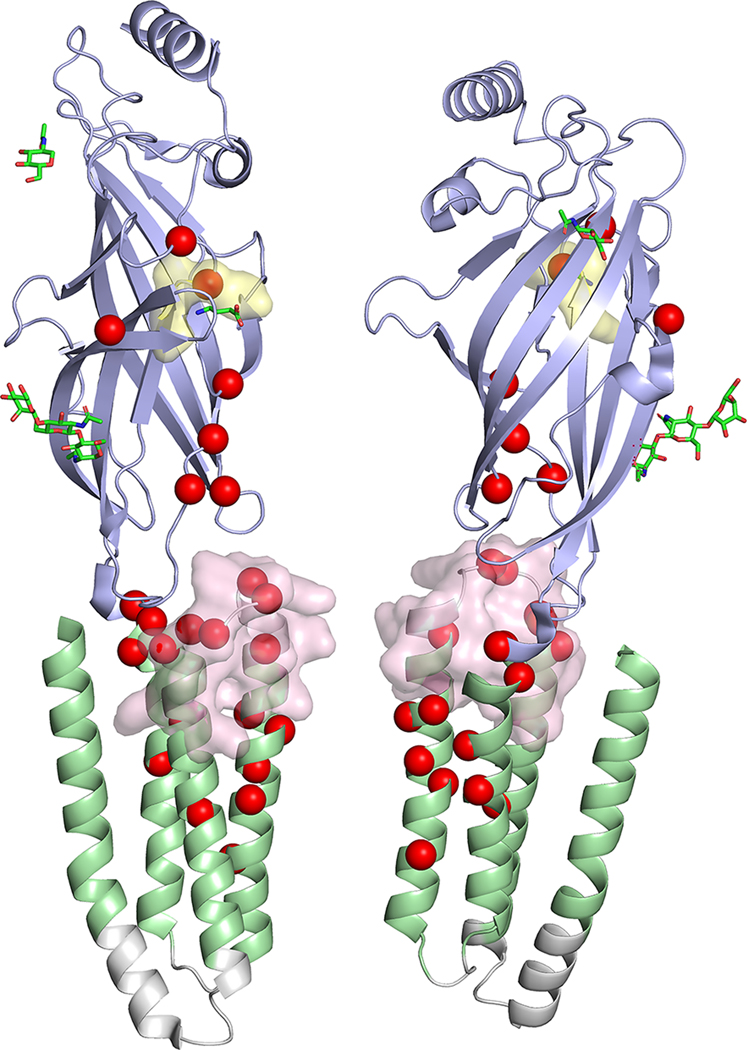
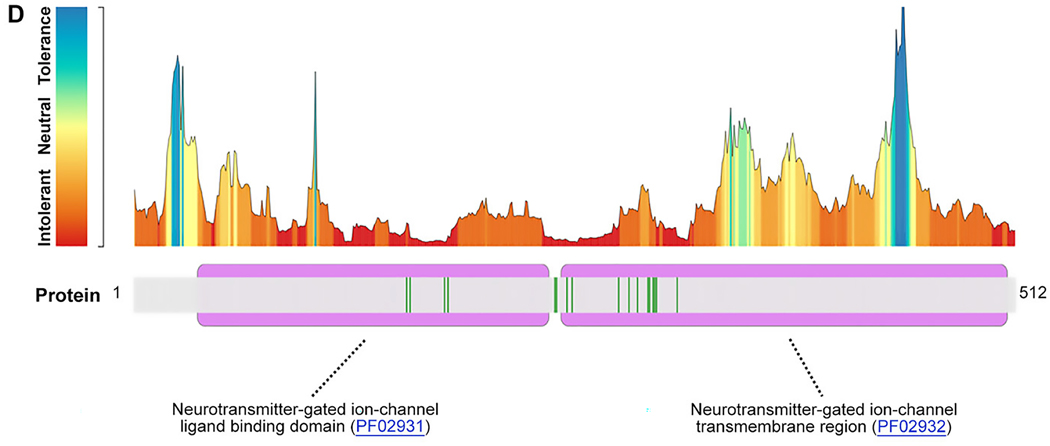
FIGURE 1:
Location of GABRB2 variants in our cohort, from the literature, and in control populations. (A) Variant location in individuals reported in this study (red text) and from the literature in a 2-dimensional model of the GABRB2 protein. Red spheres: single cases; red triangles: recurrent variants. (B) Location of missense variants in affected individuals (red spheres) and normal controls from gnomAD (blue spheres) mapped on the experimentally solved model of human β2-subunit (Protein Data Bank [PDB] ID: 6D6T, chain A and C).48 Disease-associated variants are in regions lacking variation in controls. (C) Disease-associated variant locations (red spheres) by β2-subunit protein domain (PDB ID: 6D6T, chain A and C). The extracellular and helical transmembrane domains are highlighted in purple and green, respectively. Two functionally critical regions, the agonist binding region (yellow, harbors the patient variant: p.Tyr181Phe) and the allosteric effector binding region (pink, contains 12 patient variants) are also shown in the structure. (D) GABRB2 tolerance landscape using MetaDome.49 Functional domains are highlighted in purple. Vertical green lines in the schematic representation of the GABRB2 protein indicate the locations where (likely) pathogenic missense variants were observed. From this visualization, it can be concluded that all missense variants locate to regions of GABRB2 that are intolerant to functional variation.
Missense variants were located in regions of the protein that are intolerant to functional variation (Figure 1C). The 20 unique variants occurred at 17 different amino acid positions. Thirteen amino acid positions were associated with the same recurrent or different missense variants in 2 or more unrelated individuals in this series or overlapping with reported literature within the N-terminal, transmembrane domains and the extra-cellular loop between M2-M3 (Figure 1, Table S1).
