Abstract
The distribution and abundance of sulfate-reducing bacteria (SRB) and eukaryotes within the upper 4 mm of a hypersaline cyanobacterial mat community were characterized at high resolution with group-specific hybridization probes to quantify 16S rRNA extracted from 100-μm depth intervals. This revealed a preferential localization of SRB within the region defined by the oxygen chemocline. Among the different groups of SRB quantified, including members of the provisional families “Desulfovibrionaceae” and “Desulfobacteriaceae,” Desulfonema-like populations dominated and accounted for up to 30% of total rRNA extracted from certain depth intervals of the chemocline. These data suggest that recognized genera of SRB are not necessarily restricted by high levels of oxygen in this mat community and the possibility of significant sulfur cycling within the chemocline. In marked contrast, eukaryotic populations in this community demonstrated a preference for regions of anoxia.
A central role of sulfate-reducing bacteria (SRB) in the biogeochemistry of chemically stratified marine habitats, as represented by sediments and microbial mats, is well documented (3, 4, 20). Sulfate respiration may account for as much as 50% of the total organic carbon oxidized in some marine sediments, and SRB are the dominant anaerobes in hypersaline cyanobacterial mat communities (20). Several studies have shown that they are not necessarily restricted by the presence of oxygen, and there is increasing recognition that their habitat range may extend well beyond the anoxic settings to which they have been traditionally relegated (3, 11, 13, 15, 18, 23, 24, 38, 39). Most notably, the highest rates of sulfate reduction yet documented in a natural system were observed in the highly oxic near-surface region of a cyanobacterial microbial mat similar to that examined in this study (3, 13). Thus, the contribution of SRB to biogeochemical cycling may be significantly greater than is now appreciated.
To partly address the need for more direct measures of SRB diversity and environmental distribution, we have used two molecular tools to identify and quantify them. The more established method uses group-specific DNA probes targeting the 16S rRNAs of different phylogenetic groups (phylotypes). Rather than hybridize to individual species, the probes were designed to encompass large phylogenetic groups of SRB, a method that more readily gives a phylogenetic overview of community structure (10, 35). The second method uses a general PCR primer set to directly recover gene sequences for a key enzyme in the pathway for sulfate respiration, the dissimilatory sulfite reductase (DSR) (40). We here present the use of group-specific probes to define the depth distribution of major SRB phylotypes at high resolution (ca. 100-μm depth intervals) in the near surface of a hypersaline microbial mat system from Solar Lake (Sinai, Egypt). In the accompanying study (29) we examine the use of DSR sequencing as an independent measure of SRB population distribution in this system.
This work complements a lower-resolution analysis of SRB population distribution within a similar microbial mat community from Guerrero Negro (Baja California Sur, Mexico) (35). That study revealed a clear stratification of major phylogenetic groups of SRB with depth and showed that group distribution was generally nonoverlapping, suggesting that populations representing the major phylogenetic assemblages serve specific community functions linked to carbon cycling. However, since the resolution of that analysis was at approximately 2-mm depth intervals, the distribution of populations relative to oxygen could not be resolved. We here present a much higher-resolution study of the upper 4 mm of a cyanobacterial mat from Solar Lake, Egypt (8). This has revealed a preferential localization of SRB to the highly oxic chemocline and a predominance of Desulfonema-like populations. Thus, the presence of oxygen per se does not appear to restrict the distribution of recognized lineages of SRB. The distribution of eukaryotes was also shown to differ from more conventional expectations, with the organisms showing a pronounced preference for the anoxic region of the mat.
This study more generally addressed a common information gap in microbiology: the uncertain relationship between the physiologies of organisms in culture and their environmental distribution and activities. The preferred habitats of recognized sulfate respirers, which include bacterial and archaeal lineages, are mostly unknown. Thus, the application of methods that provide for more direct measurements of natural abundance and activity should contribute to a better understanding of the general ecology of sulfate respirers.
MATERIALS AND METHODS
Mat maintenance.
Mat samples (approximately 25 by 15 by 4 cm) were collected from Solar Lake (Sinai, Egypt) and air shipped to Northwestern University (Evanston, Ill.) in sealed containers filled with brine from the lake. Upon arrival, the mats were placed in glass aquaria containing fresh 2× synthetic sea water (Instant Ocean; Aquarium Systems) and 1% (vol/vol) Solar Lake water aerated with standard aquarium pumps and kept under constant illumination with 500-W halogen lights (800 microeinsteins/s/m2) for 72 h. The mats were maintained with constant aeration on a regimen of 12 h of light followed by 12 h of darkness. The maximum water temperature during illumination was 30°C; the minimum temperature in darkness was 24°C. Evaporatively lost water was replaced daily with distilled water. A discussion of the relationship of this system to native mats is presented in the accompanying paper (29).
