Abstract
Japanese encephalitis virus (JEV), a single-stranded, enveloped RNA virus, is a health concern across Asian countries, associated with severe neurological disorders, especially in children. Primarily, pigs, bats, and birds are the natural hosts for JEV, but humans are infected incidentally. JEV requires a few host proteins for its entry and replication inside the mammalian host cell. The endoplasmic reticulum (ER) plays a significant role in JEV genome replication and assembly. During this process, the ER undergoes stress due to its remodelling and accumulation of viral particles and unfolded proteins, leading to an unfolded protein response (UPR). Here, we review the overall strategy used by JEV to infect the host cell and various cytopathic effects caused by JEV infection. We also highlight the role of JEV structural proteins (SPs) and non-structural proteins (NSPs) at various stages of the JEV life cycle that are involved in up- and downregulation of different host proteins and are potentially relevant for developing efficient therapeutic drugs.
Graphical abstract
Introduction
Japanese encephalitis virus (JEV) causes severe neurological disease in humans and horses, characterized by massive swelling in the central nervous system (CNS). JEV is a mosquito-borne arbovirus (arthropod-borne virus), belonging to the family Flaviviridae, which also includes yellow fever virus, West Nile virus (WNV), dengue virus (DENV), and Zika virus (ZIKV). JEV is the major cause of flaviviral encephalitis, especially in East and Southeast Asian countries. The first JEV infection case was observed and documented in 1871 in Japan, and the disease was later named Japanese encephalitis (JE) [1, 2]. The global number of JE cases is unknown because of poor monitoring, but the number of reported JEV infections is approximately 68,000, including approximately 20,000 fatal cases, annually [3]. JEV mainly affects children, with a fatality rate of up to 30%, and up to 50% of surviving patients experience permanent and sometimes severe neuropsychiatric sequelae [4] such as recurrent seizures [5], memory loss, convulsions [6, 7], and paralysis [8].
JEV virions contain a positive-sense single-stranded ribonucleic acid (+ssRNA) genome of approximately 11 kb encoding three structural proteins (SPs) (the capsid, membrane/premembrane, and envelope proteins) and seven non-structural proteins (NSPs) (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). Upon entering the host cell, JEV releases its genomic RNA into the cytoplasm, where it undergoes initial translation using host cell machinery to produce a polyprotein, which is cleaved by various host and viral proteases, and this is followed by replication of the JEV genome. The SPs are incorporated into virus particles, whereas the NSPs participate in the formation of the replication complex (RC) and assembly complex (AC) and the replication of the viral genomic RNA. The newly assembled virus is in an immature form, and before leaving the host cell, it must undergo a maturation process. The maturation of JEV occurs due to consecutive reductions in pH in the successive components of host cell organelles. Thus, the ER lumen (pH 6.7) serves as an ideal location for the packaging and assembly of virus particles. Budding and fusion of ER vesicles containing the immature virus results in their transport to the cis-Golgi-network (CGN, pH 6.0) and then to the trans-Golgi-network (TGN, pH 5.7). Due to the successive reduction in pH, the prM protein is cleaved by host furin protease into pr and M subunits, resulting in the maturation of the JEV virion before its release from the host cell.
JEV circulates among three different types of hosts during its life cycle, viz., amplifying hosts (major reservoir), transmission hosts (carrier), and dead-end hosts (incidental). Mosquitoes of the genus Culex, such as Culex tritaeniorhynchus, act as the primary vectors for JEV infection of different hosts and maintaining the zoonotic transmission cycle. Pigs and water birds also amplify JEV by developing high viremia titers, facilitating transmission to biting mosquitoes. Bats and migratory birds are the principal transmission and carrier hosts and are important for overwintering and dispersal of JEV to new geographical locations. Humans and horses are not natural hosts of JEV, but they become infected by chance, often due to living in proximity to domestic pigs and birds; therefore, they are known as "incidental" hosts. Incidental hosts do not develop a sufficiently high viral titer for further transmission of the virus and are therefore also known as “dead-end” hosts for JEV, but they do develop encephalitis and severe neurological disorders [9, 10]. In nature, JEV follows pig-mosquito-pig, bird-mosquito-bird [11], and bird-mosquito-pig transmission pathways [12]. However, the transmission of JEV can sometimes occur even without involving mosquitoes: from infected pigs to naïve pigs living together, through oronasal secretions [13]. Another mode of transmission involving only mosquitoes is transovarial transmission. In transovarial transmission, infected female culex or non-culex mosquitoes produce JEV-infected progeny mosquitoes [14]. Filgueira et al. have reviewed the geographical distribution and transmission of JEV involving multiple hosts [15], and this topic is therefore discussed only briefly in this review.
Japanese encephalitis is probably an underestimated disease. Several compounds have been shown in vitro and in animal models to have significant anti-JEV activity. Despite that, very few clinical trials have been conducted in the last 10 years [16]. Another reason for the unavailability of anti-JEV therapeutics may be a poor understanding of the functions of the NSPs in JEV infection and pathogenesis, with the exception of the well-studied enzymatic activities of the NS3 and NS5 proteins. As a result, no effective antiviral drugs have been approved for treatment of JE. Therefore, this review summarizes current information related to the pathogenicity and infectivity of JEV by bringing together recent findings concerning the involvement of host proteins and JEV proteins at the stages of viral entry, replication, assembly, and maturation, which will be important for the development of therapeutic drugs against JEV infection.
JEV structure and genome organisation
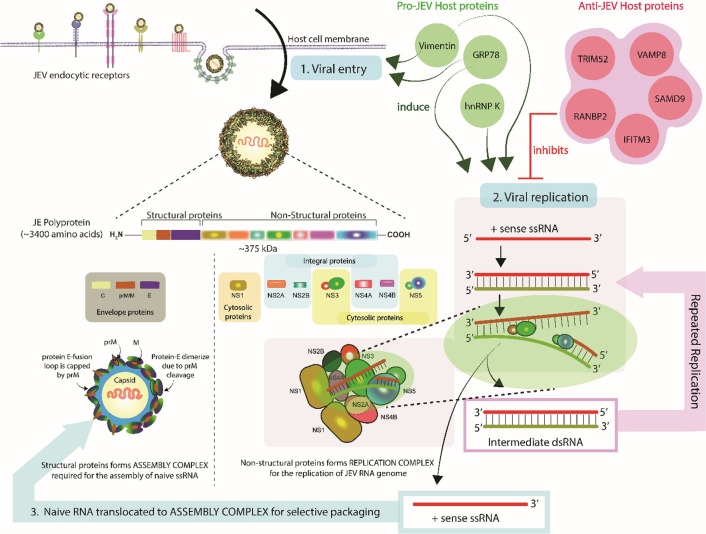
JEV, an enveloped virus, has a linear, (+) ssRNA genome enclosed by multiple copies of the capsid protein (Fig. 1). The viral nucleic acid and capsid protein form a nucleocapsid that is surrounded by a host-derived lipid bilayer. The 5′ end of the genomic RNA contains a methylated cap (m7GpppAmp), but it lacks a poly-A tail at the 3′ end [17–20]. The genomic RNA is ~11 kb in length and contains a single open reading frame (ORF) between two short non-coding regions (NCR) at 5′ and 3′ ends (Fig. 2a). The NCRs are highly conserved among all mosquito-borne flaviviruses and form secondary structures to support viral replication, transcription, and translation processes. The 5′ NCR contains functional RNA elements, such as promoters, enhancers, and putative cyclisation sequences that are required for interaction between the distantly located 5′ and 3′ NCRs of the genomic RNA. The NS5 protein interacts with the circularised RNA and initiates RNA replication at the 3′-NCR during viral infection [21–23]. The ORF encodes a polyprotein consisting of ~3400 amino acids (Fig. 2b), which is cleaved by both viral and host proteases to form three SPs – the capsid (C), membrane/pre-membrane (M/prM), and envelope (E), proteins – and seven NSPs: NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5. Notably, the JEV NS2B-NS3 protease performs proteolytic cleavages between NS2A and NS2B, between NS2B and NS3, between NS3 and NS4A, and between NS4B and NS5. The host protease signalase performs proteolytic cleavages between C and prM, between prM and E, between E and NS1, and between NS4A and NS4B in the polyprotein. During maturation in the Golgi complex, the prM protein is cleaved into the pr subunit (~10 kDa) and the mature M protein (~8 kDa) by the host protease furin (Fig. 2c). The protease that carries out the cleavage between NS1 and NS2A is still unknown and needs to be identified. The JEV NS3 protein has protease activity in its N-terminal domain and serves as binding site for the cofactor protein NS2B. NS3 also has helicase activity in its C-terminal domain that induces negative supercoiling (unwinding) of the dsRNA during viral RNA replication. NS5 is also a critical protein because its N-terminal domain (methylase domain) possesses methylase activity that is required for the 5′ capping of naïve viral RNA, and its C-terminal domain (RdRp domain) has RNA-dependent RNA polymerase (RdRp) activity that is required for viral RNA replication. The SPs are involved in the formation of the viral capsid and outer envelope, whereas the NSPs participate in the formation of the RC and the AC and replication of the viral genome [24, 25].
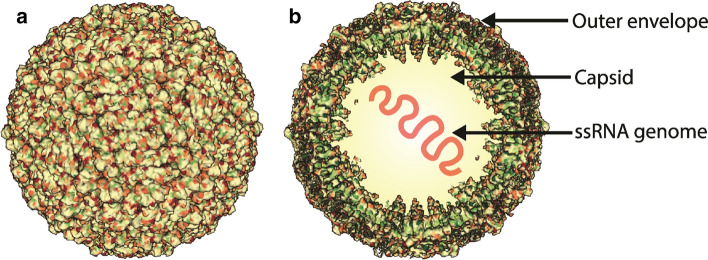
Fig. 1.
JEV structure. (a) Cryo-electron microscopy structure of a JEV virion retrieved from the protein data bank database with PDB ID: 5WSN (produced by Wang et al. (2017) [208]), and the image was created using Mol* viewer [209]. (b) The JEV virion contains a (+) ssRNA genome enclosed by a capsid and an outer envelope. The outer envelope of mature virion contains 180 copies of the envelope and membrane proteins.
Fig. 2.
Genome organization and proteolytic cleavage of JEV proteins. (a) The genome of JEV is (+) ssRNA of approximately 11 kb in length, flanked by non-coding regions (NCRs) at the 5′ and 3′ ends. The NCR at the 5′ end is shorter in length, with ~100 nucleotides, than the NCR at 3′ end, which can vary between 100 to 700 nucleotides. The NCR forms a secondary structure and participates in viral replication and transcription. (b) A singe open reading frame (ORF) in the JEV genomic RNA encodes a polyprotein of ~375 kDa, which is further cleaved by host and viral proteases to form three structural proteins (prM/M, C, and E) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). (c) JEV polyprotein cleavage. The NS2B-NS3 protease (viral protease) cleaves the NS2A-NS2B, NS2B-NS3, and NS3-NS4A junctions. Signalase (host protease) cleaves the C-prM, prM-E, E-NS1, and NS4A-NS4B junctions. Furin protease (host protease) cleaves prM into pr and M. NS1-NS2A is cleaved by an unknown protease.
Structural proteins
Envelope protein
The envelope (E) protein in JEV has a molecular mass of ~45 kDa, and is organized into three domains – domain I (central β-barrel), domain II (fusion loop), and domain III (immunoglobulin-like module) – in a homodimer [26]. The E protein of immature JEV exists in an irregular trimeric form, while it exhibits a flattened dimeric structure in mature particles. Notably, domain II, which includes the fusion loop, is capped by prM (precursor of membrane protein) in immature particles. During maturation in the TGN of Golgi bodies, the prM protein is cleaved into a pr subunit and the mature M protein, resulting in the exposure of the fusion loop of the E protein. [27–29]. In addition to membrane fusion, the E protein mediates binding to the host cell receptor and the transduction process. It is therefore the most important viral antigen for induction of neutralising antibodies (NAbs) in the host [30, 31].
Membrane protein
The JEV membrane protein is present in virions in two forms, viz., the ~24 kDa precursor membrane protein (prM) and the ~8 kDa mature membrane protein (M) [32]. The prM protein promotes the intracellular assembly of virions by acting as a chaperon as well as preventing the formation of the E dimer [33, 34]. Cleavage of the prM protein by the host furin protease results in dimerization of the envelope protein required for the maturation of JEV [27, 32]. Following the furin cleavage, the pr subunit remains attached to the E protein, forming an E-pr heterodimer until the virus is released from the host cell. The pr subunit is then released outside of the cell, leading to E homodimer formation in mature virions [35, 36].
Capsid protein
The mature JEV particle contains 180 copies of the capsid (C) protein (13 kDa) [37]. The precursor of the capsid (C) protein, i.e., the membrane-anchored capsid (anchC) protein, contains a hydrophobic moiety at its C-terminus that acts as a signal sequence for the translocation of prM into the lumen of ER, the site where virus assembly occurs [37]. The hydrophobic moiety of the anchC is cleaved by the JEV NS2B-NS3 serine protease, and this results in the folding and dimerization of the capsid protein [38, 39]. The dimerization of the capsid protein creates a moiety of basic amino acids on one side and hydrophobic amino acids on another side of the dimer, which interact with the ssRNA genome and membrane of the virus, respectively [40]. The mature capsid protein contains a nuclear localisation signal (NLS) for phosphorylation and later interacts with importin (host nuclear protein B23) [41], which is required for entry into the host nucleus [42]. The reason for the shuttling of the capsid protein between the nucleus and the cytoplasm is not clearly understood, but recently, Sarkar et al., using immunofluorescence staining, observed the presence of capsid protein in the cytoplasm as well as in the nucleus, and they suggested that transition of the capsid protein might be involved in viral assembly [43]. In addition, the capsid protein suppresses the formation of stress granules by interacting with the host caprin-1 protein to promote JEV propagation [44]. In other flavivirus infections, the capsid protein interacts with various host proteins such as the importin-α/HDM2 protein [45] and phospholipid-binding protein [46, 47], which are required for assembly and maturation of viral particles. In the case of WNV and DENV, the human Sec 3 (hSec3P) protein has been shown to have chaperonin activity toward the capsid protein, which can delay or suppress the infection [48, 49]. Thus, the capsid protein is a dynamic and multifunctional protein that is required at various stages of the viral life cycle, and hence, among the three structural proteins (E, prM/M, and C), the capsid protein is the best suited as a target of therapeutic drugs against JEV.