Epilepsy was diagnosed in 24/25 individuals (96.0%): 6 generalized, 2 focal, 15 combined, and 2 of unknown type (Table 1). Median age of onset was 9 months with a range from 3 days to 18 years, including 15 with onset before age 1 year. Of 13 individuals with DEE, median age at seizure onset was 5 months, range 3 days to 16 years, all with early developmental disabilities. Generalized seizure types included generalized tonic-clonic (GTC), tonic, spasms, myoclonic, atonic, typical and atypical absence (Table S2). Focal seizure types included motor (focal tonic, focal clonic, focal myoclonic) and non-motor. GTCs with focal or generalized onset occurred in at least 17 of 24 individuals with epilepsy (70.8%). Spasms were seen in 2 individuals and myoclonic seizures (focal or generalized) in 6. Typical absence seizures were reported in 5 individuals including three from a single family with Genetic Generalized Epilepsy (GGE) (p.Val316Ile, individuals 22–24),(29) and atypical absence seizures were reported in an additional 3 individuals. Fever sensitivity, defined as a frequent trigger for seizures, was reported in 12 of 24 individuals (50.0%). Eleven of 24 individuals with epilepsy (45.8%) had at least one episode of status epilepticus, including convulsive and non-convulsive types.
Syndrome classification included early onset epileptic encephalopathy with burst suppression including Early Myoclonic Encephalopathy (2); early onset infantile spasms (2), Febrile Seizures plus (FS+) or Dravet-like (3), Lennox-Gastaut syndrome-like (3), epileptic encephalopathy with continuous spike wave in slow-wave sleep (CSWS) (1), and temporal lobe epilepsy (1) (Table S2).
Epilepsy was medically refractory in 13 of 24 (54.1%) individuals. Seizure frequency was daily in at least 8 of 24 individuals (33.3%) (Table 3).
Table 3.
Additional neurologic characteristics for 25 individuals with GABRB2 variants.
| ID | Intractable epilepsy | SE | Epilepsy syndrome | CVI | Cognitive impairment | Movement disorder and type |
|---|---|---|---|---|---|---|
| 1 | No | Yes | Dravet-like | No | ID, mild | No |
| 2 | No | No | FS+ | No | LD | No |
| 3 | Yes | Yes | Dravet-like | No | U | No |
| 4 | Yes | Yes | No | U | ID, mild | Ataxia, asymmetric |
| 5 | No | No | No | No | ID, unspecified | No |
| 6 | Yes | Yes | LGS-like | Yes | U | No |
| 7 | Yes | No | EME | Yes | ID, severe | Severe dystonia |
| 8 | No | No | CSWS | Yes | ID, severe | No |
| 9 | Yes | Yes | EOEE, IS, LGS-like | Yes | ID, severe | Choreoathetosis, dystonia, dyskinesias |
| 10 | Yes | No | No | Yes | U | Choreoathetosis, dystonia, tremors, opsoclonus-like eye movements, paroxysmal downgaze |
| 11 | No | No | No | Yes | U | Chorea |
| 12 | Yes | Yes | LGS-like | Yes | ID, unspecified | Chorea |
| 13 | U | U | No | U | U | Hyperkinetic, chorea |
| 14a | Yes | Yes | No | No | Borderline | No |
| 14b | Yes | No | TLE | No | None | No |
| 15 | Yes | Yes | No | Yes | Unknown | Chorea, dyskinesias, myoclonus |
| 16 | Yes | No | IS | No | Unknown | Choreoathetosis |
| 17 | No | No | No | No | ID, unspecified | No |
| 18 | No | Yes | No | No | ID, unspecified | Ataxia and chorea, hyperkinetic |
| 19 | Yes | Yes | EOEE-BS | U | ID, profound | No |
| 20 | Yes | Yes | No | U | ID, unspecified | Ataxic gait |
| 21a | No | No | JAE | No | None | No |
| 21b | No | No | JAE | No | None | No |
| 21c | No | No | CAE | No | None | No |
| 22 | No | No | No | No | None | No |
Abbreviations: CAE: childhood absence epilepsy; CSWS: continuous spike wave in slow-wave sleep; CVI: cortical visual impairment, EME: early myoclonic encephalopathy; EOEE: early onset epileptic encephalopathy; EOEE-BS: early onset epileptic encephalopathy with burst suppression; FS+: febrile seizures plus; ID: intellectual disability IS: infantile spasms; JAE: juvenile absence epilepsy; LD: learning disability; LGS: Lennox Gastaut Syndrome; SE: Status Epilepticus; TLE: temporal lobe epilepsy, U:unknown
EEGs showed epileptic encephalopathy patterns in 13 of 25 individuals (52.0%, Table 1). These included severe early encephalopathy patterns of hypsarrhythmia and/or burst suppression in 5 individuals in the first 2 years of life, ESES in 2 individuals, and nonspecific slowing with generalized and/or multifocal sharp waves in 6 individuals (Supplemental Table 1). In 12 individuals, EEG background was normal but in some cases demonstrated focal (N=4) or generalized (N=8) epileptiform activity. Two individuals with generalized epilepsy had a photo-paroxysmal response.
Response to anti-seizure medications is available in detail in Table S6. Detailed analysis did not show that anti-seizure medications targeting the GABA pathway, such as benzodiazepines, vigabatrin, and phenobarbital, were either more beneficial or harmful compared to anti-seizure medications with other mechanisms. Levetiracetam and valproic acid were most commonly used but were not clearly superior to other medications in response rate. The ketogenic diet and modified Atkins diet were reported as beneficial in 3 of 4 individuals. Age at initiation and discontinuation of medications was not available in detail in most individuals.
