Abstract
The African cichlid radiations have created thousands of new cichlid species with a wide diversity of trophic morphologies, behaviors, sensory systems and pigment patterns. In addition, recent research has uncovered a surprising number of young sex chromosome systems within African cichlids. Here we refine methods to describe the differentiation of young sex chromosomes from whole genome comparisons. We identified a novel XY sex chromosome system on linkage group 14 in Oreochromis mossambicus, confirmed a linkage group 1 XY system in Coptodon zillii and also defined the limits of our methodology by examining a ZW system on linkage group 3 in Pelmatolapia mariae. These data further demonstrate that cichlids are an excellent model system for understanding the early stages of sex chromosome evolution.
Keywords: Sex chromosome evolution, Oreochromis mossambicus, Coptodon zillii, Pelmatolapia mariae
Background
Traditionally, sex chromosomes were thought to be stable features of animal karyotypes (Abbott et al., 2017). For example, the sex chromosomes of eutherian mammals arose ~181 million years ago and remain relatively similar in structure among species (Cortez et al., 2014). However, mounting evidence from other vertebrate groups suggests that sex chromosomes may turnover much more rapidly in some lineages (Lee et al., 2004; Cnaani et al., 2008; Gamble, 2010; Ser et al., 2010; Myosho et al., 2012). Transitions from one sex chromosome system to another most often begin when a mutation creates a novel sex-determination allele. Selection on linked, sexually antagonistic alleles may help drive this novel sex-determination allele to fixation. Inversions, and other mechanisms that reduce recombination between the novel sex-determination allele and nearby sexually antagonistic variation, are selectively favored (Rice, 1987; Marshall Graves, 2008). However, the loss of recombination also triggers Muller’s ratchet, which contributes to the decay of many genes linked to the new sex-determination allele (Muller, 1964; Blaser et al., 2012). This decay may gradually reduce the fitness for carriers of the new sex chromosome, allowing new mutations in the sex-determination network to begin the cycle anew, a process that has been dubbed the ‘hot-potato model’ (Blaser et al., 2014). To better understand this process, there is a need to characterize the patterns of decay during the earliest stages of sex chromosome divergence.
The adaptive radiations of African cichlids have created diversity in trophic morphology, behavior, pigmentation and sensory systems (Kocher, 2004). Recently, it has been discovered that African cichlids also harbor a diverse array of sex chromosomes. Among haplochromine cichlid species, distinct XY systems have been discovered on linkage group (LG) 7, LG18, LG20 and on a fusion of LG5 and LG14, while a ZW system has been discovered on LG5, and an XYW system found on LG13 (Roberts et al., 2009, 2016; Ser et al., 2010; Parnell & Streelman, 2013; Böhne et al., 2016; Peterson et al., 2017). Some species segregate multiple sex chromosome systems simultaneously (Ser et al., 2010). An additional study from three tribes of Lake Tanganyikan cichlids has identified a ZW system on LG7, an XY system on LG19 and a ZW system on LG5 that is possibly convergent with a ZW system on LG5 in Lake Malawi (Roberts et al., 2009; Gammerdinger et al., 2018).
Deeper lineages of the Pseudocrenilabrinae segregate several additional sex chromosome systems, including XY systems on LG1 and LG23 and a ZW system on LG3. The causative sex-determination gene on LG23 is a duplication of anti-Müllerian hormone, which is known to play an important role in the vertebrate sex-determination network (Eshel et al., 2014; Li et al., 2015). Throughout Pseudocrenilabrinae, multiple sex-determination loci sometimes segregate within a given genus or even within a given species (Lee et al., 2004; Cnaani et al., 2008; Ser et al., 2010). For example, some populations of the blue tilapia, Oreochromis aureus (Steindachner), simultaneously segregate both a LG1 XY system and a LG3 ZW system (Lee et al., 2004).
In this study, we provide a more detailed characterization of sex chromosomes from deeper lineages of the Pseudocrenilabrinae (Figure 1). We analyzed the sex chromosome systems of three species: Oreochromis mossambicus (Peters), Coptodon zillii (Gervais) and Pelmatolapia mariae (Boulenger). Previous research suggested that O. mossambicus has either a LG1 XY system or a LG3 ZW system, while C. zillii only has a LG1 XY system and P. mariae only has a LG3 ZW system (Cnaani et al., 2008). However, this previous study used a small number of microsatellite markers to characterize these systems and thus could not characterize sequence divergence between the sex chromosomes in each species (Cnaani et al., 2008).
Figure 1.
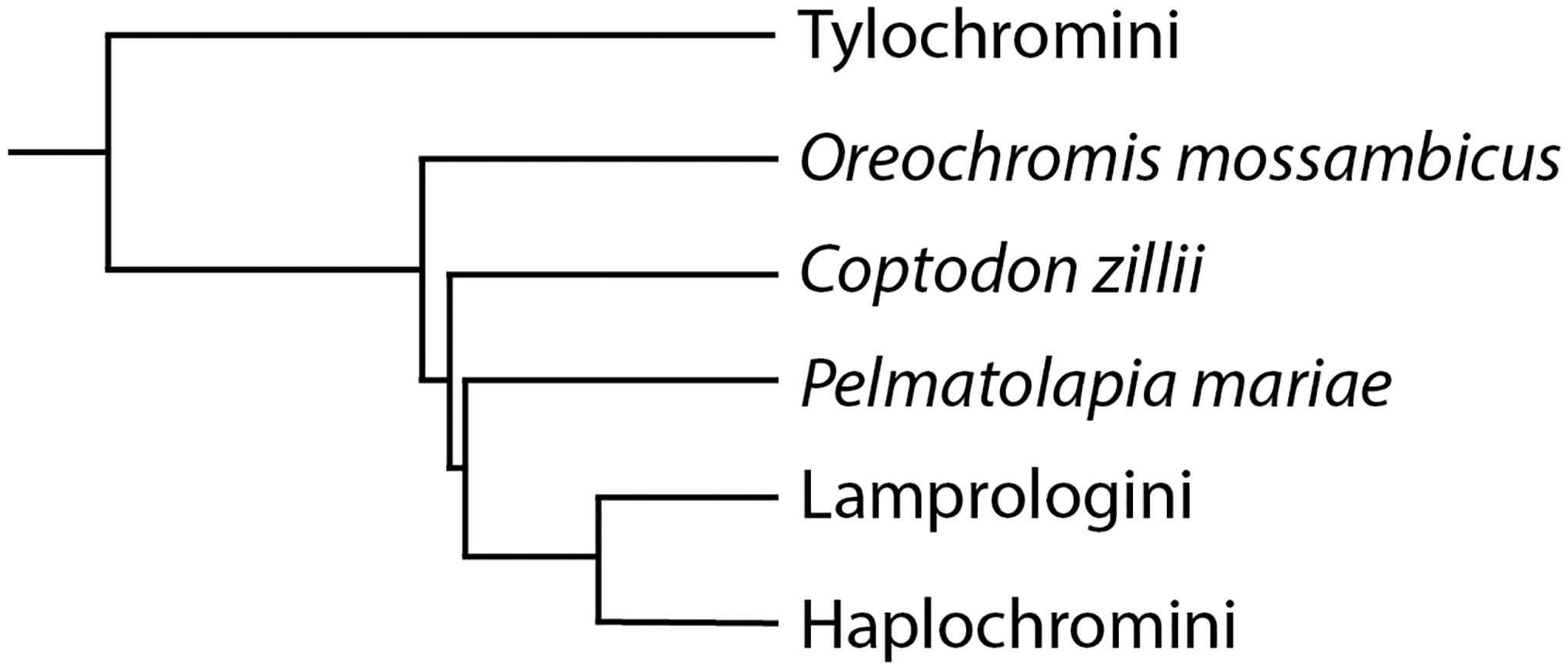
Phylogeny of the species studied showing the relationships to other members of Pseudocrenilabrinae. Phylogeny adapted from Dunz and Schliewen, 2013 (Dunz & Schliewen, 2013).