Mat sampling and RNA extraction.
Mat was sampled after 5 h of exposure to light, providing time for the mat to reach steady-state chemical and physical conditions (13). The sampling procedure was as follows. A cylindrical core (2 cm3) was collected with a Teflon-coated plastic tube for measurement of dissolved oxygen with oxygen microelectrodes (see below). Three cores from the immediate periphery of the central core were removed for rRNA analyses and kept frozen on dry ice until they were sectioned at 50-μm intervals (ca. 5-μl slice volume) with a cryomicrotome operated at −30°C. Total rRNA was extracted (as described below) from alternating 50-μm sections for the first 4 mm and every 1 mm thereafter to a depth of 9 mm. Freezing resulted in an approximate 7% vertical expansion of the mat. We have not compensated for this expansion in our presentation of population distribution but note that such a correction would serve to shift the distribution slightly closer to the surface.
rRNA was recovered by the bead-beating method previously described (26, 37). Briefly, individual 50-μm-thick sections were placed into 2.2-ml screw-cap tubes containing 500 μl of phenol saturated with buffer (50 mM sodium acetate, 10 mM EDTA [pH 5.1]), 500 μl of buffer (pH 5.1), 50 μl of 20% (wt/vol) sodium dodecyl sulfate, and 500 mg of zirconium beads. Each tube was immediately vortexed for 1 min, frozen on dry ice, and held at −80°C for further processing. The extracted rRNA was characterized by polyacrylamide gel electrophoresis to evaluate the recovery of intact high-molecular-weight species (16 and 23S rRNAs). Due to limited rRNA recovery from each section, recovered rRNAs from equivalent depths in each of the three cores were pooled for hybridization.
Hybridization of extracted rRNA.
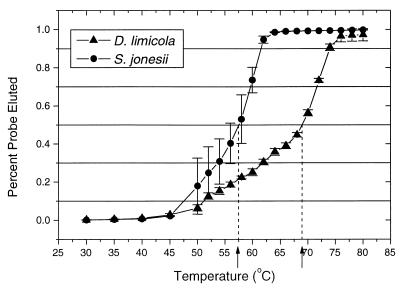
Total nucleic acids were immobilized on MagnaCharge nylon membranes (Micron Separation Inc., Westboro, Mass.) with a slot-blot apparatus (Minifold II; Schleicher & Schuell Inc., Keene, N.H.), hybridized with 32P-labeled oligonucleotide probes, and washed at the experimentally determined temperature of dissociation (Td) as previously described (26, 37). Each rRNA sample was applied to the membrane in triplicate. rRNA used as the standard for hybridization to the Desulfonema genus probe (S-G-Dsnm-0657-a-A-16) was obtained via in vitro transcription of Desulfonema limicola ribosomal DNA, cloned in the pGEM-T vector (Promega, Madison, Wis.) as previously described (30). Cloned ribosomal DNA was transcribed with SP6 RNA polymerase (Life Technologies, Inc.) via standard protocols according to the manufacturer’s suggestions. The Td for probe S-G-Dsnm-0657-a-A-16 was determined via a graded-temperature wash series (Fig. 1) as previously described (30, 33). A universal probe (S-*-Univ-1392-c-A-15) was used to quantify total rRNA abundance in the mat (41). Hybridization signals were quantified with a PhosphorImager (Molecular Dynamics Inc., Sunnyvale, Calif.). Probes and reference rRNA used in this study are listed in Table 1. We also call the reader’s attention to recent recognition that certain members of the provisional family “Geobacteriaceae” may contribute to the Desulfovibrio probe hybridization (27).
FIG. 1.
Normalized dissociation curves were obtained with the probe for Desulfonema spp. (S-G-Dsnm-0657-a-A-16) that hybridized to a target rRNA (Desulfonema limicola) and a single-mismatch nontarget rRNA (Synergistes jonesii). Curves were constructed with average values from triplicate experiments. Error bars define the standard deviation for each temperature point. The estimated Td (midpoint of probe dissociation) for each probe-rRNA duplex is indicated by an arrow on the temperature axis.
TABLE 1.