Developmental disability was reported in 21 of 25 individuals (84.0%, Table 1), of whom all except one had confirmed epilepsy. Of these, 6 were severely impaired with lack of head control and predominantly nonspecific vocalizations. Nine were moderately to severely globally impaired, and 6 had mild neurodevelopmental delays, ID or learning disability (Table S3). Severity was determined based on best attained milestones and cognitive level. Those individuals who were most severely impaired developmentally additionally had severe epilepsy. Age at seizure onset was also associated with severity of developmental impairment, median 5 months [IQR 3 months, 1.6 years] for those with moderate to severe developmental impairment compared to 1 year [IQR 9 months, 9 years] for those with normal development or mild impairment (Wilcoxon rank sum test p = 0.03). Five (20.0%) had reported developmental regression that occurred during infancy in 2 individuals, in the setting of severe movement disorder requiring prolonged hospitalization at age 3 years in 1 individual, and consisted of language regression in school age in the other 2 individuals. One of the individuals with language regression in school age had periodic improvement and decline in developmental skills with associated behavioral changes and sleep disturbance, unrelated to epileptic encephalopathy (# 5). Four individuals (16.0%) had a normal developmental profile. The behavioral and psychiatric profile consisted of 5 individuals with a notable social/happy personality, 5 with ADHD or hyperactivity, 3 with anxiety, 2 with obsessive compulsive disorder, 2 with aggressive behavior, 1 with delusions, and 1 with hallucinations and ritualistic behaviors. Seven individuals (28.0%) had autistic features including poor eye contact, motor stereotypies, repetitive behaviors and ritualistic behaviors. Cortical visual impairment was reported in 8 of 25 individuals (32.0%).
Movement disorders were reported in 11 individuals (44.0%), including chorea or choreoathetosis (8), dystonia (3), ataxia (3), hyperkinetic (2), not otherwise specified dyskinesias (2), tremors (1), non-epileptic myoclonus (1), and forced paroxysmal downgaze (1) (Table 3). In individual #10, the movement disorder was the presenting symptom and the severity resulted in chronic hospitalization culminating in placement of deep brain stimulation. Thalamic DBS resulted in substantial improvement in his dystonia, choreoathetosis, developmental progress and seizures. Movement disorders were chronic, with onset in infancy to early childhood, in all 11 individuals. We demonstrate representative movement disorders in the supplemental videos. Diagnosis of movement disorder was more common in the presence of an encephalopathy pattern on EEG (9 of 13, 69%, compared to 2 of 12, 17%, Fisher’s exact test p = 0.02) and moderate to severe cognitive/developmental impairment (10 of 15, 66% compared to 1 of 10, 10%, Fisher’s exact test p = 0.01). Median seizure onset was 6 months [IQR 3 months, 1.6 years] for those with movement disorder compared to 1 year [IQR 5 months, 9 years] for those without movement disorder, but the relationship does not reach statistical significance (Wilcoxon rank sum test, p = 0.2).
On neurological examination, strabismus was reported in 8 (32.0%, Table S4). Tone was abnormal in 18 individuals (72.0%) including hypotonia (11, axial or diffuse), hypertonia (5, spasticity, rigidity or dystonia) or mixed tone abnormalities (2). Both hypotonia and hypertonia were seen in infancy, childhood, and adolescence in different patients. Tone varied with age in some individuals from hypotonia to normal tone. None developed hypotonia after having had a normal exam or hypertonicity. Reflexes were increased in 7 individuals (28.0%) in association with hypotonia, hypertonia or mixed tone abnormalities. Two individuals had reported areflexia on physical examination. The first had hyperreflexia initially but was always described as hypotonic (#9). The second had hypertonia with no report of areflexia over time (#11). Head size was reported as normal in 15, microcephaly in 8, macrocephaly in 1 (familial), and unknown in 1 (Table 1). Microcephaly was congenital in 1 individual, acquired in 5, and subtype was unknown in the other 2. Descriptions of mild characteristic facial and limb features not fitting a specific pattern were reported in 8 of 25 individuals (32.0%) including flat facies, up-slanting or down-slanting palpebral fissures, hypertelorism, ptosis, low set dysplastic pinnae, short philtrum, micrognathia, high arched palate, and tapered fingers (Table S4).
In terms of overall outcomes, at the time of this analysis, 3 individuals had died; all 3 had had DEE with early onset seizures, refractory epilepsy, and severe developmental disability. Twelve individuals became seizure-free (including individual #3 who was initially refractory), 3 with no developmental disabilities or cognitive impairment and 8 with mild or moderate developmental disabilities. Seizure outcome data were unavailable on 1 individual (#13). The remaining 9 had refractory epilepsy, 2 with no developmental disabilities or cognitive impairments, and 7 with a range of developmental disabilities. Movement disorders were chronic and persistent in those affected (N=11), including one (#10) with high severity requiring deep brain stimulation.
Brain MRI was reported as normal in 13 of 23 individuals for whom data were available (56.5%, Table S5). Mild volume loss with prominent sulci and/or mild lateral ventriculomegaly indicating grey or white matter volume loss was observed in 5 individuals and borderline low volume was seen in an additional 2; imaging from 5 of these 7 individuals was reviewed formally by our neuroradiologist. Neuroimaging was initially normal in at least 2 of the 7 individuals with grey or white matter volume loss. Other nonspecific abnormalities each seen in a single individual included possible cortical dysplasia, hippocampal volume loss, hippocampal inversion/under-rotation, nonspecific T2 signal change or gliosis, mild hyperintensity of the splenium of the corpus callosum, thick corpus callosum, and Chiari malformation with syrinx. Neuroimaging data were not available for 1 individual and another had only a head CT scan that was normal.