Materials and Methods
Materials
The O. mossambicus and P. mariae samples were from lab-reared fish purchased through the aquarium trade. The O. mossambicus stock likely came originally from South Africa. The C. zillii samples were derived from individuals originally collected in Lake Manzala, Egypt. Finclips were collected from each individual and their gonads were visually inspected to determine their sex. For O. mossambicus, we sampled two families (Family 1: 8M, 12 F; Family 2: 15M, 15F). We also sampled two families of P. mariae (Family 1: 25M, 21F; Family 2: 14M, 18F). The single family of C. zillii consisted of 9 males and 13 females.
Sequencing
DNA was purified from fin clips by phenol-chloroform extraction using phase-lock gel tubes (5Prime, Gaithersburg, Maryland). DNA concentrations for each sample were quantified using fluorescence spectroscopy and DNA pools for each sex were constructed with equal representation of each individual. Libraries were constructed using the TruSeq DNA PCR-Free LT Kit (Illumina, San Diego, California). For O. mossambicus and P. mariae, we pooled the two families of each species separately. Males and females from C. zillii and O. mossambicus each shared a lane of 100bp paired-end Illumina sequencing. Males and females from P. mariae each received a separate lane of 100bp paired-end Illumina sequencing.
Read mapping
Raw reads were de-multiplexed and filtered using the CASAVA 1.8 filter and low-quality reads that did not meet this criterion were discarded. The read qualities for each pool were visually inspected using FastQC version 0.11.2 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were aligned to the Oreochromis niloticus (O_niloticus_UMD_1) reference genome using BWA version 0.7.12 with default parameters and utilizing the read groups option (Li & Durbin, 2009; Conte et al., 2017). The alignments were sorted, marked for duplicates, and indexed using Picard version 1.119 (http://broadinstitute.github.io/picard/). Samtools was used to filter-out reads with an alignment quality of zero (Li et al., 2009). Then, the alignments from families of the same species were merged and indexed using Picard version 1.119. Coverage estimates were obtained across the genome and for each linkage group using the INTERVALS option within the CollectWgsMetrics program for both sexes of each species with Picard version 2.17.4 (http://broadinstitute.github.io/picard/).
Variant calling
Variants were called with GATK version 3.7 software package (DePristo et al., 2011). We used the HaplotypeCaller command of GATK for each sex within each species and set the sample ploidy (--sample_ploidy) to its maximum of 10 due to the pooled nature of our datasets. We followed this up with GATK’s GenotypeGVCF to combine the sexes for each of our species into a single output VCF file.
We also converted our alignment files into mpileup files using Samtools version 0.1.18 and subsequently into sync files with a PHRED score cutoff of 20 using Popoolation2 (Li et al., 2009; Kofler et al., 2011). Next, each species was run through Sex_SNP_finder_GA.pl (https://github.com/Gammerdinger/sex-SNP-finder) in order to identify XY and ZW sex-patterned SNPs, and to calculate FST using a minimum coverage threshold of 10 and a maximum coverage threshold of 50. We also quantified the density of these sex-patterned SNPs in non-overlapping 10kb windows and looked for an elongation of the high SNP density tail in the distributions of both the XY and ZW patterned SNPs. If one tail was more elongated than the other, we ran a Mann-Whitney U-test to compare the distributions using a significance threshold of 0.05. If there was a significant difference, then we would fit a negative binomial distribution to the distribution without the elongated tail, using the fitdistr function from the MASS library in R (Venables & Ripley, 2002; R Core Team, 2016), to create a null expectation for the distribution of non-overlapping, sex-patterned 10kb windows in a given species. Next, we calculated the top 0.1% of this fitted negative binomial using the qnbinom function in R and we applied this threshold to our distribution with the elongated tail to define regions of high differentiation (R Core Team, 2016). We filtered the VCF file of each species by the list of sex-patterned SNPs generated by Sex_SNP_finder_GA.pl and used Bedtools version 2.26.0 intersect command to extract the sex-patterned SNPs within regions of high differentiation (Quinlan & Hall, 2010; Gammerdinger et al., 2014, 2016).
Functional annotation
The functional significance of each sex-patterned SNP was evaluated using SnpEff version 4.3 (Cingolani et al., 2012b). Sex-patterned SNPs classified as “high-impact” or missense mutations were extracted using SnpSift (Cingolani et al., 2012a). Missense mutations were subsequently analyzed with PROVEAN version 1.1.5 to predict the functional impact of each missense mutation on their respective protein (Choi et al., 2012). We used the recommended PROVEAN score threshold of −2.5. Substitutions lower than this threshold were considered to be deleterious.
Results
Read mapping
For P. mariae, we obtained an 80.25% alignment rate for 22.96X coverage in the merged male pool and an 82.47% alignment rate for 19.41X coverage in the merged female pool. The sequencing from C. zillii returned an alignment rate of 77.59% for a coverage of 8.84X in males and an alignment rate of 78.74% for a coverage of 17.52X in females. From O. mossambicus, we retrieved a 79.28% alignment rate for 12.89X coverage in the merged male pool and a 79.35% alignment rate for 14.91X coverage in the merged female pool.
Differentiation in Oreochromis mossambicus
In order to differentiate if the system is an XY or ZW system, and to define the regions of high differentiation, we needed to compare the XY and ZW distributions of the non-overlapping, 10kb windows with sex-patterned SNPs. The tail of the XY distribution was much larger than the tail of the ZW distribution, and the XY distribution is shifted significantly higher than the ZW distribution (Supplementary File 1; p < 0.05). The top 0.1% of the negative binomial distribution fit to the ZW distribution corresponded to 13 or more sex-patterned SNPs per non-overlapping, 10kb window. The corresponding regions of high XY differentiation encompass ~5.1Mb of the genome. Approximately 2.6Mb are found on LG14 and ~890kb on LG3b (Figure 2). No other linkage group or unanchored contig contained more than 200kb of highly differentiated regions. These regions of high differentiation overlap with the regions that show a strong signal of FST differentiation. The regions of differentiation on LG14 are tightly clustered almost entirely within the first 10Mb (Figure 3). We identified 69 genomic missense mutations, 41 on the Y-chromosome and 28 on the X-chromosome, that are responsible for 112 transcript missense mutations within the regions of high differentiation. PROVEAN predicted that 27 of these 112 transcript missense mutations are deleterious. Also, SnpEff predicted three mutations, two on the Y-chromosome and one on the X-chromosome, to be “high-impact” mutations. A complete list of all missense, predicted deleterious missense and “high-impact” SNPs along with a complete list of the genes found in the regions of high differentiation in O. mossambicus can be found in Supplementary File 2.
Figure 2.
