Abstract
Naturally-occurring variants of human cytochrome c (Cytc) that induce thrombocytopenia IV occur within Ω-loop C (residues 40 – 57). These variants enhance the peroxidase activity of human Cytc apparently by facilitating access to the heme by destabilizing Ω-loops C and D (residues 70 – 85). Given the importance of peroxidase activity in the early stages of apoptosis, we identified three sites with the EVmutation algorithm in or near Ω-loop C that coevolve and differ between yeast iso-1-Cytc and human Cytc. We prepared iso-1-Cytc variants with all possible combinations of the S40T, V57I and N63T substitutions to determine if these residues decrease the peroxidase activity of iso-1-Cytc to that of human Cytc producing an effective off state for a peroxidase signaling switch. At pH 6 and above, all variants significantly decreased peroxidase activity. However, the correlation of peroxidase activity with local and global stability, expected if cooperative unfolding of Ω-loops C and D is required for peroxidase activity, was generally poor. The m-values derived from the guanidine hydrochloride dependence of the kinetics of imidazole binding to horse Cytc, which is well-characterized by native-state hydrogen exchange methods, and K72A/K73A/K79A iso-1-Cytc show that local structural fluctuations and not subglobal cooperative unfolding of Ω-loops C and D are sufficient to permit binding of a small molecule like peroxide to the heme. A 2.46 Å structure of N63T iso-1-Cytc identifies a change to a hydrogen bond network linking Ω-loops C and D that could modulate the local fluctuations needed for the intrinsic peroxidase activity of Cytc.
Keywords: Cytochrome c, Protein dynamics, Peroxidase activity, Alkaline conformational transition, Apoptosis, Imidazole heme binding
1. Introduction
Cytochrome c (Cytc) is usually found near the mitochondrial membrane and is part of the Electron Transport Chain [1]. The heme of Cytc allows it to do redox chemistry, transporting electrons between Complex III and Complex IV. It also leaves the mitochondria as part of the intrinsic pathway of apoptosis [2,3]. Via its alternate function as a peroxidase, Cytc catalyzes the oxidation of the inner mitochondrial membrane lipid cardiolipin (CL), which causes CL to migrate to the outer mitochondrial membrane facilitating the release of Cytc from mitochondria [4]. Upon entering the cytosol, Cytc induces formation of a protein complex called the apoptosome initiating a cascade of caspases that ultimately leads to cell death [3,5].
Various mutations in Cytc are linked to thrombocytopenia type IV (THC4), which is a disease that is mainly characterized by low levels of blood platelets. Three specific mutations linked to THC4 have been discovered: G41S [6], Y48H [7], and A51V [8–10]. These mutations have been shown to promote peroxidase activity and to increase dynamics in omega loops C and D (residues 40–57 and 70–85, respectively) in the human protein [10–13]. However, recent molecular dynamics (MD) simulations on the THC4 variants indicate no correlation between the flexibility of Ω-loop C and peroxidase activity and in fact a negative correlation with the flexibility of Ω-loop D [14]. Peroxidase activity is also higher in yeast iso-1-Cytc when compared to human Cytc and other higher eukaryotes like horse [15–17]. The higher peroxidase activity of yeast iso-1-Cytc may function to convert H2O2 to H2O, leading to protection from H2O2-induced apoptosis in yeast [18].
The hypothesis driving the current work is that the increased dynamics in the omega loops of Cytc leads to a more accessible heme and therefore higher peroxidase activity seen in both variant forms of human Cytc and wild type yeast iso-1-Cytc. For a substrate like peroxide to access the heme iron, Met80 must be removed from its coordination site proximal to the heme iron to create a pentacoordinate species [19]. Previous work has shown that the oxidation of Met80 to a sulfoxide both enhances the peroxidase activity of Cytc [19–22] and allows hydrogen peroxide to access the heme more readily [20,21]. Cytc variants like those linked to THC4 in Ω-loop C more readily react with H2O2 to produce the Met80 sulfoxide and a direct correlation between the presence of Met80 sulfoxide and peroxidase activity is observed [22]. The role of Ω-loops C and D in mediating the dynamics needed to induce peroxidase activity is not surprising because they have been shown to be the two least stable substructures of horse Cytc [23,24].
Certain residues in the human Cytc omega loops appear to have coevolved (Fig. 1) [25]. For Ω-loop C, pairwise coevolution is most significant for residues Thr40 and Ile57 of human Cytc. Being at opposite ends of Ω-loop C, the side chains of these residues are in direct contact (Fig. 1). In yeast iso-1-Cytc, these positions are occupied by residues with one less methyl group (Ser40 and Val57). Equally significant pairwise coevolution occurs between Ile57 and Lys55. Other significant pairwise coevolution is observed between Ile57 and Thr63, Thr63 and Tyr74, and Thr40 and Lys55. As a group, Thr40, Lys55, Ile57, Thr63, and Tyr74 are proximal in the structure of human Cytc at the interface between Ω-loops C and D (Fig. 1). Lys55 and Tyr74 do not change between yeast iso-1-Cytc and human Cytc, although Asn63 in yeast iso-1-Cytc evolves to Thr63 in human Cytc.
Fig. 1.
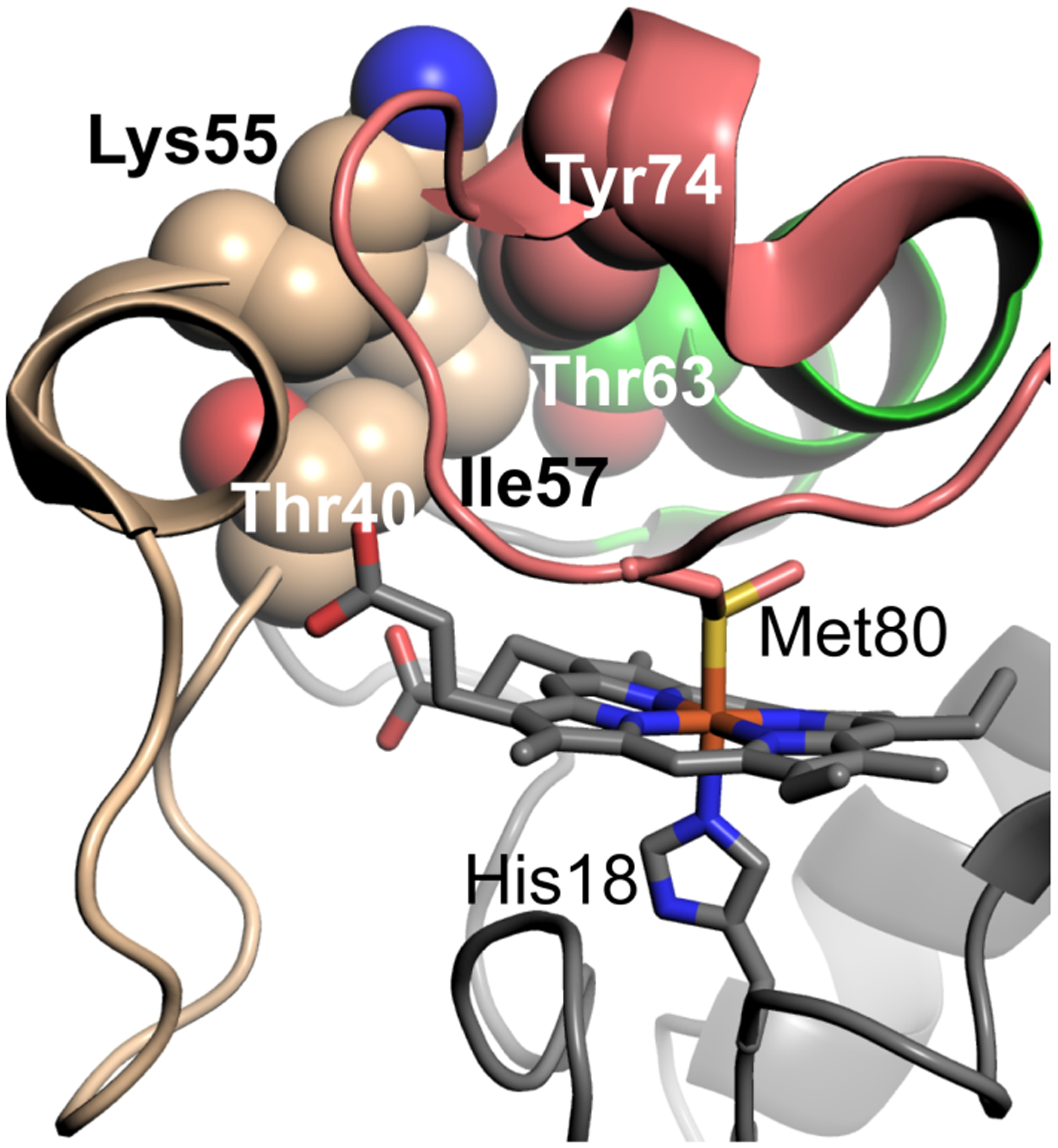
Visualization of human Cytc (PDB ID: 3ZCF) [26] residues (space-filling models) in or near Ω-loop C (shown in wheat) that have coevolved. Ω-loop D is shown in salmon and the 60s helix in green. Shown as stick models are the heme and its axial ligands Met 80 and His 18.
As mentioned previously these residues are in a part of the protein important for regulating peroxidase activity [9–13,27]. We hypothesized that as eukaryotes evolved, tighter control of the intrinsic peroxidase activity Cytc was required, so that it could act as a signaling switch to initiate apoptosis. Therefore, the residues that have changed between the yeast and human proteins may be responsible for the large differences in intrinsic peroxidase activity seen between Cytc from these two species [27]. More specifically, coevolution at the interface of Ω-loops C and D has led to residues that are bulkier and more hydrophobic in human Cytc than in yeast iso-1-Cytc, which could limit the dynamics of these omega loops and therefore hinder heme access.
In the current work, we have mutated the coevolved Ω-loop C/D interface residues of yeast iso-1-Cytc to the amino acids observed in the human protein. These substitutions are S40T, V57I, and N63T, as well as all double substitution combinations and the triple substitution variant. Our goal was to better understand how these mutations affect the peroxidase activity of Cytc and the stability of the local substructures involved, and therefore how the intrinsic peroxidase activity of Cytc decreased as eukaryotes evolved.
2. Experimental section
2.1. Site-directed mutagenesis
Mutagenesis was done using the QuikChange Lightning mutagenesis kit (Agilent Technologies). All variants of yeast iso-1-Cytc contain a background mutation of C102S (WT) [28], which prevents intermolecular disulfide dimerization. To better mimic the human protein, we have chosen not to include a K72A mutation (variant usually referred to as WT*), which is often included because Lys72 is trimethylated in its native host, Saccharomyces cerevisiae, and is not when iso-1-Cytc is expressed from Escherichia coli [29]. For imidazole binding experiments, a K72A/K73A/K79A variant was prepared using DNA containing the WT* variant [30]. The template DNA for mutagenesis to produce all iso-1-Cytc variants was the pRbs_BTR1 vector [31], a derivative of pBTR1 [29,32] carrying an optimized ribosomal binding site [33]. Sequencing to confirm mutations was done either at the University of Montana Genomics Core or by Eurofins Genomics (Louisville, KY).
2.2. Protein expression and purification
Each pRbs_BTR1 vector carrying the desired mutated iso-1-Cytc gene was used to transform competent phage T1 resistant E. coli BL21(DE3) cells (New England Biolabs). Transformed cells were inoculated into 2xYT media and grown for 30–36 hours at 37 °C with shaking at 150 rpm. Cells were pelleted and then suspended in lysis buffer consisting of 10 mM Tris, pH 7.5, 500 mM NaCl, 2 mM PMSF, and a small amount of DNAse. Cells were then lysed via sonication using a Qsonica Q700 sonicator. The lysate was centrifuged and then incubated overnight at 4 °C in 50% ammonium sulfate to precipitate out most protein impurities, which were removed via centrifugation. The supernatant was subjected to two rounds of dialysis for 8 to 24 hours each in buffer consisting of 12.5 mM sodium phosphate, pH 7.2, 1 mM disodium EDTA, and 2 mM β-mercaptoethanol (βME). The dialyzed solution was then loaded onto a CM Sepharose column equilibrated to 50 mM sodium phosphate, pH 7.2, 1 mM EDTA, and 2 mM βME. The protein was eluted using a linear gradient of the same buffer with 0.8 M NaCl added. The eluate was then exchanged into a low salt solution consisting of 50 mM sodium phosphate, pH 7.0, before flash freezing and storing in −80 °C. At most 24 hours before experiments, the protein was thawed and further purified using a HiTrap HP SP 5 mL cation exchange column attached to an AKTAprime plus chromatography system (GE Healthcare Life Sciences) and was eluted using a 50 mL 0 – 50 %B gradient with 50 mM sodium phosphate buffer, pH 7.0, as buffer A and 50 mM sodium phosphate buffer, pH 7.0, with 1 M NaCl added as buffer B. The eluate was concentrated by centrifuge ultrafiltration (Amicon Ultra-15, 10k MWCO, Millipore) and the high salt removed by several rounds of exchange into MilliQ water.
2.3. Alkaline conformational transition
To evaluate the alkaline transition, each iso-1-Cytc variant was oxidized with potassium ferricyanide at room temperature for 20–30 minutes. The protein was then separated from the potassium ferricyanide using a G25 size exclusion column (GE Healthcare) with 200 mM NaCl as the running buffer. The solution at the start of the experiment had a final protein concentration of 100 μM in 100 mM NaCl. As HCl or NaOH was added to change the pH of the solution, an equal amount of a 2-fold concentrated stock solution (200 μM protein in 200 mM NaCl) was added to maintain the concentrations of protein and NaCl in the titration solution. Absorbance measurements were collected using a Beckman Coulter DU 800 UV-Vis instrument. Wavelength scans were from 750 nm to 500 nm in a quartz suprasil cuvette. A Denver Instrument DU-10 pH meter with a Fisher Scientific Accumet Double Junction Semi-Micro pH probe was used for pH measurements. All measurements were taken at 21 ± 2 °C. Absorbance at 695 nm, A695, was used to monitor the conformational transition. Subtraction of 750 nm absorbance values, A750, was used as a baseline correction to account for instrument drift during the period of the titration. Corrected 695 nm absorbance values, A695corr (A695 – A750), were then plotted as a function of pH, and a modified form of the Henderson-Hasselbalch equation was fit to the data that allows the number of protons associated with the alkaline transition (n) to be determined as well as pKa values for the alkaline transition [27].