Localization of variants in the protein
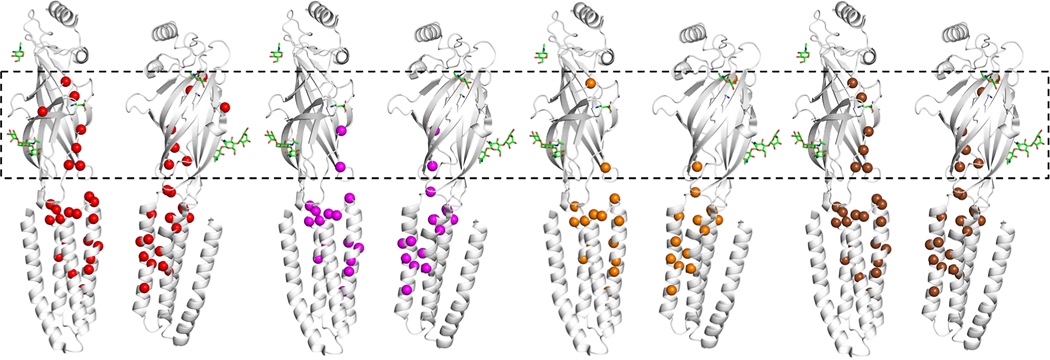
Overall, disease-associated variants in our series and the literature are clustered in the functional core of the protein, mainly the N-terminus and transmembrane regions M2 and M3 (shown in red in Fig 1B). Population-based variants derived from the database gnomAD (gnomAD browser: http://gnomad.broadinstitute.org/) are distributed differently in functionally less important regions, primarily in the outer surface of the pentameric GABA(A) receptor complex (shown in blue in Fig 1B). Variants in the extracellular N-terminus and in M3 were associated with a less severe phenotype including predominantly a milder neurodevelopmental disability and less often epileptic encephalopathy and movement disorder compared to variants in M1, M2 and the allosteric binding domain between M2 and M3 (Fig 1, Figure 3), as confirmed by Fisher’s Exact testing. Of 17 individuals with variants in M1, M2 or in the allosteric binding domain, moderate to severe developmental disability was reported in 15 (88.2%), epileptic encephalopathy patterns were reported on EEG in 13 (76.5%), and movement disorder was reported in 10 (58.8%). In contrast, of 8 individuals with variants in the N-terminus and M3, all had normal development or mild developmental disability (p<0.0001), none had an epileptic encephalopathy pattern on EEG (p = 0.0005), and movement disorder was reported in 1 (12.5%, p=0.04).
FIGURE 3:
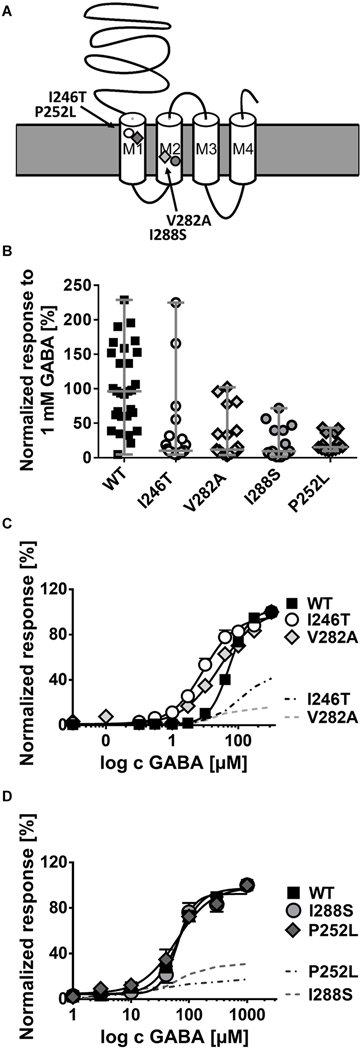
Functional analysis in Xenopus oocytes of 4 de novo GABRB2 variants associated with developmental and epileptic encephalopathies. (A) Schematic representation of the β2-subunit of the γ–aminobutyric acid type A (GABAA) receptor showing the localization of the variants. (B) Normalized current response to 1mM GABA application for α1β2γ2s wild-type (WT) receptors (n = 27), and receptors carrying the variants p.Val282Ala (V282A, n = 16), p.Pro252Leu (P252L, n = 14), p.Ile288Ser (I288S, n = 10), and p.Ile246Thr (I246T, n = 21) in the β2-subunit. Shown are all data points for each oocyte and median with range ***p<0.001, ****p<0.0001 (Kruskal-Wallis with Dunńs posthoc test). (C) Dose-response curve for WT (n = 10), I246T (n = 16) and V282A (n = 16) containing receptors after application of different GABA concentrations ranging from 0.001 μM to 1mM and normalization to the maximal GABA response for each cell. (D) Dose-response curve for WT (n = 21), p.Ile288Ser (I288S, n = 12) and p.Pro252Leu (P252L, n = 14) as in C. Shown in C and D are means ± SEM (error bars are sometimes smaller than symbol size).
Functional analysis
We performed functional analysis of 4 de novo GABRB2 variants identified in the first individuals with DEE phenotypes enrolled in this study. They are located in M1 (p.Ile246Thr, p.Pro252Leu) and M2 (p.Ile288Ser, p.Val282Ala) segments (Fig 2), all affecting conserved amino acids. Data are presented as median with range. Statistically significant differences with p values are indicated in the legend to Figure 2.