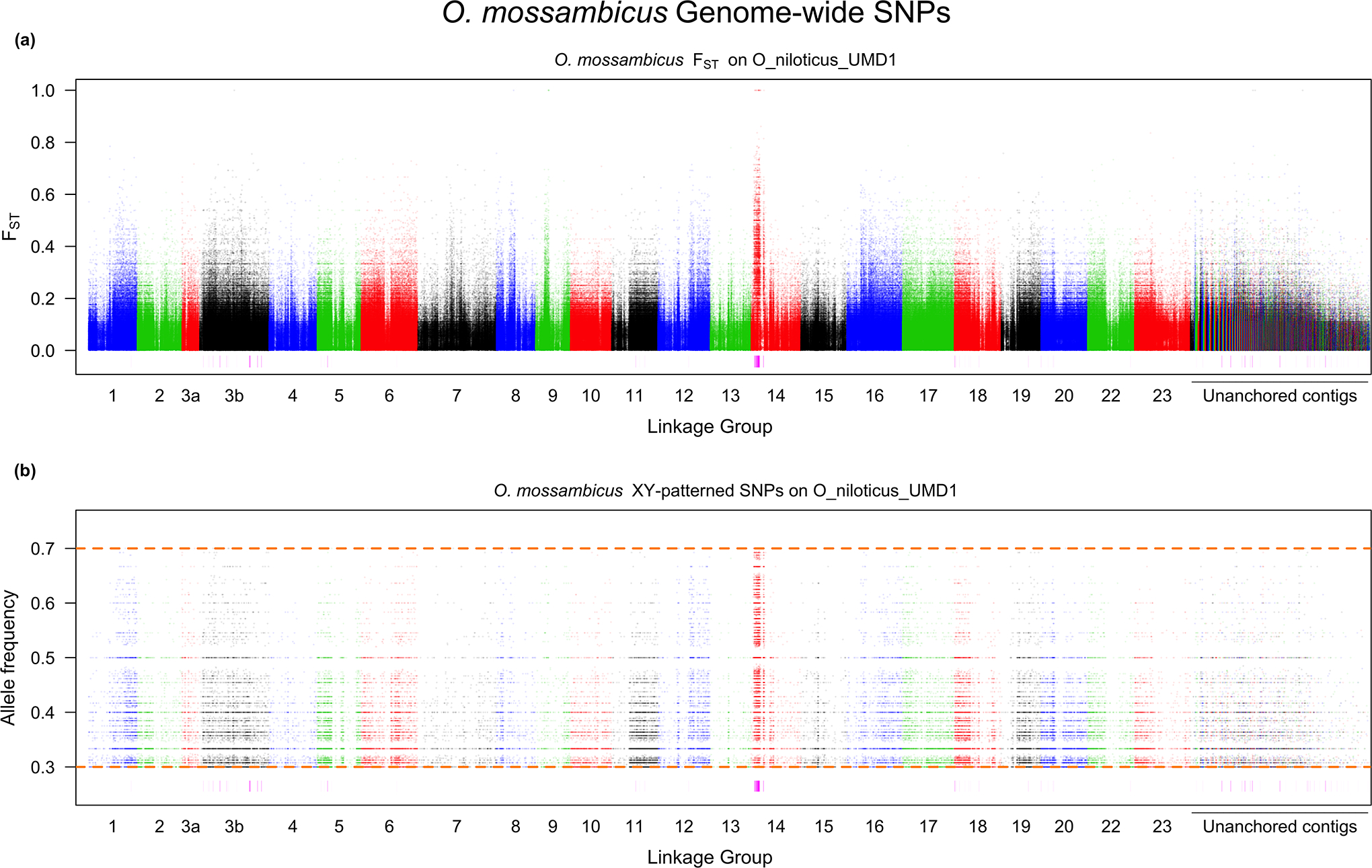
Whole genome scan of O. mossambicus for a) FST and b) the allele frequency of the Y SNP in the male pool. The rectangles underneath each figure represent regions of high differentiation.
Figure 3.
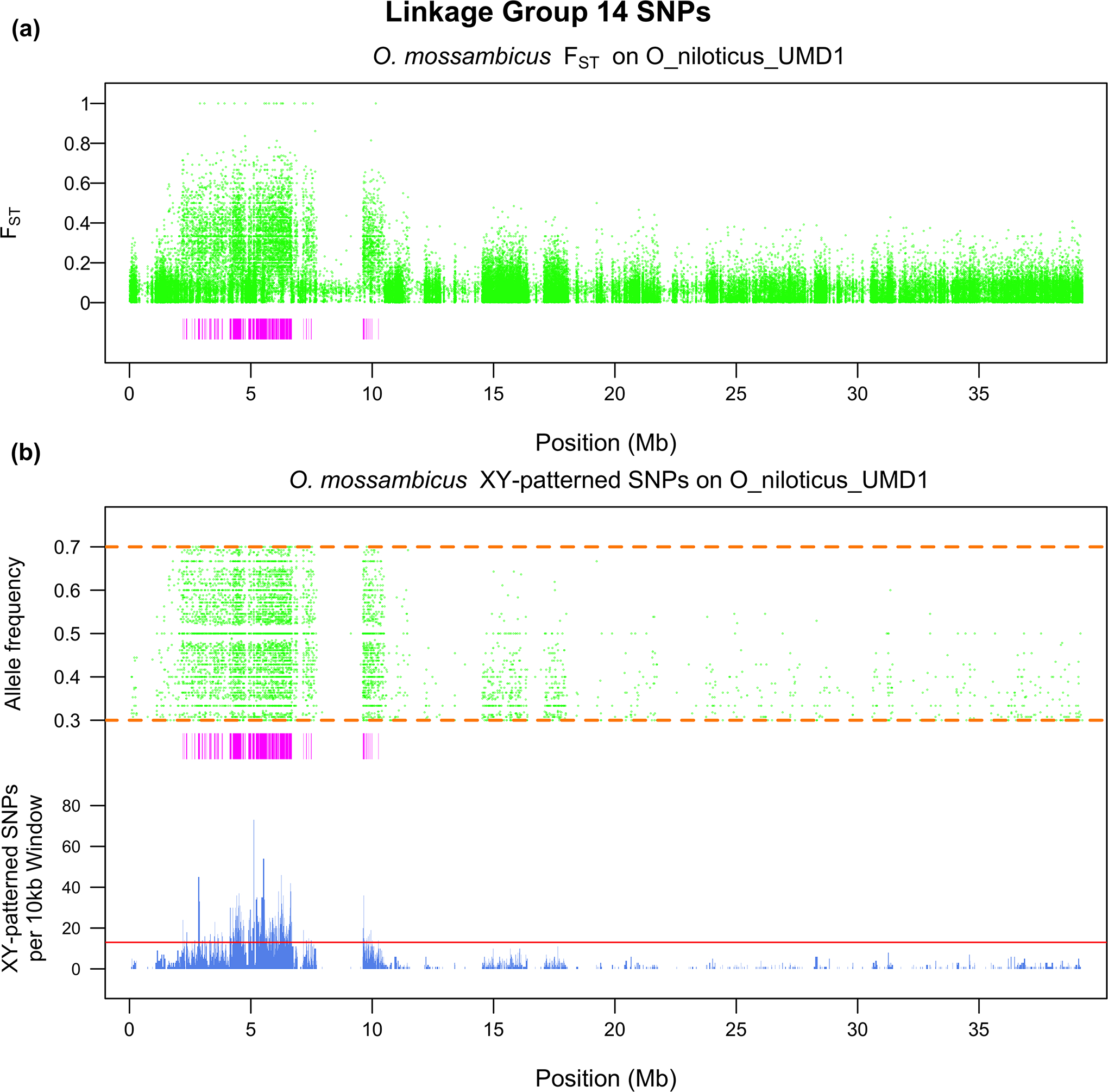
LG14 scan of O. mossambicus for a) FST and b) the allele frequency of the Y SNP in the male pool. The rectangles underneath each figure represent regions of high differentiation. The bars underneath panel (b) represent the number of XY-patterned SNPs in each non-overlapping, 10kb window and the horizontal line represents the threshold for assigning a region as a region of high differentiation.