Non-structural proteins
Non-structural protein 1 (NSP1)
Non-structural protein 1 (NSP1) is a 48-kDa glycoprotein that exists in a dimeric (~96 kDa) and a hexameric form in complex with the host ER membrane and in secretions, respectively [50]. Dimeric NSP1 is a multifunctional glycoprotein that is involved in the assembly of the replication complex [51] and replication of JEV [52, 53] by interacting with the other JEV NSPs as well as with several host proteins, including the RPL18, RPL18a, vimentin, and hnRNP K proteins [54, 55]. In contrast, it appears that the main function of the hexameric form of NSP1 is modulation of the host immune system to support JEV propagation [56, 57]. Furthermore, an extended form of the NSP1 protein, NSP1' (~53 kDa) of JEV has been reported to suppress the activity of interferon (IFN) type I [58], a crucial component of the host innate and adaptive immune response to viral infections. Hence, as an early marker of infection, the NS1 protein can be targeted to develop drugs, vaccines, and monoclonal antibodies to inhibit JEV pathogenesis. Notably, an NS1-based vaccine was developed recently by fusing a truncated NS1 protein with the Escherichia coli heat-labile enterotoxin subunit [59].
NS2A
NS2A is a 22-kDa membrane-associated hydrophobic protein located in the RC that interacts with the 3′-UTR of the JEV (+) sense ssRNA with high affinity and with other NSPs required for the replication process [60, 61]. In addition to its involvement in replication, NS2A is also required for virion assembly, as it translocates the newly synthesised (+) sense ssRNA genome from the RC to the AC, as shown in Figure 5b [62, 63]. It has also been reported that NS2A plays a vital role in JEV infection by suppressing the antiviral response generated by host cells, where NS2A suppresses protein kinase (PKR)-induced cell death [64]. JEV requires NS1' for its survival inside the host cell by modulating the host immune response, and it has been shown that a single mutation in NS2A prevents NS1' formation. Therefore, as a regulator of NS1', NS2A protein plays an essential role in JEV infection and pathogenesis [65].
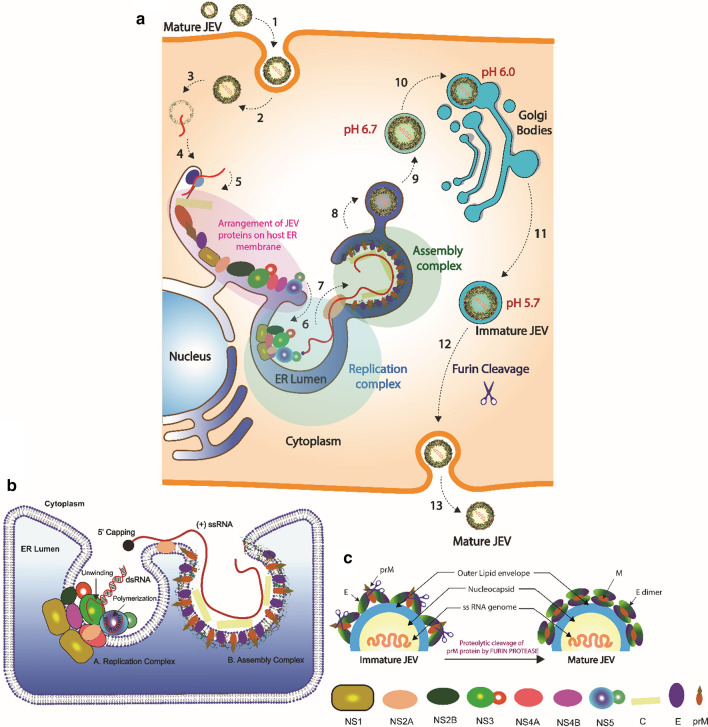
Fig. 5.
Hypothetical model of the JEV life cycle. (a) 1. Interaction of JEV with host cell receptors. 2. Entry of JEV through the endocytic pathway, which may be clathrin dependent, clathrin independent, or cholesterol dependent. 3. Once inside the host cell, JEV releases its genomic RNA into the cytosol. 4. Binding of JEV genomic RNA to the ribosome located on the ER membrane. 5. Initial translation of genomic RNA followed by polyprotein cleavage and arrangement of structural and non-structural proteins in the ER membrane, ER lumen, and cytosol as shown in Fig. 3. 6. Formation of the replication complex required for replication of JEV genomic RNA. 7. Packaging of the JEV genomic RNA into the viral capsid protein, forming the nucleocapsid. 8. Budding of the ER membrane carrying newly assembled virions. 9. Release of the newly assembled JEV virion through ER budding (pH 6.7). 10. The immature virion then enters into the Golgi body (pH 6.0) through the cis-Golgi network (CGN). 11. Release of JEV through the trans-Golgi network (TGN) (pH 5.7) 12. The furin protease site of prM gets exposed due to the decrease in pH, and prM is cleaved by the host furin protease. The conversion of prM into the M protein results in a rearrangement of the viral envelope proteins, which results in the maturation of JEV. 13. Mature JEV virions are released through exocytosis. (b) JEV replication complex and assembly complex. The formation of the replication complex occurs at vesicle packets, which are an extended and modified network of the ER. Non-structural proteins participate in the formation of the replication complex and replication of JEV RNA. NS1 proteins induce curvature into the ER membrane, which is required for the recruitment of other non-structural proteins (NS2, NS3, NS4, NS5). The NS3 protein, which has helicase activity, and the NS5 protein, which has RdRp activity, interact with the viral RNA and participate in replication. NS2B acts as a cofactor for the NS3 protein. NS4A and NS4B interact with NS1 and help in the formation of the viral replication complex. The assembly complex is formed by JEV structural proteins (C, prM, and E), where newly synthesized RNA is incorporated into a virion particle. (c) Proteolytic cleavage of the prM protein by furin protease is required for maturation of the virion.
NS2B-NS3 protease
The 14-kDa NS2B protein is the smallest NSP of JEV. It contains a conserved hydrophilic domain of 40 amino acid residues that interacts with the N-terminal protease domain of the NS3 protein (69 kDa) to form the NS2B-NS3 protease complex, in which NS2B acts as a cofactor [66, 67]. NS3 is the best studied protein of JEV, and its three-dimensional structure has been determined by X-ray crystallography at a resolution of 1.80 Å, showing that it contains an N-terminal protease domain and a C-terminal helicase domain [68]. The NS2B-NS3 protease performs proteolytic cleavage specifically at dibasic amino acid motifs (K-R, R-R, R-K, or, rarely, Q-R) between NS2A and NS2B, between NS2B and NS3, NS3-NS4A, and between NS4B and NS5, as shown in Figure 2 [69, 70]. The helicase and NTPase domain at the C-terminal end of NS is required for negative supercoiling of the dsRNA intermediate during JEV replication and transcription [71, 72]. A recent study by Xie et al. showed that the NS2B-NS3 protein complex induces cell apoptosis by degrading the AXL membrane protein through the ubiquitin-proteasome pathway [73]. Therefore, NS2B and NS3 are suitable therapeutic targets, as they play a central role in JEV replication and post-translational processing of the polyprotein.
NS4A and NS4B
NS4A and NS4B are integral membrane proteins with a molecular mass of 17 kDa and 27 kDa, respectively, that play multiple roles in JEV infection, including assembly of the replication complex and viral replication. NS4A regulates the NTPase activity of the NS3 helicase, while NS4B acts as a cofactor for the NS3 helicase activity [74].
NS5
NS5 is the largest viral protein (100 kDa), and like NS3, it is also a well-studied protein whose three-dimensional structure has been determined by X-ray crystallography and shown to contain a methyltransferase domain and an RdRp domain at its N- and C-terminus, respectively [75]. Due to the polymerase activity of its RdRp domain and the 5′-capping enzymatic activity of its methyltransferase domain, NS5 is of central importance for JEV replication. The RdRp domain of NS5 contains three subdomains: palm, thumb, and finger. The palm subdomain contains conserved aspartic acid residues and forms the active site for the binding of RNA [76, 77], metal ions, and nucleotides and is also involved in the transfer of phosphate groups. The finger subdomain escorts the template RNA to the active site by forming a tunnel [78, 79], and the thumb subdomain is required to form an RNA synthesis complex and regulates RNA synthesis [77, 79]. Due to its central role in JEV replication, NS5 is a promising target for the development of therapeutic drugs.
Host proteins involved in JEV infection
JEV infection alters the expression of various host proteins, which can have either anti-viral or pro-viral activity. The role of host proteins in JEV infection has not yet been studied precisely. However, several studies have shown that some host proteins, such as IFITM3, RANBP2, SAMD9, VMP8, and TRIM52, are upregulated in JEV-infected cells, and knocking down these genes significantly enhances JEV replication. These same proteins are also involved in inhibiting other viral infections, such as HIV, influenza A virus type H1N1, hepatitis C virus, Sendai virus, dengue virus, and West Nile virus (WNV) [80, 81]. Recent studies have also shown that some host proteins, including GRP78, vimentin, and hnRNP K, assist in JEV entry and replication in the host cell [55, 82].
Host proteins with anti-JEV activity
JEV diagnosis involves the observation of clinical signs and serological tests; therefore, the host proteins upregulated during JEV infection may serve as biomarkers for the detection of infection and evaluation of its severity. There are several problems associated with the traditional diagnosis of JEV infection, as has been discussed previously by Roberts and Gandhi [83]. Therefore, detection of the upregulated host proteins discussed below may help in the diagnosis of JEV infection. The concentrations of these biomarkers may also be associated with the severity of JEV infection and therefore potentially useful for choosing appropriate clinical measures.
IFITM3
Brass et al. reported that interferon-induced transmembrane protein 3 (IFITM3), a host factor of the IFITM family, inhibits WNV infection by blocking the entry of the virus into the cytoplasm [80]. Later, Zhang et al. reported that this transmembrane protein is upregulated during JEV infection and that a loss of IFITM3 gene expression results in enhanced JEV replication [84]. Chesarino et al. reported that post-translational modification regulates the antiviral activity of IFITM3. This was supported by the findings of Wang et al., who reported that the tumour suppressor protein p53 promotes palmitoylation of IFITM3, which is required to inhibit JEV replication, but the mechanism by which this occurs is still unclear [85]. IFITM3 shows antiviral activity against several other viruses, such as SARS-CoV-2 [86] and human metapneumovirus (hMPV) [87]. Zani and Yount have summarised the various in vivo studies in which IFITM3-knockout mice experience severe disease when exposed to different viruses in comparison to wild-type mice [88]. Therefore, the IFITM3 protein can be considered one of the vital biomarkers for diagnosing JEV infection.
RANBP2
RAN binding protein 2 (RANBP2) belongs to the nucleoprotein family and forms a nuclear pore complex that acts as a shuttle for the translocation of proteins between the nucleus and cytoplasm [89]. Zhang et al. also reported that knockdown of the RANBP2 gene greatly enhanced JEV replication and also showed that there is an increase in expression and accumulation of the RANBP2 protein inside the nucleus during JEV infection [84]. Maarifi et al. also reported that RANBP2 regulates anti-retroviral activity by sumoylation of tripartite-motif-containing protein 5 (TRIM5α), which is known to block retroviral infection in the post-entry phase [90].
SAMD9
Zhang et al. found that sterile alpha motif domain-containing protein 9 (SAMD9) was upregulated by ~2.5-fold in JEV infection, whereas IFITM3 and RANBP2 were upregulated by ~1.7-fold [84]. Like IFITM3 and RANBP2, SAMD9 also inhibits JEV replication and is present in large amounts in the cytoplasm only. Previously, SAMD9 was shown to have antiviral activity against a poxvirus [91]. There have been few studies related to the antiviral activity of the SAMD9 protein, but the role of this protein in the JEV life cycle should not be neglected and needs to be studied further.
VAMP8
Vesicle-associated membrane protein 8 (VAMP8) is an integral membrane protein that gets upregulated along with IFITM3, RANBP2, and SAMD9 during JEV infection, and HeLa cells lacking VAMP8 have shown enhanced JEV replication [84]. Van Tol et al. reported VAMP8 to be a novel regulator of an interferon-I signalling system that generates an antiviral response during West Nile virus infection [92].
TRIM52
Human TRIM52 is a tripartite-motif-containing protein that possesses antiviral activity. Fan et al. reported that TRIM52 interacts with and degrades the NS2A protein of JEV in a ubiquitin-proteasome-dependent manner, resulting in inhibition of JEV replication and infection [93].
Host proteins with pro-JEV activity
Protein-protein interactions play a crucial role in pathogenesis, whether viral-viral or viral-host protein interactions. Lv et al. have reported that the various host proteins discussed below interact mainly with the proteins E, M/prM, C, and NS1, which play a crucial role in the JEV life cycle, especially in neuroinvasion. Therefore, they can also be used as targets for developing anti-JEV therapeutics and inhibiting JEV infection.
GRP78
Glucose-regulated protein 78 (GRP78) is an ER chaperon belonging to the 70-kilodalton heat shock protein (HSP70) family that is expressed on epithelial and neuronal cell membranes. GRP78 interacts with domain III of the JEV envelope protein, which is required for entry of JEV into the host cell. It is also involved in the replication of the viral genomic RNA and viral protein synthesis. GRP78 assists JEV in entry and replication, which are critical stages in any virus's life cycle [82]. Therefore, GRP78 is considered an important factor that can be targeted in JEV infection.
Vimentin
Vimentin is an intermediate filament protein that is found on the cell surface and intracellularly. Like GRP78, surface vimentin interacts with the JEV envelope protein and is involved in viral entry [94, 95], while intracellular vimentin assists in JEV replication by interacting with NS1 [55]. It is still obscure how vimentin interacts with NS1 and helps JEV replication.
hnRNP K
Heterogenous nuclear ribonucleoprotein K (hnRNP K) is a crucial pre-mRNA binding protein that is involved in hnRNA metabolism in the nucleus. Like GRP78 and vimentin, hnRNP K also affects JEV replication by interacting with the NS1 protein [55]. hnRNP K also assists dengue virus and Junin virus in their replication [96]. In the case of JEV, hnRNP K may be involved in the translocation of the NS1 protein within the host cell [96]. It also interacts with the JEV NS5 protein [97], which has polymerase activity and plays a central role in viral replication. Therefore, hnRNP K can be considered an important intracellular target due to interaction with the critical proteins NS1 and NS5 of JEV.