FIGURE 2:
Location of all disease-associated variants (red spheres) mapped on the experimentally solved model of human β2-subunit (Protein Data Bank ID: 6D6T, chain A and C),48 and variants associated with electroencephalographic encephalopathy (magenta spheres), movement disorder (orange spheres), and developmental disorder (brown spheres). Spatial distributions of variants by phenotypes show that patient variants located in the extracellular domain (in the dashed box) are mostly associated with developmental delay and less often with epileptic encephalopathy or movement disorder.
GABA-evoked ionic currents were recorded with an automated 2-microelectrode voltage clamp technique from oocytes expressing wild-type (WT) or mutant β2-subunits together with WT α1- and γ2-subunits. After application of 1 mM GABA, we observed a significant reduction of amplitudes of GABA-induced anion currents for all 4 variants in comparison to the WT (Fig 2B). The GABA sensitivity of the altered receptors was studied by applying different GABA concentrations. Dose-response curves of variants, p.Ile246Thr and p.Val282Ala, indicated a higher GABA sensitivity upon application of very low GABA concentrations of 0.01 to 10 μM (Fig 2C). However, taking into account the observed reduction in current amplitude (Fig 2B), an overall effect of these variants within the whole range of GABA applications, including synaptically relevant high concentrations, is a loss of function (Fig 2C, dashed lines). For p.Ile288Ser and p.Pro252Leu variants we observed no change in GABA sensitivity (Fig 2D). The EC50 values and the statistical difference between mean GABA sensitivity expressed as EC50 for each variant compared to wild type determined with the Kruskal-Wallis and Dunn’s multiple comparison test were as follows (means ± SEM: WT 80 ± 10 μM; p.I246T 11 ± 3 μM (p<0.0001); p.V282A 27 ± 5 μM (p<0.001); p.I288S 57 ± 6 μM (p>0.99); p.P252L 47 ± 9 μM (p=0.6).
Discussion
Through our analysis of 25 individuals and the literature, we delineate a range of neurodevelopmental phenotypes associated with GABRB2 variants, including various epilepsy phenotypes, developmental disabilities, and movement disorders. We newly report movement disorders with onset in infancy, at times severe enough to consider deep brain stimulation, and in one case as the presenting feature. We further describe preliminary differences in phenotypic severity based on location of variants within the protein. Functional analysis of 4 missense variants in transmembrane domains 1 and 2, all associated with severe neurological phenotypes, demonstrates a loss of function of GABA-induced anion currents.
All but 1 individual in our cohort had confirmed epilepsy, and the 1 individual without epilepsy had events that were considered suspicious for seizures. This high rate of epilepsy may represent an ascertainment bias, however, as many individuals were investigated because of their diagnosis of epilepsy. We demonstrated a wide range of epilepsy types and syndromes from GGE to DEEs as well as a wide range of developmental disabilities, overlapping closely with those reported in association with variants in other GABAA receptor subunit genes. (1–21) Distinct fever sensitivity is noted in half of our cohort, as in prior reports of GABAA receptor subunit variation in association with syndromes including Genetic Epilepsy with Febrile Seizures plus spectrum and Dravet syndrome.(5, 7, 13, 17, 20) A recent series also demonstrated fever sensitivity in 13 out of 15 individuals.(34) We newly report association of age of seizure onset with severity of developmental impairment, a relationship that is not unexpected but not previously assessed by studies of GABAA receptor-associated neurodevelopmental disorders. The effect of GABAergic medications on seizures was mixed for patients with the 2 variants associated with higher receptor sensitivity to GABA (p.Ile246Thr and p.Val282Ala) (Table S6). Retrospective assessment of treatment responses did not suggest a difference in response to anti-seizure medications targeting GABA pathways compared to other mechanisms. Prior studies of epilepsy related to GABAA receptor subunit variants did not specifically evaluate treatment response or report distinct patterns. Retrospective data collection and cohort size limit our ability to assess response to medications with regards to age or particular seizure types.
Movement disorders were observed in 44% of this cohort, including choreoathetosis, dystonia, ataxia, and tremors, and demonstrated association with encephalopathy on EEG and moderate to severe developmental impairment. Similarly, 5 of the 11 individuals reported by Hamdan et al. with GABRB2 variants (45%) had movement disorders including incoordination, ataxia, dystonia, dyskinesia and choreoathetosis.(27) Prominent infantile onset movement disorder, as seen in 5 of the 11 individuals with movement disorders in this series, were not previously described in individuals with variants in other GABAA receptor genes. Ataxia is reported in 4 and dyskinesias in 1 of 22 individuals with GABRB3 variants in the largest reported series and in scattered additional cases.(2, 7, 32) Choreoathetosis and dystonia are reported in 2 individuals with GABRA1 variants.(14) The overlap of movement disorders and epilepsy is well-described in a number of other genetic disorders, including FOXG1, FRRS1L, GNAO1, PRRT2, SLC2A1, STXBP1 and others.(42–44) The spectrum of movement disorders seen in the setting of variants in GABAA receptor subunit genes and phenotype-genotype correlations warrants further study. A key feature of this cohort design is that individuals were enrolled at various ages, and it is possible that the phenotypic developmental delay and movement disorders may develop or be diagnosed later for some of the individuals enrolled in this study.
