Abstract
The death fold domain-containing protein PIDD1 has recently attracted renewed attention as a regulator of the orphan cell death-related protease, Caspase-2. Caspase-2 can activate p53 to promote cell cycle arrest in response to centrosome aberrations, and its activation requires formation of the PIDDosome multi-protein complex containing multimers of PIDD1 and the adapter RAIDD/CRADD at its core. However, PIDD1 appears to be able to engage with multiple client proteins to promote an even broader range of biological responses, such as NF-κB activation, translesion DNA synthesis or cell death. PIDD1 shows features of inteins, a class of self-cleaving proteins, to create different polypeptides from a common precursor protein that allow it to serve these diverse functions. This review summarizes structural information and molecular features as well as recent experimental advances that highlight the potential pathophysiological roles of this unique death fold protein to highlight its drug-target potential.
Keywords: caspases, cell cycle, cell death, p53
Death fold-containing proteins in cell death and inflammation
Cell death is an essential part of life in multi-cellular organisms and is critical for development, tissue homeostasis and host defence. Caspases are a class of cysteine-dependent and aspartate-specific endopeptidases that constitute major regulators of cell death and inflammation [1,2]. The activation of caspases can be induced by their dimerization in large signaling complexes such as the apoptosome (Caspase-9), the PIDDosome (Caspase-2) or different types of inflammasomes (CASP1/4/5) [3,4]. Once fully active, these apical caspases are primed to cleave key-substrates that also include but are not limited to distal effector proteins of the same family, such as Caspase-3 or Caspase-7 [5]. The assembly of these platforms is marked by homotypic interactions between conserved structural motifs containing so-called death folds (DF). DFs are six alpha-helical bundle-containing globular domains that form the structural core of death domains (DD), death effector domains (DED), caspase activation and recruitment domains (CARD), and pyrin domains (PYD) [3].
Structural and genomic analysis suggests that many of the networks involved in the innate immune system have evolved from early cell death inducers that contained such DFs [3,6]. During host-pathogen interaction, a plethora of pathways can trigger either cell death, cytokine release or the activation of inflammatory target gene expression [3]. Examples of those are pathogen-associated molecular patterns (PAMPs) converging on inflammasome-driven Caspase-1 activation, which promotes local inflammation through activation of the interleukins IL-1β and IL-18, as well as Gasdermin D (GSDMD)-mediated pore-formation and pyroptotic cell death [7,8]. Cytochrome c release into the cytoplasm is needed to activate Caspase-9 in the apoptosome, which is still mainly considered anti-inflammatory [9]. Another prominent example that underlines the close relationship between inflammatory signaling and cell death is the signaling cascade downstream of tumor necrosis factor receptor 1 (TNFR1) [10]. Ligand-binding first promotes the formation of the membrane-bound complex I to activate NF-κB signaling through TRADD, TRAF2 and RIPK1. Sustained signaling leads to endocytosis of the multimeric receptor complex and formation of the cytoplasmic complex II which recruits Caspase-8 through FADD to induce apoptosis. Simultaneously, complex II suppresses the inflammatory cell death modality known as necroptosis [3,10]. Finally, the death domain-containing protein PIDD1 [11,12], was long believed to either induce apoptosis by activation of Caspase-2 or cell survival via NEMO-dependent activation of NF-κB. Yet, selective triggers of PIDDosome activation remained elusive until recently [13].
The p53-induced death domain protein 1 (PIDD1)
Two groups described PIDD1 (aka, Leucin-Rich Repeat and Death Domain-Containing Protein, LRDD) as a pro-apoptotic protein in the year 2000 [11,12]. Lin et al. [12] reported that PIDD1 is involved in p53-mediated growth suppression in response to ionizing radiation. Similarly, Telliez et al. [11] identified PIDD1 in a bioinformatics screen for DD interactors of RIPK1. Its pro-apoptotic function in response to disturbances of cellular fitness was assigned to the death domain found at its C-terminal end [11]. Early studies suggested that upon genotoxic stress, PIDD1 kick-starts pro-inflammatory signaling via activation of the NF-κB pathway, before initiating mitochondrial cell death by activating Caspase-2 [14,15]. However, the fact that PIDD1 knockout mice failed to show a clear phenotype in response to DNA damage suggested that the pathophysiological context in which PIDD1 becomes relevant was not fully understood [16–18]. Indeed, recent evidence defines PIDD1 as a sensor of centrosome number and regulator of cellular ploidy [19–21].
Transcriptional control of PIDD1 expression
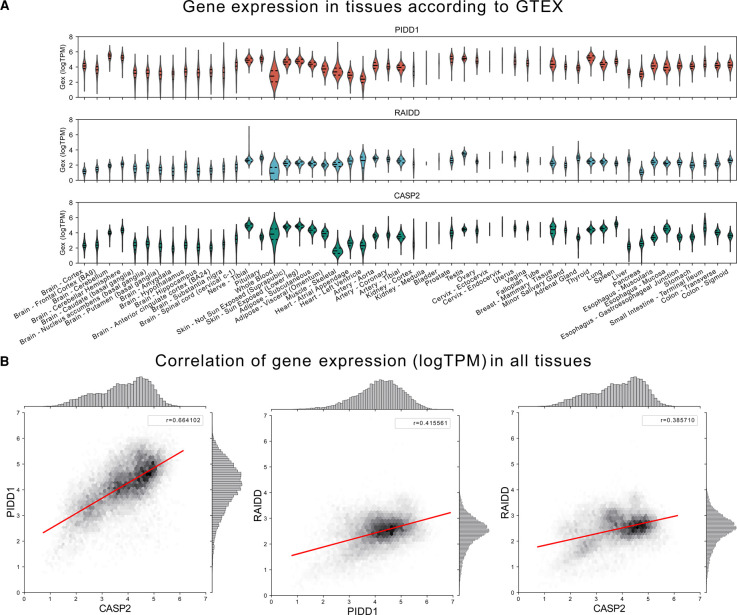
The human PIDD1 gene is located on chromosome 11p15.5. There are five potential transcript variants of which three have been confirmed to be expressed experimentally [22]. The PIDD1 gene promoter contains a p53 consensus sequence. In fact, PIDD1 expression is directly regulated by p53 and is strongly induced in response to γ-irradiation [12]. Furthermore, the E2F transcription factor family controls PIDD1 expression in mouse hepatocytes, and E2F1 overexpression induces PIDD1 mRNA levels in HeLa cells [23]. In neonatal mouse hepatocytes, the onset of weaning coincides with reduced E2F1/2 activity and increased expression of atypical and repressive E2F7/8, which promote hepatocyte polyploidization by interfering with cytokinesis [24,25]. The switch in transcription factor activity results in decreased PIDD1 and CASP2 expression [23]. Whether this regulatory mechanism is also linked to cell cycle progression, during which E2F levels oscillate significantly or if this regulation is specific to hepatocytes is not yet known. Generally, PIDD1 seems to be expressed weakly in most adult tissues in the mouse, with the highest expression levels in the kidneys, lung and spleen [12]. What controls base line expression of PIDD1 is currently unknown but protein is also detectable in the absence of p53 [22]. Interestingly, more recent GTEx datasets suggest that PIDD1, together with CASP2, is most strongly expressed in the brain, skin, small intestine, thyroid and testis, in addition to the aforementioned tissues (Figure 1A). Expression of RAIDD, the adapter protein linking PIDD1 and Caspase-2 via its DD and CARD, respectively, appears to be regulated differently, as its expression levels do not correlate well with the two other genes across tissues [23] (Figure 1B). So far, the PIDD1- and Caspase-2-independent functions of RAIDD have been rarely reported in the literature. Although initially suggested to be involved in TNF signaling [26], a critical role in this inflammatory pathway was not firmly established. However, others have suggested inhibitory roles in NF-kB signaling downstream of antigen receptors in T cells by interference with the CARMA1 signalosome [27].