MAP1LC3
Microtubule-associated protein 1 light chain 3 (MAP1LC3) is a host protein that plays an essential role in autophagy. Sarkar et al. showed that MAP1LC3 interacts with the JEV capsid protein and promotes viral replication and pathogenesis [43].
JEV life cycle
The life cycle of JEV starts with the bite of a carrier mosquito, which releases virus particles that interact with receptors present on the surface of host cells, such as pericytes [98], fibroblasts [99], endothelial cells [100–102], dendritic cells [103], and myeloid cells [10], which are susceptible to JEV infection and are the primary sites of propagation of the virus. After replication in these cells, the newly assembled virion particles now migrate towards brain cells such as neuronal cells [104] and microglial cells [105] by using infected dendritic cells and T cells for their transport [98]. The early stages of JEV infection, which include internalization of virions and membrane fusion, have already been reviewed by Yun and Lee [10] and are therefore not included in this review. The mechanisms by which JEV infects brain tissues and crosses the blood brain barrier as well as the consequences of these events have been reviewed by Filgueira and Lannes [15] and by Hsieh and John [106]. Before entering the brain tissues, the propagation of JEV is necessary, which takes place in cells present at the primary site of infection, such as endothelial cells, fibroblasts, pericytes, macrophages, and dendritic cells [106, 107]. This review mainly discusses the role of JEV structural and non-structural proteins required for entry, genome replication, protein synthesis, and virion assembly in all types of cells that are susceptible to JEV, which was not the focus of the reviews by Yun and Lee [10] and Filgueira and Lannes [15].
Entry and fusion with the host endosomal membrane
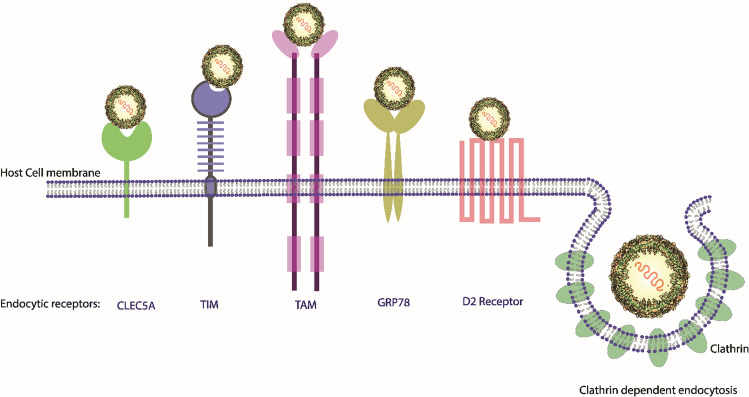
The binding of JEV to the host cell occurs in two stages: (i) Initial binding to attachment factors and (ii) specific binding to endocytic receptors. The initial interaction is nonspecific, occurring between the JEV envelope glycoprotein E and heparan sulfate glycosaminoglycans present on the surface of the host cells [26]. The interaction involves electrostatic interaction between positively charged amino acid residues of the E protein and negatively charged glycosaminoglycans [28], which helps to concentrate the virions over the host cell surface and induce interaction of the virus with the endocytic receptors present on the surface of the host cell. The receptor-mediated interaction between JEV and the host cell is not fully understood. However, some studies have shown that receptors such as HSP70 (N2a cell receptor) [27, 29], CLEC5A (C-type lectin receptor) [30], TIM/TAM phosphatidylserine receptor [31–33], GRP78 [82], D2 receptor [82], and αvß3 (a glycoprotein of the integrin family) [33] may assist in the entry of JEV into the host cell via endocytosis, like other flaviviruses (Fig. 3). Once the virus interacts with endocytic receptors, it can enter the host cell either through clathrin-dependent endocytosis [27], clathrin-independent endocytosis [36], or cholesterol-dependent endocytosis [35]. Due to acidification inside the endosome, protein E undergoes irreversible conformational changes and oligomerization, leading to exposure of a hydrophobic domain that interacts with the host endosomal membrane and driving fusion of the viral and endosomal membranes [37, 38, 108]. After membrane fusion, viral genomic RNA is uncoated and released into the host cytoplasm, where it has multiple fates. It can undergo replication [40, 41], serve as an mRNA for translation of the single open reading frame to produce the precursor polyprotein [42, 44], or be encapsidated and incorporated into immature viral particles.
Fig. 3.
Schematic representation of endocytic receptors and clathrin-mediated endocytosis for the entry of JEV into the host cell
Initial translation of viral genomic RNA
Once the viral RNA is in the host cytoplasm, it is transported to the host ER membrane for initiation of translation. The viral RNA lacks a poly-A tail at its 3′ end [47], but, like the majority of eukaryotic mRNAs, it has type 1 cap (m7GpppAmp) at its 5′ end [46], and viral RNA is therefore translated in a cap-dependent manner. The viral RNA encodes a polyprotein of ~375 kDa, which is cleaved by viral and host proteases into three SPs and seven NSPs [49]. The arrangement of these proteins on ER membrane is still unclear and needs to be studied more, but some biochemical and interaction studies with other flaviviruses have suggested that NS3 and NS5 are cytosolic proteins and that NS1 is found in the ER lumen; however, the remaining NSPs, as well as the SPs E, C, and prM are integral proteins (Fig. 4). The NSPs are involved in virus replication, where NS1, NS2A, NS2B, NS4A, and NS4B are involved in remodelling the ER to form the RC, NS3 in unwinding, and NS5 in the polymerisation of JEV genomic RNA. NS2A interacts with the newly synthesized RNA and is translocated from the RC to AC. NS2A acts a bridge between the modified ER structures. The SPs play a vital role in the assembly of viral particles, where the newly synthesised RNA interacts with the capsid proteins and undergoes encapsidation. The prM holds and stabilises the E protein arranged in the outer envelope, which is derived from the host membrane [48].
Fig. 4.
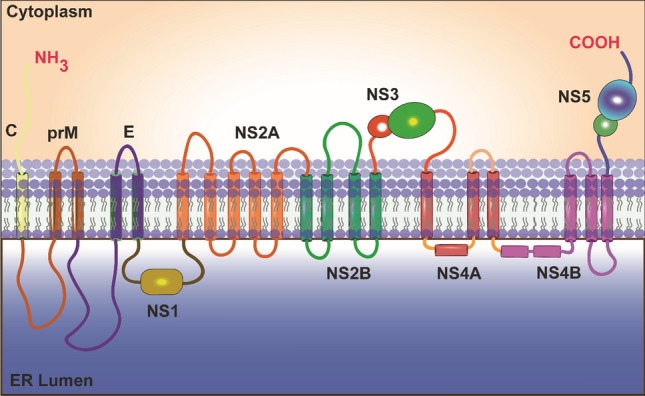
Schematic representation of the JEV polyprotein arrangement in the host endoplasmic reticulum (ER) membrane. The topology of JEV structural and non-structural proteins with respect to the cytosol and ER lumen is shown. The JEV proteins are distributed into the ER lumen (NS1), cytoplasm (NS3 and NS5), and in ER membrane (C, prM/M, E, NS2A, NS2B, NS4A, and NS4B).
Remodelling the host’s endoplasmic reticulum membrane
Flavivirus genome replication and transcription occur on extended and modified ER membrane regions, known as vesicle packets (VPs), while translation and polyprotein processing occur at convoluted membranes (CM) derived from the ER membrane [67, 109]. Based on genetic studies of various flaviviruses, NSPs are predicted to interact inside the VPs to form the RC on the modified network of the ER membrane (Fig. 5a and b). NS1 is a 48-kDa protein that mainly exists as a homodimer [66] that interacts with the envelope (E) protein [52, 68], NS4A protein [59], and NS5 protein [69] and is co-localised with the viral genomic RNA [52]. It is therefore essential for stabilising the RC by creating a scaffold on the ER membrane. Among the NSPs present in the RC, NS3 and NS5 are the two most important and best-studied proteins, and both have essential enzymatic activity. NS3 is located on the cytoplasmic side but remains attached to the ER membrane due to its interaction with polar residues of the integral NS2B protein, which acts as a cofactor and binds to the N-terminal domain of NS3 to form the NS3-NS2B serine protease [110]. Binding assays have suggested that NS2A interacts strongly with NS3 and NS5 as well as the untranslated region (UTR) of the viral RNA in both the RC and the virus assembly complex. These interactions support a hypothetical model in which NS2A acts as carrier protein and transports the newly replicated RNA from the RC to the adjacent assembly complex [70, 71, 111]. Wen et al. reported that NS4B of JEV interacts with the helicase domain of NS3 and facilitates the unwinding of dsRNA and, ultimately, the replication of the genomic RNA [72]. NS4A is considered a crucial protein for the organisation and assembly of the RC [61, 63], and like NS2B, which is a cofactor for the NS3 protease domain, it also serves as a cofactor for NS3. NS4A interacts with the C-terminal helicase domain of the NS3 protein to regulate its ATPase activity and helps in the unwinding of dsRNA and replication of viral genome [62]. The C-terminal ‘2K’ fragment of NS4A translocates the NS4B protein into the ER lumen by acting as a signal sequence [112]. NS5 is the most conserved and the largest protein (~100 kDa) in all flaviviruses, and it interacts strongly with NS3 and other NSPs as well as the genomic RNA in the RC [73–75]. The N-terminal domain (~30 kDa) of NS5 exhibits methyltransferase activity and is responsible for the addition of a methylated cap to the viral RNA at 5′ end [76, 77]. The C-terminal region (~70 kDa) of NS5 comprises the RdRp domain, which is required for replication of the viral genomic RNA [78, 79].
Replication of JEV genomic RNA
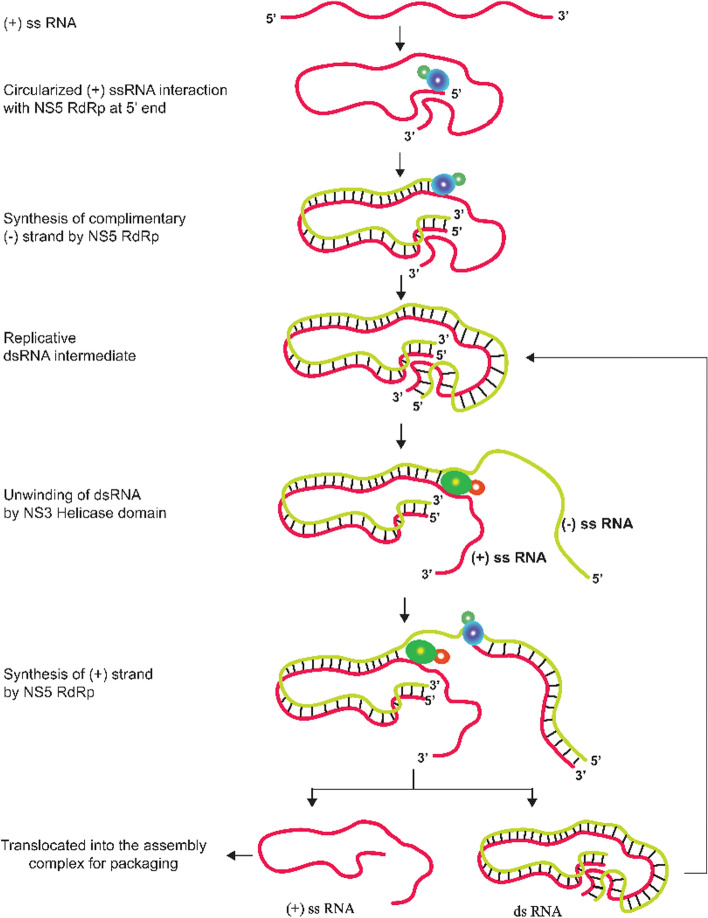
Replication starts when viral RNA, NSPs, and host factors come together to form the RC inside the virus-induced infoldings of the ER membrane termed "vesicle packets" (VPs) [113]. The host cytoplasm and VPs are connected through pores, which are used to translocate nucleotides and other host factors required for replication [113, 114]. In the RC, NS5 interacts with the circularised (+) sense ssRNA, and through the polymerase activity of the RdRp domain, it synthesises a complementary (-) strand RNA, which remains base-paired with the (+) strand and forms a replicative double-stranded (ds) RNA intermediate, which undergoes unwinding due to the helicase activity of NS3, and the (-) strand RNA serves as a template for generating a new (+) sense RNA. Immediately after synthesising the (+) ssRNA, a methylated cap is added to its 5′ end by the NS5 methylase domain, and the RNA is translocated from the RC to the AC. In the RC, NS1 is colocalised with NS5 and the double-stranded RNA intermediate and is involved in the initial replication of negative RNA strands [52, 115]. The synthesis of dsRNA intermediates and (+) sense ssRNA is asymmetric, with the dsRNA intermediate being present at a lower copy number and entering again into the replication machinery for the synthesis of new (+) sense ssRNA. The (+) sense ssRNA are synthesised in high copy numbers and are transported to the assembly complex. The (+) ssRNA in the assembly complex interacts with the capsid (C) protein for selective packaging (Fig. 6) [80, 116].
Fig. 6.
Replication of JEV genomic RNA. The RdRp domain of NS5 protein binds to the 5′ end of circularized (+) ssRNA and synthesizes the (-) strand. Once the dsRNA is fully synthesized, the NS3 protein with its helicase domain binds and starts unwinding the RNA. NS5 then uses the (-) strand as a template and starts synthesis of the (+) strand. The final products of this replication cycle are the replicative dsRNA intermediate and (+) ssRNA. dsRNA then enters another replication cycle, while (+) ssRNA is incorporated into virions.