Probes and target groups
| Probe | Target group | Reference rRNA | Reference |
|---|---|---|---|
| S-*-Univ-1392-c-A-15 | All organisms | Escherichia coli | 41 |
| S-D-Euca-1379-a-A-16 | Eukaryotes | Saccharomyces cerevisiae | 17 |
| S-F-Dsv-0687-a-A-16 | “Desulfovibrionaceae” | Desulfovibrio salexigens | 10 |
| S-*-Dsb-0804-a-A-18 | Desulfobacter group | Desulfococcus multivorans | 10 |
| S-*-Dscoc-0814-a-A-18 | Desulfococcus group | Desulfococcus multivorans | 10 |
| S-G-Dsnm-0657-a-A-16a | Desulfonema limicola and Desulfonema magnum | Desulfonema limicola | 38 |
This probe has one nucleotide difference from the sequence of Desulfonema ishimotei (C at position 6).
The probes used in this study and their target-group coverages are listed in Table 1. We present the abundance of each SRB target group as the amount of rRNA recovered from each depth interval. We also discuss population abundance in terms of fractional contribution, the group-specific fraction of total rRNA recovered from each depth interval. Total rRNA was quantified with a well-characterized universal probe that hybridizes with comparable efficiencies to virtually all small-subunit rRNAs under optimized hybridization conditions (41). The universal probe also provided an overview of general microbial population distribution with depth in the mat community.
Dissolved-oxygen measurements.
Profiles of dissolved oxygen were obtained with a Clark-type oxygen microelectrode (22, 34). The references for oxygen saturation and zero oxygen were oxygen- and nitrogen-saturated deionized room-temperature water, respectively. The oxygen profiles within and between mat specimens were very similar, as shown by the ranges and median values for half-maximum and minimum concentrations of oxygen determined from six independent microelectrode measurements (Fig. 2). The range and median values were calculated with the central reference core from this study (one profile) and three well-separated cores from a different mat specimen, including three profiles from different regions of the same core.
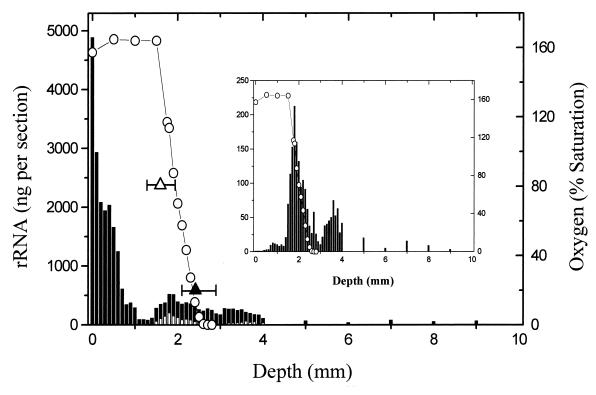
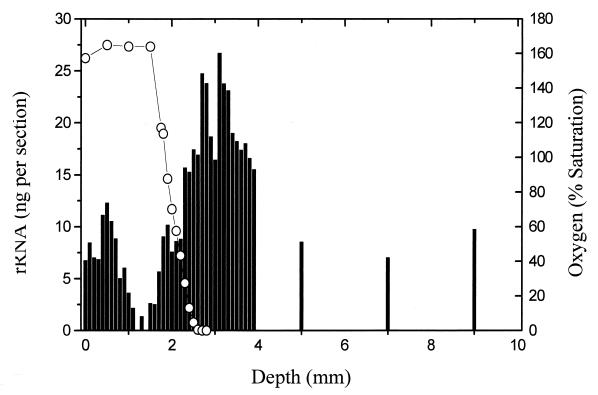
FIG. 2.
Distribution of total and Desulfonema-like rRNAs at 100-μm depth intervals in relation to oxygen. The abundance of Desulfonema is represented by the unshaded region in the total-abundance plot and individually in the inset plot. The medians and ranges for the one-half maximum-oxygen (open triangle) and minimum-oxygen (filled triangle) concentrations were calculated from six independent microelectrode measurements as discussed in the text.
RESULTS
General population distribution.
The most general feature revealed by quantifying total rRNA was a pronounced separation of surface and subsurface populations. A major surface population was separated from deeper populations by a region of very low rRNA recovery (Fig. 2). The near-surface maximum was anticipated, corresponding to cyanobacterial primary production and growth of associated bacteria (25). The secondary peak was coincident with the midpoint of the oxygen chemocline and was shown in this study (discussed below) to be comprised of a significant population of SRB. The interpeak minimum was positioned near the beginning of the chemocline, marking the depth of an initial significant decrease in O2 concentration. This feature has not been reported from previous mat studies and we further consider its significance in the discussion.
The presence of one or two minor peaks below the base of the oxygen chemocline suggested additional stratification of populations. Fine structure was more evident with specific probes. For example, the Desulfonema probe identified a second well-resolved population peak in the region immediately below the oxygen chemocline (discussed below).
Distribution of SRB.