2.4. Guanidine hydrochloride denaturation
Iso-1-Cytc variants were oxidized as described for the alkaline transition, except that the buffer for G25 chromatography was 20 mM Tris, 40 mM NaCl and 1 mM EDTA at pH 7.5. Denatured (D) and Native (N) stocks were prepared with final concentrations of 4 μM protein in 20 mM Tris, 40 mM NaCl, and 1 mM EDTA at pH 7.5. The D stock also contained 6 M guanidine hydrochloride (GdnHCl). For circular dichroism (CD) measurements, 2 mL N stock was placed in a 1 cm square quartz cuvette with a stir bar and tubing for withdrawing N stock and adding D stock, and with masking to ensure that light does not scatter off the stir bar or tubing. The titration was carried out by increasing GdnHCl concentration in 0.1 M increments with a Hamilton titrator interfaced to the CD spectrometer. Ellipticity measurements were taken using an Applied Photophysics Chirascan spectrometer at 25 °C. Data were collected at wavelengths of 250 nm and 222 nm, with 250 nm ellipticity values subtracted from the 222 nm values as a baseline correction, θ222corr. A two-state model assuming a linear relationship between the free energy of unfolding, ΔGu, and GdnHCl concentration (eq 1) was fit to plots of θ222corr vs GdnHCl concentration using nonlinear least squares methods.
| (1) |
In eq 1, θN is the native state baseline, θD and mD are the intercept and slope of the denatured state baseline, m is the rate of change of ΔGu with respect to GdnHCl concentration and ΔGuo′(H2O) is the free energy of unfolding in the absence of GdnHCl.
2.5. Peroxidase activity measurements
Iso-1-Cytc variants were oxidized as described for the alkaline transition. Peroxidase activity was measured by the oxidation of guaiacol to tetraguaiacol [34], which absorbs strongly at 470 nm. Different 50 mM buffers were used for different pH values: sodium acetate for pH 5, 2-(N-morpholino)ethanesulfonic acid (MES) for pH 6, sodium phosphate for pH 7, and tris(hydroxymethyl)aminomethane (Tris) for pH 8. Each buffer was adjusted to the desired pH using HCl or NaOH. The solutions were degassed under argon for 30–45 minutes to limit non-enzymatic oxidation of guaiacol by O2. The degassed solutions were then used to make three separate solutions of 4 μM protein, 400 μM guaiacol, and 100 ± 5 mM hydrogen peroxide such that H2O2 is above the Michaelis constant, KM, with respect to H2O2 for peroxidase activity (~37 mM for human Cytc) [35,36]. The concentrations of guaiacol and hydrogen peroxide were measured by absorbance using ε274 = 2150 M−1 cm−1 [37], and ε240 = 41.5 M−1 cm−1 [38,39], respectively. The protein solution, the degassed buffer, and differing amounts of the guaiacol solution were mixed into a single solution immediately prior to measurements. This solution and the hydrogen peroxide solution were mixed 1:1 with an Applied Photophysics SX20 stopped-flow spectrometer and time-dependent changes in absorbance at 470 nm (A470) were recorded. Final protein concentration was 1 μM, final hydrogen peroxide concentration was 50 mM in 50 mM buffer. Data were collected at 25 ± 0.1 °C. For each trial, A470 was plotted as a function of time and the linear region of the resulting graph with the highest slope was used to determine the initial enzyme velocity, ν. Five runs were done for each guaiacol concentration, and the slopes were averaged then multiplied by 4 (oxidation of guaiacol requires removal of 4 electrons [34,40], divided by the protein concentration and the reported extinction coefficient [34,40,41] of 26.6 mM−1 cm−1 for tetraguaiacol (3,3’-dimethoxy-4,4’-biphenoquinone) [42] to obtain ν/[Cytc] values at each guaiacol concentration. These ν/[Cytc] values were plotted as a function of guaiacol concentration and the Michaelis-Menten equation was fit to the data to obtain the Michaelis constant, KM, and the catalytic rate constant, kcat.
2.6. Equilibrium and kinetics of imidazole binding
Imidazole binding experiments were carried out either by equilibrium titration monitored by UV-Vis spectroscopy (Beckman DU 800 UV-Vis Spectrometer) or by stopped-flow mixing methods (Applied Photophysics SX20 Stopped-flow Spectrometer). Horse heart Cytc (Sigma used without further purification) and K72A (WT*) and K72A/K73A/K79A variants of yeast iso-1-Cytc were used for these experiments. All experiments were done in 20 mM 3-(N-morpholino)propanesulfonic acid (MOPS), pH 7 at constant ionic strength. NaCl was used to maintain constant ionic strength as the concentration of imidazole was varied.
For equilibrium experiments, individual samples were prepared for each data point by mixing together two solutions at the same ionic strength, both containing 10 μM Cytc, one containing imidazole and one not containing imidazole. The solutions were mixed in proportions to produce a set of solutions covering the desired range of imidazole concentrations. Samples were equilibrated at room temperature (21 ± 2 °C) for at least two hours before measuring UV-Vis spectra.
Stopped-flow experiments were carried out by 1:1 mixing of Cytc with imidazole. Both solutions contained 20 mM MOPS, pH 7. The concentration of the imidazole solution was varied. The ionic strengths of both the Cytc and imidazole solutions were adjusted to the same value with NaCl. The final concentration of Cytc after mixing was 10 μM. Stopped-flow experiments in the presence of GdnHCl were carried out at an ionic strength of 1.25 M with GdnHCl replacing NaCl to keep ionic strength constant in the protein and imidazole containing solutions. Binding of imidazole was followed at 405 nm, A405, as a function of time. Exponential functions were fit to plots of A405 versus time.
2.7. Crystallization and structure determination of N63T iso-1-Cytc
The N63T variant of iso-1-Cytc was purified as described above, oxidized with 5 mg of K3[Fe(CN)6] per mg of protein, separated from K3[Fe(CN)6] using a sephadex G25 column equilibrated to and run with 50 mM sodium phosphate pH 6.0, and then concentrated to 37.2 mg/mL using an Amicon Ultra-15 (10k MWCO) centrifuge ultrafiltration device. Qiagen’s PEGs I Suite, PEGs II Suite, JCSG Core III Suite, and JCSG Core IV Suite were used to screen for crystallization conditions. Needle-like crystals were obtained from 1:1 sitting drops incubated at 20 °C for several screening conditions including: PEGs II-16 (0.1 M Tris pH 8.5, 30% w/v polyethylene glycol (PEG) 1000), JCSG III-33 (0.1 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.5, 30% v/v PEG 400, 5% v/v PEG 3000, 10% glycerol), JCSG III-61 (0.1 M MES pH 6.0, 40% v/v PEG 400, 5% PEG 3000) and JCSG IV-40 (0.1 M HEPES pH 7.5, 50% v/v PEG 200). Crystals were harvested directly from the crystal trial plates and cryoprotected with 20% glycerol. Crystals that diffracted to 2.46 Å resolution were obtained from the PEGs II-16 screening condition. X-ray diffraction data were collected at the Stanford Synchrotron Radiation Lightsource beamline 12–2 with a DECTRIS PILATUS 6M detector. The data were indexed, integrated and scaled in the P41 space group using iMosflm [43] and Aimless [44]. The structure was solved by molecular replacement using Molrep [45], integrated into the CCP4i2 software suite [46]. Model building was accomplished in REFMAC [47] and PHENIX [48] and the structure was refined through iterative cycles of manual adjustment in Coot [49] and refinement of atomic positions, real space, occupancy, and thermal parameters in PHENIX, yielding Rwork = 0.165 and Rfree = 0.217. Data collection and refinement statistics are provided in Table S1. Coordinates have been deposited at the Protein Data Bank (www.rcsb.org) under PDB code: 7MRI. All figures were produced with PyMol [50].
3. Results
3.1. Stability of iso-1-Cytc variants
Fig. 2 shows typical GdnHCl denaturation data for several iso-1-Cytc variants with humanlike mutations. It is evident from the data in Fig. 2 that the effects of humanlike substitutions at positions 40, 57 and 63 are modest. In these plots, the open data points were not used in data fitting because of the initial downward curvature of ellipicity at 222 nm at low GdnHCl concentrations. This effect is a result of the high charge density of iso-1-Cytc, which causes initial additions of GdnHCl to stabilize helical structure [51,52]. To further mitigate the effects of this behavior, a native state baseline that is invariant with GdnHCl concentration was used.
Fig. 2.
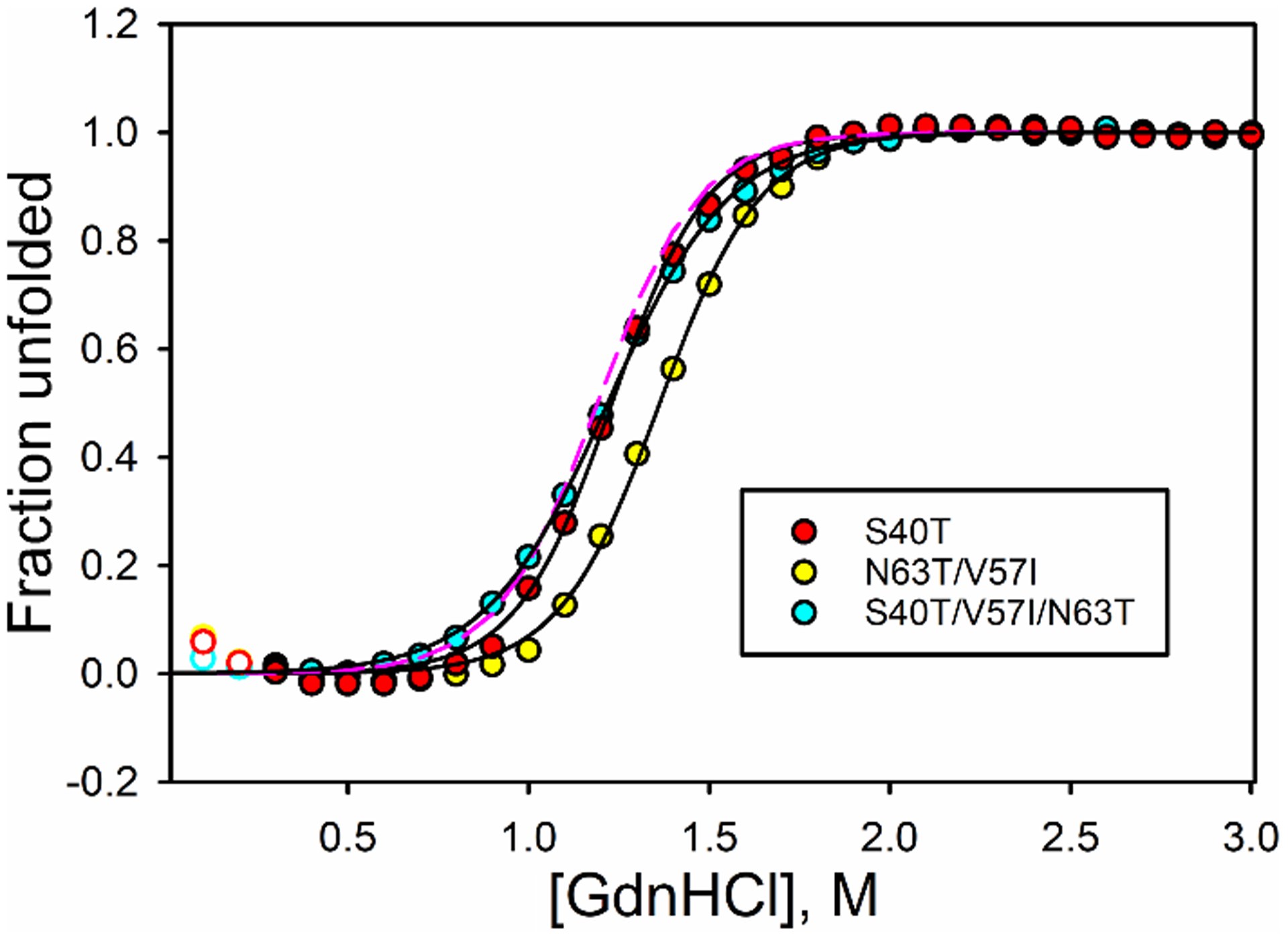
Plots of fraction of protein unfolded for selected iso-1-Cytc variants as a function of GdnHCl concentration. Fraction unfolded was calculated from fits of eq 1 to θ222corr vs GdnHCl concentration data. Open circles are data points that were not used in the fit. Solid curves are fits of a two-state model (eq 1) to the data. Data are shown for the S40T (red circles), N63T/V57I (yellow circles) and S40T/V57I/N63T (cyan circles) variants. The dashed magenta line is fraction unfolded for WT iso-1-Cytc using the parameters in Table 1.
As can be seen from the parameters obtained from fitting eq 1 to the data in this manner (Table 1), the differences in global stability are small. Only the V57I, S40T/V57I, andS40T/V57I/N63T have significant changes in ΔGuo′(H2O) relative to WT iso-1-Cytc. The m values for the variants are significantly smaller than that of WT iso-1-Cytc, decreasing to values similar to human Cytc (3.5 – 3.7 kcal/mol×M) [15]. The GdnHCl concentration at the midpoint of unfolding, Cm, increases for some of the single and double mutant variants, but is the same for WT iso-1-Cytc and the final S40T/V57I/N63T variant.
Table 1.
Thermodynamic parameters for GdnHCl unfolding of iso-1-Cytc variants at 25 °C.
| Variant | ΔGuo′(H2O) (kcal/mol) | m (kcal/mol×M) | Cm (M) |
|---|---|---|---|
| WTa | 5.05 ± 0.30 | 4.24 ± 0.13 | 1.19 ± 0.04 |
| V57I | 4.22 ± 0.13 | 3.54 ± 0.12 | 1.193 ± 0.007 |
| S40T | 4.99 ± 0.09 | 3.94 ± 0.16 | 1.27 ± 0.03 |
| N63T | 5.38 ± 0.12 | 4.01 ± 0.10 | 1.343 ± 0.005 |
| N63T/V57I | 5.40 ± 0.16 | 3.87 ± 0.12 | 1.40 ± 0.03 |
| S40T/V57I | 4.23 ± 0.07 | 3.66 ± 0.22 | 1.16 ± 0.06 |
| S40T/N63T | 4.50 ± 0.12 | 3.85 ± 0.07 | 1.17 ± 0.01 |
| S40T/V57I/N63T | 4.197 ± 0.004 | 3.49 ± 0.05 | 1.20 ± 0.02 |
Parameters from Duncan et al. 2009 [31]
The GdnHCl denaturation results overall indicate that the global stability of the variants is not affected strongly with the exception of the V57I, S40T/V57I, and S40T/V57I/N63T variants. All of these variants contain the V57I mutation, which appears to be important for decreasing the m-value (compare S40T data to S40T/V57I/N63T data in Fig. 2). In general, all variants unfold less cooperatively than WT iso-1-Cytc, indicating that the differences between the human and yeast proteins may have more to do with changes in substructure dynamics and stability than with global stability.