Figure 1. Expression analysis of PIDDosome components, PIDD1, RAIDD, CASP2.
(A) Tissue-specific expression of H. sapiens PIDD1, RAIDD and CASP2 obtained from the Genotype-Tissue Expression (GTEx v8) project (https://www.gtexportal.org/). mRNA expression profiles are shown as log-transformed transcript per million (log(TPM)). (B) Correlation of PIDD1, RAIDD, and CASP2 gene expression (logTPM) in human tissues based on data from GTEx v8. Pearson's correlation coefficient shown as r-value.
PIDD1 structure, protein domain organization and auto-processing
Structurally, PIDD1 is defined by a C-terminal DD that exhibits a typical six alpha-helix bundle comparable to that found in other DD proteins [3,28]. In the years following its discovery, several additional structural features of PIDD1 were defined. On its N-terminus, the protein possesses seven leucine-rich repeats (LRRs). LRRs are often associated with pattern recognition of PAMPs or DAMPs, however, no ligand for PIDD1 has been identified to date. Following the LRRs, PIDD1 contains two ZU-5 domains and a poorly characterized UPA domain (domain found in UNC5, PIDD1 and ANKYRIN) [11,29].
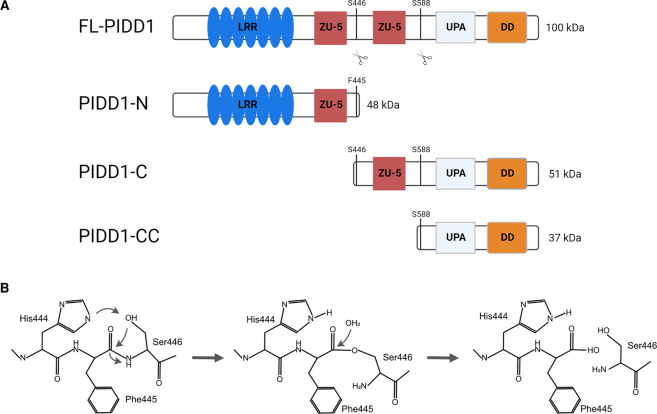
To become activated, the full-length precursor of PIDD1, FL-PIDD1, is post-translationally processed at two serine residues, generating an N-terminal fragment (48 kDa PIDD1-N) and two C-terminal fragments (51 kDa PIDD1-C and the 37 kDa PIDD1-CC) [11,30] (Figure 2A). This process depends on a tripeptide motif, His-Phe-Ser, and occurs in an autocatalytic manner, similar to what has been reported for the nuclear pore protein, NUP98 [30,31]. The respective serine — where cleavage occurs — initiates a nucleophilic attack on the preceding scissile amide bond. This process is initiated by the proximity of the side chains in His and Ser. Subsequently, the hydroxyl group induces a reversible N → O acyl shift, forming an intermediate with an ester, replacing the Phe-Ser peptide bond [32]. The ester intermediate is then attacked by a water molecule, and hydrolyzed (Figure 2B). Such autocatalytic processing has also been found in several other proteins with diverse functions such as inteins, Hedgehog, EMR2, N-terminal nucleophile family enzymes (NTNs) or, more recently, two proteins of the NLR-family, NLRP1 and CARD8 [30,33]. In all of these, the side chains of a Ser, Cys or Thr residue act as the nucleophile to initiate self-processing. Interestingly, while the histidine in position -2 in respect to the cleavage site is essential to initiate the second nucleophilic attack in PIDD1 and NUP98, it is exchanged for another serine in NLRP1 and CARD8 without impacting auto-processing [34]. Studies on NUP98 have shown that the conserved tripeptide motif lies within a hydrophobic pocket which is created by the amino acids upstream of the sequence motif. Thus, a longer N-terminal stretch is necessary to allow for cleavage, which may also apply for PIDD1 [35]. Notably, the N-terminus of PIDD1 is required for proper processing, as a Δ1–379 deletion mutant is unable to do so efficiently [30].
Figure 2. PIDD1 auto-processing.
(A) Full length PIDD1 (FL-PIDD1) is processed into three different fragments. Auto-proteolytic cleavage between F445 and S446 produces PIDD1-N (48 kDa) and PIDD1-C (51 kDa). Further cleavage at S588 forms the PIDD1-CC (37 kDa) fragment. (B) Outline of the chemical transitions needed for PIDD1 auto-proteolysis. The same mechanism is supposed to occur to generate PIDD1-C and PIDD1-CC, in a sequential manner (please see main text for further details).