Differentiation in Coptodon zillii
The tail of the distribution of non-overlapping, 10kb windows containing XY sex-patterned SNPs was significantly larger than the tail of the ZW distribution in C. zillii (p < 0.05) (Supplementary File 3). The threshold derived from the top 0.1% of the negative binomial distribution fit to the ZW dataset was 14 or more sex-patterned SNPs per non-overlapping, 10kb window. Using this threshold, ~13.8 Mb of the genome falls within a region of high XY differentiation. Approximately 7.9Mb of this corresponds to LG1, with an additional ~1.1Mb on LG22 and ~728kb on LG3b (Figure 4). While there were many other small regions of differentiation scattered across the genome, no other linkage group or unanchored contig contained more than 350kb of highly differentiated regions. These regions of high differentiation explain the elevated FST on LG1 relative to the rest of the genome, and this differentiation corresponds to one end of the linkage group (Figure 5). There are 265 genomic sex-patterned mutations, 132 on the Y-chromosome and 133 on the X-chromosome, creating 498 transcript missense mutations within the regions of high differentiation. PROVEAN predicts that 92 of the 498 missense mutations are deleterious. Lastly, a single mutation on the X-chromosome caused a “high-impact” mutation within this region of high differentiation. This mutation, along with a complete list of the missense mutations, predicted deleterious missense mutations and the genes in the regions of high differentiation in C. zillii, can be found in Supplementary File 2.
Figure 4.
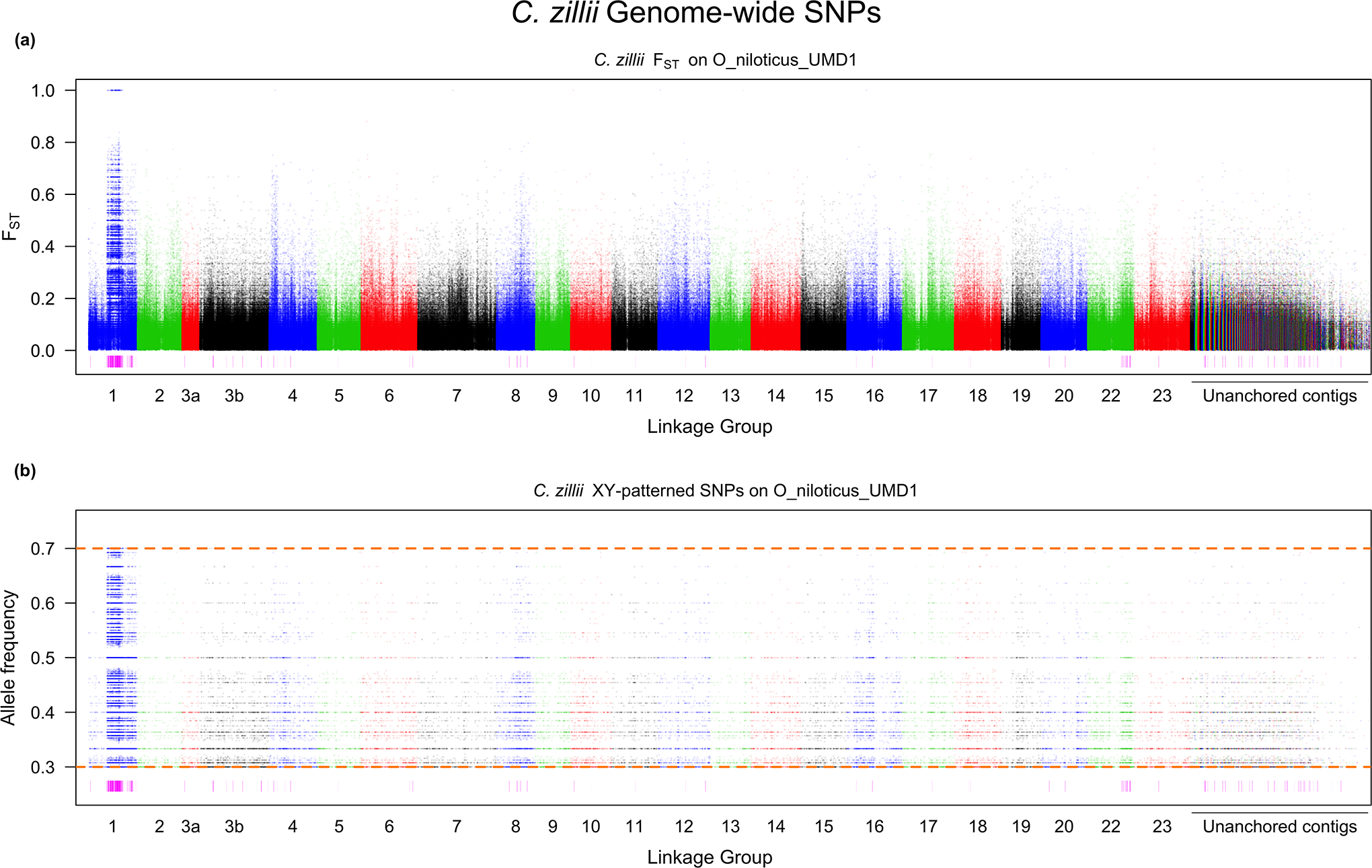
Whole genome scan of C. zillii for a) FST and b) the allele frequency of the Y SNP in the male pool. The rectangles underneath each figure represent regions of high differentiation.
Figure 5.