3.2. Alkaline conformational transition of iso-1-Cytc variants
The alkaline transition of Cytc happens under mildly basic conditions when one of the lysine residues in Ω-loop D replaces the methionine that interacts with the iron of the heme. The pKa of the alkaline transition can provide information about the local stability of Ω-loop D (residues 70 – 85) [53,54]. Fig. 3 shows typical data for the alkaline conformational transition of the iso-1-Cytc variants with humanlike substitutions at residues 40, 57 and 63, derived from spectra measured from 500 to 750 nm (Fig. S1). As with the global unfolding of these variants, the effects on the local stability of Ω-loop D as measured by the alkaline transition appear to be small. Fits of the Henderson-Hasselbalch equation to alkaline transition data yield pKa values similar to the WT protein (Table 2). The N63T/V57I variant shows a significant decrease in the pKa of the alkaline transition relative to WT iso-1-Cytc, whereas the V57I and S40T/N63T variants show a small increase and a small decrease, respectively, in the pKa of the alkaline transition. The n values (Table 2) in all cases are approximately 1, as is expected based on the net deprotonation of a single lysine. Overall, the alkaline transition data indicate that the stability of the Ω-loop that contains Met80 and the nearby lysine residues is similar for WT iso-1-Cytc and all variants.
Fig. 3.
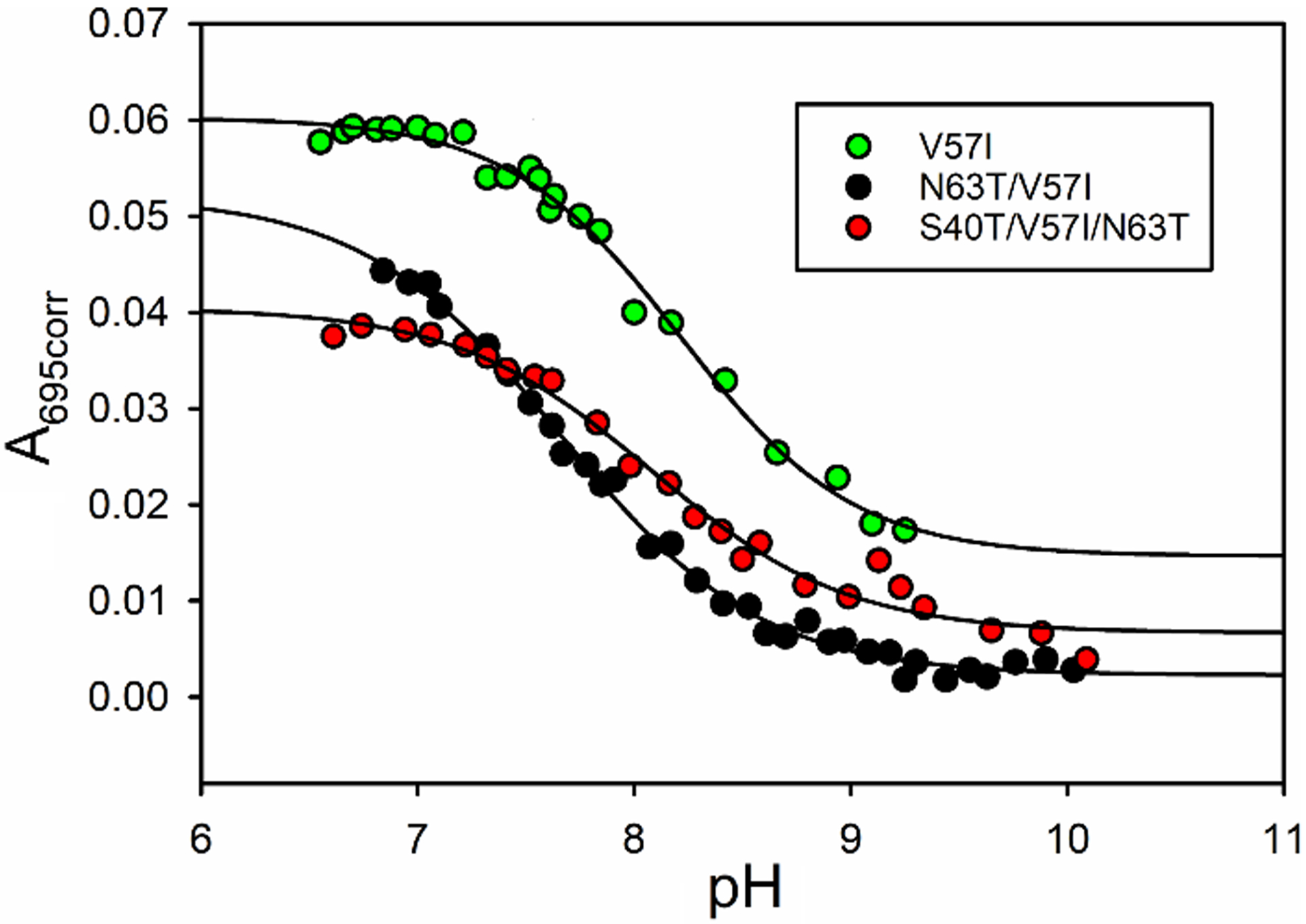
Plot of corrected absorbance at 695nm, A695corr, versus pH for iso-1-Cytc variants. Black lines are fits of a modified form of the Henderson-Hasselbalch equation to the data. Parameters from these fits are provided in Table 2. Data for the V57I (green circles), N63T/V57I (black circles), and S40T/V57I/N63T (red circles) are shown.
Table 2.
Alkaline transition parameters of iso-1-Cytc variants at 25 °C.
| Variant | pKa | n |
|---|---|---|
| WTa | 8.00 ± 0.05 | 0.98 ± 0.01 |
| V57I | 8.17 ± 0.06 | 1.13 ± 0.12 |
| S40T | 7.99 ± 0.06 | 0.98 ± 0.17 |
| N63T | 8.01 ± 0.05 | 0.95 ± 0.06 |
| N63T/V57I | 7.64 ± 0.04 | 0.96 ± 0.05 |
| S40T/V57I | 7.94 ± 0.16 | 1.05 ± 0.13 |
| S40T/N63T | 7.84 ± 0.11 | 0.98 ± 0.12 |
| S40T/V57I/N63T | 7.96 ± 0.10 | 0.89 ± 0.08 |
Parameters from Duncan et al. 2009 [31]
3.3. Peroxidase activity of iso-1-Cytc variants
The peroxidase activities of the variants of iso-1-Cytc were measured from pH 5 to 8 using guaiacol as substrate [15,16,27,34,55,56]. Michaelis-Menten plots for peroxidase activity at pH 8 are shown in Fig. 4 for theV57I, N63T/V57I, and S40T/V57I/N63T variants. More significant changes in kcat are evident in Fig. 4 than might be expected based on the changes in the global and local stability described above. The KM and kcat values from fits of the Michaelis-Menten equation to peroxidase activity data are provided in Table S2 and plotted in Fig. 5 as a function of pH for WT and variant forms of iso-1-Cytc and for human Cytc. The KM values do not differ much among the WT and variant forms of yeast iso-1-Cytc. The KM values for all variants and WT are higher at more acidic pH values, decrease as the pH becomes more alkaline until pH 8, where most variants show an increase relative to pH 7. With regard to kcat, the WT iso-1-Cytc value is in the middle of the range observed for the variants at pH 5, but at higher pH, the variants show significantly lower kcat values relative to WT iso-1-Cytc. However, the decrease in kcat for the variants does not approach that observed for human Cytc. Interestingly at pH 8, peroxidase activity as measured by kcat becomes much lower for all the variants. Peroxidase measurements at pH 9 were attempted, but the peroxidase activity for most variants was so low that it became difficult to distinguish signal from noise in the data. The pH 9 results are likely due to the increasing presence of the alkaline form of iso-1-Cytc, which cannot perform peroxidase reactions due to binding of the strong field ligand lysine to the heme [34].
Fig. 4.
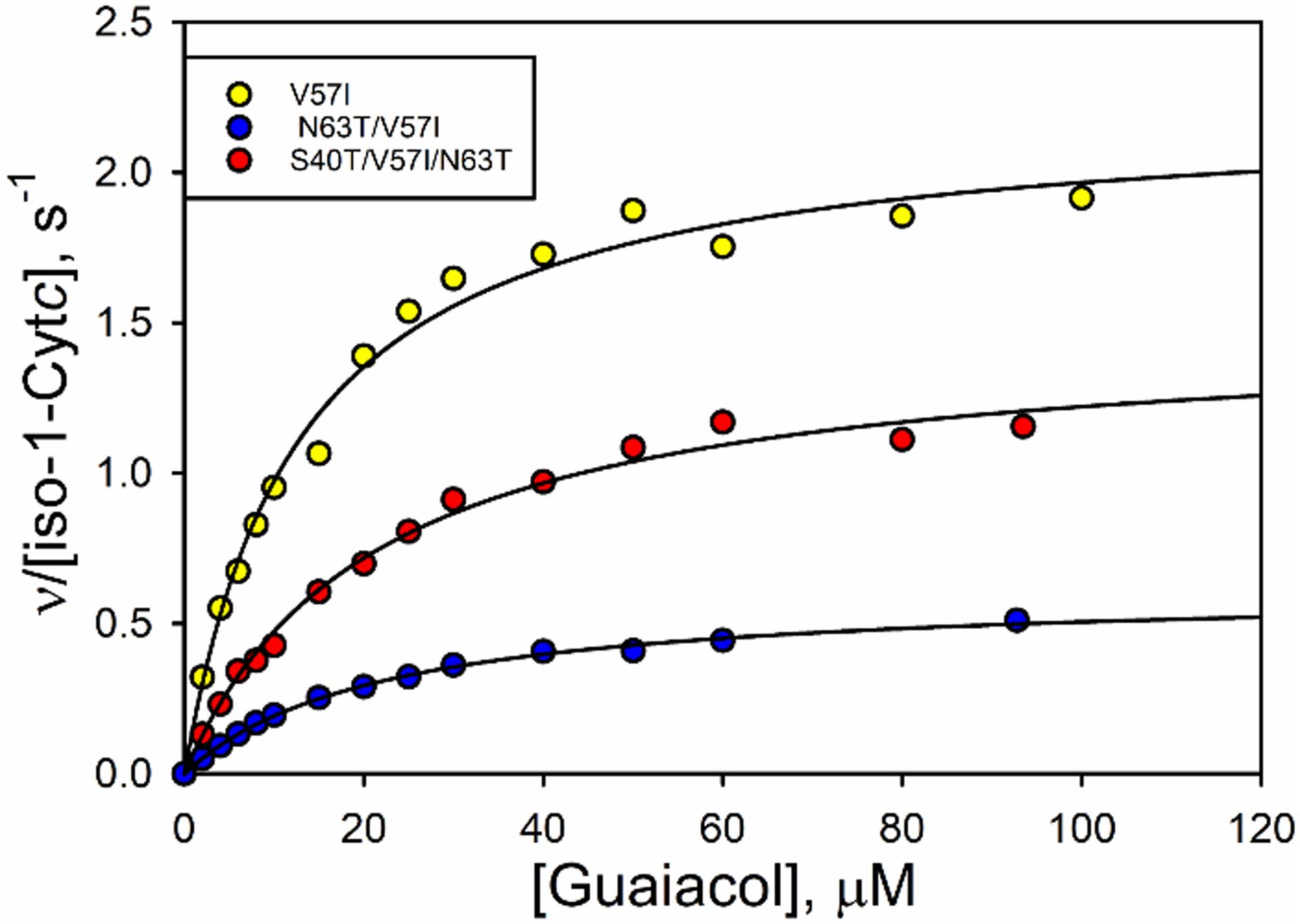
Michaelis-Menten plots at pH 8 for variants V57I (yellow), N63T/V57I (blue) and S40T/V57I/N63T (red). Black lines are fits of the Michaelis-Menten equation to the data.
Fig. 5.
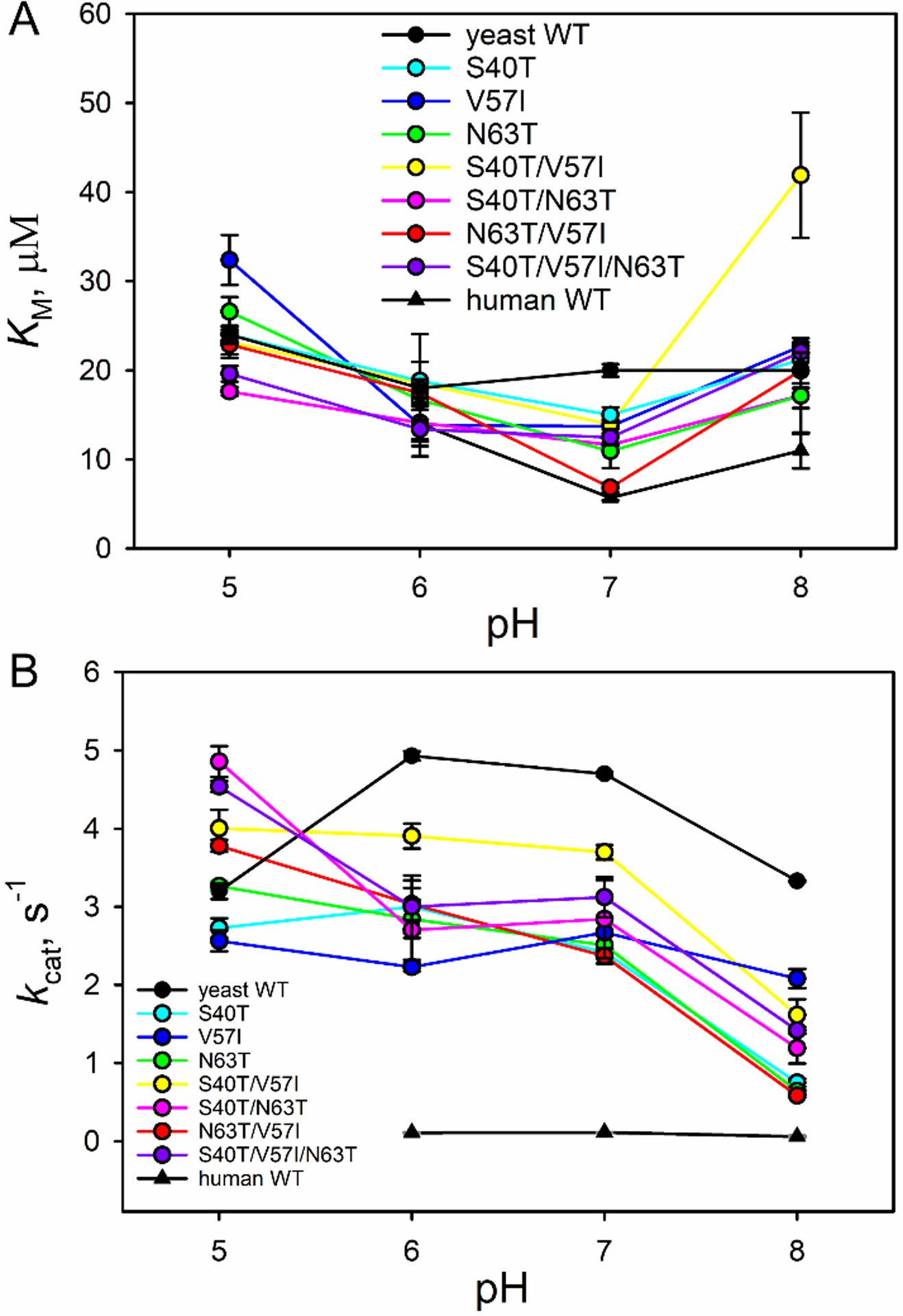
Plots of (A) KM and (B) kcat versus pH for all variants. For comparison, KM and kcat values for WT yeast iso-1-Cytc (black circles) and WT human Cytc (black triangles) are provided.