Lessons from other death fold protein-containing complexes
Structurally, the PIDD1 C-terminus is closely related to two proteins of the NOD-like receptors (NLR), NLRP1 and CARD8, which both contain a C-terminal FIIND (function to find) domain that resembles the ZU-5 — UPA motif in PIDD1 [36,37]. NLRP1-mediated Caspase-1 activation depends on homotypic CARD — CARD interaction between the NLRP1 C-terminus and the adapter protein ASC which recruits Caspase-1 into the assembling inflammasome [8,33]. Interestingly, NLRP1, similar to PIDD1, is constitutively auto-processed in its FIIND domain to release the biologically active C-terminus [37], raising the question how sustained caspase activation is prevented. It seems that the N- and the C-terminal fragments of NLRP1 remain associated after cleavage, thereby sequestering the CARD domain in an inhibitory complex that prevents the initiation of inflammasome assembly [38,39]. Elegant studies have established that triggers of the NLRP1-inflammasome, including the B. subtilis toxin Anthrax Lethal Factor and the DPP8/9 inhibitor Val-boro-pro, act by the disruption of this inhibitory complex and subsequent release of the active NLRP1 C-terminus [40,41].
In addition, the domain organization of the PIDD1 C-terminus is closely related to the UNC5 family of netrin-receptors and to ankyrins such as ANK2. UNC5 proteins are a family of transmembrane receptors which can bind netrin- and netrin-related proteins. Their intracellular domain is composed of a ZU-5 — UPA — DD (ZUD) module, similar to the one found on the PIDD1 C-terminus. UNC5 receptors are widely expressed during neural development where they are involved in axonal guidance and cell migration [42] and function as dependence receptors with additional roles in apoptosis and tumorigenesis [43]. Interestingly, Wang et al. [29] established that within this ZUD module, the UPA and DD fold back onto the ZU-5 domain, thereby sequestering the DD in an inhibitory state and preventing the induction of apoptosis. ANK2, which contains a similar ZU-5 — ZU-5 — UPA — DD (ZZUD) structure, forms the same supra-module that sequesters its DD [44]. In ANK2, the UPA — DD interact with the first of the two ZU-5 domains, which corresponds to the ZU-5 domain present in PIDD1-N.
The high similarity of the PIDD1 C-terminus suggests, that such a mechanism might be relevant for PIDDosome activation, especially, since a dominant negative effect of the LRR in PIDD1-N on NF-κB activation has been reported [30]. This is further substantiated by co-immunoprecipitation experiments showing a direct interaction between PIDD1-N and PIDD1-C [20,30], which coincidentally impairs binding to other interaction partner. Although the mechanism through which the LRR in PIDD1 exerts its inhibitory effect is unknown, the features mentioned here are clearly reminiscent of the inhibitory complexes described earlier for NLRP1/CARD8, UNC5 or ANK2.
It therefore appears plausible that a PIDD1-C or -CC molecule is held in an inactive state by binding to the LRR of PIDD1-N until a specific trigger disrupts this interaction. One such trigger might be the phosphorylation of PIDD1 in its DD, which is required for RAIDD binding upon DNA damage [45,46]. However, a specific trigger for PIDDosome formation, e.g. in response to supernumerary centrosomes is still elusive.
PIDD1-containing protein complexes
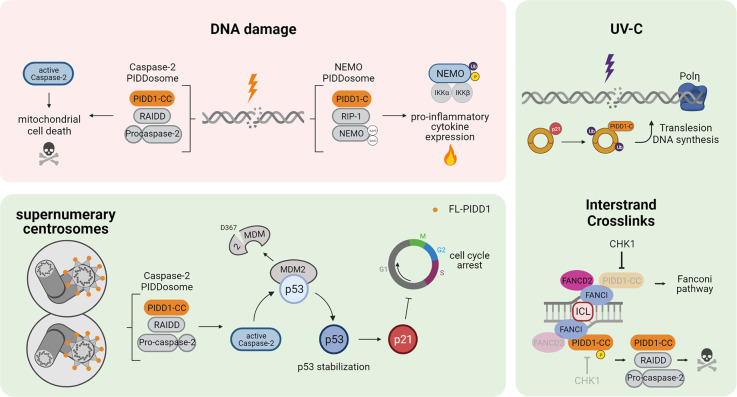
The most prominent functions of PIDD1 are associated with its ability to nucleate protein complexes through its DD. Initial studies speculated about a host of different DD-containing interaction partners of PIDD1, of which only RAIDD/CRADD and the kinase RIPK1, both carrying a DD, were ultimately confirmed [11,15]. In support of their integration into multi-protein signaling complexes, they co-elute with PIDD1 in high-molecular weight fractions which have been dubbed to correspond to two distinct PIDDosome complexes, namely a complex consisting of PIDD1-CC, RAIDD and Caspase-2 (the ‘Caspase-2-PIDDosome’) and a complex consisting of PIDD1-C, RIPK1 and NEMO/IKKγ (the ‘NEMO-PIDDosome’) [11,13–15]. In addition to these two complexes several PIDD1 interactors have been identified in mass spectrometry analyses, including PCNA [47] and FANCI [46], which engage with PIDD1 in different settings based on other structural features, namely the distal ZU-5 and the distal ZU-5 and UPA domain, respectively (Figure 3). Furthermore, PIDD1 has also been detected in the nucleolus, where it interacts with nucleophosmin 1 (NPM1) via its N-terminal LRRs [48].
Figure 3. Biological functions of PIDD1-containing protein complexes.
DNA damage is reported to lead to activation of the Caspase-2 — and NEMO — PIDDosome, resulting in either mitochondrial cell death or pro-inflammatory NF-kB activation and cytokine expression. In response to supernumerary centrosomes, PIDDosome formation leads to p53 stabilization and p21-dependent cell cycle arrest. Under specific circumstances, e.g. after UV irradiation, PIDD1 associates with PCNA to promote Polη-mediated translesion DNA synthesis. Finally, interstrand DNA crosslinks (ICLs) that cannot be resolved in time trigger the formation of the Caspase-2 — PIDDosome via recruitment of PIDD1-CC to FANCI.