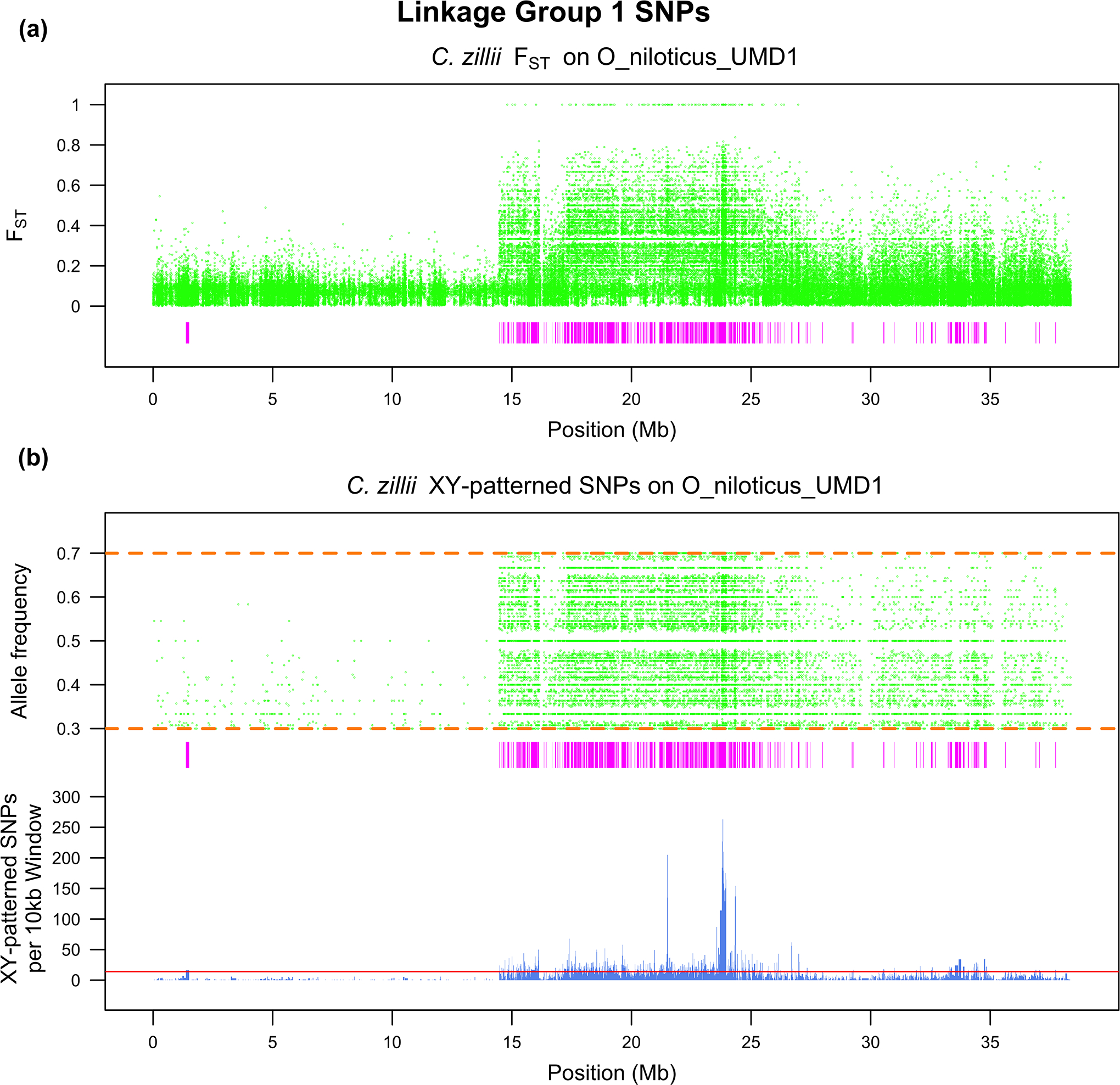
LG1 scan of C. zillii for a) FST and b) the allele frequency of the Y SNP in the male pool. The rectangles underneath each figure represent regions of high differentiation. The bars underneath panel (b) represent the number of XY-patterned SNPs in each non-overlapping, 10kb window and the horizontal line represents the threshold for assigning a region as a region of high differentiation.
Differentiation in Pelmatolapia mariae
Analysis of P. mariae revealed a high level of both XY and ZW signal. We detected 82,937 sex-patterned SNPs in the XY direction and 80,945 in the ZW direction. Neither tail for the distributions of non-overlapping, sex-patterned, 10kb windows appeared more elongated than the other (Supplementary File 4). Figure 6 and 7 show that LG3b has a high level of FST differentiation. There are 18,974 sex-patterned XY SNPs and 20,718 sex-patterned ZW SNPs on LG3b. We did not attempt to identify regions of high differentiation in this species for two reasons: (1) neither tail appeared to be elongated relative to the other and (2) there was a large excess of sex-patterned XY SNPs relative to what would have been expected given the results of a previous microsatellite study (Cnaani et al., 2008).
Figure 6.
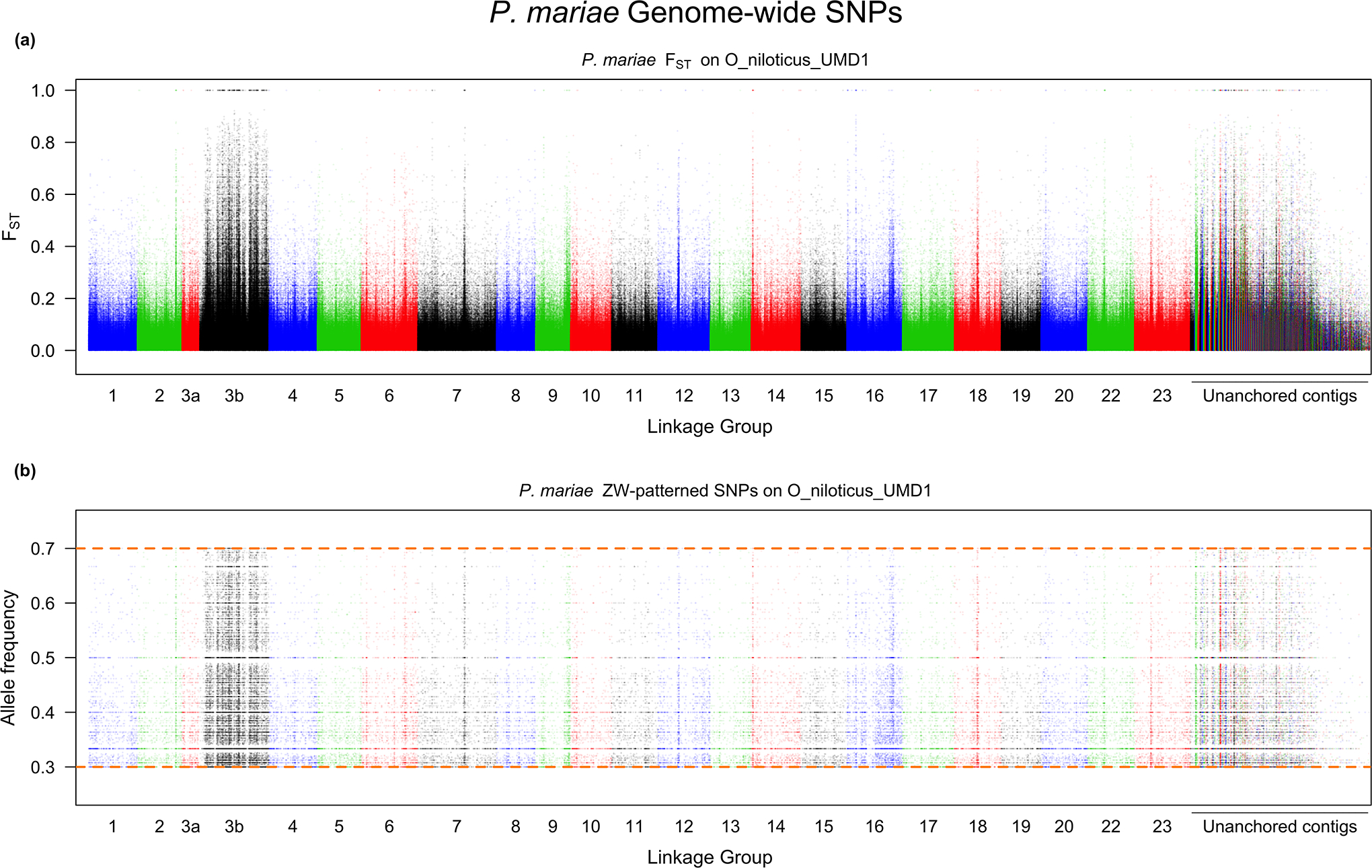
Whole genome scan of P. mariae for a) FST and b) the allele frequency of the W SNP in the female pool.