3.4. Imidazole binding as a function of GdnHCl concentration
In hydrogen-deuterium exchange (HX) experiments, it is known that exchange can occur via subglobal unfolding or by lower energy local structural fluctuations [57]. Recent work on equine Cytc has shown that some protein dynamics may not be detectable by the HX methods used to study the dynamics of Ω-loops C and D [58]. The observation that the effects of the Ω-loop C mutations on peroxidase activity were more pronounced than their effects on local and global stability led us to test the extent of structural rearrangement necessary for a small ligand like peroxide to bind to the heme. In analogy to HX experiments on horse heart Cytc [23,24], the extent of unfolding of Ω-loop D can be assessed by measuring the GdnHCl-dependence of the free energy of binding (m-value) for the small exogenous ligand imidazole (Im). For these studies, we used horse heart Cytc to allow direct comparison with the m-value for opening of the Ω-loop D substructure as measured by HX methods [23,24]. We also used two variants of the yeast protein, K72A (WT*) and K72A/K73A/K79A. The latter variant was used to eliminate interference from alkaline conformers on the dynamics of Ω-loop D that mediate Im binding. The equilibria and kinetics of binding of Im as a function of ionic strength, I, were measured first to find optimal conditions for measurements as a function of GdnHCl concentration.
Equilibrium binding of Im to the WT* and the K72A/K73A/K79A iso-1-Cytc variants was measured by absorbance spectroscopy (Fig. S2). Data were analyzed as a function of total imidazole concentration to allow direct comparison with values reported, previously [59,60]. Because it is deprotonated Im that binds to the heme of Cytc, and Im is only about 50% ionized at pH 7, all reported equilibrium constants are apparent equilibrium constants for Im binding. Measurements at ionic strengths of 0.05 to 1.25 M show that the apparent equilibrium constant for Im binding, KIm,equil, increases slowly up to I = 1 M for WT* and then levels off (Fig. S3). The magnitude of KIm,equil for the K72A/K73A/K79A variant initially decreased as ionic strength increased, but then increased again leveling off at I = 1 M (Fig. S3). At I = 1.0 M, KIm,equil is 313 ± 35 M−1 for WT* iso-1-Cytc and 503 ± 56 M−1 for K72A/K73A/K79A iso-1-Cytc. These values are 20-fold and 30-fold larger, respectively, than the previously reported value of 15.3 ± 0.5 M−1 for the binding constant of Im for horse heart Cytc at I = 1.0 M [59].
A kinetic scheme involving an initial rearrangement of Cytc to a conformer competent for Im binding, Cytc†, has been found to be consistent with the dependence of the observed rate constant, kobs, on Im concentration [59,60]. In this mechanism (Scheme 1), kobs levels off at high Im concentration because the observed rate constant, kobs, for Im binding (eq 2) becomes limited
| (2) |
by the rate constant, k1, for formation of Cytc†. We investigated the kinetics of Im binding to WT* and K72A/K73A/K79A iso-1-Cytc and horse heart Cytc at 1.25 M ionic strength. Fits of the kinetic data are consistent with a major fast phase due to Im binding and a slow phase that is on the time scale expected for proline isomerization (Fig. S4 and Table S3) [61]. Proline isomerization phases are also observed near neutral pH for the alkaline transition of iso-1-Cytc mediated by His73 in Ω-loop D for a K73H variant of iso-1-Cytc [62]. Therefore, we believe the slow phase is due to proline isomerization within the protein following imidazole binding. Plots of total amplitude at 405 nm versus total Im concentration show that the affinity of Im for yeast iso-1-Cytc is considerably higher than for horse heart Cytc (Fig. S5). The data for horse heart Cytc yield apparent binding constants similar to those reported by Sutin and Yandell at 1.0 M ionic strength (Table S4) [59]. Apparent binding constants from amplitude data for the yeast iso-1-Cytc variants are consistent with values from our equilibrium titrations (Table S4).
Scheme 1.
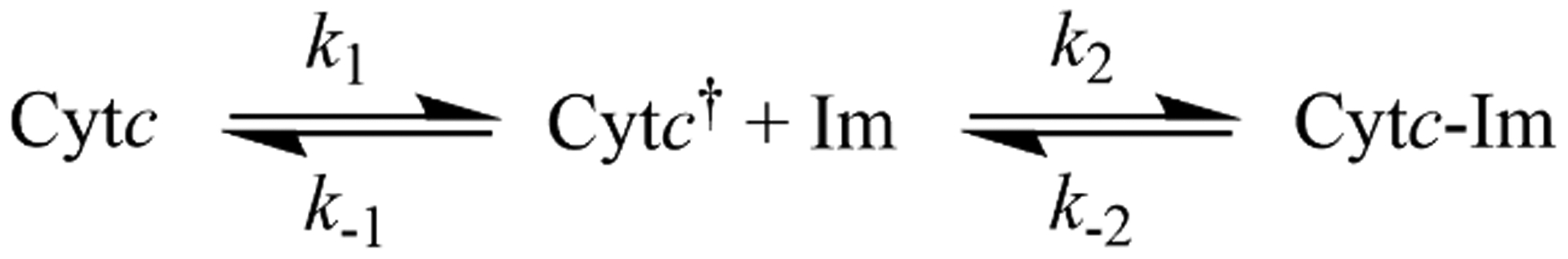
Kinetics mechanism of Im binding to Cytc.
Plots of kobs versus Im concentration for K72A/K73A/K79A iso-1-Cytc and horse heart Cytc at 1.25 M ionic strength fit well to eq 2 (Fig. S6, Fig. S7 and Table S5). However, the data for WT* iso-1-Cytc do not deviate detectably from linearity. Therefore, we used horse heart Cytc and K72A/K73A/K79A iso-1-Cytc to measure the GdnHCl dependence of the binding of Im to Cytc. Interestingly, in line with the ionic strength dependence of KIm,equil observed for the yeast iso-1-Cytc variants, comparison of our kinetic data with previous data for horse heart Cytc suggest that the kinetics of Im binding to Cytc depend on ionic strength (Fig. S6). Therefore, measurements of the kinetics of Im binding as a function of GdnHCl concentration were carried out at a constant ionic strength of 1.25 M.
Fig. 6 shows plots of kobs versus total Im concentration at 0, 0.25 and 0.50 M GdnHCl for K72A/K73A/K79A iso-1-Cytc and horse heart Cytc. It is evident that kobs at each Im concentration increases as GdnHCl concentration increases and that overall the kobs values are larger for the yeast protein compared to the horse protein. From plots of amplitude data as a function of total Im concentration, the affinity of Im for horse Cytc can be seen to increase as GdnHCl concentration increases (Fig. S8). Fits of the data to a single site binding model confirm this observation for both horse heart Cytc and K72A/K73A/K79A iso-1-Cytc, although the error bars are large for the yeast variant (Table 3). Plots of the apparent free energy of Im binding, ΔG°′Im (Table 3), as a function of GdnHCl concentration are shown in Fig. 7A. The plots are linear yielding m-values in the range of 0.4 – 0.7 kcal mol−1 M−1 for both proteins (Table 4) indicating that Im binding causes only a small structural rearrangement. By comparison, unfolding of the Ω-loop D substructure as detected by hydrogen exchange [24,63] or the alkaline transition [53,64] yield GdnHCl m-values of 1.5 – 1.7 kcal mol−1 M−1.
Fig. 6.
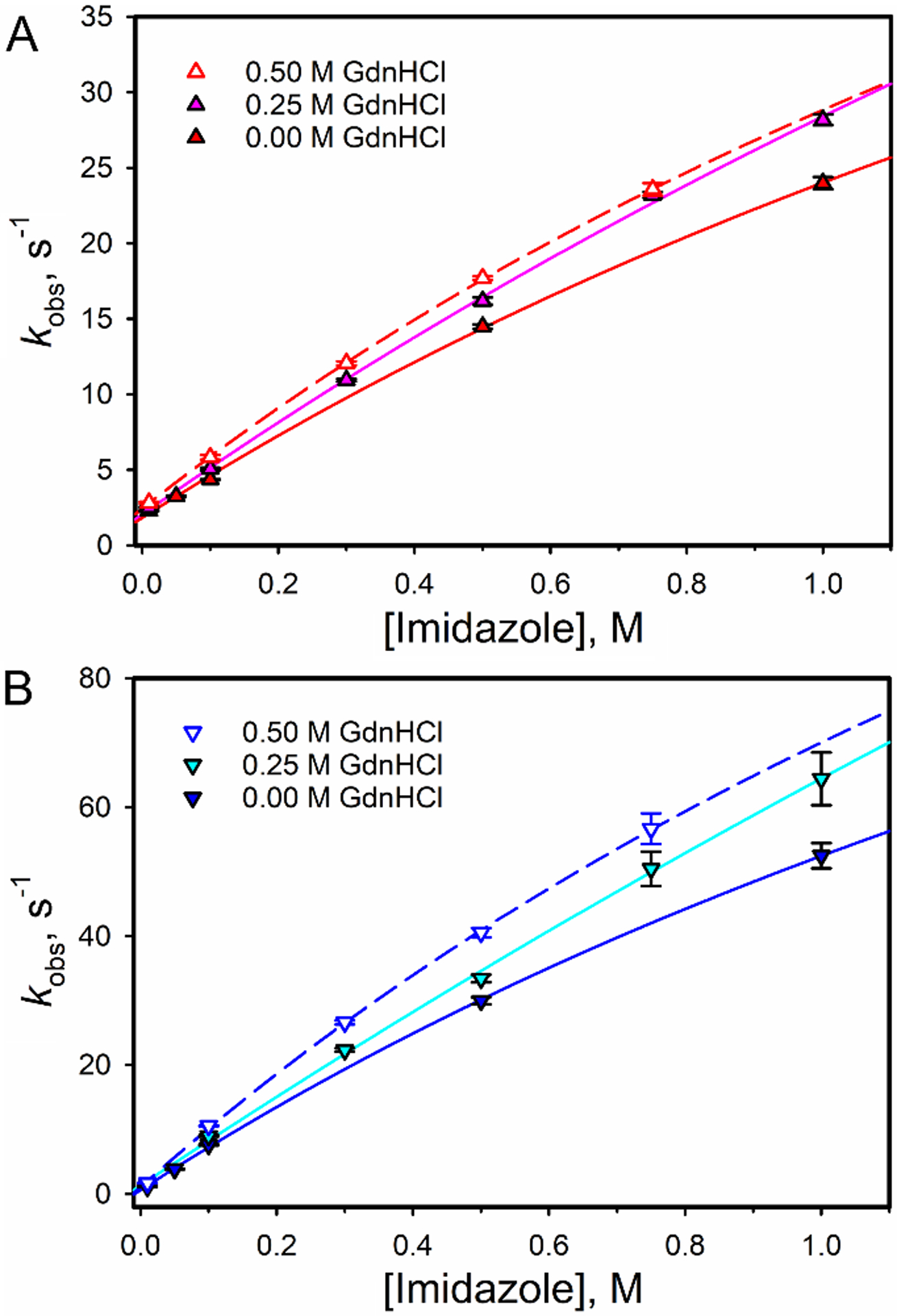
Plots of kobs versus Im concentration at 0, 0.25 and 0.50 M GdnHCl for (A) horse heart Cytc and (B) K72A/K73A/K79A iso-1-Cytc. All data were measured at a constant ionic strength of 1.25 M. Solid curves are fits of eq 2 to the data.
Table 3.
Thermodynamic parameters for imidazole binding to Cytc as a function of GdnHCl at I = 1.25 M, pH 7 and 25 °C.
Fig. 7.