Assembly and packaging of JEV genomic RNA
The packaging of viral particles is precise, as only (+) RNA is encapsidated and assembled into new virions; however, the molecular mechanism behind the encapsidation and assembly is still unclear. In some viruses, selective packaging is due to packaging signals, which may be secondary structures or specific sequences to which the capsid protein interacts and binds to promote encapsidation [117, 118]. However, in flaviviruses, the specific interactions involved are not known. Since no packaging signal has been identified, the interaction with the C protein is thought to be nonspecific [119, 120], despite the fact that only viral genomic RNA gets incorporated into virions. As mentioned above, in comparison to (-) ssRNAs, the (+) sense RNAs are synthesised asymmetrically in high copy numbers and selectively packaged, suggesting that the viral RNA replication and assembly processes are closely associated [81]. The replication of viral RNA takes place inside the lumen of the VPs, and the components required for replication enter through the pores. The viral RNA synthesised inside the VPs is released and, for packaging, subsequently enters budded regions of the ER membrane created just opposite of the VPs (Fig. 5b) [67, 113]. It is still obscure how the viral RNA is transported from VPs to the assembly site and how the viral NSPs and host proteins function in the assembly of virus particles.
Maturation and release of JEV particles
The mature virus particle contains a nucleocapsid surrounded by an outer lipid bilayer derived from the host cell membrane. The E and M/prM proteins are embedded in the outer envelope. Intracellular immature virion particles contain the prM (premembrane) protein, whereas extracellular fully matured virion particles contain the M (membrane) protein (Fig. 5c) [52]. The maturation of the virus takes place during transport of virus particles from the assembly site in ER membrane invaginations to the extracellular region via the trans-Golgi network (TGN). The reduction in pH of exocytic compartments triggers a structural rearrangement in the prM proteins, resulting in exposure of the furin protease site, where the furin protease cleaves the prM protein into pr and M protein subunits (Fig. 5c). The pr subunits remain bound to the viral surface during the maturation process and are dissociated only when the fully matured virus is released outside the host cell. Therefore, the intracellular immature virus appears spiky and uneven due to pr subunits all over the surface, while the fully mature infectious virus in the extracellular region has a smooth spherical surface [82]. The spiky appearance of the immature virus is due to the presence of 60 spikes on the outer envelope of JEV interacting with each other, and each spike contains three prM-E heterocomplexes. The JEV virion contains 180 copies of prM/M (prM in immature and M in mature JEV) and protein E on its surface [84].
Host metabolic pathways affected by JEV infection
JEV infection induces various cytopathic effects by inducing cellular apoptosis, which has been observed in neuronal cells and astrocytes [121–124], mediated mainly by endoplasmic reticulum stress (ER stress) [125], oxidative stress [126], and mitochondrion-dependent activation of caspases [127]. The JEV-NS5 protein has also been reported to affect the JAK-STAT pathway by inhibiting the phosphorylation of the STAT protein [128]. It has also been shown that JEV induces apoptosis by inhibiting the Foxo signalling pathway [129].
Apoptotic pathway
ER-tress-mediated apoptosis
The ER lumen in JEV-infected cells contains a large number of viral proteins involved in viral RNA replication and virion assembly. Due to the accumulation of virus particles, the ER membrane becomes hypertrophic, resulting in extensive proliferation, as has also been observed in other flavivirus infections [130]. The eukaryotic ER serves as the site for protein synthesis, post-translational modification, folding, and oligomerisation and also plays a significant role in cellular signal-transduction pathways. Therefore, the excessive accumulation of viral proteins in and around the ER lumen results in the modification of ER membrane. The modified ER membrane serves as a central site for genomic RNA replication, assembly, and maturation of the newly synthesised JEV viral particles [24, 131].
Consequently, the ER experiences alterations in its homeostasis to which it is extremely sensitive, and ER stress results in an unfolded protein response (UPR) [125]. ER stress triggers increased expression of death signals such as C/EBP homologous protein (CHOP) and p38 MAP kinase, which is transduced from the ER to the nucleus, resulting in cellular apoptosis mediated by the UPR signalling pathway. The p38 MAP kinase belongs to the serine/threonine protein kinase family and is overexpressed in stress-activated pathways [132]. The CHOP protein, also known as GADD153, is a transcriptional regulator and has been noted for its high expression during ER stress, causing cell-cycle arrest and apoptosis by inhibiting the wnt signalling pathway [125, 132–134].
Oxidative-stress-mediated apoptosis
Like ER stress, oxidative stress is also a cause of JEV-induced apoptosis, which occurs due to the uncontrolled release of intracellular reactive oxygen species (ROS) and several other free radicals such as hydrogen peroxide and hydroxyl radicals from the mitochondria [135, 136]. The massive release of ROS results in the oxidation of lipids, proteins, and nucleic acids, which can cause permeabilization of the mitochondrial membrane, ultimately leading to cellular apoptosis [137, 138]. It has been reported that the intracellular accumulation of ROS results in the downregulation of superoxide dismutase and thioredoxin, leading to apoptosis in JEV-infected human neuronal cells. ROS-induced apoptosis occurs in JEV-infected cells through the activation of the AKS1-p38 MAPK and ASK1-ERK1/2 signalling pathways [139].
Mitochondrion-dependent caspase-mediated apoptosis
The role of mitochondria in JEV-induced apoptosis was first reported by Liao et al. in 1997 [140]. The same group later found that mitochondrion-dependent apoptosis occurs due to altered mitochondrial homeostasis caused by caspase-9 activation during JEV infection [127]. Due to the high level of JEV replication at the ER membrane, infected cells experience ER stress and generate a UPR, which alters the ER homeostasis, depletes Ca2+ in the ER lumen, and results in accumulation of proteins in the cytoplasm up to an immoderate level. The Ca2+ imbalance alters the mitochondrial membrane potential and affects modulation of mitochondrial permeability pores [125, 141–143]. Consequently, cytochrome c (Cyt-c) is released from mitochondria and interacts with monomeric inactive Apaf-1 in the cytoplasm to form a multimeric functional apoptosome complex. Procaspase 9 (precursor of caspase 9) bound to the N-terminal caspase recruitment domain (CARD) of Apaf-1 is converted to active caspase 9 initiator caspase, which remains bound to the apoptosome complex and cleaves caspase 3 (effector caspase) between its smaller and larger subunits to activate it. The maturation and activation of caspase 3 results in various cytopathic effects and apoptosis due to degradation of various regulatory proteins, such as poly(ADP-ribose) polymerase (PARP), which is involved in DNA replication, repair, and recombination as well as programmed cell death [144, 145], gelsolin, which is an actin-modulating protein [146], and the anti-apoptotic bcl-2 [127, 147, 148].
JAK-STAT pathway
This pathway involves three principal factors – (1) a cell-membrane-bound receptor, (2) a receptor-bound enzyme, Janus kinase (JAK), and (3) a transcription factor, signal transducer and activator of transcription (STAT); therefore, it is known as the JAK-STAT pathway [149]. The JAK-STAT pathway plays an essential role in the division, activation, and recruitment of immune cells. Interferon (IFN-α and IFN-β) signalling pathways play a significant role in recovery from infections caused by flaviviruses, but in JEV infection, the functions of IFNs get aborted due to inhibition of the JAK-STAT pathway.
The two crucial enzymes, Jak1 and Jak2, have phosphorylase activity, which plays an essential role in IFN-α and IFN-β signalling. Once the IFNs bind to the receptor, Jak1 and Jak2 phosphorylate each other, and this is followed by phosphorylation of a tyrosine residue in the intracellular domain of the IFN receptor. Due to phosphorylation, the receptor undergoes conformational changes and binds to the STAT proteins, which are then phosphorylated and dimerized. The STAT protein dimers are then translocated into the nucleus and bind specifically to the promoter regions of IFN-stimulated genes (ISGs) on the DNA sequence. As a result, the ISGs are transcribed, leading to the production of IFNs with antiviral properties, inhibiting viral replication and the production of new virus particles. In JEV infection, NS5 blocks the phosphorylation of STAT proteins, resulting in inactivation of the STAT protein, which remains outside the nucleus, thus disrupting the JAK-STAT pathway. Hence, the NS5 protein is a crucial factor in JEV pathogenesis and therefore a potential drug target [128, 150].
Recent insights and developments: vaccines, therapeutics, and diagnostics
An underestimated disease with a 30% mortality rate and permanent neuropsychiatric sequelae in 50% of the survivors lacks effective treatment despite several in vivo, in vitro, and clinical studies. However, several vaccines are available, such as inactivated mouse brain vaccines, inactivated Vero-cell-derived vaccines, live attenuated vaccines, and live recombinant vaccines (chimeric vaccines). All of these vaccines were based on genotype III of JEV but showed cross-reactivity against genotypes I-IV [151]. The formulation of all four types of vaccines (Table 1) have been reviewed by Turtle and Solomon [16], and their immunogenicity and efficacy have been reviewed by Nagendra and Milind [152], Barzon and Palu [153], Kumar et al. [154], Satchidanandam [155], Hu and Lee [156], and Kanamori et al. [157]. In addition, Lee et al. have reviewed the applicability and usage of inactivated Vero-cell-derived vaccines, live attenuated vaccines, and live recombinant vaccines [158]. Connor et al. have reviewed the US Advisory Committee on Immunization Practices (ACIP) policy for travellers, which has recommended JEV vaccination for those who frequently travel to JEV-endemic countries or decide to stay there for an extended period of time [159]. These types of policies may help to reduce the risk of infection in healthy people and should be adopted by other countries that are geographically close to JEV-endemic countries but are not themselves affected by JEV. Recently, using reverse vaccinology and in silico approaches, Chakraborty et al. designed a peptide vaccine loaded with multiple epitopes belonging to the E, prM, NS1, NS3, and NS5 proteins of JEV [160]. This peptide vaccine is being studied through in vivo and in vitro experiments and still needs to be tested in clinical trials to examine its immunogenicity and efficacy. Keeping the disadvantages of previous vaccines in mind, Wan et al. have developed an NS1-based vaccine (LTB-NS1Δ63) by fusing the truncated NS1 protein of JEV with the heat-labile enterotoxin B subunit of E. coli. They also tested the vaccine for its immunogenicity and toxicity through in vitro and in vivo experiments and found that mice inoculated with LTB-NS1Δ63 showed a higher survival rate than those receiving the live attenuated SA14-14-2 vaccine when challenged with a lethal dose of JEV [59].
Table 1.
List of Japanese encephalitis vaccines
| Type | Generic name (Trade name) | Derived from | JEV strain | Developed/licensed in | Characteristics | References |
|---|---|---|---|---|---|---|
|
Inactivated (Two doses are recommended by WHO at an interval of four weeks in children ≥ 6 months of age) |
MB-JEV/JE-MB (JE-VAX)*† |
Mouse brain | Nakayama | BIKEN, Japan in 1954 |
Poor immunogenicity Requires multiple doses 80-100% seroconversion rate ~90% efficacy Replaced with attenuated JEV vaccines on WHO recommendation |
[152, 153, 155, 161, 162] |
|
MB-JEV/JE-MB (JE-VAX)* † |
Mouse brain | Beijing-1 | BIKEN, Japan in 1989 |
Enhanced neutralizing antibody response compared to the Nakayama strain Requires multiple doses Induces higher viral and heterologous antibody titers in immunized mice than the Nakayama strain 80-100% seroconversion rate ~90% efficacy Replaced with attenuated JEV vaccines on WHO recommendation |
[152, 153, 155, 161–163] | |
| JEV Beijing-3 strain vaccine*† | Primary hamster kidney (PHK) cells | Beijing-3 | China in 1968 | In randomized field trials, the efficacy was observed to be between 76 and 90%. | [155, 164] | |
|
IC51 vaccine * (JESPECT®, JEEV®, IXIARO®) |
Vero cell culture, alum-adjuvanted and formalin-inactivated | SA14-14-2 |
Developed by Valneva Scotland Limited in 2009. Licensed in Australia and New Zealand in 2009 and 2012, respectively as JESPECT®. Licensed in India as JEEV® in 2012. IXIARO® is the only JEV vaccine licensed in the US and Europe. |
Jelinek et al. (2013) reviewed IXIARO, showing a 100% seroconversion rate (SCR) on booster dose, but a phase III follow-up study by Taucher et al. (2020) showed a reduction of SCR with time. IXIARO is used for vaccination of travellers to Asian countries. Highly immunogenic and induces a significantly higher antibody titer than mouse brain-derived vaccine Tomas Jelinek (2013) and Christa and Jilma (2015) reviewed IXIARO, and Taucher et al. (2020) found that IXIARO has an excellent safety profile and therefore is highly recommendable. IXIARO is suggested for children as well as adults at risk of JEV infection. |
[153, 165–172] | |
| JEBIK® V* | Vero cell culture | Beijing-1 strain | Biken, Japan, 2009 |
Showed high immunogenicity 86.8% seroconversion rate on single dose in adults, and 100% seroconversion rate in children following three doses of vaccine Good safety profile with no serious adverse events More effective than licensed mouse-brain-derived JE vaccine Limited international distribution |
[153, 173–175] | |
| ENCEVAC®* | Vero cell culture | Beijing-1 strain | Kaketsutan, Japan, 2011 | |||
|
CVI-JE (JEVACTM) |
Cell culture, inactivated | Beijing P3 strain | Liaoning Chengda Biotechnology Co., and licensed in China |
89.3% seroconversion after two doses and 100% seroconversion after booster dose was observed in children vaccinated with JEVACTM Limited international distribution |
[153, 176] | |
| Kolar-821564XY (JENVAC)* | Vero cell culture | Indian JEV genotype III strain | Bharat Biotech International Limited, India, 2013 |
Phase II/III clinical trial in India showed >95% SCR on single dose of JENVAC Intramuscular administration into the deltoid region of upper arm for adults and anterolateral region of thigh for children Induced cross-neutralizing antibodies against JEV I-IV genotypes Recommended by Indian Academy of Pediatrics (IAP) to be given at a minimum of 1 year and up to 50 years of age. Limited international distribution |
[153, 175, 177–180] | |
|
Live attenuated (A single dose is recommended by WHO in children ≥ 8 months of age) |
SA 14-14-2 Vaccine (CD.JEVAX™)*† |
Primary hamster kidney (PHK) cells | Obtained through the multiple passages of the wild-type SA14 strain | Chengdu Institute of Biological Products (CDIBP), China in 1980 |
Six mutations in the E protein and three in non-structural proteins were associated with attenuation. 80% efficacy with a single dose and 98% efficacy a with double dose |
[181–184] |
|
Live chimeric (A single dose is recommended by WHO in children ≥ 9 months of age) |
JE-CV/ChimeriVAXTM-JE (IMOJEV®)* |
Vero cells | Developed by replacing the pre-membrane and envelope genes of the attenuated yellow fever strain 17D with the corresponding genes of strain SA 14-14-2 |
Sanofi Pasteur. First registered in 2010 and licensed in several countries |
Recommended for both adults and children. >99% and 95% SCR was observed in adults and children, respectively. Protection efficacy of 97-100% was observed in children after a booster dose given 12 to 24 months of first dose. Generally, for JE-CV, a booster dose is not required for up to 5 years in adults and, a few studies have reported a robust immune response upon administration of a booster dose, even after the 5-year interval. |
[185–192] |
*Licensed JEV vaccines
†Discontinued JEV vaccines
Its asymptomatic nature, short viraemia period, and cross-reactivity with other flaviviruses make diagnosis of JEV infection difficult. Therefore, a combination of diagnostic methods should be applied to detect JEV infection, such as virus isolation, plaque reduction neutralisation test (PRNT), hemagglutination test, complement fixation test, immunofluorescence test, RT-PCR, Q-PCR, ELISA, and biosensor-based diagnosis. All of these diagnostic methods and their procedures have been reviewed by Roberts and Gandhi [83]. Recently, Zhang et al. developed a dual-mode chromatography method (Sepharose 4 fast flow chromatography and CaptoTM core 700 chromatography) to purify cell-derived JEV with intact infectivity and immunogenicity. This method is rapid and straightforward compared to the traditional purification method [193]. Zhou et al. developed a real-time nucleic-acid-sequence-based amplification (RT-NASBA) method for detection of JEV RNA within 10 minutes without compromising specificity and sensitivity. The sensitivity of RT-NABSA was compared to RT-PCR and found to be 100 to 1000 times higher [194].