The Caspase-2-PIDDosome
In the Caspase-2-PIDDosome five PIDD1-CC molecules interact with seven RAIDD molecules, which in turn recruit the same number of Caspase-2 molecules [28]. Complex formation leads to the proximity-induced activation of Caspase-2. Careful evaluations of the crystal structure of the DDs of PIDD1 and RAIDD in complex imply an asymmetric assembly mechanism where all of the components have their distinct positions and interaction spectrum. This data also provides a hint to the necessity for PIDD1-CC in the PIDDosome complex since any of the other fragments is likely too bulky to allow complex formation [49–51].
In recent years, our understanding of the mechanisms that trigger PIDDosome assembly has broadened. First, Burigotto et al. [19] have shown that PIDD1 shuttles between the cytoplasm and the mother centriole at the centrosome. It is therefore optimally positioned to respond to alterations in centrosome numbers. Indeed, upon centrosome amplification, e.g. induced by cytokinesis failure or centriole overduplication, the PIDDosome assembles and promotes Caspase-2 activation [21]. The jury is still out as to how and where exactly PIDDosome formation is triggered by supernumerary centrosomes. Two recent studies have shown that knockout of the key factors responsible for the recruitment of PIDD1 to the centrosome, namely the latent subdistal appendage protein ANKRD26 or the more proximal protein SCLT1, prevent PIDDosome activation in response to cytokinesis failure. This suggests that a priming event needed for PIDDosome assembly happens at the centrosome [19,20]. Notably, processing of PIDD1 into its active fragments is not affected by disrupted centrosomal localization. Furthermore, we were so far unable to find RAIDD or Caspase-2 localizing at the centrosome, indicating that while PIDD1 has to shuttle through the centrosome at some point in order to nucleate the PIDDosome, the centrosome is neither required for PIDD1 processing, nor does it seem to be the location of PIDDosome formation. Of note, PIDDosome activation can promote cell cycle arrest in G1-cells with extra centrosomes but this does not necessarily trigger cell death [21].
Ando et al. [45] observed that forced mitotic entry via CHK1 inhibition in the presence of unresolved DNA damage induces phosphorylation of PIDD1 on T788 in the DD by ATM kinase. This phosphorylation facilitates PIDD1-RAIDD binding and BCL2-independent cell death while simultaneously preventing binding to RIPK1 for cell survival [45,46]. In a follow-up study, Shah et al. described the recruitment of PIDD1 to FANCI, which induces Caspase-2 activation and cell death when interstrand DNA crosslinks (ICLs) cannot be resolved in time by the Fanconi repair pathway [46].
Additional evidence has implicated BUBR1 and other components of the mitotic checkpoint complex (MCC) as negative regulators of Caspase-2-PIDDosome formation [52]. The authors of this study speculate that the mitotic checkpoint complex suppresses a DNA-damage dependent pro-apoptotic program which is in part mediated by the PIDDosome. However, PIDDosome activation is most often evaluated on a bulk of treated cells, as indicated by studies of Ho et al. [53] or Thompson et al. [52], due to technical constraints that prevent the analysis of PIDDosome formation at the single cell level. Therefore, we currently cannot rule out that the PIDDosome-mediated effects on cell death observed in these studies can be traced back to indirect effects of the treatments used, or activation of the PIDDosome at later stages of the cell cycle, e.g. in the next G1 phase. As such, CHK1 inhibition and MCC disruption have both been implicated in mitotic slippage or cytokinesis failure, which might create conditions in which the PIDDosome is activated by the presence of supernumerary centrosomes [54,55]. The current experimental evidence is insufficient to give a final answer whether DNA damage, mitotic arrest and centrosome amplification represent completely separate triggers for PIDDosome activation or whether they are linked. Hence, to shed new light, it would be interesting to see whether ATM- or ATR-dependent PIDD1 phosphorylation is also required for centrosome-mediated PIDDosome activation or likewise whether centrosome localization of PIDD1 presents a requirement for activation downstream of CHK1 inhibition, MCC inactivation or response to DNA interstrand crosslinks.
The NEMO-PIDDosome
It is now established that DNA damage can induce NF-κB activation and sterile inflammation. In this context, an alternative high-molecular weight complex, consisting of PIDD1, RIPK1 and NEMO, seems to be involved. Upon DNA-damage, exogenous PIDD1-C was shown to translocate to the nucleus and to interact with RIPK1 and NEMO. It is assumed that PIDD1-C and RIPK1 interact via their DDs, based on co-immunoprecipitation experiments performed by Janssens et al. [15]. Surprisingly enough, definite proof that this binding is direct and depends on DD interaction is actually lacking. Complex formation initiates a chain of posttranslational modifications of NEMO, most notably SUMOylation, which lead to the activation of NF-κB signaling [15,56]. While this NF-κB activation was initially described as an early pro-survival stimulus to prevent apoptosis immediately after DNA-damage, follow-up studies could not find that the NEMO-PIDDosome mediated a clear survival benefit [57]. Instead, DNA damage-induced NF-κB activation was shown to trigger pro-inflammatory gene expression in irradiated MEFs that relied in part on RIPK1 and PIDD1 [57].
The recent advances in our understanding of PIDDosome assembly in response to centrosome amplification pose another interesting question: Can extra centrosomes induce the formation of the NEMO-PIDDosome and therefore trigger NF-κB mediated target gene expression? Such an activation might be relevant primarily in certain myeloid cells that become physiologically polyploid during differentiation and where NF-κB activation might speed up differentiation or enhance effector function [58–60]. Additionally, we also speculate that NF-κB activation in cells with aberrantly increased centrosome numbers could trigger an inflammatory response that stimulates their removal by the innate immune system or contribute to the senescence-associated secretory phenotype (SASP), seen in cells with complex karyotypes [61].
Aside from its incorporation into multi-protein complexes mentioned above, a study by Logette et al. [47] showed that PIDD1 interacts with several DNA synthesis and DNA repair factors such as PCNA, RFC4, RFC5 and POLσ in response to UV irradiation. The interaction between PIDD1-C and PCNA is required for dissociation of p21 from PCNA and subsequent PCNA-mono-ubiquitination which leads to recruitment of the translesion DNA synthesis polymerase, POLη, to sites of DNA damage to allow error prone DNA replication, facilitating cell survival [47].