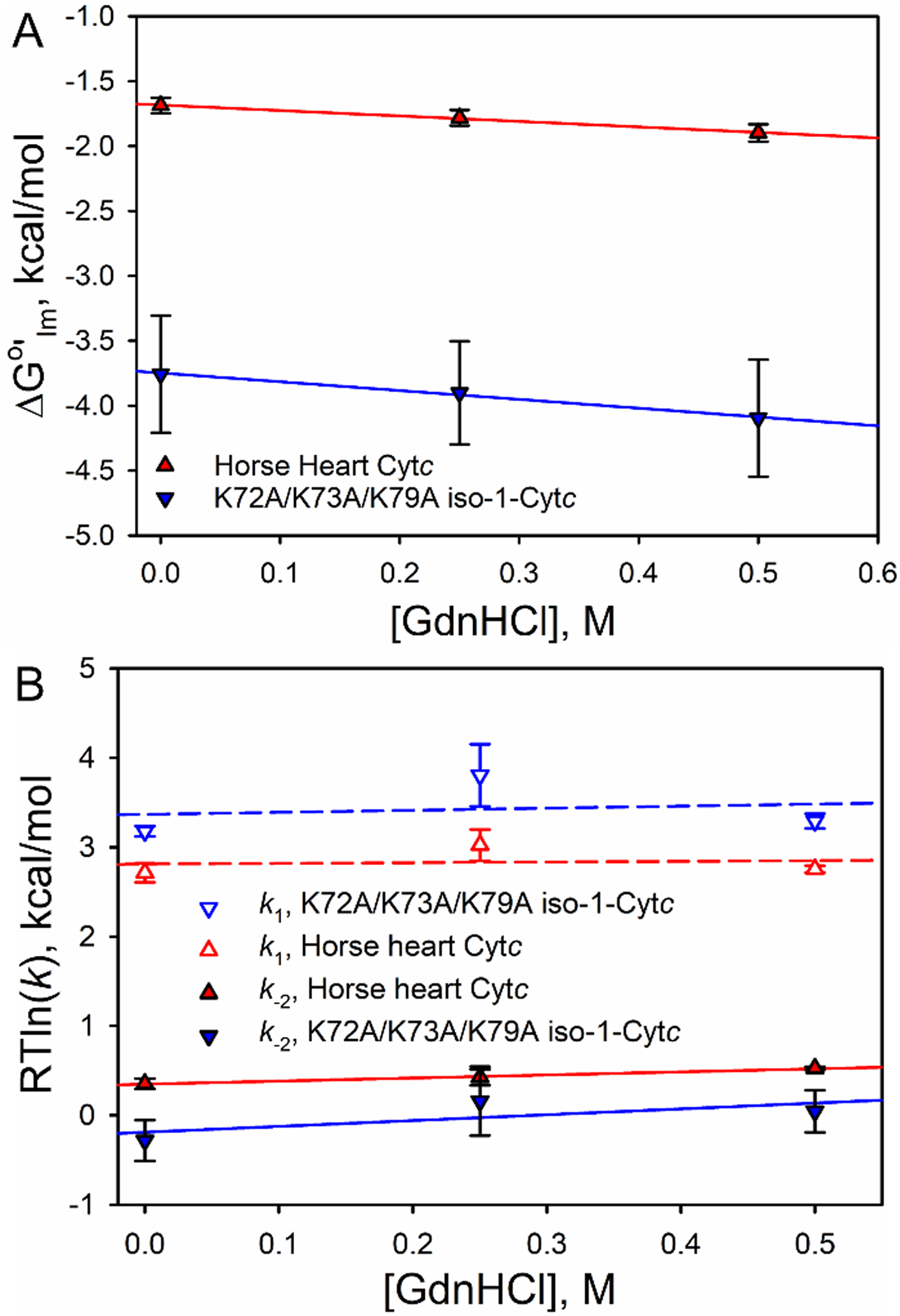
Plots of (A) ΔG°′Im and (B) RTln(k) for k1 and k−2 versus GdnHCl for horse heart Cytc and K72A/K73A/K79A iso-1-Cytc. Solid and dashed curves are fits of a linear free energy relationship to the data. The m-values from these fits are provided in Table 4.
Table 4.
GdnHCl m-values for ΔG°′Im, k1 and k−2 for imidazole binding to Cytc at I = 1.25 M, pH 7 and 25 °C.
| Protein | mIm (kcal mol−1 M−1) | (kcal mol−1 M−1) | (kcal mol−1 M−1) |
|---|---|---|---|
| Horse heart Cytc | 0.42 ± 0.02 | 0.08 ± 0.67 | 0.34 ± 0.03 |
| K72A/K73A/K79A iso-1-Cytc | 0.68 ± 0.06 | 0.2 ± 1.3 | 0.65 ± 0.64 |
The GdnHCl concentration dependence of the activation free energy can be obtained from plots of RTln(k) versus GdnHCl for the rate constants k1 and k−2 obtained from fits of eq 2 to the data in Fig. 6 (Table S6). The slopes of these plots are small (Fig. 7B). The m-value for k1, , which is a measure of the extent of the conformational change needed for Im binding is within error zero. The m-value for k−2, , which is a measure of the extent of the conformational change needed to break the Im bond to the iron of the heme is similar in magnitude to the equilibrium m-value for Im binding, mIm (Table 4).
3.5. Structure of the N63T variant
Fig. 8 shows alignments of the X-ray structure of N63T iso-1-Cytc with those of WT iso-1-Cytc and human Cytc. The backbone RMSD values obtained from PyMol show that N63T when aligned with WT yeast iso-1-Cytc has a backbone RMSD of 0.405 Å, while it is 0.323 Å when aligned with WT human Cytc, indicating that the backbone of the N63T variant aligns slightly better with that of human Cytc than with yeast iso-1-Cytc. The all atom RMSD values show the same trend: the RMSD for the N63T variant aligned to yeast iso-1-Cytc is 1.626 Å, while it is 0.871 Å for N63T iso-1-Cytc aligned to WT human Cytc.
Fig. 8.
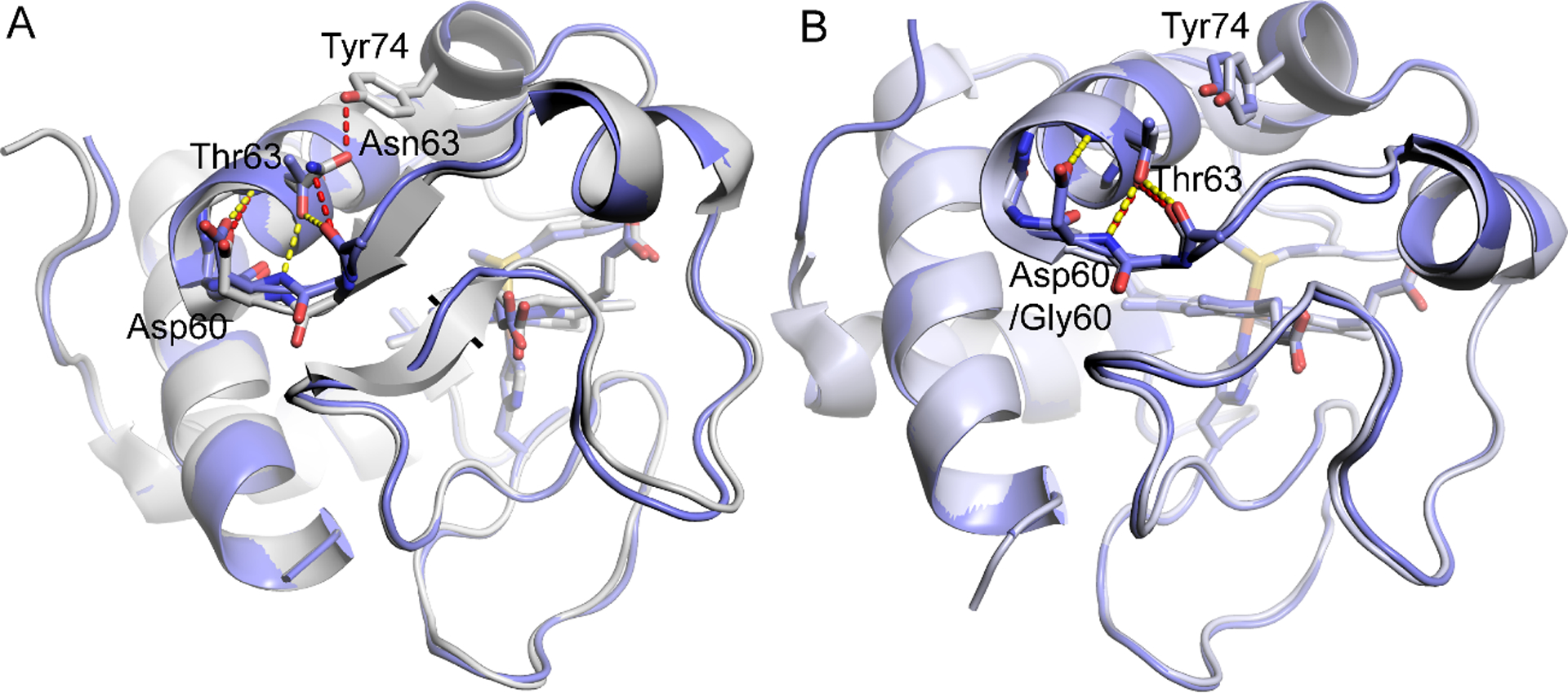
Structure of N63T iso-1-Cytc (PDB ID: 7MRI) overlaid with (A) WT iso-1-Cytc (PDB ID: 2YCC) [65] and with (B) WT human Cytc (PDB ID: 3ZCF) [26]. In both panels hydrogen bonds for N63T iso-1-Cytc are shown as yellow dashed lines and for WT yeast and human proteins as red dashed lines. In both panels, the backbone atoms from Leu58 to Thr63 (Asn63) are shown as sticks.
There are also changes in hydrogen bonding that result from the N63T mutation to iso-1-Cytc. For WT yeast iso-1-Cytc, there is a hydrogen bond from the carbonyl of the Asn63 side chain to the −OH group of Tyr74 and from the −NH2 of the Asn63 side chain to the carbonyl of Leu58 (Fig. 8A). Thus, for WT iso-1-Cytc, the N-terminal end of Ω-loop D is anchored to the short neck of β-sheet at either end of Ω-loop C. For N63T iso-1-Cytc, as for human Cytc, the side chain of Thr63 maintains a hydrogen bond to the carbonyl of Leu58. However, the hydrogen bond to the side chain −OH of Tyr 74 is lost (Fig. 8B), disrupting the connection between Ω-loop D and the β-sheet neck at the ends of Ω-loop C. The side chain of Thr63 in the N63T variant also forms a hydrogen bond to the backbone amide NH of Asp60, which is the N-cap residue of the 60s helix. The side chain of Asp60 caps the backbone amide NH of Thr63 (Fig. 8B). This reciprocal interaction constitutes a classic “capping box” motif, where the sidechains of the N3 residue of the helix (Thr63) and of the N-cap residue of the helix (Asp60) are each hydrogen bonded to the backbone amide NH of the other [66,67]. For WT human Cytc, the residue at position 60 is Gly, which is incapable of forming this “capping box” motif (Fig. 8B). WT and N63T yeast iso-1-Cytc form a hydrogen bond from the side chain of Asp60 to the amide NH of Asn62 also capping this amide NH. The hydrogen bond is stronger (2.95 Å) for the N63T variant than for WT iso-1-Cytc (3.29 Å). The N63T variant may have unique properties as a result of this “capping box” motif. However, all three proteins have the hydrophobic interaction between the 4th residue of the helix (Met64 in yeast, Leu64 in human) and the N′ residue of the helix cap (Trp59) typical of the “capping box” motif [66].
4. Discussion
4.1. Effects of coevolved mutations on local and global stability and the peroxidase activity of iso-1-Cytc.
There is considerable evidence that the effects of mutations on protein function depend upon sequence context [68]. Sites that coevolve in a protein, while distant in sequence are observed to be proximal in the three-dimensional structure [68–70], as observed for the set of three residues used for mutational analysis in this work. The coevolved sites in or near Ω-loop C of Cytc were predicted using the EVmutation algorithm [25]. EVmutation is particularly effective at accounting for the effects of mutation on enzymatic activity, yielding Spearman correlation coefficients of 0.80 and 0.87 between mutation and function for the M.Hae III DNA methyltransferase and β-lactamase, respectively [25]. EVmutation also successfully predicted the effects of mutations on function in other proteins, such as poly(A)-binding protein [25]. EVmutation analyzes the sequence of a protein in organisms that have evolved earlier and compares it to the protein sequence in organisms that evolved more recently. Given the prominent role that the dynamics of Ω-loop C plays in the peroxidase activity of Cytc [10–13], it seemed likely that the three coevolved residues studied here at positions 40, 57 and 63 might explain the large difference in peroxidase activity between yeast and human Cytc.
As is evident from our data, the effects of single, double and triple mutant variants at the coevolved positions, 40/57 and 57/63, on global and local stability, in particular, are relatively small. With regard to local stability, while individual single or double mutation variants perturb the pKa of the alkaline transition, there is no net effect of all three mutations on the local stability of Ω-loop D as measured by the pKa of the alkaline transition (Table 2). Thus, evolutionary pressure appears to act to maintain the instability of the least stable Ω-loop C and Ω-loop D substructures of Cytc. In support of the notion that evolution has preserved the properties of Ω-loop C, the stability of this least stable substructure is similar in yeast iso-1-Cytc versus equine Cytc (Fig. 9) [31,71]. By contrast, the stability of the most stable substructure increases by more than 7 kcal/mol in equine Cytc compared to yeast iso-1-Cytc. The net result is a much smaller free energy range for the partial unfolding of the substructures of yeast iso-1-Cytc compared to equine Cytc [23,24,31,72]. The broad free energy range for the substructures of horse Cytc is presumably maintained in human Cytc, which is somewhat more stable than equine Cytc [73].
Fig. 9.