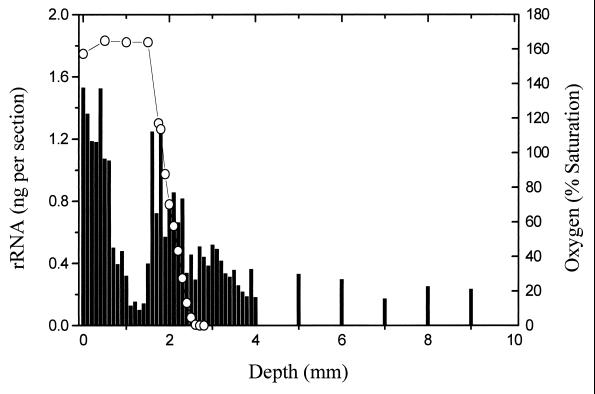
By far the most abundant population of SRB observed in this study was bacteria related to the genus Desulfonema. Peak abundance of this population was primarily restricted to the chemocline and exceeded 200 ng/section at its maximum, comprising as much as 30% of the total rRNA in this region. A similar localization within the chemocline was observed for other SRB target groups (discussed below). The only exception to this trend was observed for Desulfovibrio-like organisms. They were slightly more abundant in the upper 0.5 mm of the mat, relative to a second maximum within the oxygen chemocline (Fig. 3).
FIG. 3.
Distribution of Desulfovibrio-like rRNA at 100-μm depth intervals.
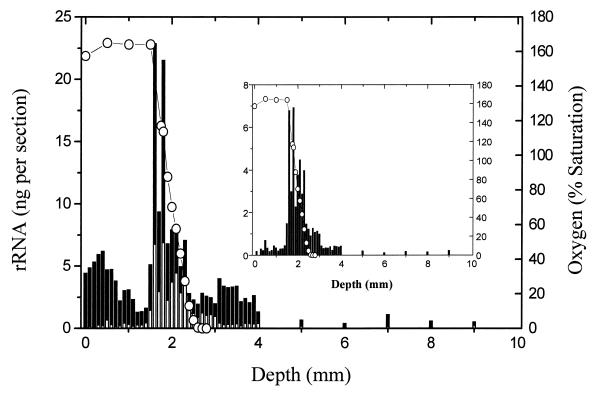
The next-most-abundant SRB population among those evaluated was comprised of members of the provisional family “Desulfobacteriaceae” (S-*-Dsb-0804-a-A-18), having a peak abundance of about 25 ng in the chemocline (Fig. 4). This approximately family-level probe was designed prior to the availability of 16S rRNA sequences for Desulfonema species. Although Desulfonema species are members of this family, the 804 probe has one mismatch in sequence from that of the 16S rRNA of the dominant Desulfonema population recognized by probe S-G-Dsnm-0657-a-A-16. All four Desulfonema isolates now available (14) contain single nucleotide differences from the sequences of both the S-*-Dsb-0804-a-A-18 and S-*-Dscoc-0814-a-A-18 probes. In addition, only one of the two characterized Desulfonema probes was used in this study and it is possible that the total abundance of this general group may be greater than that determined with a single probe. Probe S-*-Dscoc-0814-a-A-18 was designed to target a phylogenetic subgroup of probe 804, encompassing species of Desulfosarcina, Desulfococcus, and Desulfobotulus. Consistent with this design strategy, comparison of the 804 and 814 profiles shows that all values obtained with probe 814 are less than values obtained with probe 804 (Fig. 4). The maximum value for probe 814 was less than 8 ng at its highest point in the chemocline.
FIG. 4.
Distribution of populations encompassed by the general probe for the provisional family “Desulfobacteriaceae” and for the desulfococcus subgroup within this family (Table 1). The abundance of the desulfococcus group is represented by the unshaded region in the abundance plot for the family and individually in the inset plot.
Comparison of the population profiles inferred by hybridization with all three SRB probes suggests a finely stratified vertical distribution. The Desulfosarcina-Desulfococcus-Desulfobotulus population was highly localized, reflected by very low rRNA abundance above and below the chemocline. In contrast, the Desulfonema population was distributed between two peaks, a major peak within the chemocline (150- to 200-ng maximum abundance) and a minor peak below the point of oxygen depletion (50- to 75-ng maximum abundance). In addition, even though Desulfonema spp. were the most abundant target group in the system, this group was virtually absent from the upper 0.4 mm of the mat (hybridization values of less than 1 ng per depth interval).