Functional analysis indicates a loss-of-function mechanism for GABRB2, comparable to findings for variants in GABRB3 and other GABAA receptor genes.(7, 10, 29, 45, 46) Two of the variants (p.Ile246Thr and p.Val282Ala) led to a higher GABA sensitivity at low concentrations but did not impact the profound reduction of currents evoked by physiologic GABA concentrations during synaptic transmission. The suggested epileptogenic mechanism is therefore a disinhibition at GABAergic synapses, providing a target for the development of future attempts at precision medicine to treat GABRB2-related conditions. Reduced GABA-evoked peak current amplitudes and abnormal subunit trafficking are mechanisms identified for disease associated-variants in other GABAA receptor β-subunit genes.(7, 45, 46) Similarly, functional studies of DEE-associated de novo variants in GABRB3 and GABRB1 in an HEK cell model show reduced peak current amplitudes in LGS-associated variants and altered kinetics in IS-associated variants.(45)
We highlight several amino acid positions with disease-associated missense changes affecting more than 1 individual in this series or the literature.(26–28, 30, 32, 34) Disease-associated variants, unique and recurrent, are localized in regions intolerant to functional variation (lacking variation in control populations) including portions of the extracellular domain, as well as the extracellular agonist binding region, the helical transmembrane domains near the gating and pore regions, and the allosteric effector binding region. Variants detected in controls, as provided by gnomAD, are in functionally less important regions, primarily in the outer surface of the GABAA receptor complex (Fig 1B). Notably, we identified only missense variants. This may reflect that truncating variants or deletions may be either better tolerated or prenatally lethal. The pLI score (probability of being loss-of-function intolerant) in gnomAD is 0.85, suggesting that GABRB2 truncating variants or deletions are not likely lethal. A mouse model of missense knock-in versus knock-out of the γ2-subunit demonstrated absence seizures in both models but a severe phenotype with thermal seizure susceptibility in the γ2(R43Q) knock-in.(47) Further study is needed to determine the impact of loss-of-function variants compared to missense variants.
While the cohort size is small for assessment of genotype-phenotype correlations, we observed more severe phenotypes in association with variants in M1 and M2 and the allosteric binding site between M2 and M3 compared to variants in the extracellular N-terminus and M3. This finding is consistent with recent work highlighting a more severe phenotype in association with variants in the transmembrane domains.(34) Localization of 97 variants found in different GABAA receptor subunit genes in monogenic epilepsies were contrasted to variants listed in gnomAD, and significant differences were found exactly in defined and highly conserved regions of the N-terminus, M1–3 and the linker region between M2 and M3.(10) Mild and severe phenotypes were not differentiated in that review. Further studies of larger cohorts are needed to validate our genotype-phenotype correlations and identify others.
Given the overall small number of individuals, variant-specific genotype-phenotype correlation is more difficult than domain-specific genotype phenotype correlation. Some of the recurrent amino acid positions were associated with similar phenotypes (e.g. 181 with Dravet and Dravet-like Syndrome, 316 with GGE in 3 family members and generalized epilepsy with mild delays in 1 unrelated individual), whereas others showed no phenotypic similarities (e.g. 252 with CSWS in one individual and EOEE/IS in another).
In conclusion, we delineate the epilepsy and other neurodevelopmental phenotypes associated with variants in GABRB2, underscoring the wide range of epilepsy and developmental phenotypes and calling attention to movement disorders as a key feature which have been previously unrecognized. We highlight functionally important regions of the β2-subunit in which disease-associated variants, including some recurrent variants, are located, and note that these are distinct from regions in which there are benign polymorphisms in the general population. Finally, we demonstrate loss of function in a cellular model. We thus provide several lines of evidence to establish the role of GABRB2 in neurodevelopmental disease. Given the different manifestations of GABRB2, this gene should be considered as a possible cause for patients presenting with any of the above symptoms, alone or in combination. While we did not identify specific treatment responses to GABAergic or other classes of anti-seizure medication, early genetic diagnosis and future prospective studies may better identify treatments beneficial to treat the seizures and movement disorders associated with GABRB2 and other GABAA receptor encoding genes. Genetic diagnosis will also be critical in the case of future disease-modifying therapy or gene-specific clinical trials.
Supplementary Material
Acknowledgments:
We thank the individuals and families who participated in the collection of clinical data for this project and for enrolling in our research studies. We thank our colleagues who referred families to our center and our clinical colleagues who evaluated patients. HEO was supported by the National Institute of Neurologic Disorders and Stroke (K23 NS107646-02 and NINDS 5K12NS079414-02), and by the Administrative Core of Boston Children’s Hospital IDDRC, 1U54HD090255. AP was supported by the Boston Children’s Hospital Translational Research Program. RSM was supported by the Novo Nordisk Foundation (NNF19OC0058749). HL was supported by the German Research Foundation (DFG, FOR-2715, grant LE1030/16-1).
SW receives funding from FWO (FKM 1861419N) University of Antwerp (BOF-FFB180053), and the EU H2020 program (856592). PS and FZ developed this work within the framework of the DINOGMI Department of Excellence of MIUR 2018–2022 (legge 232 del 2016). KS is supported by Ministry of Health of the Czech Republic, grant nr. AZV NU20-04-00279.
Sequencing and analysis for individual #10 was provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) and was funded by the National Human Genome Research Institute, the National Eye Institute, and the National Heart, Lung and Blood Institute grant UM1 HG008900 and in part by National Human Genome Research Institute grant R01 HG009141.
Footnotes
Potential Conflicts of Interest:
ET and FZ are employees of GeneDx, Inc. GeneDx is a referring clinical laboratory for this study. The remaining authors have nothing to report.