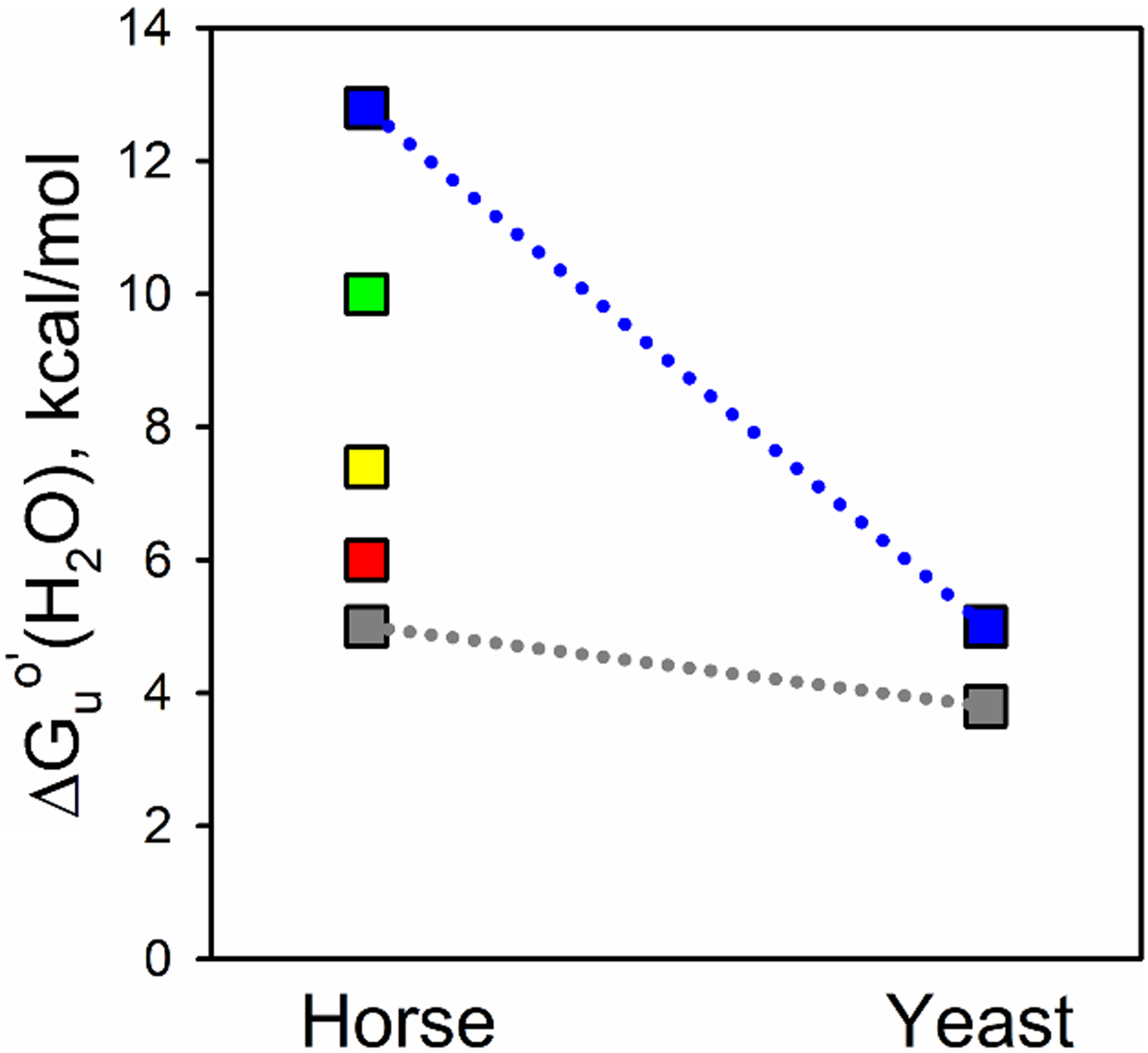
Substructure energy levels for horse Cytc and yeast iso-1-Cytc. The foldons as determined for horse Cytc, using Englander’s nomenclature, are from lowest to highest energy, the infrared (Ω-loop C, shown in gray), red (Ω-loop D), yellow (residues 37 – 39 and 58 – 60), green (residue 19 – 36 Ω-loop and 60s helix), and blue (N- and C-terminal helices) foldons [72]. Limited proteolysis was used to determine the energy of the least stable substructure of yeast iso-1-Cytc [31], which precludes measuring the energies of the other substructures. ΔGuo′(H2O) for unfolding of iso-1-Cytc by GdnHCl was used for the stability of the most stable substructure because the m-value for global unfolding of the yeast protein is similar to that for the most stable structure of horse Cytc obtained from HX experiments.
Denaturant m-values have been shown to correlate with the changes in the solvent-accessible surface area that occurs upon unfolding [74,75]. As a result, mutation-induced changes in stability have often been interpreted in terms of changes in the compactness of the denatured state [76,77], which in some cases result from a reverse hydrophobic effect [78,79]. However, mutation-induced changes in denaturant m-values for apparent two-state unfolding can also be caused by changes in the population of intermediates [80,81]. For horse Cytc, it has been shown that the large difference in substructure stabilities leads to low populations of intermediates during the GdnHCl unfolding transition. The result is a decreases in the cooperativity of the apparent two-state transition as measured by the m-value of unfolding of horse Cytc when monitored by CD or fluorescence relative to the true two-state m-value of 4.6 kcal/mol×M measured by HX methods [82]. Denaturant m-values monitored by standard optical methods yield lower m-values for GdnHCl unfolding of horse (3.0 kcal/mol×M) [83] and human Cytc (3.5 – 3.7 kcal/mol×M) [15] relative to yeast iso-1-Cytc (4.2 – 5.1 kcal/mol×M) [31,84,85]. The m-value of WT iso-1-Cytc is closer to that of the m-value for true two-state unfolding of Cytc measured by HX, consistent with the tight spacing of the substructure stabilities leading to GdnHCl unfolding that is more closely two-state (more cooperative, larger m-value). The decrease in the m-values for the substitution of the human residues at positions 40, 57 and 63 of yeast iso-1-Cytc suggests that unfolding of these variants is less cooperative. The loss of the Asn63 to Tyr74 hydrogen bond in the X-ray structure of the N63T variant (see section 3.5) is consistent with the observed loss of cooperativity.
At pH 6 and above, the peroxidase activity of iso-1-Cytc is affected significantly by the substitutions at positions 40, 57 and 63. However, kcat is not decreased to the level expected if the coevolved residues were solely responsible for the differences in intrinsic peroxidase activity between the yeast and human proteins. It is possible that pairwise coevolution revealed by the EVmutation algorithm [25] is insufficient to detect a broader set of coevolving residues necessary to decrease the intrinsic peroxidase reactivity to the level observed for human Cytc. In the highly conserved Ω-loop D, the I81A mutation in human Cytc [86], which substitutes the yeast residue alanine for the human residue isoleucine, had much larger effects on peroxidase activity than the A81I mutation in yeast iso-1-Cytc [27]. This observation indicates that a broader context of sequence differences between the yeast and human proteins are needed to change the dynamics of the two least stable substructures in a manner that limits the intrinsic peroxidase activity of Cytc. Mutations in Ω-loop C, may have smaller effects on stability and function in the context of the yeast iso-1-Cytc sequence than they do in equine or human Cytc because of the smaller energy differences in the substructures of the yeast protein [31,71].
4.2. Correlation between Peroxidase Activity and Local and Global Stability
Plots of the change in the activation free energy for kcat, ΔΔG‡(kcat), versus ΔGu°′(H2O) from GdnHCl denaturation or the change in the free energy of the alkaline transition, ΔΔG°′alk, show very little correlation, except at pH 8 (Fig. 10) and for ΔGu°′(H2O) at pH 7 (Fig. S9). A plot of ΔΔG‡(kcat) at pH 8 versus ΔGu°′(H2O) (Fig. 10A) that excludes WT iso-1-Cytc gives a correlation coefficient, R, of 0.96. This correlation suggests that this set of substitutions affects the properties of iso-1-Cytc so that they are distinct from WT iso-1-Cytc. The slope of the correlation is only 0.5 at pH 8 and even lower at pH 7 (~0.1) indicating that the change in ΔGuo′(H2O) is not the only modulator of peroxidase activity.
Fig. 10. (.
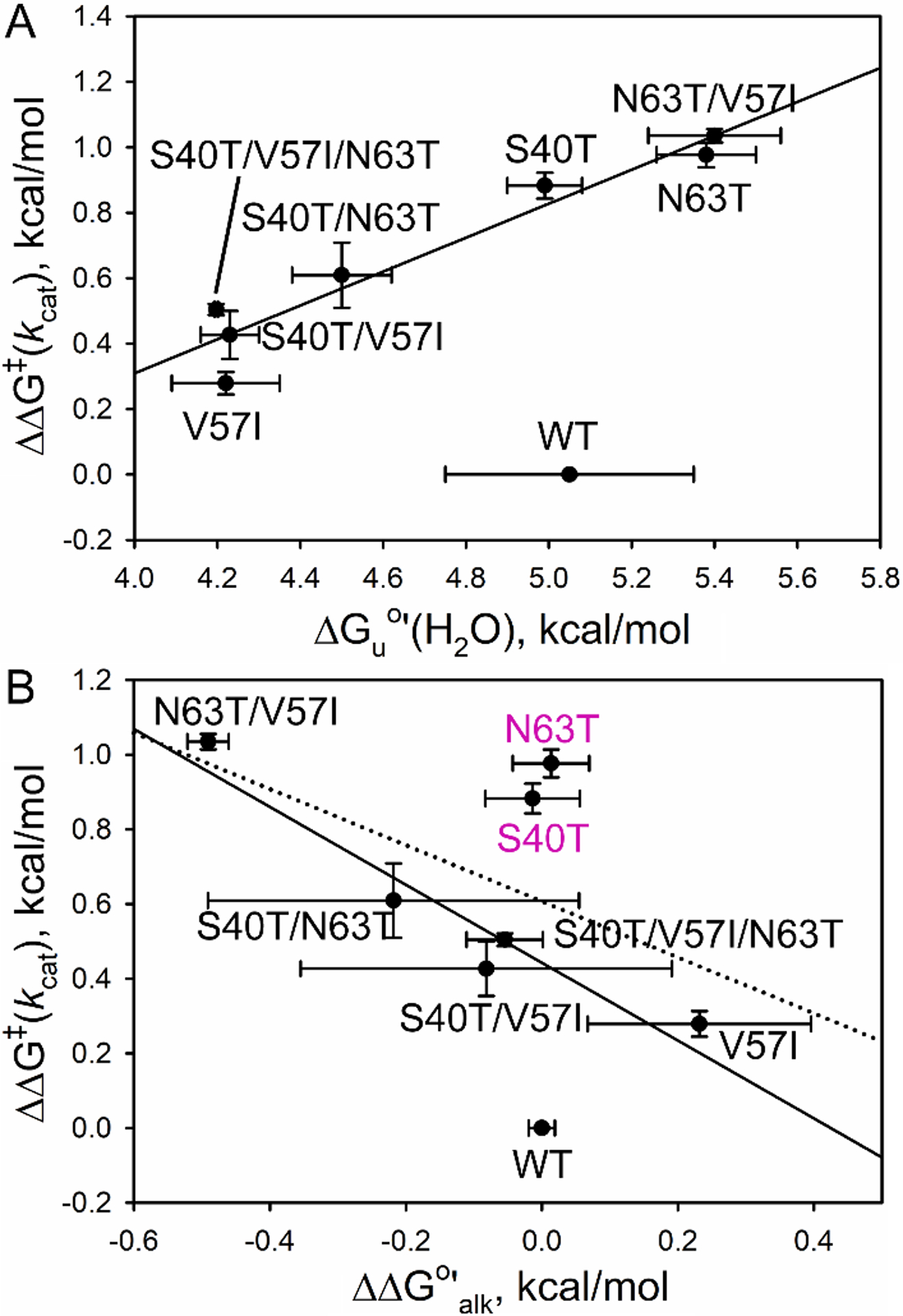
A) Plot of ΔΔG‡(kcat) at pH 8 versus ΔGuo′(H2O) for all variants. The solid line is the correlation without WT iso-1-Cytc (R = 0.96). (B) Plot of ΔΔG‡(kcat) at pH 8 versus ΔΔG°′alk. The dotted line is the correlation without WT iso-1-Cytc (R = 0.57). The solid correlation line (R = 0.96) also does not include the S40T and N63T variants. ΔΔG‡(kcat) = −RTln(kcat,mut/kcat,WT). The error bars are propagated from the errors in kcat. ΔΔG°′alk = 2.3RT(pKa,mut – pKa,WT). The error bars are propagated from the errors in the pKa for the alkaline transition.
A plot of ΔΔG‡(kcat) at pH 8 versus the change in the free energy of the alkaline transition, ΔΔG°′alk, (Fig. 10B) that excludes WT iso-1-Cytc gives a correlation coefficient, R, of 0.57 (dotted line). A significantly better correlation coefficient (0.96) can be obtained if the S40T and N63T variants are not included in the correlation. This observation suggests that N63T and S40T variants may differ from the WT protein and from the other variants in some manner that affects peroxidase activity. The inverse correlation observed between ΔΔG‡(kcat) and ΔΔGalk at pH 8 is also consistent with Lys-heme binding near the midpoint of the alkaline transition for these variants slowing formation of the open coordination site required for peroxide binding [34]. Recent studies showing that H2O2-induced carbonylation of lysines involved in the alkaline transition may be important for activating the peroxidase activity of Cytc [19] is also consistent with this correlation. The observation that the correlation is not absolute indicates that other aspects of the dynamics of Ω-loops C and D are important in regulating the activation of peroxidase activity.
4.3. Relationship between structure and dynamics
Hydrogen-exchange (HX) data from previous work shows that human wild type Cytc [11] has larger protection factors at most residues than yeast iso-1-Cytc [87]. The protection factors, P, for Ω-loop D of yeast iso-1-Cytc measured at pD = 4.6 are generally low or unassigned, except for trimethyllysine 72, Lys73 and Tyr74 with LogP values of 3 – 4 and Thr78 and Phe82 which have LogP around 2 [87]. The protection factors for human Cytc in this same region measured at pD = 6.5 are also low, however residues Tyr74 and Ile75 have LogP values of almost 3, while residues Lys73, Lys79, Met80, and Ile85 have LogP values around 1 [11]. Although the measurements were carried out at different pD values, unlike Ω-loop C, the protection factors for Ω-loop D are not strongly dependent on pD [63]. Thus, the data suggest that the dynamics of Ω-loop D near Lys 73 and Lys79 are similar for human Cytc, whereas for yeast iso-1-Cytc Ω-loop D is more dynamic near Lys79 than Lys73. Equilibrium data for the alkaline transition of horse Cytc indicate that Lys79 predominates over Lys73 in the alkaline state and that Lys72 does not contribute [54]. Studies comparing WT and K72A human Cytc also indicate that Lys72 is not a significant contributor to the alkaline state but are consistent with Lys73 rather than Lys79 being the predominant ligand for the kinetics of the alkaline transition. Equilibrium data for the alkaline transition of yeast iso-1-Cytc suggest that Lys73 is a somewhat stronger ligand for the alkaline state than Lys79, in the presence of trimetyllysine72 [88]. Lysines 72, 73 and 79 all contribute to the alkaline transition when Lys72 of iso-1-Cytc is not trimethylated [29] with mutational studies indicating that Lys72 has the lowest alkaline transition pKa when only a single Lys is available in Ω-loop D [89]. However, kinetic data point to Lys79 as the predominate ligand for the kinetics of the alkaline transition of iso-1-Cytc [27]. The dynamics of Ω-loop D as measured by the alkaline transition and HX methods are often equated [53,54]. The comparison is not straightforward if the GdnHCl-dependence of HX is not measured because HX can occur by local structural fluctuations rather than cooperative unfolding of a foldon like Ω-loop D. However, the hydrogen exchange data are qualitatively consistent with kinetic data for alkaline transition which indicate that for yeast iso-1-Cytc, Lys79 dominates the kinetics [27], while for human Cytc Lys73 is dominant in the kinetics of the alkaline transition [15].