Despite the emergence of JEV in non-endemic countries, it is surprising to see that effective treatments and anti-JEV drugs are still unavailable, and in the last ten years (2011 to 2020), only four clinical trials have been conducted [195–198]. Turtle and Solomon have reviewed all of the drugs tested in clinical trials, including dexamethasone [199], interferon [200], ribavirin [201], IVIG [198], and minocycline [195, 197], and further updates about the drugs in clinical trials, together with their methodology, outcomes, and limitations have been published by Ajibowo et al. Only minocycline showed promising results in all the clinical trials drugs [202]. Turtle and Solomon also reviewed the drugs showing protective efficacy in animal models and significant anti-JEV activities in vitro. Testing these drugs in clinical trials may result in effective anti-JEV drugs [16]. Recently, various inhibitors were reported to inhibit JEV entry and replication (Table 2), and these can also be considered for further studies in animal models and tested in clinical trials.
Table 2.
List of recently identified anti-JEV compounds that can be tested for their safety and protective efficacy
| Compounds | Type | Method | Target | Reference |
|---|---|---|---|---|
| Ouabain and digoxin | FDA-approved drug | Identification through high-content screening (HCS) and validation through in vivo and in vitro assays |
Ouabain and digoxin block JEV RNA synthesis by inhibiting the Na+/K+ ATPase. Ouabain significantly reduces the morbidity and mortality in BALB/c mice caused by JEV infection. |
[203] |
| Manidipine | FDA-approved drug | Identification through high-throughput screening (HTS) and validation through in vitro and in vivo assays | Inhibitor of intracellular calcium, which is required for JEV entry, replication, and budding | [204] |
| P1 Inhibitor | Peptide inhibitor | Identification through screening of phage-displayed peptides and validated through in vitro and in vivo assays. | A non-cytotoxic inhibitor, P1 interacts with the JEV envelope protein and blocks its entry into the host cell. | [205] |
| Gedunine, nimbolide, ohchinin acetate, and kulactone | Natural product | Identification through structure-based virtual screening. The affinity and stability were validated through binding free energy calculation and molecular dynamics simulation | Inhibitors of JEV RNA-dependent RNA polymerase (RdRp). Being a natural product, they would be less toxic and therefore can be considered further for the development of anti-JEV drugs. | [206] |
| Berbamine | Natural product | Validated through in vitro and in vivo assays | Berbamine blocks viral entry by lowering the amount of low-density lipoprotein receptor (LDLR), which acts as a receptor for JEV and interacts with the envelope protein. | [207] |
Concluding remarks
Despite being a major cause of viral encephalitis, JEV has received less attention than some other flaviviruses and remains an important disease for which there is no effective specific treatment. Despite the availability of vaccines, outbreaks of Japanese encephalitis occur annually in Asian countries. Therefore, continuing efforts are needed to develop drugs that inhibit the functions of JEV proteins. Although recent advances in JEV research have helped us to understand the basic mechanisms involved in pathogenesis, more detailed information is still needed. This review sheds light on a few host proteins possessing anti-JEV and pro-JEV activity, as well as the importance of several SPs and NSPs of JEV involved at various stages of its pathogenesis, such as viral entry, modification of the host ER membrane required for RNA replication and virion assembly a>nd maturation. Eventually, the insights into the pathogenicity and infection caused by JEV will provide a lead that will help identify prominent targets and develop a strategy to inhibit their functions at the molecular level.
Acknowledgements
Author Sanjay Kumar and Akanksha Verma acknowledge the Council of Scientific & Industrial Research (CSIR), Govt. of India, and the Department of Science & Technology-INSPIRE, Govt. of India, respectively, for providing Senior Research Fellowship (SRF) during PhD. The authors would like to acknowledge Jawaharlal Nehru University, New Delhi, India, for providing access to the Grammarly Business version through which the English language of the manuscript was improved.
Author contributions
SSM, VDD, EIA, and SK conceptualised the study and hypotheses. SK, PY, SKD, and AV performed literature search. SK and VDD drew the schemes, drafted the artwork and wrote the manuscript. All authors contributed significantly in editing the manuscript. All authors read, edited, and approved the manuscript.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Esam Ibraheem Azhar, Email: eazhar@kau.edu.sa.
S. S. Maitra, Email: ssm2100@mail.jnu.ac.in
Vivek Dhar Dwivedi, Email: vivek_bioinformatics@yahoo.com.
References
- 1.Solomon T. Control of Japanese encephalitis—within our grasp? N Engl J Med. 2006;355:869–871. doi: 10.1056/NEJMp058263. [DOI] [PubMed] [Google Scholar]
- 2.Misra UK, Kalita J. Overview: Japanese encephalitis. Prog Neurobiol. 2010;91:108–120. doi: 10.1016/j.pneurobio.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 3.WHO (2020) 1–4.
- 4.Sarkari NBS, Thacker AK, Barthwal SP, et al. Japanese encephalitis (JE) part II: 14 years’ follow-up of survivors. J Neurol. 2012;259:58–69. doi: 10.1007/s00415-011-6131-9. [DOI] [PubMed] [Google Scholar]
- 5.Solomon T, Dung NM, Kneen R, et al. Seizures and raised intracranial pressure in Vietnamese patients with Japanese encephalitis. Brain. 2002;125:1084–1093. doi: 10.1093/brain/awf116. [DOI] [PubMed] [Google Scholar]
- 6.Dickerson RB, Newton JR, Hansen JE. Diagnosis and immediate prognosis of Japanese B encephalitis; observations based on more than 200 patients with detailed analysis of 65 serologically confirmed cases. Am J Med. 1952;12:277–288. doi: 10.1016/0002-9343(52)90356-2. [DOI] [PubMed] [Google Scholar]
- 7.Kumar R, Tripathi P, Singh S, Bannerji G. Clinical Features in Children Hospitalized during the 2005 Epidemic of Japanese Encephalitis in Uttar Pradesh, India. Clin Infect Dis. 2006;43:123–131. doi: 10.1086/505121. [DOI] [PubMed] [Google Scholar]
- 8.Solomon T, Kneen R, Dung NM, et al. Pollomyelitis-like illness due to Japanese encephalitis virus. Lancet. 1998;351:1094–1097. doi: 10.1016/S0140-6736(97)07509-0. [DOI] [PubMed] [Google Scholar]
- 9.Unni SK, Růžek D, Chhatbar C, et al. Japanese encephalitis virus: from genome to infectome. Microbes Infect. 2011;13:312–321. doi: 10.1016/j.micinf.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Yun S-I, Lee Y-M. Early events in Japanese encephalitis virus infection: viral entry. Pathogens (Basel, Switzerland) 2018;7:68. doi: 10.3390/pathogens7030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van den Hurk AF, Ritchie SA, Mackenzie JS. Ecology and geographical expansion of Japanese encephalitis virus. Annu Rev Entomol. 2009;54:17–35. doi: 10.1146/annurev.ento.54.110807.090510. [DOI] [PubMed] [Google Scholar]
- 12.Erlanger TE, Weiss S, Keiser J, et al. Past, present, and future of Japanese encephalitis. Emerg Infect Dis. 2009;15:1–7. doi: 10.3201/eid1501.080311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ricklin ME, Garcìa-Nicolàs O, Brechbühl D, et al. Japanese encephalitis virus tropism in experimentally infected pigs. Vet Res. 2016;47:34. doi: 10.1186/s13567-016-0319-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosen L, Shroyer DA, Lien JC. Transovarial transmission of Japanese encephalitis virus by Culex tritaeniorhynchus mosquitoes. Am J Trop Med Hyg. 1980;29:711–712. doi: 10.4269/ajtmh.1980.29.711. [DOI] [PubMed] [Google Scholar]
- 15.Filgueira L, Lannes N. Review of emerging Japanese encephalitis virus: new aspects and concepts about entry into the brain and inter-cellular spreading. Pathogens (Basel, Switzerland) 2019 doi: 10.3390/pathogens8030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turtle L, Solomon T. Japanese encephalitis - the prospects for new treatments. Nat Rev Neurol. 2018;14:298–313. doi: 10.1038/nrneurol.2018.30. [DOI] [PubMed] [Google Scholar]
- 17.Harris E, Holden KL, Edgil D, et al. Molecular biology of flaviviruses. Novartis Found Symp. 2006;277:23–39. [PubMed] [Google Scholar]
- 18.Dong H, Fink K, Züst R, et al. Flavivirus RNA methylation. J Gen Virol. 2014;95:763–778. doi: 10.1099/vir.0.062208-0. [DOI] [PubMed] [Google Scholar]
- 19.Klema VJ, Padmanabhan R, Choi KH. Flaviviral replication complex: coordination between RNA synthesis and 5′-RNA capping. Viruses. 2015;7:4640–4656. doi: 10.3390/v7082837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu B, Qin Y, Li B, et al. Full-length genome sequence of Japanese encephalitis virus strain FC792, isolated from Guangxi, China. Genome Announc. 2017;5:e01054–e1117. doi: 10.1128/genomeA.01054-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alvarez DE, Lodeiro MF, Ludueña SJ, et al. Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol. 2005;79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lodeiro MF, Filomatori CV, Gamarnik AV. Structural and functional studies of the promoter element for dengue virus RNA replication. J Virol. 2009;83:993–1008. doi: 10.1128/JVI.01647-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gebhard LG, Filomatori CV, Gamarnik AV. Functional RNA elements in the dengue virus genome. Viruses. 2011;3:1739–1756. doi: 10.3390/v3091739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinz-J BDL, Rice CM (2007) Flaviviridae: the viruses and their replication
- 25.Kim JK, Kim JM, Song BH, Yun SI, Yun GN, Byun SJLY. Profiling of viral proteins expressed from the genomic RNA of Japanese encephalitis virus using a panel of 15 region-specific polyclonal rabbit antisera implications for viral gene expression. PLoS ONE. 2015 doi: 10.1371/journal.pone.0124318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luca VC, AbiMansour J, Nelson CA, Fremont DH. Crystal structure of the Japanese encephalitis virus envelope protein. J Virol. 2012;86:2337–2346. doi: 10.1128/JVI.06072-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stadler K, Allison SL, Schalich J, Heinz FX. Proteolytic activation of tick-borne encephalitis virus by furin. J Virol. 1997;71:8475–8481. doi: 10.1128/jvi.71.11.8475-8481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elshuber S, Allison SL, Heinz FX, Mandl CW. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J Gen Virol. 2003;84:183–191. doi: 10.1099/vir.0.18723-0. [DOI] [PubMed] [Google Scholar]
- 29.Yu I-M, Zhang W, Holdaway HA, et al. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science (80-) 2008;319:1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 30.McMinn PC. The molecular basis of virulence of the encephalitogenic flaviviruses. J Gen Virol. 1997;78(Pt 11):2711–2722. doi: 10.1099/0022-1317-78-11-2711. [DOI] [PubMed] [Google Scholar]
- 31.Konishi E, Yamaoka M, Kurane I, et al. The anamnestic neutralizing antibody response is critical for protection of mice from challenge following vaccination with a plasmid encoding the Japanese encephalitis virus premembrane and envelope genes. J Virol. 1999;73:5527–5534. doi: 10.1128/JVI.73.7.5527-5534.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keelapang P, Sriburi R, Supasa S, et al. Alterations of pr-M cleavage and virus export in pr-M junction chimeric dengue viruses. J Virol. 2004;78:2367–2381. doi: 10.1128/jvi.78.5.2367-2381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guirakhoo F, Bolin RA, Roehrig JT. The Murray Valley encephalitis virus prM protein confers acid resistance to virus particles and alters the expression of epitopes within the R2 domain of E glycoprotein. Virology. 1992;191:921–931. doi: 10.1016/0042-6822(92)90267-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinz FX, Stiasny K, Püschner-Auer G, et al. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology. 1994;198:109–117. doi: 10.1006/viro.1994.1013. [DOI] [PubMed] [Google Scholar]
- 35.Wengler G, Wengler G. Cell-associated West Nile flavivirus is covered with E+pre-M protein heterodimers which are destroyed and reorganized by proteolytic cleavage during virus release. J Virol. 1989;63:2521–2526. doi: 10.1128/jvi.63.6.2521-2526.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stiasny K, Allison SL, Marchler-Bauer A, et al. Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. J Virol. 1996;70:8142–8147. doi: 10.1128/JVI.70.11.8142-8147.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freire JM, Santos NC, Veiga AS, et al. Rethinking the capsid proteins of enveloped viruses: multifunctionality from genome packaging to genome transfection. FEBS J. 2015;282:2267–2278. doi: 10.1111/febs.13274. [DOI] [PubMed] [Google Scholar]