Emerging pathologicial and physiolocial roles of PIDD1
PIDD1 as an initiator and effector of p53 function
Early findings about the role of PIDD1 in the p53-response to DNA-damage have been documented in certain human cell lines and zebrafish experiments, they were, however, difficult to reconstruct in primary cell lines derived from PIDDosome mutant mice or universally reproducible across human cell lines. Pidd1−/− mouse hematopoietic cells or embryonic fibroblasts do not show decreased apoptosis or increased clonogenic potential upon induction of DNA damage with etoposide or ionizing radiation, even though in vitro assays in some human cell lines suggest a direct pro-apoptotic role of the PIDDosome [57,62]. Moreover, in vivo evidence of an anti-apoptotic phenotype of Pidd1−/− mice comes from skin keratinocytes, which display higher levels of DNA damage after UV irradiation [47].
This prompts the question whether early findings could in fact be partly explained by accidental cellular perturbations that might accompany treatment with DNA damage inducing agents. In line with this, Caspase-2 has been implicated in apoptosis upon treatment with drugs that impair actin filaments or microtubules or promote mitotic errors [53,63,64]. Since the microtubule targeting agents used for the study mentioned above [53] are known to induce mitotic slippage in certain cell types, such as U2OS or A549, we speculate that PIDDosome activation after cytoskeleton disruption may be a result of tetraploidization and the resulting centrosome amplification in a subset of cells. However, the mechanisms that govern whether PIDDosome activation triggers cell death (as observed in the studies of Ho et al. and others [53,63]) or an arrest in G1 phase (as observed by Fava et al. [21]) are currently unknown. Oliver et al. [65] have identified MDM2 as a second bona fide Caspase-2 target, whose cleavage leads to p53 stabilization in the context of Doxorubicin treatment. Yet, p53-induced transcriptional activation of PIDD1 after DNA-damage appears to suffice to allow PIDDosome activation, independent of centrosome amplification and promote secondary Caspase-2-mediated cell death in some settings [66,67].
P53 stabilization does not inevitably induce cell death, but can also contribute to cell cycle arrest and senescence through the transcriptional up-regulation of p21 in certain cell types [68] and subsequently to the secretion of senescence-associated cytokines [69,70]. Accordingly, loss of p53 facilitates survival and cell growth in cells that carry extra centrosomes, leading to CIN and eventually aneuploidy [71,72]. As such, it was tempting to postulate that loss of the PIDDosome should — similarly to loss of p53 — increase CIN and foster aneuploidy. While loss of PIDD1 could indeed facilitate proliferation and growth of cells overexpressing PLK4 to induce centrosome amplification [19,20], our analysis of PIDDosome-defective hepatocytes did not provide evidence for increased levels of aneuploidy in primary hepatocytes or DEN-driven liver cancers [23,73]. This suggests that secondary events that are associated with CIN or aneuploidy, such as DNA damage or proteotoxic stress will eventually activate p53 in a PIDDosome-independent manner to stop the outgrowth of PIDD1-deficient cells.
PIDD1 as a regulator of organogenesis and tissue regeneration
The physiological role of the centrosome-PIDDosome-p53 axis has been explored in vivo in naturally polyploid tissues, which have acquired supernumerary centrosomes through different mechanisms [74]. For example, liver cells become polyploid through a process of incomplete cytokinesis, which is triggered around the time of weaning by changes in insulin signaling [75,76]. Hepatocytes of Pidd1, Raidd or Casp2 knockout mice exhibit a substantially higher ploidy compared with wild-type animals [23] (reviewed in Sladky et al. [74,77]). Increased ploidy does not significantly affect liver function in steady state, but is associated with a higher capacity to regenerate liver tissue after partial hepatectomy [73]. Surprisingly, increased ploidy protects hepatocytes from transformation, and deletion of all three PIDDosome components reduced tumor burden [74].
One interesting aspect of PIDDosome function, which has received some attention in recent years, is associated with the marked expression of Caspase-2-PIDDosome components in the brain. Genetic evidence clearly links perturbations of each of the PIDDosome components to neurodevelopmental or neurodegenerative phenotypes. Loss-of-function mutations in RAIDD and PIDD1 which prevent PIDDosome activation are associated with lissencephaly [78–82], a form of impaired cortical development, and intellectual disability [83,84]. Even though the circumstances which lead to the activation of the PIDDosome in the brain or the cell type where this becomes relevant are not known, the authors speculate that apoptosis induced by the PIDDosome might be critical for proper cortical development [85,86]. Strikingly, however, a number of patients with intellectual disability have been identified that carry mutations in the DD of PIDD1 which abrogate its ability to activate Caspase-2 [84]. Moreover, the activation of Caspase-2 is implicated in the pathology of Alzheimer's disease (AD) [87,88]. There is evidence that Aβ-mediated apoptosis is dependent on Caspase-2 and partially also on RAIDD and PIDD1. However, there are also cases where Caspase-2 activation occurs independently of PIDD1, which implies that there might be alternative ways to activate Caspase-2 [48,89].
Open questions
On the one hand, PIDDosome-mediated Caspase-2 activation appears potentially relevant when the DNA repair response is impaired, where it can promote cell death, e.g. upon defective ICL-resolution or prolonged p53 activation [45,46,62,67]. The molecular pathway governing the PIDDosome's response to ICLs has been nicely delineated recently, however, the molecular details concerning execution of Caspase 2-PIDDosome-mediated cell death are still undefined. On the other hand, PIDDosome activation in response to supernumerary centrosomes has put it on the map of cell cycle control, CIN and cancer biology but the relevance of the PIDDosome in tumorigenesis, cancer evolution or response to therapy is far from clear. Moreover, several tissues become polyploid during development and thereby acquire extra centrosomes, which activates the PIDDosome. Whether this is indeed the case in polyploid tissues other than the liver will hopefully be answered in the coming years. Finally, polyploidization is also a feature often observed under inflammatory conditions, most prominently viral infections or bacteria-driven granulomas [90]. There, the NEMO-PIDDosome might be required to coordinate inflammation and immunity in a manner similar to other inflammasomes, and studies addressing these possibilities are eagerly awaited.
Perspectives
Death fold (DF)-containing proteins are key regulators of cell death and inflammation.
The DF protein PIDD1 exerts additional effector functions by regulating p53 and cell cycle arrest in response to extra centrosomes.