For human Cytc (PDB: 3ZCF), the Thr63 side chain hydrogen bonds to main chain atoms of Gly60 and Ile58, whereas in yeast iso-1-Cytc (PDB: 2YCC), the Asn63 side chain hydrogen bonds to the side chain hydroxyl of Tyr74 and the main chain of Leu58. Because Tyr74 is in Ω-loop D, this residue could be a link between the two loops that modulates the dynamics that control opening of the heme crevice. The structure of N63T iso-1-Cytc shows that the hydrogen bond to the side −OH of Tyr74 is lost (Fig. 8) such that the side chain at position 63 no longer bridges Ω-loops C and D. If this hydrogen bond causes the larger protection factor at Lys73 compared to near Lys79 for iso-1-Cytc, one might expect that the N63T variant would have similar dynamics near both Lys73 and Lys 79, as indicated by the similar protection factors for these residues in human Cytc. Mutation-induced switching in the preferred ligand for the alkaline conformer may explain the poor correlation between the pKa of the alkaline transition and peroxidase activity (Fig. 10 and Fig. S9). The formation of a “capping box” motif in the N63T iso-1-Cytc variant (Fig. 8) not present in WT iso-1-Cytc or human Cytc (Fig. 8) could further confound this correlation.
Our previous results for an I81A variant in Ω-loop D of human Cytc showed that the effect on the kinetics of the alkaline transition was selective for one of two detectable alkaline conformers [86]. Selective effects on the kinetics of one alkaline conformer were also observed for the naturally-occurring A51V variant of human Cytc [9]. Based on the effects of A81I and G83V variants in Ω-loop D of iso-1-Cytc on the kinetics of the alkaline transition [27], we tentatively assigned the Lys79 alkaline conformer as the one that was preferentially affected by Ω-loop D substitutions. For iso-1-Cytc, a good correlation between the rate constant for forming the Lys79 alkaline conformer and the kcat for peroxidase activity of the Ω-loop D variants was observed [27] suggesting that backbone dynamics near Lys79 might correlate with the dynamics needed for peroxidase activity. The decreased peroxidase activity for the Ω-loop C variants also may result from a redistribution of the relative backbone dynamics near Lys79 versus Lys 73. Recent MD simulations show a somewhat significant (R = 0.67) correlation between the flexibility of Ω-loop D and the pKa of the alkaline transition of THC4 variants of human Cytc [14]. The fact that this correlation is imperfect may reflect the tendency of mutations in Ω-loops C and D to affect the dynamics of different parts of Ω-loop D.
4.4. Imidazole binding as a function of GdnHCl concentration
The effect of mutations on the thermodynamics of the alkaline transition correlate well with the thermodynamics of the cooperative unfolding of the Ω-loop D substructure of Cytc as measured by HX methods [53,54]. However, all substructures of Cytc have amides that exchange via lower energy local fluctuations [57]. Thus, it is possible that the dynamics required for peroxide binding to the heme go through lower energy pathways that do not require cooperative unfolding of the Ω-loop C and D substructures. In Scheme 1, formation of Cytc† is the limiting step in the reaction for Cytc with small molecules. We used imidazole in this work, but similar behavior is expected for the binding of hydrogen peroxide as it relates to the peroxidase activity of human Cytc. Plots of both ΔG°′Im and the activation free energy for forming Cytc† as a function of GdnHCl concentration (Fig. 7), yield m-values much less than observed for cooperative unfolding of the Ω-loop D substructure (Table 4). Thus, it can be concluded that Ω-loop D does not need to fully unfold for small molecules to bind to the heme. The GdnHCl dependence of the activation free energy for forming Cytc† (Fig. 10B) is zero within error (Table 4) for both the horse and yeast proteins indicating that the conformational change to allow a small ligand to bind to the heme is minimal. These results are consistent with the small perturbation to the structure of horse Cytc with imidazole bound [90], and the minimal perturbation to the structure of yeast iso-1-Cytc when Met80 is displaced by hydroxide at moderately alkaline pH [16]. Thus, it is likely that mutation-induced effects on activation of the peroxidase activity of Cytc will correlate better with local backbone fluctuations than with the pKa of the alkaline transition or with global stability.
5. Conclusion
Decreases in the pKa of the alkaline transition of Cytc often occur for mutations that increase the intrinsic peroxidase activity of Cytc. This observation has led to the assumption that cooperative unfolding of Ω-loop D linked to the dynamics of the adjacent Ω-loop C is needed to permit Cytc to act as a peroxidase. Substituting human residues into yeast iso-1-Cytc at three coevolved sites in or near Ω-loop C, we observed poor correlations between peroxidase activity and both the pKa of the alkaline transition and global stability. The GdnHCl dependence of the kinetics of Im binding to horse and yeast cytochromes c confirmed that small local structural fluctuations of the backbone of Cytc are sufficient to allow small ligands like peroxide to bind to the heme. These results suggest that subtle mutation-induced changes in backbone dynamics may be sufficient to decrease the susceptibility of Cytc to pre-activation by reactive oxygen species so as to maintain the off state of the peroxidase signaling switch for apoptosis, particularly since the stability of Ω-loop D appears to be robust to mutations at coevolved sites in and near Ω-loop C. In summary, previous work has focused on the unfolding of the entire Ω-loop D as being necessary for the binding of hydrogen peroxide and other small molecules, but our work indicates that the unfolding of the entire substructure is not necessary to activate the peroxidase activity of Cytc.
Supplementary Material
Acknowledgements
This work was supported by grants from the NSF [CHE-1609720 and CHE-1904895 (B.E.B)]. The Integrated Structural Biology Core at the University of Montana was supported by a CoBRE grant from the National Institute of General Medical Sciences [P20GM103546].
Abbreviations
- βME
β-mercaptoethanol
- CD
circular dichroism
- CL
cardiolipin
- Cytc
cytochrome c
- ΔG°′Im
Apparent free energy of binding of imidazole to Cytc
- GdnHCl
guanidine hydrochloride
- HX
hydrogen/deuterium exchange
- Im
imidazole
- iso-1-Cytc
yeast iso-1-cytochrome c
- K Im,equil
apparent equilibrium constant for Im binding to Cytc
- MD
molecular dynamics
- THC4
thrombocytopenia type IV
- WT*
variant of yeast iso-1-Cytc with a K72A substitution
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at
References
- [1].Dickerson RE, Timkovich R, in: Boyer PD (Ed.), The Enzymes, vol. 11, Academic Press, New York, 1975, pp. 397–547. [Google Scholar]
- [2].Okada M, Smith NI, Palonpon AF, Endo H, Kawata S, Sodeoka M, Fujita K. Label-free Raman observation of cytochrome c dynamics during apoptosis, Proc. Natl. Acad. Sci. U.S.A 109 (2012) 28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c, Cell 86 (1996) 147–157. [DOI] [PubMed] [Google Scholar]
- [4].Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors, Nat. Chem. Biol 1 (2005) 223–232. [DOI] [PubMed] [Google Scholar]
- [5].Zou H, Li Y, Liu X, Wang X. An APAF-1·cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9, J. Biol. Chem 274 (1999) 11549–11556. [DOI] [PubMed] [Google Scholar]
- [6].Morison IM, Cramer Bordé EM, Cheesman EJ, Cheong PL, Holyoake AJ, Fichelson S, Weeks RJ, Lo A, Davies SMK, Wilbanks SM, Fagerlund RD, Ludgate MW, da Silva Tatley FM, Coker MSA, Bockett NA, Hughes G, Pippig DA, Smith MP, Capron C, Ledgerwood EC. A mutation of human cytochrome c enhances the intrinsic apoptotic pathway but causes only thrombocytopenia, Nat. Genet 40 (2008) 387–389. [DOI] [PubMed] [Google Scholar]
- [7].De Rocco D, Cerqua C, Goffrini P, Russo G, Pastore A, Meloni F, Nicchia E, Moraes CT, Pecci A, Salviati L, Savoia A. Mutations of cytochrome c identified in patients with thrombocytopenia THC4 affect both apoptosis and cellular bioenergetics, Biochim. Biophys. Acta 1842 (2014) 269–274. [DOI] [PubMed] [Google Scholar]
- [8].Johnson B, Lowe GC, Futterer J, Lordkipanidzé M, MacDonald D, Simpson MA, Sanchez-Guiú I, Drake S, Bem D, Leo V, Fletcher SJ, Dawood B, Rivera J, Allsup D, Biss T, Bolton-Maggs PHB, Collins P, Curry N, Grimley C, James B, Makris M, Motwani J, Pavord S, Talks K, Thachil J, Wilde J, Williams M, Harrison P, Gissen P, Mundell S, Mumford A, Daly ME, Watson SP, Morgan NV. Whole exome sequencing identifies genetic variants in inherited thrombocytopenia with secondary qualitative function defects, Haematologica 101 (2016) 1170–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lei H, Bowler BE. The naturally occurring A51V variant of human cytochrome c destabilizes the native state and enhances peroxidase activity, J. Phys. Chem. B 123 (2019) 8939–8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Deacon OM, White RW, Moore GR, Wilson MT, Worrall JAR. Comparison of the structural dynamic and mitochondrial electron-transfer properties of the proapoptotic human cytochrome c variants, G41S, Y48H and A51V, J. Inorg. Biochem 203 (2020) 110924. [DOI] [PubMed] [Google Scholar]
- [11].Karsisiotis AI, Deacon OM, Wilson MT, Macdonald C, Blumenschein TMA, Moore GR, Worrall JAR. Increased dynamics in the 40–57 Ω-loop of the G41S variant of human cytochrome c promote its pro-apoptotic conformation, Sci. Rep 6 (2016) 30447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Deacon OM, Karsisiotis AI, Moreno-Chicano T, Hough MA, Macdonald C, Blumenschein TMA, Wilson MT, Moore GR, Worrall JAR. Heightened dynamics of the oxidized Y48H variant of human cytochrome c increases its peroxidatic activity, Biochemistry 56 (2017) 6111–6124. [DOI] [PubMed] [Google Scholar]
- [13].Deacon OM, Svistunenko DA, Moore GR, Wilson MT, Worrall JAR. Naturally occurring disease-related mutations in the 40−57 Ω-loop of human cytochrome c control triggering of the alkaline isomerization, Biochemistry 57 (2018) 4276–4288. [DOI] [PubMed] [Google Scholar]
- [14].Fellner M, Parakra R, McDonald KO, Kass I, Jameson GNL, Wilbanks SM, Ledgerwood EC. Altered structure and dynamics of pathogenic cytochrome c variants correlate with increased apoptotic activity, Biochem. J 478 (2021) 669–684. [DOI] [PubMed] [Google Scholar]
- [15].Nold SM, Lei H, Mou T-C, Bowler BE. Effect of a K72A mutation on the structure, stability, dynamics and peroxidase activity of human cytochrome c, Biochemistry 56 (2017) 3358–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].McClelland LJ, Mou T-C, Jeakins-Cooley ME, Sprang SR, Bowler BE. Structure of a mitochondrial cytochrome c conformer competent for peroxidase activity, Proc. Natl. Acad. Sci. U.S.A 111 (2014) 6648–6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hirota S, Hattori Y, Nagao S, Taketa M, Komori H, Kamikubo H, Wang Z, Takahashi I, Negi S, Sugiura Y, Kataoka M, Higuchi Y. Cytochrome c polymerization by successive domain swapping at the C-terminal helix, Proc. Natl. Acad. Sci. U.S.A 107 (2010) 12854–12859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang C.-q., Li X, Wang M.-q., Qian J, Zheng K, Bian H.-w., Han N, Wang J.-h., Pan J.-w., Zhu M.-y.. Protective effects of ETC complex III and cytochrome c against hydrogen peroxide-induced apoptosis in yeast, Free Radical Res 48 (2014) 435–444. [DOI] [PubMed] [Google Scholar]
- [19].Yin V, Shaw GS, Konermann L. Cytochrome c as a peroxidase: activation of the precatalytic native state by H2O2-induced covalent modifications, J. Am. Chem. Soc 139 (2017) 15701–15709. [DOI] [PubMed] [Google Scholar]
- [20].Chen Y-R, Deterding LJ, Sturgeon BE, Tomer KB, Mason RP. Protein oxidation of cytochrome c by reactive halogen species enhances Its peroxidase activity, J. Biol. Chem 277 (2002) 29781–29791. [DOI] [PubMed] [Google Scholar]
- [21].Capdevila DA, Marmisollé WA, Tomasina F, Demicheli V, Portela M, Radi R, Murgida DH. Specific methionine oxidation of cytochrome c in complexes with zwitterionic lipids by hydrogen peroxide: potential implications for apoptosis, Chem Sci 6 (2015) 705–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Parakra RD, Kleffmann T, Jameson GNL, Ledgerwood EC. The proportion of Met80-sulfoxide dictates peroxidase activity of human cytochrome c, Dalton Trans. 47 (2018) 9128–9135. [DOI] [PubMed] [Google Scholar]
- [23].Krishna MM, Lin Y, Rumbley JN, Englander SW. Cooperative omega loops in cytochrome c: role in folding and function, J. Mol. Biol 331 (2003) 29–36. [DOI] [PubMed] [Google Scholar]
- [24].Bai Y, Sosnick TR, Mayne L, Englander SW. Protein folding intermediates: native-state hydrogen exchange, Science 269 (1995) 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hopf TA, Ingraham JB, Poelwijk FJ, Schärfe CPI, Springer M, Sander C, Marks DS. Mutation effects predicted from sequence co-variation, Nat. Biotechnol 35 (2017) 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rajagopal BS, Edzuma AN, Hough MA, Blundell KLIM, Kagan VE, Kapralov AA, Fraser LA, Butt JN, Silkstone GG, Wilson MT, Svistunenko DA, Worrall JAR. The hydrogen-peroxide-induced radical behaviour in human cytochrome c–phospholipid complexes: implications for the enhanced pro-apoptotic activity of the G41S mutant, Biochem. J 456 (2013) 441–452. [DOI] [PubMed] [Google Scholar]