Figure 7.
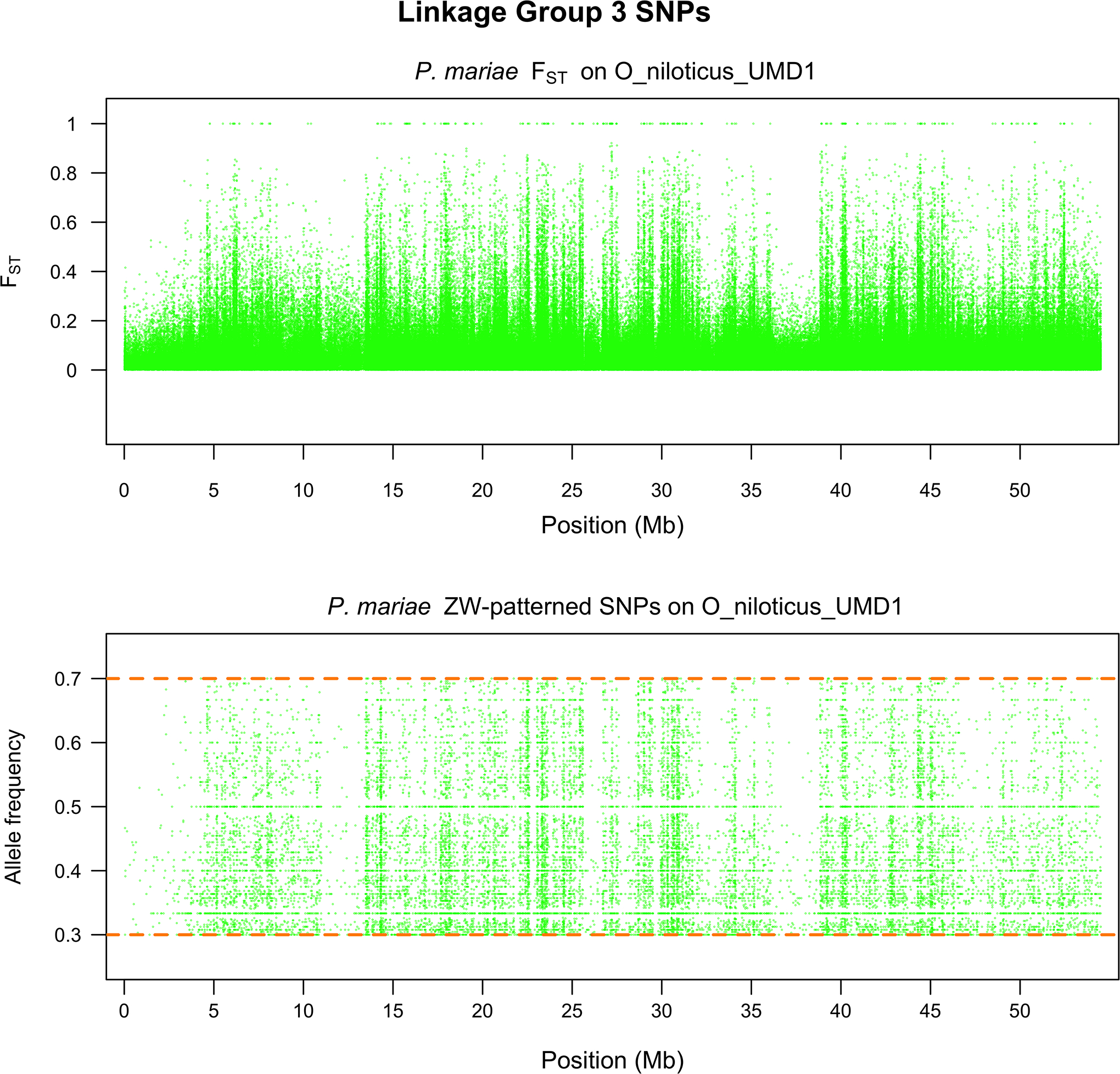
LG3b scan of P. mariae for a) FST and b) the allele frequency of the W SNP in the female pool.
Additionally, our analysis of coverage from each linkage group revealed that LG3a and LG3b consistently contained the lowest mean coverage, and the highest standard deviation in coverage, in both sexes for all of the species analyzed (Supplementary File 5). An example of this alignment problem is visualized in Supplementary File 6, which compares a region on LG3b to a similarly sized region on LG6 and illustrates how reads align much better to LG6 than LG3b.
Discussion
In this study, we used whole genome sequencing of pools of males and females to identify regions harboring a high density of sex-patterned SNPs in deeply divergent species of Pseudocrenilabrinae. Our results provide additional support to the narrative that sex chromosomes transitions occur at a rapid rate in cichlid fishes. However, they also illustrate the limits of our methodology and we propose solutions for moving forward.
LG14 in Oreochromis mossambicus
The high density of XY sex-patterned SNPs on LG14 likely indicates that this region of the genome harbors a novel sex-determination locus. Additionally, the lack of a bimodal distribution for the non-overlapping sex-patterned 10kb windows, as was observed in a recent study of Lake Tanganyikan cichlids, suggests that this novel sex chromosome system may be younger than its Lake Tanganyikan counterparts (Gammerdinger et al., 2018). This is further corroborated by its small size, which suggests it has not had sufficient time to accumulate structural rearrangements that expand the regions of high differentiation, a characteristic of older sex chromosome systems. Furthermore, there are relatively few genes that have accumulated predicted deleterious missense and “high-impact” mutations as would be expected in an established sex chromosome. Taken together, this evidence suggests this is a young sex chromosome.
Since LG14 has not been previously described as a sex chromosome in cichlid fishes, it could be that this system is epistatically recessive to the previously reported LG1 XY and LG3 ZW systems in other species of Oreochromini. Studies of O. aureus demonstrated a LG3 W that was epistastically dominant to a LG1 Y, and individuals who were ZZ at LG3 and XX at LG1 were a mix between males and females, suggesting at least one additional factor for controlling sex in Oreochromini (Lee et al., 2004). It could be that LG14 represents this additional factor, which may have been revealed in our strain if the LG3 W and LG1 Y were lost after several generations of lab rearing. Alternatively, there may be geographic variation in sex-determination in this species.
A candidate gene for sex-determination within this region on LG14 is AHNAK, which has a “high-impact” SNP altering a splice acceptor site on the X-chromosome. AHNAK is involved in a multitude of cellular processes including the regulation of calcium channels, cell signaling and membrane repair. But it also interacts with the TGF-β pathway as a tumor suppressor gene (Davis et al., 2014; Lee et al., 2014). The TGF-β pathway has been previously implicated to play a critical role in sex-determination by modulating the germ cell count in the bipotential gonad (Kikuchi & Hamaguchi, 2013). It has been observed that bipotential gonads with fewer germ cells develop into testes, while bipotential gonads with more germ cells develop into ovaries (Kikuchi & Hamaguchi, 2013). Because AHNAK acts as a tumor suppressor for the TGF-β pathway, a SNP that disrupts this function on the X-chromosome could promote more germ cell growth in XX versus XY individuals. If this is true, the XX individuals with two mutant copies of AHNAK would develop into females and XY individuals with only one mutant copy of AHNAK would develop into males. However, the pleiotropic effects of such a mutation might impose large fitness costs on individuals.