Understanding PIDD1 auto-processing will allow selective targeting of its versatile actions to treat diseases associated with deregulated cellular or nuclear ploidy.
Acknowledgements
Work on the PIDDosome in our lab is currently supported by the European Research Council, ERC-AdG ‘POLICE’ (#787171) and the Austrian Academy of Sciences (ÖAW). Figures were created in part using biorender.com
Abbreviations
- CARD
caspase activation and recruitment domains
- DD
death domains
- DF
death folds
- ICLs
interstrand DNA crosslinks
- LRRs
leucine-rich repeats
- MCC
mitotic checkpoint complex
- PAMPs
pathogen-associated molecular patterns
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Author Contributions
E.S.W. wrote manuscript and prepared figures, T.G.S. provided expression data analysis, I.G.C. edited manuscript, A.V. wrote and edited manuscript and figures.
References
- 1.van Opdenbosch, N. and Lamkanfi, M. (2019) Caspases in cell death, inflammation, and disease. Immunity 50, 1352–1364 10.1016/j.immuni.2019.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuan, J., Shaham, S., Ledoux, S., Ellis, H.M. and Horvitz, H.R. (1993) The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell 75, 641–652 10.1016/0092-8674(93)90485-9 [DOI] [PubMed] [Google Scholar]
- 3.Park, H.H., Lo, Y.-C., Lin, S.-C., Wang, L., Yang, J.K. and Wu, H. (2007) The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu. Rev. Immunol. 25, 561–586 10.1146/annurev.immunol.25.022106.141656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sohn, J. and Hur, S. (2016) Filament assemblies in foreign nucleic acid sensors. Curr. Opin. Struct. Biol. 37, 134–144 10.1016/j.sbi.2016.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramirez, M.L.G. and Salvesen, G.S. (2018) A primer on caspase mechanisms. Semin. Cell Dev. Biol. 82, 79–85 10.1016/j.semcdb.2018.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reed, J.C., Doctor, K.S. and Godzik, A. (2004) The domains of apoptosis: a genomics perspective. Sci. STKE 2004, re9 10.1126/stke.2392004re9 [DOI] [PubMed] [Google Scholar]
- 7.Shi, J., Gao, W. and Shao, F. (2017) Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem. Sci. 42, 245–254 10.1016/j.tibs.2016.10.004 [DOI] [PubMed] [Google Scholar]
- 8.Martinon, F., Burns, K. and Tschopp, J. (2002) The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10, 417–426 10.1016/S1097-2765(02)00599-3 [DOI] [PubMed] [Google Scholar]
- 9.Zou, H., Henzel, W.J., Liu, X., Lutschg, A. and Wang, X. (1997) Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90, 405–413 10.1016/S0092-8674(00)80501-2 [DOI] [PubMed] [Google Scholar]
- 10.Ting, A.T. and Bertrand, M.J.M. (2016) More to life than NF-κB in TNFR1 signaling. Trends Immunol. 37, 535–545 10.1016/j.it.2016.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Telliez, J.-B., Bean, K.M. and Lin, L.-L. (2000) LRDD, a novel leucine rich repeat and death domain containing protein. Biochim. Biophys. Acta 1478, 280–288 10.1016/S0167-4838(00)00029-7 [DOI] [PubMed] [Google Scholar]
- 12.Lin, Y., Ma, W. and Benchimol, S. (2000) Pidd, a new death-domain–containing protein, is induced by p53 and promotes apoptosis. Nat. Genet. 26, 122–127 10.1038/79102 [DOI] [PubMed] [Google Scholar]
- 13.Sladky, V., Schuler, F., Fava, L.L. and Villunger, A. (2017) The resurrection of the PIDDosome - emerging roles in the DNA-damage response and centrosome surveillance. J. Cell Sci. 130, 3779–3787 10.1242/jcs.203448 [DOI] [PubMed] [Google Scholar]
- 14.Tinel, A. and Tschopp, J. (2004) The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 304, 843–846 10.1126/science.1095432 [DOI] [PubMed] [Google Scholar]
- 15.Janssens, S., Tinel, A., Lippens, S. and Tschopp, J. (2005) PIDD mediates NF-κB activation in response to DNA damage. Cell 123, 1079–1092 10.1016/j.cell.2005.09.036 [DOI] [PubMed] [Google Scholar]
- 16.Berube, C., Boucher, L.-M., Ma, W., Wakeham, A., Salmena, L., Hakem, R.et al. (2005) Apoptosis caused by p53-induced protein with death domain (PIDD) depends on the death adapter protein RAIDD. Proc. Natl Acad. Sci. U.S.A. 102, 14314–14320 10.1073/pnas.0506475102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manzl, C., Krumschnabel, G., Bock, F., Sohm, B., Labi, V., Baumgartner, F.et al. (2009) Caspase-2 activation in the absence of PIDDosome formation. J. Cell Biol. 185, 291–303 10.1083/jcb.200811105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, I.R., Murakami, K., Chen, N.-J., Saibil, S.D., Matysiak-Zablocki, E., Elford, A.R.et al. (2009) DNA damage- and stress-induced apoptosis occurs independently of PIDD. Apoptosis 14, 1039–1049 10.1007/s10495-009-0375-1 [DOI] [PubMed] [Google Scholar]
- 19.Burigotto, M., Mattivi, A., Migliorati, D., Magnani, G., Valentini, C., Roccuzzo, M.et al. (2021) Centriolar distal appendages activate the centrosome-PIDDosome-p53 signalling axis via ANKRD26. EMBO J. 40, e104844 10.15252/embj.2020104844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans, L.T., Anglen, T., Scott, P., Lukasik, K., Loncarek, J. and Holland, A.J. (2021) ANKRD26 recruits PIDD1 to centriolar distal appendages to activate the PIDDosome following centrosome amplification. EMBO J. 40, e105106 10.15252/embj.2020105106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fava, L.L., Schuler, F., Sladky, V., Haschka, M.D., Soratroi, C., Eiterer, L.et al. (2017) The PIDDosome activates p53 in response to supernumerary centrosomes. Genes Dev. 31, 34–45 10.1101/gad.289728.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuenin, S., Tinel, A., Janssens, S. and Tschopp, J. (2008) p53-induced protein with a death domain (PIDD) isoforms differentially activate nuclear factor-kappaB and Caspase-2 in response to genotoxic stress. Oncogene 27, 387–396 10.1038/sj.onc.1210635 [DOI] [PubMed] [Google Scholar]