- 38.Jones CT, Ma L, Burgner JW, et al. Flavivirus capsid is a dimeric alpha-helical protein. J Virol. 2003;77:7143–7149. doi: 10.1128/jvi.77.12.7143-7149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dokland T, Walsh M, Mackenzie JM, et al. West Nile virus core protein: tetramer structure and ribbon formation. Structure. 2004;12:1157–1163. doi: 10.1016/j.str.2004.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma L, Jones CT, Groesch TD, et al. Solution structure of dengue virus capsid protein reveals another fold. Proc Natl Acad Sci USA. 2004;101:3414–3419. doi: 10.1073/pnas.0305892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsuda Y, Mori Y, Abe T, et al. Nucleolar protein B23 interacts with Japanese encephalitis virus core protein and participates in viral replication. Microbiol Immunol. 2006;50:225–234. doi: 10.1111/j.1348-0421.2006.tb03789.x. [DOI] [PubMed] [Google Scholar]
- 42.Mori Y, Okabayashi T, Yamashita T, et al. Nuclear localization of Japanese encephalitis virus core protein enhances viral replication. J Virol. 2005;79:3448–3458. doi: 10.1128/JVI.79.6.3448-3458.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarkar R, Sharma KB, Kumari A, et al. Japanese encephalitis virus capsid protein interacts with non-lipidated MAP1LC3 on replication membranes and lipid droplets. J Gen Virol. 2021 doi: 10.1099/jgv.0.001508. [DOI] [PubMed] [Google Scholar]
- 44.Katoh H, Okamoto T, Fukuhara T, et al. Japanese encephalitis virus core protein inhibits stress granule formation through an interaction with caprin-1 and facilitates viral propagation. J Virol. 2013;87:489–502. doi: 10.1128/JVI.02186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhuvanakantham R, Cheong YK, Ng ML. West Nile virus capsid protein interaction with importin and HDM2 protein is regulated by protein kinase C-mediated phosphorylation. Microbes Infect. 2010;12:615–625. doi: 10.1016/j.micinf.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Faustino AF, Carvalho FA, Martins IC, et al. Dengue virus capsid protein interacts specifically with very low-density lipoproteins. Nanomed Nanotechnol Biol Med. 2014;10:247–255. doi: 10.1016/j.nano.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 47.Guevara J, Romo J, McWhorter T, Guevara NV. Analogs of LDL receptor ligand motifs in dengue envelope and capsid proteins as potential codes for cell entry. J. Viruses. 2015;2015:1–15. doi: 10.1155/2015/646303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhuvanakantham R, Li J, Tan TTT, Ng M-L. Human Sec3 protein is a novel transcriptional and translational repressor of flavivirus. Cell Microbiol. 2010;12:453–472. doi: 10.1111/j.1462-5822.2009.01407.x. [DOI] [PubMed] [Google Scholar]
- 49.Bhuvanakantham R, Ng M-L. West Nile virus and dengue virus capsid protein negates the antiviral activity of human Sec3 protein through the proteasome pathway. Cell Microbiol. 2013;15:1688–1706. doi: 10.1111/cmi.12143. [DOI] [PubMed] [Google Scholar]
- 50.Alcalá AC, Palomares LA, Ludert JE. Secretion of nonstructural protein 1 of dengue virus from infected mosquito cells: facts and speculations. J Virol. 2018 doi: 10.1128/jvi.00275-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Youn S, Ambrose RL, MacKenzie JM, Diamond MS. Non-structural protein-1 is required for West Nile virus replication complex formation and viral RNA synthesis. Virol J. 2013;10:1–14. doi: 10.1186/1743-422X-10-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindenbach BD, Rice CM. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71:9608–9617. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fan J, Liu Y, Yuan Z. Critical role of dengue virus NS1 protein in viral replication. Virol Sin. 2014;29:162–169. doi: 10.1007/s12250-014-3459-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cervantes-Salazar M, Angel-Ambrocio AH, Soto-Acosta R, et al. Dengue virus NS1 protein interacts with the ribosomal protein RPL18: This interaction is required for viral translation and replication in Huh-7 cells. Virology. 2015;484:113–126. doi: 10.1016/j.virol.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 55.Wang P, Liu X, Li Q, et al. Proteomic analyses identify intracellular targets for Japanese encephalitis virus nonstructural protein 1 (NS1) Virus Res. 2021;302:198495. doi: 10.1016/j.virusres.2021.198495. [DOI] [PubMed] [Google Scholar]
- 56.Chung KM, Thompson BS, Fremont DH, Diamond MS. Antibody recognition of cell surface-associated NS1 triggers Fc-gamma receptor-mediated phagocytosis and clearance of West Nile virus-infected cells. J Virol. 2007;81:9551–9555. doi: 10.1128/JVI.00879-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson JR, de Sessions PF, Leon MA, Scholle F. West Nile virus nonstructural protein 1 inhibits TLR3 signal transduction. J Virol. 2008;82:8262–8271. doi: 10.1128/JVI.00226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou D, Li Q, Jia F, et al. The Japanese encephalitis virus NS1′ protein inhibits type I IFN production by targeting MAVS. J Immunol. 2020;204:1287–1298. doi: 10.4049/jimmunol.1900946. [DOI] [PubMed] [Google Scholar]
- 59.Wan J, Wang T, Xu J, et al. Novel Japanese encephalitis virus NS1-based vaccine: truncated NS1 fused with E. coli heat labile enterotoxin B subunit. EBioMedicine. 2021;67:103353. doi: 10.1016/j.ebiom.2021.103353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chambers TJ, McCourt DW, Rice CM. Yellow fever virus proteins NS2A, NS213, and NS4B: identification and partial N-terminal amino acid sequence analysis. Virology. 1989;169:100–109. doi: 10.1016/0042-6822(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 61.Mackenzie JM, Khromykh AA, Jones MK, Westaway EG. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology. 1998;245:203–215. doi: 10.1006/viro.1998.9156. [DOI] [PubMed] [Google Scholar]
- 62.Khromykh AA, Varnavski AN, Sedlak PL. Coupling between replication and packaging of flavivirus RNA: evidence derived from the use of DNA-based full-length cDNA clones of Kunjin virus. J Virol. 2001;75:4633–4640. doi: 10.1128/JVI.75.10.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gonzaleza ME, Carrasco L. Virporins. FEBS Lett. 2003;552:28–34. doi: 10.1016/S0014-5793(03)00780-4. [DOI] [PubMed] [Google Scholar]
- 64.Tu Y-C, Yu C-Y, Liang J-J, et al. Blocking double-stranded RNA-activated protein kinase PKR by Japanese encephalitis virus nonstructural protein 2A. J Virol. 2012;86:10347–10358. doi: 10.1128/JVI.00525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ye Q, Li X-F, Zhao H, et al. A single nucleotide mutation in NS2A of Japanese encephalitis-live vaccine virus (SA14-14-2) ablates NS1’ formation and contributes to attenuation. J Gen Virol. 2012;93:1959–1964. doi: 10.1099/vir.0.043844-0. [DOI] [PubMed] [Google Scholar]
- 66.Yusof R, Clum S, Wetzel M, et al. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J Biol Chem. 2000;275:9963–9969. doi: 10.1074/jbc.275.14.9963. [DOI] [PubMed] [Google Scholar]
- 67.Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. Mutagenesis of the West Nile virus NS2B cofactor domain reveals two regions essential for protease activity. J Gen Virol. 2008;89:1010–1014. doi: 10.1099/vir.0.83447-0. [DOI] [PubMed] [Google Scholar]
- 68.Yamashita T, Unno H, Mori Y, et al. Crystal structure of the catalytic domain of Japanese encephalitis virus NS3 helicase/nucleoside triphosphatase at a resolution of 1.8 Å. Virology. 2008;373:426–436. doi: 10.1016/j.virol.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 69.Nestorowicz A, Chambers TJ, Rice CM. Mutagenesis of the yellow fever virus NS2A/2B cleavage site: effects on proteolytic processing, viral replication, and evidence for alternative processing of the NS2A protein. Virology. 1994;199:114–123. doi: 10.1006/viro.1994.1103. [DOI] [PubMed] [Google Scholar]
- 70.Teo KF, Wright PJ. Internal proteolysis of the NS3 protein specified by dengue virus 2. J Gen Virol. 1997;78:337–341. doi: 10.1099/0022-1317-78-2-337. [DOI] [PubMed] [Google Scholar]
- 71.Warrener P, Tamura JK, Collett MS. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J Virol. 1993;67:989–996. doi: 10.1128/JVI.67.2.989-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wen G, Chen C, Luo X, et al. Identification and characterization of the NTPase activity of classical swine fever virus (CSFV) nonstructural protein 3 (NS3) expressed in bacteria. Arch Virol. 2007;152:1565–1573. doi: 10.1007/s00705-007-0969-2. [DOI] [PubMed] [Google Scholar]
- 73.Xie S, Liang Z, Yang X, et al. Japanese encephalitis virus NS2B-3 protein complex promotes cell apoptosis and viral particle release by down-regulating the expression of AXL. Virol Sin. 2021;36:1503–1519. doi: 10.1007/s12250-021-00442-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shiryaev SA, Chernov AV, Aleshin AE, et al. NS4A regulates the ATPase activity of the NS3 helicase: a novel cofactor role of the non-structural protein NS4A from West Nile virus. J Gen Virol. 2009;90:2081–2085. doi: 10.1099/vir.0.012864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lu G, Gong P. Crystal structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog. 2013;9:e1003549. doi: 10.1371/journal.ppat.1003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Joyce CM. Polymerase structures and function: variations on a theme? J Bacteriol. 1995;177:6321–6329. doi: 10.1128/jb.177.22.6321-6329.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ng KKS, Arnold JJ, Cameron CE. Structure-function relationships among RNA-dependent RNA polymerases. Curr Top Microbiol Immunol. 2008;320:137–156. doi: 10.1007/978-3-540-75157-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bressanelli S, Tomei L, Roussel A, et al. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc Natl Acad Sci USA. 1999;96:13034–13039. doi: 10.1073/pnas.96.23.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choi KH, Groarke JM, Young DC, et al. The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation. Proc Natl Acad Sci USA. 2004;101:4425–4430. doi: 10.1073/pnas.0400660101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brass AL, Huang I-C, Benita Y, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu J, Pan Q, Rong L, et al. The IFITM proteins inhibit HIV-1 infection. J Virol. 2011;85:2126–2137. doi: 10.1128/JVI.01531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nain M, Mukherjee S, Karmakar SP, et al. GRP78 is an important host factor for Japanese encephalitis virus entry and replication in mammalian cells. J Virol. 2017 doi: 10.1128/JVI.02274-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roberts A, Gandhi S. Japanese encephalitis virus: a review on emerging diagnostic techniques. Front Biosci (Landmark Ed) 2020;25:1875–1893. doi: 10.2741/4882. [DOI] [PubMed] [Google Scholar]
- 84.Zhang L-K, Chai F, Li H-Y, et al. Identification of host proteins involved in Japanese encephalitis virus infection by quantitative proteomics analysis. J Proteome Res. 2013;12:2666–2678. doi: 10.1021/pr400011k. [DOI] [PubMed] [Google Scholar]
- 85.Wang X, Wu Z, Li Y, et al. p53 promotes ZDHHC1-mediated IFITM3 palmitoylation to inhibit Japanese encephalitis virus replication. PLoS Pathog. 2020;16:e1009035. doi: 10.1371/journal.ppat.1009035. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 86.Shi G, Kenney AD, Kudryashova E, et al. Opposing activities of IFITM proteins in SARS-CoV-2 infection. EMBO J. 2021;40:e106501. doi: 10.15252/embj.2020106501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McMichael TM, Zhang Y, Kenney AD, et al. IFITM3 restricts human metapneumovirus infection. J Infect Dis. 2018;218:1582–1591. doi: 10.1093/infdis/jiy361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zani A, Yount JS. Antiviral protection by IFITM3 in vivo. Curr Clin Microbiol Rep. 2018;5:229–237. doi: 10.1007/s40588-018-0103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beddow AL, Richards SA, Orem NR, Macara IG. The Ran/TC4 GTPase-binding domain: identification by expression cloning and characterization of a conserved sequence motif. Proc Natl Acad Sci USA. 1995;92:3328–3332. doi: 10.1073/pnas.92.8.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maarifi G, Fernandez J, Portilho DM, et al. RanBP2 regulates the anti-retroviral activity of TRIM5α by SUMOylation at a predicted phosphorylated SUMOylation motif. Commun Biol. 2018;1:193. doi: 10.1038/s42003-018-0198-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang J, Dupuis C, Tyring SK, Underbrink MP. Sterile α motif domain containing 9 is a novel cellular interacting partner to low-risk type human papillomavirus e6 proteins. PLoS ONE. 2016;11:e0149859. doi: 10.1371/journal.pone.0149859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van Tol S, Atkins C, Bharaj P, et al. VAMP8 contributes to the TRIM6-mediated type I interferon antiviral response during West Nile virus infection. J Virol. 2020 doi: 10.1128/JVI.01454-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fan W, Wu M, Qian S, et al. TRIM52 inhibits Japanese Encephalitis Virus replication by degrading the viral NS2A. Sci Rep. 2016;6:33698. doi: 10.1038/srep33698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Das S, Ravi V, Desai A. Japanese encephalitis virus interacts with vimentin to facilitate its entry into porcine kidney cell line. Virus Res. 2011;160:404–408. doi: 10.1016/j.virusres.