- [27].Lei H, Bowler BE. Humanlike substitutions to Ω-loop D of yeast iso-1-cytochrome c only modestly affect dynamics and peroxidase activity, J. Inorg. Biochem 183 (2018) 146–156. [DOI] [PubMed] [Google Scholar]
- [28].Elmer-Dixon MM, Bowler BE. Site A-mediated partial unfolding of cytochrome c on cardiolipin vesicles is species-dependent and does not require Lys72, Biochemistry 56 (2017) 4830–4839. [DOI] [PubMed] [Google Scholar]
- [29].Pollock WB, Rosell FI, Twitchett MB, Dumont ME, Mauk AG. Bacterial expression of a mitochondrial cytochrome c. Trimethylation of Lys72 in yeast iso-1-cytochrome c and the alkaline conformational transition, Biochemistry 37 (1998) 6124–6131. [DOI] [PubMed] [Google Scholar]
- [30].McClelland LJ, Seagraves SM, Khan MKA, Cherney MM, Bandi S, Culbertson JE, Bowler BE. The response of Ω-loop D dynamics to truncation of trimethyllysine 72 of yeast iso-1-cytochrome c depends on the nature of loop deformation, J. Biol. Inorg. Chem 20 (2015) 805–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Duncan MG, Williams MD, Bowler BE. Compressing the free energy range of substructure stabilities in iso-1-cytochrome c, Protein Sci. 18 (2009) 1155–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rosell FI, Mauk AG. Spectroscopic properties of a mitochondrial cytochrome c with a single thioether bond to the heme prosthetic group, Biochemistry 41 (2002) 7811–7818. [DOI] [PubMed] [Google Scholar]
- [33].Rumbley JN, Hoang L, Englander SW. Recombinant equine cytochrome c in Escherichia coli: high-level expression, characterization, and folding and assembly mutants, Biochemistry 41 (2002) 13894–13901. [DOI] [PubMed] [Google Scholar]
- [34].Diederix REM, Ubbink M, Canters GW. The peroxidase activity of cytochrome c-550 from Paracoccus versutus, Eur. J. Biochem 268 (2001) 4207–4216. [DOI] [PubMed] [Google Scholar]
- [35].Samsri S, Pornsuwan S. Influence of cysteine-directed mutations at the Ω-loops on peroxidase activity of human cytochrome c, Arch. Biochem. Biophys 709 (2021) 108980. [DOI] [PubMed] [Google Scholar]
- [36].Radi R, Thomson L, Rubbo H, Prodanov E. Cytochrome c-catalyzed oxidation of organic molecules by hydrogen peroxide, Arch. Biochem. Biophys 288 (1991) 112–117. [DOI] [PubMed] [Google Scholar]
- [37].Goldschmid O. The effect of alkali and strong acid on the ultraviolet absorption spectrum of lignin and related compounds, J. Am. Chem. Soc 75 (1953) 3780–3783. [Google Scholar]
- [38].Noble RW, Gibson QH. The reaction of ferrous horseradish peroxidase with hydrogen peroxide, J. Biol. Chem 245 (1970) 2409–2413. [PubMed] [Google Scholar]
- [39].Nelson DP, Kiesow LA. Enthalpy of decomposition of hydrogen peroxide by catalase at 25 °C (with molar extinction coefficients of H2O2 solutions in the UV), Anal. Biochem 49 (1972) 474–478. [DOI] [PubMed] [Google Scholar]
- [40].Diederix REM, Ubbink M, Canters GW. Peroxidase activity as a tool for studying the folding of c-type cytochromes, Biochemistry 41 (2002) 13067–13077. [DOI] [PubMed] [Google Scholar]
- [41].de Azevedo Neto AD, Prisco JT, Enéas-Filho J, Abreu CEBD, Gomes-Filho E. Effect of salt stress on antioxidative enzymes and lipid peroxidation in leaves and roots of salt-tolerant and salt-sensitive maize genotypes, Environ. Exp. Bot 56 (2006) 87–94. [Google Scholar]
- [42].Doerge DR, Divi RL, Churchwell MI. Identification of the colored guaiacol oxidation product produced by peroxidases, Anal. Biochem 250 (1997) 10–17. [DOI] [PubMed] [Google Scholar]
- [43].Powell HR, Johnson O, Leslie AGW. Autoindexing diffraction images with iMosflm, Acta Crystallogr., Sect. D: Biol. Crystallogr 69 (2013) 1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Evans PR, Murshudov GN. How good are my data and what is the resolution?, Acta Crystallogr., Sect. D: Biol. Crystallogr 69 (2013) 1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Vagin A, Teplyakov A. Molecular replacement with MOLREP, Acta Crystallogr., Sect. D: Biol. Crystallogr 66 (2010) 22–25. [DOI] [PubMed] [Google Scholar]
- [46].Potterton L, Agirre J, Ballard C, Cowtan K, Dodson E, Evans PR, Jenkins HT, Keegan R, Krissinel E, Stevenson K, Lebedev A, McNicholas SJ, Nicholls RA, Noble M, Pannu NS, Roth C, Sheldrick G, Skubak P, Turkenburg J, Uski V, von Delft F, Waterman D, Wilson K, Winn M, Wojdyr M. CCP4i2: the new graphical user interface to the CCP4 program suite, Acta Crystallogr., Sect. D: Biol. Crystallogr 74 (2018) 68–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC 5 for the refinement of macromolecular crystal structures, Acta Crystallogr., Sect. D: Biol. Crystallogr 67 (2011) 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution, Acta Crystallogr., Sect. D: Biol. Crystallogr 66 (2010) 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot, Acta Crystallogr., Sect. D: Biol. Crystallogr 66 (2010) 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].DeLano WL. The PyMOL molecular graphics system, Version 1, DeLano Scientific, LLC, San Carlos, CA, USA: (2006) [Google Scholar]
- [51].Hagihara Y, Aimoto S, Fink A, Goto Y. Guanidine hydrochloride-induced folding of proteins, J. Mol. Biol 231 (1993) 180–184. [DOI] [PubMed] [Google Scholar]
- [52].Hagihara Y, Tan Y, Goto Y. Comparison of the conformational stability of the molten globule and native states of horse cytochrome c, J. Mol. Biol 237 (1994) 336–348. [DOI] [PubMed] [Google Scholar]
- [53].Kristinsson R, Bowler BE. Communication of stabilizing energy between substructures of a protein, Biochemistry 44 (2005) 2349–2359. [DOI] [PubMed] [Google Scholar]
- [54].Maity H, Rumbley JN, Englander SW. Functional role of a protein foldon - an Ω-loop foldon controls the alkaline transition in ferricytochrome c, Proteins: Struct., Funct., Genet 63 (2006) 349–355. [DOI] [PubMed] [Google Scholar]
- [55].Wang Z, Matsuo T, Nagao S, Hirota S. Peroxidase activity enhancement of horse cytochrome c by dimerization, Org. Biomol. Chem 9 (2011) 4766–4769. [DOI] [PubMed] [Google Scholar]
- [56].Steele HBB, Elmer-Dixon MM, Rogan JT, Ross JBA, Bowler BE. The human cytochrome c domain-swapped dimer facilitates tight regulation of intrinsic apoptosis, Biochemistry 59 (2020) 2055–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Englander SW, Mayne L, Kan Z-Y, Hu W. Protein folding—how and why: by hydrogen exchange, fragment separation, and mass spectrometry, Annu. Rev. Biophys 45 (2016) 135–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Scrosati PM, Yin V, Konermann L. Hydrogen/deuterium exchange measurements may provide an incomplete view of protein dynamics: a case study on cytochrome c, Anal. Chem 93 (2021) 14121–14129. [DOI] [PubMed] [Google Scholar]
- [59].Sutin N, Yandell JK. Mechanisms of the reactions of cytochrome c. Rate and equilibrium constants for ligand binding to horse heart ferricytochrome c, J. Biol. Chem 247 (1972) 6932–6936. [PubMed] [Google Scholar]
- [60].Dumortier C, Meyer TE, Cusanovich MA. Protein dynamics: Imidazole binding to class I c-type cytochromes, Arch. Biochem. Biophys 371 (1999) 142–148. [DOI] [PubMed] [Google Scholar]
- [61].Fersht A, W. H. Freeman and Company, New York, 1998. [Google Scholar]
- [62].Martinez RE, Bowler BE. Proton-mediated dynamics of the alkaline conformational transition of yeast iso-1-cytochrome c, J. Am. Chem. Soc 126 (2004) 6751–6758. [DOI] [PubMed] [Google Scholar]
- [63].Krishna MM, Maity H, Rumbley JN, Lin Y, Englander SW. Order of steps in the cytochrome c folding pathway: evidence for a sequential stabilization mechanism, J. Mol. Biol 359 (2006) 1410–1419. [DOI] [PubMed] [Google Scholar]
- [64].Nelson CJ, Bowler BE. pH dependence of formation of a partially unfolded state of a Lys 73 -> His variant of iso-1-cytochrome c: implications for the alkaline conformational transition of cytochrome c, Biochemistry 39 (2000) 13584–13594. [DOI] [PubMed] [Google Scholar]
- [65].Berghuis AM, Brayer GD. Oxidation state-dependent conformational changes in cytochrome c, J. Mol. Biol 223 (1992) 959–976. [DOI] [PubMed] [Google Scholar]
- [66].Seale JW, Srinivasan R, Rose GD. Sequence determinants of the capping box, a stabilizing motif at the N-termini of α-helices, Protein Sci. 3 (1994) 1741–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Harper ET, Rose GD. Helix stop signals in proteins and peptides: the capping box, Biochemistry 32 (1993) 7605–7609. [DOI] [PubMed] [Google Scholar]
- [68].Starr TN, Thornton JW. Exploring protein sequence-function landscapes, Nat. Biotechnol 35 (2017) 125–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Morcos F, Pagnani A, Lunt B, Bertolino A, Marks DS, Sander C, Zecchina R, Onuchic JN, Hwa T, Weigt M. Direct-coupling analysis of residue coevolution captures native contacts across many protein families, Proc. Natl. Acad. Sci. U.S.A 108 (2011) E1293–E1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Marks DS, Colwell LJ, Sheridan R, Hopf TA, Pagnani A, Zecchina R, Sander C. Protein 3D structure computed from evolutionary sequence variation, PLoS ONE 6 (2011) e28766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Cherney MM, Bowler BE. Protein dynamics and function: making new strides with an old warhorse, the alkaline conformational transition of cytochrome c, Coord. Chem. Rev 255 (2011) 664–677. [Google Scholar]
- [72].Maity H, Maity M, Englander SW. How cytochrome c folds, and why: submolecular foldon units and their stepwise sequential stabilization, J. Mol. Biol 343 (2004) 223–233. [DOI] [PubMed] [Google Scholar]
- [73].Goldes ME, Jeakins-Cooley ME, McClelland LJ, Mou T-C, Bowler BE. Disruption of a hydrogen bond network in human versus spider monkey cytochrome c affects heme crevice stability, J. Inorg. Biochem 158 (2016) 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding, Protein Sci. 4 (1995) 2138–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Schellman JA. Solvent denaturation, Biopolymers 17 (1978) 1305–1322. [DOI] [PubMed] [Google Scholar]
- [76].Scholtz JM, Grimsley GR, Pace CN. Solvent denaturation of proteins and interpretations of the m value, Methods Enzymol. 466 (2009) 549–565. [DOI] [PubMed] [Google Scholar]
- [77].Shortle D. Staphylococcal nuclease: a showcase of m-value effects, Adv. Protein Chem 46 (1995) 217–245. [DOI] [PubMed] [Google Scholar]
- [78].Bowler BE, May K, Zaragoza T, York P, Dong A, Caughey WS. Destabilizing effects of replacing a surface lysine of cytochrome c with aromatic amino acids: implications for the denatured state, Biochemistry 32 (1993) 183–190. [DOI] [PubMed] [Google Scholar]
- [79].Herrmann L, Bowler BE, Dong A, Caughey WS. The effects of hydrophilic to hydrophobic surface mutations on the denatured state of iso-1-cytochrome c: investigation of aliphatic residues, Biochemistry 34 (1995) 3040–3047. [DOI] [PubMed] [Google Scholar]
- [80].Pace CN, Hermans J. The stability of globular proteins, CRC Crit. Rev. Biochem 3 (1975) 1–43. [DOI] [PubMed] [Google Scholar]
- [81].Carra JH, Privalov PL. Thermodynamics of denaturation of staphylococcal nuclease mutants: an intermediate state in protein folding, FASEB J. 10 (1996) 67–74. [DOI] [PubMed] [Google Scholar]
- [82].Mayne L, Englander SW. Two-state vs. multistate protein unfolding studied by optical melting and hydrogen exchange, Protein Sci. 9 (2000) 1873–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Knapp JA, Pace CN. Guanidine hydrochloride and acid denaturation of horse, cow, and Candida krusei cytochromes c, Biochemistry 13 (1974) 1289–1294. [DOI] [PubMed] [Google Scholar]
- [84].Godbole S, Hammack B, Bowler BE. Measuring denatured state energetics: deviations from random coil behavior and implications for the folding of iso-1-cytochrome c, J. Mol. Biol 296 (2000) 217–228. [DOI] [PubMed] [Google Scholar]
- [85].Betz SF, Pielak GJ. Introduction of a disulfide bond into cytochrome c stabilizes a compact denatured state, Biochemistry 31 (1992) 12337–12344. [DOI] [PubMed] [Google Scholar]
- [86].Lei H, Nold SM, Jung Motta L, Bowler BE. Effect of V83G and I81A substitutions to human cytochrome c on acid unfolding and peroxidase activity below neutral pH, Biochemistry 58 (2019) 2921–2933. [DOI] [PubMed] [Google Scholar]
- [87].Baxter SM, Fetrow JS. Hydrogen exchange behavior of [U-15N]-labeled oxidized and reduced iso-1-cytochrome c, Biochemistry 38 (1999) 4493–4503. [DOI] [PubMed] [Google Scholar]
- [88].Rosell FI, Ferrer JC, Mauk AG. Proton-linked protein conformational switching: definition of the alkaline conformational transition of yeast iso-1-ferricytochrome c, J. Am. Chem. Soc 120 (1998) 11234–11245. [Google Scholar]
- [89].Battistuzzi G, Borsari M, De Rienzo F, Di Rocco G, Ranieri A, Sola M. Free energy of transition for the individual alkaline conformers of yeast iso-1-cytochrome c, Biochemistry 46 (2007) 1694–1702. [DOI] [PubMed] [Google Scholar]
- [90].Banci L, Bertini I, Liu G, Lu J, Reddig T, Tang W, Wu Y, Yao Y, Zhu D. Effects of extrinsic imidazole ligation on the molecular and electronic structure of cytochrome c, J. Biol. Inorg. Chem 6 (2001) 628–637. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