Those populations targeted by the more general probe for the “Desulfobacteriaceae” had a more complex distribution, comprising at least three peaks: a major peak within the chemocline, a minor peak in the surficial 1 mm, and a second minor peak below the point of oxygen depletion. Thus, the aggregate hybridization data reveal considerable fine-scale vertical zonation of sulfate-reducing populations. Since these probes encompass phylogenetic groups that are almost certainly composed of related, but genetically distinct, populations, additional structure will almost certainly be revealed with more specific probes. An initial view of the full structural complexity of this community has been provided by our analysis of DSR gene sequences (reported in the accompanying paper [29]).
Distribution of eukaryotes.
Probes for eukaryotes and archaea were used to provide general information about their contribution to this community. The archaea were a very minor population at all depths examined, as inferred by using a well-characterized probe for this domain (33). Members of this domain comprised significantly less than 1% of total rRNA abundance throughout the system, generally 1 to 2 ng per depth interval (data not shown). A more detailed characterization of archaeal distribution and diversity within this system will be presented elsewhere (28a). In contrast to the archaea, eukaryotes were a significant population at some depth intervals, showing a minor peak in the upper 1 mm and a second more abundant peak at and below the point of oxygen depletion (Fig. 5). The eukaryotic population in the anoxic region accounted for as much as 10% of total rRNA abundance.
FIG. 5.
Distribution of eukaryotic rRNAs at 100-μm depth intervals.
DISCUSSION
The separation of surface and subsurface populations by an interval of very low biomass is the most general structural feature observed in this study. Although this population structure has not been previously reported, it is consistent with the results of studies of other stratified biological systems. For example, reduced chemical species generated in the anoxic bottom layers of stratified water bodies are rapidly oxidized by populations localized at the oxic-anoxic interface (12). The population distribution in this mat community most likely reflects a similar phenomenon; reduced substrates diffusing from the permanently anoxic region provide electron donors to support an active respiratory population within the chemocline. The low-biomass region between surface and subsurface peaks may reflect a depletion of electron donors; substrates diffusing from below (e.g., reduced sulfur species and organic fermentation products) and from above (originating directly or indirectly from cyanobacterial primary production) may be fully consumed before reaching this intermediate depth interval. Although oxygen is not limiting, electron donors may be. Other considerations include the effect of elevated pH in this region, either directly inhibiting microbial growth or promoting abiotic “growth” of inorganic precipitates (e.g., of carbonates or sulfides). Previous studies have measured pH values of greater than 9.5 in this region of the mat (22). In considering these data, we also note that these mats were maintained under relatively low-light conditions, approximating early-morning sunlight. Although the general features of the mat are consistent with the results of studies of a more native context, it is possible that full-daylight illumination may alter the observed population distributions.
In contrast to conventional expectation, SRB comprised a major fraction of populations localized to the oxygen chemocline and the eukaryotic population distribution was highly skewed to the permanently anoxic region. A significant presence of eukaryotes in this region was unanticipated, and at this time we have no additional information concerning their identities. Single-cell eukaryotes have been observed previously by microscopic examination, but their diversity and contribution to system processes are mostly unexplored (3, 6, 8). Studies of eukaryotic population structure are ongoing, and we restrict further discussion to the distribution of SRB.
Several previous studies of mats in their native context or maintained in outdoor aquaria have also suggested a significant presence of SRB near the chemocline (5, 13, 21, 35, 36, 38). For example, an earlier study of a specimen from Guerrero Negro showed that populations related to Desulfonema were most abundant in the upper 2 mm and were also the most abundant SRB population quantified within any depth interval (35). A recent study by Teske et al. (38) examining population distribution at 2-mm depth intervals reported the highest most-probable-number counts of Desulfonema species within the 2- to 4-mm depth interval of a Solar Lake mat specimen maintained in outdoor ponds. Desulfonema population abundance was between 105 and 106 cells/ml near the chemocline and generally between 104 and 105 cells/ml in other depth intervals examined. Additionally, these investigators recovered a partial 16S rRNA sequence related to the phylogenetic group composed of Desulfonema, Desulfococcus, and Desulfobotulus species. This sequence was more abundant near the chemocline than the surface (upper 1 mm), as assessed by PCR amplification.
Our high-resolution hybridization studies have more fully delineated Desulfonema distribution within the chemocline. Remarkably, the peak distribution of these organisms in this region spanned only an approximately 1-mm depth interval. Since this profile was obtained by pooling rRNAs recovered from three adjacent core samples, it is possible that their distribution is even more localized than here indicated. We also call attention to a secondary peak of Desulfonema immediately below the point of oxygen depletion. Although this anoxic localization is more consistent with conventional expectation, it represents only about 25% of the total Desulfonema rRNA quantified in the upper 4 mm of the mat.