- 23.Sladky, V.C., Knapp, K., Soratroi, C., Heppke, J., Eichin, F., Rocamora-Reverte, L.et al. (2020) E2F-family members engage the PIDDosome to limit hepatocyte ploidy in liver development and regeneration. Dev. Cell 52, 335–349.e7 10.1016/j.devcel.2019.12.016 [DOI] [PubMed] [Google Scholar]
- 24.Chen, H.-Z., Ouseph, M.M., Li, J., Pécot, T., Chokshi, V., Kent, L.et al. (2012) Canonical and atypical E2Fs regulate the mammalian endocycle. Nat. Cell Biol. 14, 1192–1202 10.1038/ncb2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pandit, S.K., Westendorp, B. and de Bruin, A. (2013) Physiological significance of polyploidization in mammalian cells. Trends Cell Biol. 23, 556–566 10.1016/j.tcb.2013.06.002 [DOI] [PubMed] [Google Scholar]
- 26.Duan, H. and Dixit, V.M. (1997) RAIDD is a new ‘death’ adaptor molecule. Nature 385, 86–89 10.1038/385086a0 [DOI] [PubMed] [Google Scholar]
- 27.Lin, Q., Liu, Y., Moore, D.J., Elizer, S.K., Veach, R.A., Hawiger, J.et al. (2012) Cutting edge: the “death” adaptor CRADD/RAIDD targets BCL10 and suppresses agonist-induced cytokine expression in T lymphocytes. J. Immunol. 188, 2493–2497 10.4049/jimmunol.1101502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park, H.H. and Wu, H. (2007) Crystallization and preliminary X-ray crystallographic studies of the oligomeric death-domain complex between PIDD and RAIDD. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 229–232 10.1107/S1744309107007889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, R., Wei, Z., Jin, H., Wu, H., Yu, C., Wen, W.et al. (2009) Autoinhibition of UNC5b revealed by the cytoplasmic domain structure of the receptor. Mol. Cell 33, 692–703 10.1016/j.molcel.2009.02.016 [DOI] [PubMed] [Google Scholar]
- 30.Tinel, A., Janssens, S., Lippens, S., Cuenin, S., Logette, E., Jaccard, B.et al. (2006) Autoproteolysis of PIDD marks the bifurcation between pro-death Caspase-2 and pro-survival NF-κB pathway. EMBO J. 26, 197–208 10.1038/sj.emboj.7601473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fontoura, B.M., Blobel, G. and Matunis, M.J. (1999) A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol. 144, 1097–1112 10.1083/jcb.144.6.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenblum, J.S. and Blobel, G. (1999) Autoproteolysis in nucleoporin biogenesis. Proc. Natl Acad. Sci. U.S.A. 96, 11370–5 10.1073/pnas.96.20.11370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robert Hollingsworth, L., David, L., Li, Y., Griswold, A.R., Ruan, J., Sharif, H.et al. (2021) Mechanism of filament formation in UPA-promoted CARD8 and NLRP1 inflammasomes. Nat. Commun. 12, 189 10.1038/s41467-020-20320-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D'Osualdo, A., Weichenberger, C.X., Wagner, R.N., Godzik, A., Wooley, J. and Reed, J.C. (2011) CARD8 and NLRP1 undergo autoproteolytic processing through a ZU5-like domain. PLoS ONE 6, e27396 10.1371/journal.pone.0027396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hodel, A.E., Hodel, M.R., Griffis, E.R., Hennig, K.A., Ratner, G.A., Xu, S.et al. (2002) The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin Nup98. Mol. Cell 10, 347–358 10.1016/S1097-2765(02)00589-0 [DOI] [PubMed] [Google Scholar]
- 36.Tschopp, J., Martinon, F. and Burns, K. (2003) NALPs: a novel protein family involved in inflammation. Nat. Rev. Mol. Cell Biol. 4, 95–104 10.1038/nrm1019 [DOI] [PubMed] [Google Scholar]
- 37.Sandstrom, A., Mitchell, P.S., Goers, L., Mu, E.W., Lesser, C.F. and Vance, R.E. (2019) Functional degradation: a mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 364, eaau1330 10.1126/science.aau1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hollingsworth, L.R., Sharif, H., Griswold, A.R., Fontana, P., Mintseris, J., Dagbay, K.B.et al. (2021) DPP9 sequesters the C terminus of NLRP1 to repress inflammasome activation. Nature 592, 778–783 10.1038/s41586-021-03350-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang, M., Zhang, X., Toh, G.A., Gong, Q., Wang, J., Han, Z.et al. (2021) Structural and biochemical mechanisms of NLRP1 inhibition by DPP9. Nature 592, 773–777 10.1038/s41586-021-03320-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taabazuing, C.Y., Griswold, A.R. and Bachovchin, D.A. (2020) The NLRP1 and CARD8 inflammasomes. Immunol. Rev. 297, 13–25 10.1111/imr.12884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong, F.L., Robinson, K., Teo, D.E.T., Tan, K.-Y., Lim, C., Harapas, C.R.et al. (2018) Human DPP9 represses NLRP1 inflammasome and protects against autoinflammatory diseases via both peptidase activity and FIIND domain binding. J. Biol. Chem. 293, 18864–18878 10.1074/jbc.RA118.004350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubina, K.A. and Tkachuk, V.A. (2015) Guidance receptors in the nervous and cardiovascular systems. Biochemistry (Mosc) 80, 1235–1253 10.1134/S0006297915100041 [DOI] [PubMed] [Google Scholar]
- 43.Zhu, Y., Li, Y. and Nakagawara, A. (2021) UNC5 dependence receptor family in human cancer: a controllable double-edged sword. Cancer Lett. 516, 28–35 10.1016/j.canlet.2021.05.034 [DOI] [PubMed] [Google Scholar]