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 95.Shen S, Shen C, Lin H, et al. Susceptibility of human embryonic stem cell-derived neural cells to Japanese encephalitis virus infection. PLoS ONE. 2014 doi: 10.1371/journal.pone.0114990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brunetti JE, Scolaro LA, Castilla V. The heterogeneous nuclear ribonucleoprotein K (hnRNP K) is a host factor required for dengue virus and Junín virus multiplication. Virus Res. 2015;203:84–91. doi: 10.1016/j.virusres.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 97.Katoh H, Mori Y, Kambara H, et al. Heterogeneous nuclear ribonucleoprotein A2 participates in the replication of Japanese encephalitis virus through an interaction with viral proteins and RNA. J Virol. 2011;85:10976–10988. doi: 10.1128/JVI.00846-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen C-J, Ou Y-C, Li J-R, et al. Infection of pericytes in vitro by Japanese encephalitis virus disrupts the integrity of the endothelial barrier. J Virol. 2014;88:1150–1161. doi: 10.1128/JVI.02738-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang K, Deubel V. Mice with different susceptibility to Japanese encephalitis virus infection show selective neutralizing antibody response and myeloid cell infectivity. PLoS ONE. 2011;6:e24744. doi: 10.1371/journal.pone.0024744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shwetank DOS, Carbone E, Manjunath R. Inhibition of ERK and proliferation in NK cell lines by soluble HLA-E released from Japanese encephalitis virus infected cells. Immunol Lett. 2014;162:94–100. doi: 10.1016/j.imlet.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 101.Shimojima M, Takenouchi A, Shimoda H, et al. Distinct usage of three C-type lectins by Japanese encephalitis virus: DC-SIGN, DC-SIGNR, and LSECtin. Arch Virol. 2014;159:2023–2031. doi: 10.1007/s00705-014-2042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kumar G, Date OS, Kim KS, Manjunath R. Infection of human amniotic and endothelial cells by Japanese encephalitis virus: increased expression of HLA-F. Virology. 2014;471–473:29–37. doi: 10.1016/j.virol.2014.09.022. [DOI] [PubMed] [Google Scholar]
- 103.Chauhan S, Rathore DK, Sachan S, et al. Japanese encephalitis virus infected human monocyte-derived dendritic cells activate a transcriptional network leading to an antiviral inflammatory response. Front Immunol. 2021;12:1880. doi: 10.3389/fimmu.2021.638694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kalia M, Khasa R, Sharma M, et al. Japanese encephalitis virus infects neuronal cells through a clathrin-independent endocytic mechanism. J Virol. 2013;87:148–162. doi: 10.1128/JVI.01399-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lannes N, Neuhaus V, Scolari B, et al. Interactions of human microglia cells with Japanese encephalitis virus. Virol J. 2017;14:8. doi: 10.1186/s12985-016-0675-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hsieh JT, St John AL. Japanese encephalitis virus and its mechanisms of neuroinvasion. PLoS Pathog. 2020;16:e1008260. doi: 10.1371/journal.ppat.1008260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Filgueira L, Lannes N. Review of emerging Japanese encephalitis virus: new aspects and concepts about entry into the brain and inter-cellular spreading. Pathogens. 2019 doi: 10.3390/pathogens8030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rana J, Slon Campos JL, Leccese G, et al. Role of capsid anchor in the morphogenesis of zika virus. J Virol. 2018;92:1–15. doi: 10.1128/jvi.01174-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Phong WY, Moreland NJ, Lim SP, et al. Dengue protease activity: The structural integrity and interaction of NS2B with NS3 protease and its potential as a drug target. Biosci Rep. 2011;31:399–409. doi: 10.1042/BSR20100142. [DOI] [PubMed] [Google Scholar]
- 110.Arias CF, Preugschat F, Strauss JH. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology. 1993;193:888–899. doi: 10.1006/viro.1993.1198. [DOI] [PubMed] [Google Scholar]
- 111.Cui T, Sugrue RJ, Xu Q, et al. Recombinant dengue virus type 1 NS3 protein exhibits specific viral RNA binding and NTPase activity regulated by the NS5 protein. Virology. 1998;246:409–417. doi: 10.1006/viro.1998.9213. [DOI] [PubMed] [Google Scholar]
- 112.Liu WJ, Chen HB, Khromykh AA. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for ns2a in virus assembly and for a nonconservative residue in NS3 in RNA replication. J Virol. 2003;77:7804–7813. doi: 10.1128/JVI.77.14.7804-7813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wu CF, Wang SH, Sun CM, et al. Activation of dengue protease autocleavage at the NS2B-NS3 junction by recombinant NS3 and GST-NS2B fusion proteins. J Virol Methods. 2003;114:45–54. doi: 10.1016/j.jviromet.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 114.van Dijk AA, Makeyev EV, Bamford DH. Initiation of viral RNA-dependent RNA polymerization. J Gen Virol. 2004;85:1077–1093. doi: 10.1099/vir.0.19731-0. [DOI] [PubMed] [Google Scholar]
- 115.Mackenzie JM, Jones MK, Young PR. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996;240:232–240. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- 116.Apcher GS, Heink S, Zantopf D, et al. Human immunodeficiency virus-1 Tat protein interacts with distinct proteasomal alpha and beta subunits. FEBS Lett. 2003;553:200–204. doi: 10.1016/s0014-5793(03)01025-1. [DOI] [PubMed] [Google Scholar]
- 117.Jiang D, Weidner JM, Qing M, et al. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J Virol. 2010;84:8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schaller T, Ocwieja KE, Rasaiyaah J, et al. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 2011;7:e1002439. doi: 10.1371/journal.ppat.1002439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yao L, Dong H, Zhu H, et al. Identification of the IFITM3 gene as an inhibitor of hepatitis C viral translation in a stable STAT1 cell line. J Viral Hepat. 2011;18:e523–e529. doi: 10.1111/j.1365-2893.2011.01452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu J, Wennier S, Zhang L, McFadden G. M062 is a host range factor essential for myxoma virus pathogenesis and functions as an antagonist of host SAMD9 in human cells. J Virol. 2011;85:3270–3282. doi: 10.1128/JVI.02243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen Y, Maguire T, Hileman RE, et al. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997;3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 122.Lee E, Pavy M, Young N, et al. Antiviral effect of the heparan sulfate mimetic, PI-88, against dengue and encephalitic flaviviruses. Antiviral Res. 2006;69:31–38. doi: 10.1016/j.antiviral.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 123.Das S, Laxminarayana SV, Chandra N, et al. Heat shock protein 70 on Neuro2a cells is a putative receptor for Japanese encephalitis virus. Virology. 2009;385:47–57. doi: 10.1016/j.virol.2008.10.025. [DOI] [PubMed] [Google Scholar]
- 124.Zhu Y-Z, Cao M-M, Wang W-B, et al. Association of heat-shock protein 70 with lipid rafts is required for Japanese encephalitis virus infection in Huh7 cells. J Gen Virol. 2012;93:61–71. doi: 10.1099/vir.0.034637-0. [DOI] [PubMed] [Google Scholar]
- 125.Su H-L, Liao C-L, Lin Y-L. Japanese encephalitis virus infection initiates endoplasmic reticulum stress and an unfolded protein response. J Virol. 2002;76:4162–4171. doi: 10.1128/jvi.76.9.4162-4171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lin RJ, Liao CL, Lin YL. Replication-incompetent virions of Japanese encephalitis virus trigger neuronal cell death by oxidative stress in a culture system. J Gen Virol. 2004;85:521–533. doi: 10.1099/vir.0.19496-0. [DOI] [PubMed] [Google Scholar]
- 127.Tsao C-H, Su H-L, Lin Y-L, et al. Japanese encephalitis virus infection activates caspase-8 and -9 in a FADD-independent and mitochondrion-dependent manner. J Gen Virol. 2008;89:1930–1941. doi: 10.1099/vir.0.2008/000182-0. [DOI] [PubMed] [Google Scholar]
- 128.Lin R-J, Chang B-L, Yu H-P, et al. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J Virol. 2006;80:5908–5918. doi: 10.1128/JVI.02714-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Guo F, Yu X, Xu A, et al. Japanese encephalitis virus induces apoptosis by inhibiting Foxo signaling pathway. Vet Microbiol. 2018;220:73–82. doi: 10.1016/j.vetmic.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 130.Hase T, Summers PL, Ray P, Asafo-Adjei E. Cytopathology of PC12 cells infected with Japanese encephalitis virus. Virchows Arch B Cell Pathol Incl Mol Pathol. 1992;63:25–36. doi: 10.1007/BF02899241. [DOI] [PubMed] [Google Scholar]
- 131.Chambers TJ, Hahn CS, Galler R, Rice CM. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 132.Kumar S, Boehm J, Lee JC. p38 MAP kinases: key signalling molecules as therapeutic targets for inflammatory diseases. Nat Rev Drug Discov. 2003;2:717–726. doi: 10.1038/nrd1177. [DOI] [PubMed] [Google Scholar]
- 133.Horndasch M, Lienkamp S, Springer E, et al. The C/EBP homologous protein CHOP (GADD153) is an inhibitor of Wnt/TCF signals. Oncogene. 2006;25:3397–3407. doi: 10.1038/sj.onc.1209380. [DOI] [PubMed] [Google Scholar]
- 134.Zhao A, Zhang Z, Zhou Y, et al. β-Elemonic acid inhibits the growth of human Osteosarcoma through endoplasmic reticulum (ER) stress-mediated PERK/eIF2α/ATF4/CHOP activation and Wnt/β-catenin signal suppression. Phytomedicine. 2020;69:153183. doi: 10.1016/j.phymed.2020.153183. [DOI] [PubMed] [Google Scholar]
- 135.Yu BP. Cellular defenses against damage from reactive oxygen species. Physiol Rev. 1994;74:139–162. doi: 10.1152/physrev.1994.74.1.139. [DOI] [PubMed] [Google Scholar]
- 136.Raung SL, Der KM, Wang YM, Chen CJ. Role of reactive oxygen intermediates in Japanese encephalitis virus infection in murine neuroblastoma cells. Neurosci Lett. 2001;315:9–12. doi: 10.1016/S0304-3940(01)02300-X. [DOI] [PubMed] [Google Scholar]
- 137.Richter C, Gogvadze V, Laffranchi R, et al. Oxidants in mitochondria: from physiology to diseases. BBA - Mol Basis Dis. 1995;1271:67–74. doi: 10.1016/0925-4439(95)00012-S. [DOI] [PubMed] [Google Scholar]
- 138.Li Y, Li Z. Mitochondria and apoptosis. Zhonghua Yu Fang Yi Xue Za Zhi. 2000;34:183–184. [PubMed] [Google Scholar]
- 139.Yang T-C, Lai C-C, Shiu S-L, et al. Japanese encephalitis virus down-regulates thioredoxin and induces ROS-mediated ASK1-ERK/p38 MAPK activation in human promonocyte cells. Microbes Infect. 2010;12:643–651. doi: 10.1016/j.micinf.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 140.Liao CL, Lin YL, Wang JJ, et al. Effect of enforced expression of human bcl-2 on Japanese encephalitis virus-induced apoptosis in cultured cells. J Virol. 1997;71:5963–5971. doi: 10.1128/JVI.71.8.5963-5971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cai J, Jones DP. Superoxide in apoptosis. Mitochondrial generation triggered by cytochrome c loss. J Biol Chem. 1998;273:11401–11404. doi: 10.1074/jbc.273.19.11401. [DOI] [PubMed] [Google Scholar]
- 142.Green DR, Kroemer G. The pathophysiology of mitochondrial cell death. Science. 2004;305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 143.Tan S, Sagara Y, Liu Y, et al. The regulation of reactive oxygen species production during programmed cell death. J Cell Biol. 1998;141:1423–1432. doi: 10.1083/jcb.141.6.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chen A. PARP inhibitors: its role in treatment of cancer. Chin J Cancer. 2011;30:463–471. doi: 10.5732/cjc.011.10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Morales J, Li L, Fattah FJ, et al. Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases. Crit Rev Eukaryot Gene Expr. 2014;24:15–28. doi: 10.1615/critreveukaryotgeneexpr.2013006875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Burtnick LD, Robinson RC, Choe S. Structure and function of gelsolin. Results Probl Cell Differ. 2001;32:201–211. doi: 10.1007/978-3-540-46560-7_14. [DOI] [PubMed] [Google Scholar]
- 147.Hu Q, Wu D, Chen W, et al. Molecular determinants of caspase-9 activation by the Apaf-1 apoptosome. Proc Natl Acad Sci USA. 2014;111:16254–16261. doi: 10.1073/pnas.1418000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li Y, Zhou M, Hu Q, et al. Mechanistic insights into caspase-9 activation by the structure of the apoptosome holoenzyme. Proc Natl Acad Sci USA. 2017;114:1542–1547. doi: 10.1073/pnas.1620626114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene. 2002;285:1–24. doi: 10.1016/s0378-1119(02)00398-0. [DOI] [PubMed] [Google Scholar]
- 150.Best SM, Morris KL, Shannon JG, et al. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J Virol. 2005;79:12828–12839. doi: 10.1128/JVI.79.20.12828-12839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Treatment JE (2019) Japanese encephalitis treatment & management. p 4–7
- 152.Hegde NR, Gore MM. Japanese encephalitis vaccines: immunogenicity, protective efficacy, effectiveness, and impact on the burden of disease. Hum Vaccin Immunother. 2017;13:1–18. doi: 10.1080/21645515.2017.1285472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Barzon L, Palù G. Recent developments in vaccines and biological therapies against Japanese encephalitis virus. Expert Opin Biol Ther. 2018;18:851–864. doi: 10.1080/14712598.2018.1499721. [DOI] [PubMed] [Google Scholar]