Since reduced sulfur species generated by SRB in the chemocline would be rapidly reoxidized by chemolithotrophs, the contribution of sulfate respiration to sulfur and carbon cycling has almost certainly been greatly underestimated in past studies of this and similar systems. Although we cannot rule out oxygen consumption by these SRB, available data more generally suggest a close association with sulfur-oxidizing bacteria, such as the morphologically conspicuous Beggiatoa species observed in this mat. This kind of association is supported by recent observations of an intimate physical association between Desulfonema species and sulfur-oxidizing Thioploca species reported by Fukui et al. (14) and Teske et al. (38). Also, initial comparisons of night-and-day patterns of distribution suggest that certain members of the “Desulfobacteriaceae” migrate towards the surface of the mat during the night, providing additional support for an active participation in mat processes during both the night and day (29a). A significant presence of SRB in the chemocline of the day mat would allow the sulfur oxidizers, both chemotrophic and phototrophic, to occupy regions where more light and higher concentrations of oxygen are available. However, this hypothesized association begs the question of competition between the SRB and aerobic heterotrophs for electron donors in this system. Although microelectrode studies have not revealed the presence of reducing microniches in this community or similar communities (19, 32), tips are several microns in diameter and there is no consensus on the possible existence of microniches at near-micrometer scales. Thus, additional evidence for aerotolerance among SRB and intimate juxtapositioning of cells may require a reevaluation of the meaning of a microniche in this community.
A close association between SRB and cyanobacteria was also previously indicated. For example, sulfate reduction in the oxic zone of a Solar Lake mat specimen was shown to be stimulated by the addition of glycolate (5, 13), a cyanobacterial photosynthate excreted by CO2-limited cyanobacteria (2, 16, 31). However, if our hypothesis that depletion of electron donors accounts for the low-biomass interval directly above the chemocline is correct, it is unlikely that cyanobacterial photosynthate directly nourishes the dominant population of SRB within the chemocline. Rather, as for other stratified systems, the turnover of biomass in deeper depth intervals likely provides reduced organic and inorganic species that are oxidized at the oxic-anoxic interface.
These data do not directly address the issue of significant reduction or respiration of oxygen by SRB. We therefore make only brief note of many recent studies showing aerotolerance or a limited capacity for oxygen consumption among SRB, including Desulfovibrio species and relatives of Desulfonema species (1, 7, 9, 11, 18, 23, 28, 39). For example, a strain of Desulfococcus multivorans was shown to have the capacity to oxidize hydrogen, lactate, formate, and sulfite by using oxygen as an electron acceptor (9). However, there has been no unequivocal demonstration that any SRB can grow with oxygen as the sole electron acceptor (18).
We have not drawn extensive comparisons between the results of this study and those of a previous lower-resolution study of a similar mat system from the Exportadora de Sal salt works (Guerrero Negro, Baja California Sur, Mexico). The present study focused on the upper 4-mm depth interval, whereas our previous investigation surveyed a depth of several centimeters at 2-mm sectioning intervals. The most significant difference in population patterns was observed for the probe targeting Desulfovibrio species. Desulfovibrio represented a significant population in the upper 2 mm of the Guerrero Negro mat and in a broad region below 7 mm. In contrast, these organisms were a relatively minor population in the Solar Lake community, which might reflect intrinsic differences between these geographically well-separated systems. Additional comparative studies are needed to more fully resolve these apparent differences.
An unresolved question is whether we have yet fully accounted for the dominant SRB in this mat community. As presented in the accompanying paper (29), we have completed initial experiments using an independent and more explicit measure of SRB diversity based on comparative sequencing of DSR, a highly conserved enzyme common to all SRB. Many of the DSR sequences recovered from the oxic region (0 to 2.5 mm) were closely related to Desulfococcus and Desulfonema (29). This finding is consistent with the major presence of Desulfonema-like species revealed in this study by rRNA-targeted probes. However, the recovery of at least one apparently novel DSR lineage from this region also suggests the presence of additional unidentified SRB populations in the highly oxic near-surface region. The identification of all key participants is a necessary prelude to fully defining the contribution of SRB to carbon and energy flow in this system.
ACKNOWLEDGMENTS
This work was supported by the Office of Naval Research (grant ONR N00014-95-1-00887). This research was partially supported by a grant from the German-Israeli Foundation for Scientific Research and Development and a grant of the BMBF to Y.C.