References
- 1.Baulac S, Huberfeld G, Gourfinkel-An I, Mitropoulou G, Beranger A, Prud’homme JF, et al. First genetic evidence of GABA(A) receptor dysfunction in epilepsy: a mutation in the gamma2-subunit gene. Nat Genet. 2001;28(1):46–8. [DOI] [PubMed] [Google Scholar]
- 2.Burgess R, Wang S, McTague A, Boysen KE, Yang X, Zeng Q, et al. The Genetic Landscape of Epilepsy of Infancy with Migrating Focal Seizures. Ann Neurol. 2019;86(6):821–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Epi KC, Epilepsy Phenome/Genome P, Allen AS, Berkovic SF, Cossette P, Delanty N, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501(7466):217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirose S Mutant GABA(A) receptor subunits in genetic (idiopathic) epilepsy. Prog Brain Res. 2014;213:55–85. [DOI] [PubMed] [Google Scholar]
- 5.Kang JQ, Macdonald RL. Molecular Pathogenic Basis for GABRG2 Mutations Associated With a Spectrum of Epilepsy Syndromes, From Generalized Absence Epilepsy to Dravet Syndrome. JAMA Neurol. 2016;73(8):1009–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macdonald RL, Kang JQ, Gallagher MJ. GABAA Receptor Subunit Mutations and Genetic Epilepsies. In: th, Noebels JL, Avoli M, Rogawski MA, Olsen RW, Delgado-Escueta AV, editors. Jasper’s Basic Mechanisms of the Epilepsies. Bethesda (MD)2012. [PubMed] [Google Scholar]
- 7.Moller RS, Wuttke TV, Helbig I, Marini C, Johannesen KM, Brilstra EH, et al. Mutations in GABRB3: From febrile seizures to epileptic encephalopathies. Neurology. 2017;88(5):483–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papandreou A, McTague A, Trump N, Ambegaonkar G, Ngoh A, Meyer E, et al. GABRB3 mutations: a new and emerging cause of early infantile epileptic encephalopathy. Dev Med Child Neurol. 2016;58(4):416–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rochtus A, Olson HE, Smith L, Keith LG, El Achkar C, Taylor A, et al. Genetic diagnoses in epilepsy: The impact of dynamic exome analysis in a pediatric cohort. Epilepsia. 2020. [DOI] [PMC free article] [PubMed]
- 10.Maljevic S, Moller RS, Reid CA, Perez-Palma E, Lal D, May P, et al. Spectrum of GABAA receptor variants in epilepsy. Curr Opin Neurol. 2019;32(2):183–90. [DOI] [PubMed] [Google Scholar]
- 11.Dibbens LM, Feng HJ, Richards MC, Harkin LA, Hodgson BL, Scott D, et al. GABRD encoding a protein for extra- or peri-synaptic GABAA receptors is a susceptibility locus for generalized epilepsies. Hum Mol Genet. 2004;13(13):1315–9. [DOI] [PubMed] [Google Scholar]
- 12.Dibbens LM, Harkin LA, Richards M, Hodgson BL, Clarke AL, Petrou S, et al. The role of neuronal GABA(A) receptor subunit mutations in idiopathic generalized epilepsies. Neurosci Lett. 2009;453(3):162–5. [DOI] [PubMed] [Google Scholar]
- 13.Johnston AJ, Kang JQ, Shen W, Pickrell WO, Cushion TD, Davies JS, et al. A novel GABRG2 mutation, p.R136*, in a family with GEFS+ and extended phenotypes. Neurobiol Dis. 2014;64:131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kodera H, Ohba C, Kato M, Maeda T, Araki K, Tajima D, et al. De novo GABRA1 mutations in Ohtahara and West syndromes. Epilepsia. 2016;57(4):566–73. [DOI] [PubMed] [Google Scholar]
- 15.Lachance-Touchette P, Brown P, Meloche C, Kinirons P, Lapointe L, Lacasse H, et al. Novel alpha1 and gamma2 GABAA receptor subunit mutations in families with idiopathic generalized epilepsy. Eur J Neurosci. 2011;34(2):237–49. [DOI] [PubMed] [Google Scholar]
- 16.Maljevic S, Krampfl K, Cobilanschi J, Tilgen N, Beyer S, Weber YG, et al. A mutation in the GABA(A) receptor alpha(1)-subunit is associated with absence epilepsy. Ann Neurol. 2006;59(6):983–7. [DOI] [PubMed] [Google Scholar]
- 17.Sun H, Zhang Y, Liang J, Liu X, Ma X, Wu H, et al. SCN1A, SCN1B, and GABRG2 gene mutation analysis in Chinese families with generalized epilepsy with febrile seizures plus. J Hum Genet. 2008;53(8):769–74. [DOI] [PubMed] [Google Scholar]
- 18.Tian M, Mei D, Freri E, Hernandez CC, Granata T, Shen W, et al. Impaired surface alphabetagamma GABA(A) receptor expression in familial epilepsy due to a GABRG2 frameshift mutation. Neurobiol Dis. 2013;50:135–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urak L, Feucht M, Fathi N, Hornik K, Fuchs K. A GABRB3 promoter haplotype associated with childhood absence epilepsy impairs transcriptional activity. Hum Mol Genet. 2006;15(16):2533–41. [DOI] [PubMed] [Google Scholar]
- 20.Carvill GL, Weckhuysen S, McMahon JM, Hartmann C, Moller RS, Hjalgrim H, et al. GABRA1 and STXBP1: novel genetic causes of Dravet syndrome. Neurology. 2014;82(14):1245–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka M, Olsen RW, Medina MT, Schwartz E, Alonso ME, Duron RM, et al. Hyperglycosylation and reduced GABA currents of mutated GABRB3 polypeptide in remitting childhood absence epilepsy. Am J Hum Genet. 2008;82(6):1249–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baumann SW, Baur R, Sigel E. Forced subunit assembly in alpha1beta2gamma2 GABAA receptors. Insight into the absolute arrangement. J Biol Chem. 2002;277(48):46020–5. [DOI] [PubMed] [Google Scholar]
- 23.Grabenstatter HL, Russek SJ, Brooks-Kayal AR. Molecular pathways controlling inhibitory receptor expression. Epilepsia. 2012;53 Suppl 9:71–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brooks-Kayal AR, Pritchett DB. Developmental changes in human gamma-aminobutyric acidA receptor subunit composition. Ann Neurol. 1993;34(5):687–93. [DOI] [PubMed] [Google Scholar]