LG1 in Coptodon zillii
The high density of XY sex-patterned SNPs on LG1 is consistent with a previous report of an XY system on LG1 in C. zillii (Cnaani et al., 2008). Similar to O. mossambicus, we did not detect a bimodal distribution of 10kb windows with sex-patterned SNPs, but rather we observed an extended tail in the XY distribution, suggesting that the LG1 system could be younger than the sex chromosome systems found in Lake Tanganyikan cichlids (Gammerdinger et al., 2018). Additionally, it appears that during the sequencing process we did not sample equal proportions of male and female DNA and thus we under-sampled the male pool. Because we continued to maintain the minimum coverage threshold of 10 for our analysis, we have likely underestimated the number of sites that show a sex-patterned profile in this species. However, we do not think the true level of divergence between the X and Y in C. zillii is close to the divergence between the apparently older but still young sex chromosome systems we identified in Lake Tanganyikan cichlids (Gammerdinger et al., 2018).
Phylogenetic evidence suggests that the divergence between C. zillii and the composite group of O. niloticus and Sarotherodon melanotheron (Rüppell), which share a common LG1 XY system, predates the emergence of the Lake Tanganyikan cichlids (Meyer et al., 2015; Gammerdinger et al., 2016). The simplest explanation is that this system could represent multiple evolutions of LG1 XY sex-determination systems. Alternatively, this pattern could be explained if the LG1 system in these species is accumulating new mutations on the sex chromosomes at a slower rate than the sex chromosomes of some Lake Tanganyikan cichlids. It could be that the mechanisms that have reduced recombination in the Oreochromini and Coptodonini LG1 systems have emerged more recently than the mechanisms that reduced recombination in the Lake Tanganyikan cichlid sex chromosomes. As a result, the Lake Tanganyikan sex chromosomes systems would have accumulated sex-patterned, deleterious SNPs at a much more rapid rate and thus appear older than the LG1 XY system in the Oreochromini and Coptodini.
Since the C. zillii system overlaps with the region previously shown to be associated with sex in O. niloticus and S. melanotheron, the candidate genes for controlling sex from this region are the same as we discussed in previous work: Ras-related protein R-Ras2, Suppression of tumorigenicity 5 protein, Ras association domain-containing protein 10, AFG3-like protein 1, Wilms tumor protein homolog, Estrogen-related receptor gamma and Growth regulation by estrogen in breast cancer 1 (Gammerdinger et al., 2014, 2016).
Interestingly, a second, slightly less divergent region of differentiation centered around ~30Mb in O. niloticus is not present in C. zillii (Conte et al., 2017). Similarly, C. zillii seems to have a second, slightly less divergent block of differentiation around ~34Mb. We suggest that these regions represent independent evolutionary strata in each lineage. Additionally, S. melanotheron shows widespread differentiation across this region (Gammerdinger et al., 2016). Theory suggests that a region harboring hypothetical sexually antagonistic alleles can remain in linkage disequilibrium with a sex-determination locus, and that the intervening region can show less differentiation than the differentiation at either locus (Kirkpatrick & Guerrero, 2014). However, given the distance between the sex-determination region and the hypothetical sexually antagonistic locus in C. zillii, there would need to be very high levels of sexual antagonism to keep these regions in linkage disequilibrium. Therefore, it is more likely that the region around ~34Mb represents a structural rearrangement that has captured a new region of LG1 and created a less differentiated stratum in C. zillii.
LG3 in Pelmatolapia mariae
Previous research suggested that P. mariae has a ZW sex chromosome system on LG3 (Cnaani et al., 2008). Our FST data (Figure 6) also clearly indicate the presence of a sex chromosome system on LG3. However, our data also show a large number of XY patterned SNPs which make it difficult to confidently call regions of high differentiation. This ‘noise’ is likely to be the result of several factors. First, LG3b harbors many repetitive blocks that make alignment of short reads difficult. While our read alignments were relatively even on other linkage groups, we had relatively poor alignments in both sexes on LG3b, with reads piling up in some locations and absent in others (Supplementary Files 5 and 6). It could be that repetitive reads originating from non-homologous regions are piling up in this alignment create an artifact that resembles an XY signal. Second, it could be that the Z-chromosome in P. mariae harbors a high level of polymorphism and thus creates spurious XY signal. Third, and likely most importantly, it could be that our LG3 sequence is simply too diverged from the reference genome to effectively align short reads. We were able to align these short reads to the rest of the genome as seen in Supplementary Files 5 and 6, but the reference sequence for this linkage group appears to be particularly problematic. It seems that our approach for finding sex-patterned SNPs has limitations that prevent us from determining whether it represents an XY or ZW signal; however, we are confident that our samples have a sex determination system on LG3b because of the high level of FST divergence between males and females on LG3b. The approach we have developed in this and previous studies seems effective at defining sex chromosomes in their earliest stages, but breaks down when sequences diverge strongly from the reference sequence. Longer sequencing read technologies and a better reference sequence will likely facilitate future studies of the LG3 ZW system on this sex chromosome pair. Lastly, this sex chromosome system, along with the other sex chromosome systems described here, could be strain-specific.
Conclusions
We report and quantify the decay of a novel XY sex chromosome system on LG14 in O. mossambicus. The discovery of this new sex chromosome system continues to illustrate the incredible diversity of cichlid sex chromosomes and argues for more studies to quantify their rich variety. Additionally, we confirm a previous study that reported a XY system on LG1 in C. zillii and we quantified the level of decay on this chromosome. We were unable to distinguish the presumed ZW system from an XY system in P. mariae, likely due to both technical and biological issues. We suggest refinements to our approach, such as longer reads and a more closely related reference assembly of LG3, which should allow for the characterization of this sex chromosome in future studies.
Supplementary Material
Supplementary File 2. A list of all sex-patterned missense mutations, deleterious missense mutations, “high-impact” mutations and genes within the regions of high differentiation in both O. mossambicus and C. zillii.
Supplementary File 1. Distribution of sex-patterned windows both the XY and ZW direction for O. mossambicus. The vertical red line denotes the threshold of 13 sex-patterned SNPs per non-overlapping 10kb window.
Supplementary File 3. Distribution of sex-patterned windows both the XY and ZW direction for C. zillii. The vertical red line denotes the threshold of 14 sex-patterned SNPs per non-overlapping 10kb window.
Supplementary File 4. Distribution of sex-patterned windows both the XY and ZW direction for P. mariae.
Supplementary File 5. Mean and standard deviations for coverage on each linkage group in both sexes for each species.
Supplementary File 6. Coverage in P. mariae from males and females on (a) LG3 and (b) LG6.
Acknowledgements
The authors acknowledge the University of Maryland supercomputing resources (http://www.it.umd.edu/hpcc) made available for conducting the research reported in this paper. Samples were processed in accordance with University of Maryland IACUC protocol #R-10-73.
Funding
This work was supported by the National Science Foundation under Grant Number DEB-1143920; U.S. Department of Education Graduate Assistance in Areas of National Need fellowship offered through the University of Maryland Biology Department under Award Number P200A150160; and the American Genetics Association through an Ecological, Evolutionary and Conservation Genomics Award.