REFERENCES
- 1.Abdollahi H, Wimpenny J W T. Effects of oxygen on the growth of Desulfovibrio desulfuricans. J Gen Microbiol. 1990;136:1025–1030. [Google Scholar]
- 2.Bateson M M, Ward D M. Photoexcretion and fate of glycolate in a hot spring cyanobacterial mat. Appl Environ Microbiol. 1988;54:1738–1743. doi: 10.1128/aem.54.7.1738-1743.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canfield D E, Des Marais D J. Aerobic sulfate reduction in microbial mats. Science. 1991;251:1471–1473. doi: 10.1126/science.11538266. [DOI] [PubMed] [Google Scholar]
- 4.Cohen Y. Oxygenic photosynthesis, anoxygenic photosynthesis and sulfate reduction in cyanobacterial mats. In: Klug M J, Reddy C A, editors. Current perspectives in microbial ecology. Washington, D.C.: ASM Press; 1984. pp. 435–441. [Google Scholar]
- 5.Cohen Y, Helman Y, Sigalevich P. Light-driven sulfate reduction and methane emission in hypersaline cyanobacterial mats. NATO ASI Ser G Ecol Sci. 1994;35:421–427. [Google Scholar]
- 6.Cohen Y, Krumbein W E, Shilo M. Solar Lake (Sinai). 3. Bacterial distribution and production. Limnol Oceanogr. 1977;22:621–634. [Google Scholar]
- 7.Cypionka H, Widdel F, Pfennig N. Survival of sulfate-reducing bacteria after oxygen stress growth in sulfate-free oxygen-sulfide gradients. FEMS Microbiol Ecol. 1985;31:39–45. [Google Scholar]
- 8.D’Amelio E D, Cohen Y, Des Marais D J. Comparative functional ultrastructure of two hypersaline submerged microbial mats: Guerrero Negro, Baja California Sur, Mexico, and Solar Lake, Sinai, Egypt. In: Cohen Y, Rosenberg E, editors. Microbial mats: physiological ecology of benthic microbial communities. Washington, D.C.: American Society for Microbiology; 1989. pp. 97–113. [Google Scholar]
- 9.Dannenberg S, Kroder M, Dilling W, Cypionka H. Oxidation of H2, organic compounds and inorganic sulfur compounds coupled to reduction of O2 or nitrate by sulfate-reducing bacteria. Arch Microbiol. 1992;158:93–99. [Google Scholar]
- 10.Devereux R, Kane M D, Winfrey J, Stahl D A. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst Appl Microbiol. 1992;15:610–619. [Google Scholar]
- 11.Dilling W, Cypionka H. Aerobic respiration in sulfate-reducing bacteria. FEMS Microbiol Lett. 1990;71:123–128. [Google Scholar]
- 12.Fenchel T, Finlay B J. Ecology and evolution in anoxic worlds. 1st ed. Oxford, United Kingdom: Oxford University Press; 1995. [Google Scholar]
- 13.Fründ C, Cohen Y. Diurnal cycles of sulfate reduction under oxic conditions in cyanobacterial mats. Appl Environ Microbiol. 1992;58:70–77. doi: 10.1128/aem.58.1.70-77.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukui, M., A. Teske, B. Assmus, G. Muyzer, and F. Widdel. Isolation, physiological characteristics, natural relationships, and 16S rRNA-targeted in situ detection of filamentous, gliding sulfate-reducing bacteria, genus Desulfonema. Submitted for publication.
- 15.Hastings D, Emerson S. Sulfate reduction in the presence of low oxygen levels in the water column of the Cariaco Trench. Limnol Oceanogr. 1988;33:391–396. [Google Scholar]
- 16.Heyer H, Krumbein W E. Excretion of fermentation products in dark and anaerobically incubated cyanobacteria. Arch Microbiol. 1991;155:284–287. [Google Scholar]
- 17.Hicks R E, Amann R I, Stahl D A. Dual staining of natural bacterioplankton with 4′,6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992;58:2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson M S, Zhulin I B, Gapuzan M E, Taylor B L. Oxygen-dependent growth of the obligate anaerobe Desulfovibrio vulgaris Hildenborough. J Bacteriol. 1997;179:5598–5601. doi: 10.1128/jb.179.17.5598-5601.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jørgensen B B. Bacterial sulfate reduction within reduced micro-niches of oxidized marine sediments. Mar Biol. 1977;41:7–17. [Google Scholar]
- 20.Jørgensen B B. Mineralization of organic matter in the sea bed—the role of sulfate reduction. Nature. 1982;296:643–645. [Google Scholar]
- 21.Jørgensen B B, Bak F. Pathways and microbiology of thiosulfate transformations and sulfate reduction in a marine sediment (Kattegat, Denmark) Appl Environ Microbiol. 1991;57:847–856. doi: 10.1128/aem.57.3.847-856.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jørgensen B B, Revsbech N P, Cohen Y. Photosynthesis and structure of benthic microbial mats: microelectrode and SEM studies of four cyanobacterial communities. Limnol Oceanogr. 1983;28:1075–1093. [Google Scholar]