- 25.Laurie DJ, Seeburg PH, Wisden W. The distribution of 13 GABAA receptor subunit mRNAs in the rat brain. II. Olfactory bulb and cerebellum. J Neurosci. 1992;12(3):1063–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bosch DG, Boonstra FN, de Leeuw N, Pfundt R, Nillesen WM, de Ligt J, et al. Novel genetic causes for cerebral visual impairment. Eur J Hum Genet. 2016;24(5):660–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamdan FF, Myers CT, Cossette P, Lemay P, Spiegelman D, Laporte AD, et al. High Rate of Recurrent De Novo Mutations in Developmental and Epileptic Encephalopathies. Am J Hum Genet. 2017;101(5):664–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heyne HO, Singh T, Stamberger H, Abou Jamra R, Caglayan H, Craiu D, et al. De novo variants in neurodevelopmental disorders with epilepsy. Nat Genet. 2018;50(7):1048–53. [DOI] [PubMed] [Google Scholar]
- 29.May P, Girard S, Harrer M, Bobbili DR, Schubert J, Wolking S, et al. Rare coding variants in genes encoding GABAA receptors in genetic generalised epilepsies: an exome-based case-control study. Lancet Neurol. 2018;17(8):699–708. [DOI] [PubMed] [Google Scholar]
- 30.Srivastava S, Cohen J, Pevsner J, Aradhya S, McKnight D, Butler E, et al. A novel variant in GABRB2 associated with intellectual disability and epilepsy. Am J Med Genet A. 2014;164A(11):2914–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishii A, Kang JQ, Schornak CC, Hernandez CC, Shen W, Watkins JC, et al. A de novo missense mutation of GABRB2 causes early myoclonic encephalopathy. J Med Genet. 2017;54(3):202–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cogliati F, Giorgini V, Masciadri M, Bonati MT, Marchi M, Cracco I, et al. Pathogenic Variants in STXBP1 and in Genes for GABAa Receptor Subunities Cause Atypical Rett/Rett-like Phenotypes. Int J Mol Sci. 2019;20(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baldridge D, Heeley J, Vineyard M, Manwaring L, Toler TL, Fassi E, et al. The Exome Clinic and the role of medical genetics expertise in the interpretation of exome sequencing results. Genet Med. 2017;19(9):1040–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y, Xiangwei W, Zhang X, Xiao J, Chen J, Yang X, et al. Phenotypic spectrum of patients with GABRB2 variants: from mild febrile seizures to severe epileptic encephalopathy. Dev Med Child Neurol. 2020. [DOI] [PubMed]
- 35.Nishikawa A, Otani Y, Ito S, Nagata S, Shiota M, Takanashi JI, et al. A de novo GABRB2 variant associated with myoclonic status epilepticus and rhythmic high-amplitude delta with superimposed (poly) spikes (RHADS). Epileptic Disord. 2020;22(4):476–81. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, Zhang YH, Chen JY, Zhang J, Yang XL, Chen Y, et al. [Clinical features of epilepsies associated with GABRB2 variants]. Zhonghua Er Ke Za Zhi. 2019;57(7):532–7. [DOI] [PubMed] [Google Scholar]
- 37.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36(10):928–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scheffer IE, Berkovic S, Capovilla G, Connolly MB, French J, Guilhoto L, et al. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia. 2017;58(4):512–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwan P, Arzimanoglou A, Berg AT, Brodie MJ, Allen Hauser W, Mathern G, et al. Definition of drug resistant epilepsy: consensus proposal by the ad hoc Task Force of the ILAE Commission on Therapeutic Strategies. Epilepsia. 2010;51(6):1069–77. [DOI] [PubMed] [Google Scholar]
- 40.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelly M, Park M, Mihalek I, Rochtus A, Gramm M, Perez-Palma E, et al. Spectrum of neurodevelopmental disease associated with the GNAO1 guanosine triphosphate-binding region. Epilepsia. 2019;60(3):406–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olson HE, Poduri A, Pearl PL. Genetic forms of epilepsies and other paroxysmal disorders. Semin Neurol. 2014;34(3):266–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carecchio M, Mencacci NE. Emerging Monogenic Complex Hyperkinetic Disorders. Curr Neurol Neurosci Rep. 2017;17(12):97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Janve VS, Hernandez CC, Verdier KM, Hu N, Macdonald RL. Epileptic encephalopathy de novo GABRB mutations impair gamma-aminobutyric acid type A receptor function. Ann Neurol. 2016;79(5):806–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi YW, Zhang Q, Cai K, Poliquin S, Shen W, Winters N, et al. Synaptic clustering differences due to different GABRB3 mutations cause variable epilepsy syndromes. Brain. 2019;142(10):3028–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reid CA, Kim T, Phillips AM, Low J, Berkovic SF, Luscher B, et al. Multiple molecular mechanisms for a single GABAA mutation in epilepsy. Neurology. 2013;80(11):1003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu S, Noviello CM, Teng J, Walsh RM, Jr., Kim JJ, Hibbs RE. Structure of a human synaptic GABAA receptor. Nature. 2018;559(7712):67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiel L, Baakman C, Gilissen D, Veltman JA, Vriend G, Gilissen C. MetaDome: Pathogenicity analysis of genetic variants through aggregation of homologous human protein domains. Hum Mutat. 2019;40(8):1030–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.