Footnotes
This paper has not been submitted elsewhere in identical or similar form, nor will it be during the first three months after its submission to Hydrobiologia.
Data Availability
All reads have been deposited in the NCBI SRA under Bioproject accession number PRJNA432420.
References
- Abbott JK, Nordén AK & Hansson B, 2017. Sex chromosome evolution: historical insights and future perspectives. Proceedings of the Royal Society B: Biological Sciences 284:20162806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser O, Grossen C, Neuenschwander S & Perrin N, 2012. Sex-chromsome turnovers induced by deleterious mutation load. Evolution 67:635–645. [DOI] [PubMed] [Google Scholar]
- Blaser O, Neuenschwander S & Perrin N, 2014. Sex-chromosome turnovers: the hot-potato model. The American Naturalist 183:140–146. [DOI] [PubMed] [Google Scholar]
- Böhne A, Wilson CA, Postlethwait JH & Salzburger W, 2016. Variations on a theme: Genomics of sex determination in the cichlid fish Astatotilapia burtoni. BMC Genomics 17:883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S et al. , 2012. Predicting the functional effect of amino acid substitutions and indels. PloS One 7:e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Patel VM, Coon M et al. , 2012.a. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Frontiers in Genetics 3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL et al. , 2012.b. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnaani A, Lee B-Y, Zilberman N et al. , 2008. Genetics of sex determination in tilapiine species. Sexual Development 2:43–54. [DOI] [PubMed] [Google Scholar]
- Conte MA, Gammerdinger WJ, Bartie KL et al. , 2017. A high quality assembly of the Nile tilapia (Oreochromis niloticus) genome reveals the structure of two sex determination regions. BMC Genomics 18:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Marin R, Toledo-Flores D et al. , 2014. Origins and functional evolution of Y chromosomes across mammals. Nature 508:488–493. [DOI] [PubMed] [Google Scholar]
- Davis TA, Loos B & Engelbrecht AM, 2014. AHNAK: The giant jack of all trades. Cellular Signalling 26:2683–2693. [DOI] [PubMed] [Google Scholar]
- DePristo M, Banks E, Poplin R et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunz AR & Schliewen UK, 2013. Molecular phylogeny and revised classification of the haplotilapiine cichlid fishes formerly referred to as “Tilapia.” Molecular Phylogenetics and Evolution 68:64–80. [DOI] [PubMed] [Google Scholar]
- Eshel O, Shirak A, Dor L et al. , 2014. Identification of male-specific amh duplication, sexually differentially expressed genes and microRNAs at early embryonic development of Nile tilapia (Oreochromis niloticus). BMC Genomics 15:774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble T, 2010. A review of sex determining mechanisms in geckos (Gekkota: Squamata). Sexual Development 4:88–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammerdinger WJ, Conte MA, Acquah EA et al. , 2014. Structure and decay of a proto-Y region in tilapia, Oreochromis niloticus. BMC Genomics 15:975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammerdinger WJ, Conte MA, Baroiller J-F et al. , 2016. Comparative analysis of a sex chromosome from the blackchin tilapia, Sarotherodon melanotheron. BMC Genomics 17:808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammerdinger WJ, Conte MA, Sandkam BA et al. , 2018. Novel sex chromosomes in three cichlid fishes from Lake Tanganyika. Journal of Heredity 1:12. [DOI] [PubMed] [Google Scholar]
- Kikuchi K & Hamaguchi S, 2013. Novel sex-determining genes in fish and sex chromosome evolution. Developmental Dynamics 242:339–353. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M & Guerrero R, 2014. Signatures of sex-antagonistic selection on recombining sex chromosomes. Genetics 197:531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher TD, 2004. Adaptive evolution and explosive speciation: the cichlid fish model. Nature Reviews Genetics 5:288–298. [DOI] [PubMed] [Google Scholar]
- Kofler R, Pandey RV & Schlötterer C, 2011. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics 27:3435–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B-Y, Hulata G & Kocher TD, 2004. Two unlinked loci controlling the sex of blue tilapia (Oreochromis aureus). Heredity 92:543–549. [DOI] [PubMed] [Google Scholar]
- Lee IH, Sohn M, Lim HJ et al. , 2014. Ahnak functions as a tumor suppressor via modulation of TGFβ/Smad signaling pathway. Oncogene 33:4675–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H & Durbin R, 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Sun Y, Zhao J et al. , 2015. A tandem duplicate of anti-müllerian hormone with a missense SNP on the Y Chromosome is essential for male sex determination in Nile Tilapia, Oreochromis niloticus. PLoS Genetics 11:e1005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall Graves JA, 2008. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annual Review of Genetics 42:565–586. [DOI] [PubMed] [Google Scholar]
- Meyer BS, Matschiner M & Salzburger W, 2015. A tribal level phylogeny of Lake Tanganyika cichlid fishes based on a genomic multi-marker approach. Molecular Phylogenetics and Evolution 83:56–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, 1964. The relation of recombination to mutational advance. Mutation Research 1:2–9. [DOI] [PubMed] [Google Scholar]
- Myosho T, Otake H, Masuyama H et al. , 2012. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191:163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell NF & Streelman JT, 2013. Genetic interactions controlling sex and color establish the potential for sexual conflict in Lake Malawi cichlid fishes. Heredity 110:239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson EN, Cline ME, Moore EC et al. , 2017. Genetic sex determination in Astatotilapia calliptera, a prototype species for the Lake Malawi cichlid radiation. The Science of Nature 104:41. [DOI] [PubMed] [Google Scholar]
- Quinlan AR & Hall IM, 2010. Genome analysis BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2016. A language and environment for statistical computing. R Foundation for Statistical Computing [Google Scholar]
- Rice WR, 1987. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution 41:911–914. [DOI] [PubMed] [Google Scholar]
- Roberts NB, Juntti SA, Coyle KP et al. , 2016. Polygenic sex determination in the cichlid fish, Astatotilapia burtoni. BMC Genomics 17:835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RB, Ser JR & Kocher TD, 2009. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science 326:998–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ser JR, Roberts RB & Kocher TD, 2010. Multiple interacting loci control sex determination in Lake Malawi cichlid fish. Evolution 64:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables WN & Ripley BD, 2002. Modern Applied Statistics with S, Fourth. Springer, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 2. A list of all sex-patterned missense mutations, deleterious missense mutations, “high-impact” mutations and genes within the regions of high differentiation in both O. mossambicus and C. zillii.
Supplementary File 1. Distribution of sex-patterned windows both the XY and ZW direction for O. mossambicus. The vertical red line denotes the threshold of 13 sex-patterned SNPs per non-overlapping 10kb window.
Supplementary File 3. Distribution of sex-patterned windows both the XY and ZW direction for C. zillii. The vertical red line denotes the threshold of 14 sex-patterned SNPs per non-overlapping 10kb window.
Supplementary File 4. Distribution of sex-patterned windows both the XY and ZW direction for P. mariae.
Supplementary File 5. Mean and standard deviations for coverage on each linkage group in both sexes for each species.
Supplementary File 6. Coverage in P. mariae from males and females on (a) LG3 and (b) LG6.
Data Availability Statement
All reads have been deposited in the NCBI SRA under Bioproject accession number PRJNA432420.