- 23.Krekeler D, Sigalevich P, Teske A, Cypionka H, Cohen Y. A sulfate-reducing bacterium from the oxic layer of a microbial mat from Solar Lake (Sinai), Desulfovibrio oxyclinae sp. nov. Arch Microbiol. 1997;167:369–375. [Google Scholar]
- 24.Krekeler D, Teske A, Cypionka H. Strategies of sulfate-reducing bacteria to escape oxygen stress in a cyanobacterial mat. FEMS Microbiol Ecol. 1998;25:89–96. [Google Scholar]
- 25.Kühl M, Jørgensen B B. Spectral light measurements in microbenthic phototrophic communities with a fiber-optic microprobe coupled to a sensitive diode array detector. Limnol Oceanogr. 1992;37:1813–1823. [Google Scholar]
- 26.Lin C, Stahl D A. Taxon-specific probes for the cellulolytic genus Fibrobacter reveal abundant and novel equine-associated populations. Appl Environ Microbiol. 1995;61:1348–1351. doi: 10.1128/aem.61.4.1348-1351.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lonergan D J, Jenter H I, Coates J D, Phillips E J P, Schmidt T M, Lovley D R. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J Bacteriol. 1996;178:2402–2408. doi: 10.1128/jb.178.8.2402-2408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marschall C, Frenzel P, Cypionka H. Influence of oxygen on sulfate reduction and growth of sulfate-reducing bacteria. Arch Microbiol. 1993;159:168–173. [Google Scholar]
- 28a.Minz, D. Unpublished data.
- 29.Minz D, Flax J L, Green S J, Muyzer G, Cohen Y, Wagner M, Rittmann B E, Stahl D A. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl Environ Microbiol. 1999;65:4666–4671. doi: 10.1128/aem.65.10.4666-4671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Minz, D., et al. Unpublished data.
- 30.Mobarry B K, Wagner M, Urbain V, Rittmann B E, Stahl D A. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microbiol. 1996;62:2156–2162. doi: 10.1128/aem.62.6.2156-2162.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nold S C, Ward D M. Photosynthate partitioning and fermentation in hot spring microbial mat communities. Appl Environ Microbiol. 1996;62:4598–4607. doi: 10.1128/aem.62.12.4598-4607.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramsing N B, Kühl M, Jørgensen B B. Distribution of sulfate-reducing bacteria, O2, and H2S in photosynthetic biofilms by oligonucleotide probes and microelectrodes. Appl Environ Microbiol. 1993;59:3840–3849. doi: 10.1128/aem.59.11.3840-3849.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raskin L, Rittmann B E, Stahl D A. Competition and coexistance of sulfate-reducing and methanogenic populations in anaerobic biofilms. Appl Environ Microbiol. 1996;62:3847–3857. doi: 10.1128/aem.62.10.3847-3857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Revsbech N P, Jørgensen B B, Blackburn T H, Cohen Y. Microelectrode studies of the photosynthesis and O2, H2S, and pH profile of a microbial mat. Limnol Oceanogr. 1983;28:1062–1074. [Google Scholar]
- 35.Risatti J B, Capman W C, Stahl D A. Community structure of a microbial mat: the phylogenetic dimension. Proc Natl Acad Sci USA. 1994;91:10173–10177. doi: 10.1073/pnas.91.21.10173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sass H, Cypionka H, Babenzien H-D. Vertical distribution of sulfate-reducing bacteria at the oxic-anoxic interface in sediments of the oligotrophic Lake Stechlin. FEMS Microbiol Ecol. 1997;22:245–255. [Google Scholar]
- 37.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teske A, Ramsing N B, Habicht K, Fukui M, Küver J, Jørgensen B B, Cohen Y. Sulfate-reducing bacteria and their activities in cyanobacterial mats of Solar Lake (Sinai, Egypt) Appl Environ Microbiol. 1998;64:2943–2951. doi: 10.1128/aem.64.8.2943-2951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Niel E W J, Gomes T M P, Willems A, Collins M D, Prins R A, Gottschal J C. The role of polyglucose in oxygen-dependent respiration by a new strain of Desulfovibrio salexigens. FEMS Microbiol Ecol. 1996;21:243–253. [Google Scholar]
- 40.Wagner M, Roger A J, Flax J L, Brusseau G A, Stahl D A. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J Bacteriol. 1998;180:2975–2982. doi: 10.1128/jb.180.11.2975-2982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng D, Raskin L, Alm E W, Stahl D A. Characterization of small-subunit rRNA-targeted universal probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol. 1996;62:4504–4513. doi: 10.1128/aem.62.12.4504-4513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]