- 154.Kumar A, Sharma P, Shukla KK, et al. Japanese encephalitis virus: associated immune response and recent progress in vaccine development. Microb Pathog. 2019;136:103678. doi: 10.1016/j.micpath.2019.103678. [DOI] [PubMed] [Google Scholar]
- 155.Satchidanandam V. Japanese encephalitis vaccines. Curr Treat Options Infect Dis. 2020 doi: 10.1007/s40506-020-00242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hu Y-L, Lee P-I. Safety of Japanese encephalitis vaccines. Hum Vaccin Immunother. 2021 doi: 10.1080/21645515.2021.1969852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Furuya-Kanamori L, Xu C, Doi SAR, et al. Comparison of immunogenicity and safety of licensed Japanese encephalitis vaccines: a systematic review and network meta-analysis. Vaccine. 2021;39:4429–4436. doi: 10.1016/j.vaccine.2021.06.023. [DOI] [PubMed] [Google Scholar]
- 158.Lee P-I, Huang Y-C, Hwang K-P, et al. Recommendations for the use of Japanese encephalitis vaccines. Pediatr Neonatol. 2020;61:3–8. doi: 10.1016/j.pedneo.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 159.Connor BA, Hamer DH, Kozarsky P, et al. Japanese encephalitis vaccine for travelers: risk-benefit reconsidered. J Travel Med. 2019 doi: 10.1093/jtm/taz037. [DOI] [PubMed] [Google Scholar]
- 160.Chakraborty S, Barman A, Deb B. Japanese encephalitis virus: a multi-epitope loaded peptide vaccine formulation using reverse vaccinology approach. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2020;78:104106. doi: 10.1016/j.meegid.2019.104106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Poland JD, Cropp CB, Craven RB, Monath TP. Evaluation of the potency and safety of inactivated Japanese encephalitis vaccine in US inhabitants. J Infect Dis. 1990;161:878–882. doi: 10.1093/infdis/161.5.878. [DOI] [PubMed] [Google Scholar]
- 162.Hoke CH, Nisalak A, Sangawhipa N, et al. Protection against Japanese encephalitis by inactivated vaccines. N Engl J Med. 1988;319:608–614. doi: 10.1056/NEJM198809083191004. [DOI] [PubMed] [Google Scholar]
- 163.Halstead SB, Jacobson J (2008) Chapter 17—Japanese encephalitis vaccines, 5th ed. In: Plotkin SA, Orenstein WA, Offit Pabt V (eds). W.B. Saunders, Elesivier, Edinburgh, p 311–352
- 164.WHO (2014) WHO JE vaccine information sheet. p 1–5
- 165.Jelinek T. Ixiaro: a new vaccine against Japanese encephalitis. Expert Rev Vaccines. 2009;8:1501–1511. doi: 10.1586/erv.09.112. [DOI] [PubMed] [Google Scholar]
- 166.Firbas C, Jilma B. Product review on the JE vaccine IXIARO. Hum Vaccin Immunother. 2015;11:411–420. doi: 10.4161/21645515.2014.983412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Taucher C, Barnett ED, Cramer JP, et al. Neutralizing antibody persistence in pediatric travelers from non-JE-endemic countries following vaccination with IXIARO® Japanese encephalitis vaccine: an uncontrolled, open-label phase 3 follow-up study. Travel Med Infect Dis. 2020;34:101616. doi: 10.1016/j.tmaid.2020.101616. [DOI] [PubMed] [Google Scholar]
- 168.Jelinek T. IXIARO updated: overview of clinical trials and developments with the inactivated vaccine against Japanese encephalitis. Expert Rev Vaccines. 2013;12:859–869. doi: 10.1586/14760584.2013.835638. [DOI] [PubMed] [Google Scholar]
- 169.Jelinek T, Cromer MA, Cramer JP, et al. Safety and immunogenicity of an inactivated Vero cell_derived Japanese encephalitis vaccine (IXIARO(®), JESPECT(®)) in a pediatric population in JE non-endemic countries: an uncontrolled, open-label phase 3 study. Travel Med Infect Dis. 2018;22:18–24. doi: 10.1016/j.tmaid.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 170.(2020) Australian Product Information—JESPECT. p 1–21
- 171.(2021) JESPECT New Zealand data sheet. p 45–51. 10.4324/9780203025574-11
- 172.Japanese I, Vaccine E, Medicine C Jespec t ®. p 1–4
- 173.Okada K, Iwasa T, Namazue J, et al. Safety and immunogenicity of a freeze-dried, cell culture-derived Japanese encephalitis vaccine (inactivated) (JEBIK(®)V) in children. Vaccine. 2012;30:5967–5972. doi: 10.1016/j.vaccine.2012.07.034. [DOI] [PubMed] [Google Scholar]
- 174.Yun KW, Lee HJ, Kang JH, et al. Safety and immunogenicity of a freeze-dried, Vero cell culture-derived, inactivated Japanese encephalitis vaccine (KD-287, ENCEVAC®) versus a mouse brain-derived inactivated Japanese encephalitis vaccine in children: a phase III, multicenter, double-blinded, randomized trial. BMC Infect Dis. 2015;15:7. doi: 10.1186/s12879-014-0744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jenvac-Brochure.pdf
- 176.Chanthavanich P, Limkittikul K, Sirivichayakul C, et al. Immunogenicity and safety of inactivated chromatographically purified Vero cell-derived Japanese encephalitis vaccine in Thai children. Hum Vaccin Immunother. 2018;14:900–905. doi: 10.1080/21645515.2017.1414763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Vashishtha VM, Kalra A, Bose A, et al. Indian Academy of Pediatrics (IAP) recommended immunization schedule for children aged 0 through 18 years, India, 2013 and updates on immunization. Indian Pediatr. 2013;50:1095–1108. doi: 10.1007/s13312-013-0292-9. [DOI] [PubMed] [Google Scholar]
- 178.Aggarwal A, Garg N. newer vaccines against mosquito-borne diseases. Indian J Pediatr. 2018;85:117–123. doi: 10.1007/s12098-017-2383-4. [DOI] [PubMed] [Google Scholar]
- 179.Patient Product Information JENVAC®. p 2–3
- 180.Singh A, Mitra M, Sampath G, et al. A Japanese encephalitis vaccine from India induces durable and cross-protective immunity against temporally and spatially wide-ranging global field strains. J Infect Dis. 2015;212:715–725. doi: 10.1093/infdis/jiv023. [DOI] [PubMed] [Google Scholar]
- 181.Yu Y (2013) Development of japanese encephalitis attenuated live vaccine virus SA14-14-2 and its characteristics. 10.5772/52980
- 182.Xin YY, Ming ZG, Peng GY, et al. Safety of a live-attenuated Japanese encephalitis virus vaccine (SA14-14-2) for children. Am J Trop Med Hyg. 1988;39:214–217. doi: 10.4269/ajtmh.1988.39.214. [DOI] [PubMed] [Google Scholar]
- 183.Ni H, Burns NJ, Chang GJ, et al. Comparison of nucleotide and deduced amino acid sequence of the 5′ non-coding region and structural protein genes of the wild-type Japanese encephalitis virus strain SA14 and its attenuated vaccine derivatives. J Gen Virol. 1994;75(Pt 6):1505–1510. doi: 10.1099/0022-1317-75-6-1505. [DOI] [PubMed] [Google Scholar]
- 184.Hennessy S, Liu Z, Tsai TF, et al. Effectiveness of live-attenuated Japanese encephalitis vaccine (SA14-14-2): a case-control study. Lancet (London, England) 1996;347:1583–1586. doi: 10.1016/s0140-6736(96)91075-2. [DOI] [PubMed] [Google Scholar]
- 185.Torresi J, McCarthy K, Feroldi E, Méric C. Immunogenicity, safety and tolerability in adults of a new single-dose, live-attenuated vaccine against Japanese encephalitis: randomised controlled phase 3 trials. Vaccine. 2010;28:7993–8000. doi: 10.1016/j.vaccine.2010.09.035. [DOI] [PubMed] [Google Scholar]
- 186.Chokephaibulkit K, Sirivichayakul C, Thisyakorn U, et al. Safety and immunogenicity of a single administration of live-attenuated Japanese encephalitis vaccine in previously primed 2- to 5-year-olds and naive 12- to 24-month-olds: multicenter randomized controlled trial. Pediatr Infect Dis J. 2010;29:1111–1117. doi: 10.1097/INF.0b013e3181f68e9c. [DOI] [PubMed] [Google Scholar]
- 187.Chokephaibulkit K, Houillon G, Feroldi E, Bouckenooghe A. Safety and immunogenicity of a live attenuated Japanese encephalitis chimeric virus vaccine (IMOJEV®) in children. Expert Rev Vaccines. 2016;15:153–166. doi: 10.1586/14760584.2016.1123097. [DOI] [PubMed] [Google Scholar]
- 188.Nasveld PE, Ebringer A, Elmes N, et al. Long term immunity to live attenuated Japanese encephalitis chimeric virus vaccine: randomized, double-blind, 5-year phase II study in healthy adults. Hum Vaccin. 2010;6:1038–1046. doi: 10.4161/hv.6.12.13057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Feroldi E, Capeding MR, Boaz M, et al. Memory immune response and safety of a booster dose of Japanese encephalitis chimeric virus vaccine (JE-CV) in JE-CV-primed children. Hum Vaccin Immunother. 2013;9:889–897. doi: 10.4161/hv.23087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kosalaraksa P, Watanaveeradej V, Pancharoen C, et al. Long-term immunogenicity of a single dose of japanese encephalitis chimeric virus vaccine in toddlers and booster response 5 years after primary immunization. Pediatr Infect Dis J. 2017;36:e108–e113. doi: 10.1097/INF.0000000000001494. [DOI] [PubMed] [Google Scholar]
- 191.Capeding MR, Alberto ER, Bouckenooghe A, et al. Five-year antibody persistence following a Japanese encephalitis chimeric virus vaccine (JE-CV) booster in JE-CV-primed children in the Philippines. J Infect Dis. 2018;217:567–571. doi: 10.1093/infdis/jix601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Feroldi E, Pancharoen C, Kosalaraksa P, et al. Single-dose, live-attenuated Japanese encephalitis vaccine in children aged 12–18 months: randomized, controlled phase 3 immunogenicity and safety trial. Hum Vaccin Immunother. 2012;8:929–937. doi: 10.4161/hv.20071. [DOI] [PubMed] [Google Scholar]
- 193.Zhang F, Luo J, Teng M, et al. Purification of cell-derived Japanese encephalitis virus by dual-mode chromatography. Biotechnol Appl Biochem. 2021;68:547–553. doi: 10.1002/bab.1960. [DOI] [PubMed] [Google Scholar]
- 194.Zhou D, Wang S, Yang K, et al. Rapid and simultaneous detection of Japanese encephalitis virus by real-time nucleic acid sequence-based amplification. Microb Pathog. 2021;150:104724. doi: 10.1016/j.micpath.2020.104724. [DOI] [PubMed] [Google Scholar]
- 195.Singh DAK, Mehta DA, Kushwaha DKP, et al. Minocycline trial in Japanese encephalitis: a double blind, randomized placebo study. Pediatr Rev Int J Pediatr Res. 2016;3:371–377. doi: 10.17511/ijpr.2016.i05.18. [DOI] [Google Scholar]
- 196.Zhao J, Chen F, Lu L, et al. Japanese encephalitis (JE) mimicking acute ischemic stroke: a case report. Medicine (Baltimore) 2020;99:e23071. doi: 10.1097/MD.0000000000023071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kumar R, Basu A, Sinha S, et al. Role of oral minocycline in acute encephalitis syndrome in India—a randomized controlled trial. BMC Infect Dis. 2016;16:67. doi: 10.1186/s12879-016-1385-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Rayamajhi A, Nightingale S, Bhatta NK, et al. A preliminary randomized double blind placebo-controlled trial of intravenous immunoglobulin for Japanese encephalitis in Nepal. PLoS ONE. 2015;10:e0122608. doi: 10.1371/journal.pone.0122608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hoke CHJ, Vaughn DW, Nisalak A, et al. Effect of high-dose dexamethasone on the outcome of acute encephalitis due to Japanese encephalitis virus. J Infect Dis. 1992;165:631–637. doi: 10.1093/infdis/165.4.631. [DOI] [PubMed] [Google Scholar]
- 200.Solomon T, Dung NM, Wills B, et al. Interferon alfa-2a in Japanese encephalitis: a randomised double-blind placebo-controlled trial. Lancet (London, England) 2003;361:821–826. doi: 10.1016/s0140-6736(03)12709-2. [DOI] [PubMed] [Google Scholar]
- 201.Kumar R, Tripathi P, Baranwal M, et al. Randomized, controlled trial of oral ribavirin for Japanese encephalitis in children in Uttar Pradesh, India. Clin Infect Dis an Off Publ Infect Dis Soc Am. 2009;48:400–406. doi: 10.1086/596309. [DOI] [PubMed] [Google Scholar]
- 202.Ajibowo AO, Ortiz JF, Alli A, et al. Management of Japanese Encephalitis: A Current Update. Cureus. 2021;13:e14579. doi: 10.7759/cureus.14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Guo J, Jia X, Liu Y, et al. Screening of natural extracts for inhibitors against Japanese encephalitis virus infection. Antimicrob Agents Chemother. 2020 doi: 10.1128/AAC.02373-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wang S, Liu Y, Guo J, et al. Screening of FDA-approved drugs for inhibitors of Japanese encephalitis virus infection. J Virol. 2017 doi: 10.1128/JVI.01055-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wei J, Hameed M, Wang X, et al. Antiviral activity of phage display-selected peptides against Japanese encephalitis virus infection in vitro and in vivo. Antiviral Res. 2020;174:104673. doi: 10.1016/j.antiviral.2019.104673. [DOI] [PubMed] [Google Scholar]
- 206.Dwivedi VD, Singh A, El-Kafraway SA, et al. Mechanistic insights into the Japanese encephalitis virus RNA dependent RNA polymerase protein inhibition by bioflavonoids from Azadirachta indica. Sci Rep. 2021;11:18125. doi: 10.1038/s41598-021-96917-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Huang L, Li H, Ye Z, et al. Berbamine inhibits Japanese encephalitis virus (JEV) infection by compromising TPRMLs-mediated endolysosomal trafficking of low-density lipoprotein receptor (LDLR) Emerg Microbes Infect. 2021;10:1257–1271. doi: 10.1080/22221751.2021.1941276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Xiangxi WS-H, Ling L, Qing-Gong Z et al (2017) Near-atomic structure of Japanese encephalitis virus reveals critical determinants of virulence and stability. Nat Commun 8(1):14. 10.1038/s41467-017-00024-6 [DOI] [PMC free article] [PubMed]
- 209.David SS, Mandar B, Radka D et al (2021) Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acid Res 49(W1):W431–W437. 10.1093/nar/gkab314 [DOI] [PMC free article] [PubMed]