- 44.Wang, C., Yu, C., Ye, F., Wei, Z. and Zhang, M. (2012) Structure of the ZU5-ZU5-UPA-DD tandem of ankyrin-B reveals interaction surfaces necessary for ankyrin function. Proc. Natl Acad. Sci. U.S.A. 109, 4822–4827 10.1073/pnas.1200613109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ando, K., Kernan, J.L., Liu, P.H., Sanda, T., Logette, E., Tschopp, J.et al. (2012) PIDD death-domain phosphorylation by ATM controls prodeath versus prosurvival PIDDosome signaling. Mol. Cell 47, 681–693 10.1016/j.molcel.2012.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah, R.B., Kernan, J.L., van Hoogstraten, A., Ando, K., Li, Y., Belcher, A.L.et al. (2021) FANCI functions as a repair/apoptosis switch in response to DNA crosslinks. Dev. Cell 56, 2207–2222.e7 10.1016/j.devcel.2021.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Logette, E., Schuepbach-Mallepell, S., Eckert, M.J., Leo, X.H., Jaccard, B., Manzl, C.et al. (2011) PIDD orchestrates translesion DNA synthesis in response to UV irradiation. Cell Death Differ. 18, 1036–1045 10.1038/cdd.2011.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ando, K., Parsons, M.J., Shah, R.B., Charendoff, C.I., Paris, S.L., Liu, P.H.et al. (2017) NPM1 directs PIDDosome-dependent caspase-2 activation in the nucleolus. J. Cell Biol. 216, 1795–1810 10.1083/jcb.201608095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jang, T.H. and Park, H.H. (2013) PIDD mediates and stabilizes the interaction between RAIDD and Caspase-2 for the PIDDosome assembly. BMB Rep. 46, 471–476 10.5483/BMBRep.2013.46.9.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jang, T.H., Zheng, C., Wu, H., Jeon, J.-H. and Park, H.H. (2010) In vitro reconstitution of the interactions in the PIDDosome. Apoptosis 15, 1444–1452 10.1007/s10495-010-0544-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jang, T.H., Seo, E.K. and Park, H.H. (2013) Analysis of mutation effects on PIDDosome core complex. Appl. Biochem. Biotechnol. 170, 210–218 10.1007/s12010-013-0184-4 [DOI] [PubMed] [Google Scholar]
- 52.Thompson, R., Shah, R.B., Liu, P.H., Gupta, Y.K., Ando, K., Aggarwal, A.K.et al. (2015) An inhibitor of PIDDosome formation. Mol. Cell 58, 767–779 10.1016/j.molcel.2015.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ho, L.H., Read, S.H., Dorstyn, L., Lambrusco, L. and Kumar, S. (2008) Caspase-2 is required for cell death induced by cytoskeletal disruption. Oncogene 27, 3393–3404 10.1038/sj.onc.1211005 [DOI] [PubMed] [Google Scholar]
- 54.Poehlmann, A., Habold, C., Walluscheck, D., Reissig, K., Bajbouj, K., Ullrich, O.et al. (2011) Cutting edge: Chk1 directs senescence and mitotic catastrophe in recovery from G₂ checkpoint arrest. J. Cell. Mol. Med. 15, 1528–1541 10.1111/j.1582-4934.2010.01143.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stevens, F.E., Beamish, H., Warrener, R. and Gabrielli, B. (2008) Histone deacetylase inhibitors induce mitotic slippage. Oncogene 27, 1345–1354 10.1038/sj.onc.1210779 [DOI] [PubMed] [Google Scholar]
- 56.Huang, T.T., Wuerzberger-Davis, S.M., Wu, Z.-H. and Miyamoto, S. (2003) Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell 115, 565–576 10.1016/S0092-8674(03)00895-X [DOI] [PubMed] [Google Scholar]
- 57.Bock, F.J., Krumschnabel, G., Manzl, C., Peintner, L., Tanzer, M.C., Hermann-Kleiter, N.et al. (2013) Loss of PIDD limits NF-κB activation and cytokine production but not cell survival or transformation after DNA damage. Cell Death Differ. 20, 546–557 10.1038/cdd.2012.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McNally, A.K. and Anderson, J.M. (2011) Macrophage fusion and multinucleated giant cells of inflammation. Adv. Exp. Med. Biol. 713, 97–111 10.1007/978-94-007-0763-4_7 [DOI] [PubMed] [Google Scholar]
- 59.Hornik, T.C., Neniskyte, U. and Brown, G.C. (2014) Inflammation induces multinucleation of microglia via PKC inhibition of cytokinesis, generating highly phagocytic multinucleated giant cells. J. Neurochem. 128, 650–661 10.1111/jnc.12477 [DOI] [PubMed] [Google Scholar]
- 60.Yagi, M., Miyamoto, T., Sawatani, Y., Iwamoto, K., Hosogane, N., Fujita, N.et al. (2005) DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells. J. Exp. Med. 202, 345–351 10.1084/jem.20050645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santaguida, S., Richardson, A., Iyer, D.R., M'Saad, O., Zasadil, L., Knouse, K.A.et al. (2017) Chromosome mis-segregation generates cell-cycle-arrested cells with complex karyotypes that are eliminated by the immune system. Dev. Cell 41, 638–651.e5 10.1016/j.devcel.2017.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manzl, C., Peintner, L., Krumschnabel, G., Bock, F., Labi, V., Drach, M.et al. (2012) PIDDosome-independent tumor suppression by Caspase-2. Cell Death Differ. 19, 1722–1732 10.1038/cdd.2012.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bonzon, C., Bouchier-Hayes, L., Pagliari, L.J., Green, D.R. and Newmeyer, D.D. (2006) Caspase-2-induced apoptosis requires bid cleavage: a physiological role for bid in heat shock-induced death. Mol. Biol. Cell 17, 2150–2157 10.1091/mbc.e05-12-1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dawar, S., Lim, Y., Puccini, J., White, M., Thomas, P., Bouchier-Hayes, L.et al. (2017) Caspase-2-mediated cell death is required for deleting aneuploid cells. Oncogene 36, 2704–2714 10.1038/onc.2016.423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oliver, T.G., Meylan, E., Chang, G.P., Xue, W., Burke, J.R., Humpton, T.J.et al. (2011) Caspase-2-mediated cleavage of Mdm2 creates a p53-induced positive feedback loop. Mol. Cell 43, 57–71 10.1016/j.molcel.2011.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fava, L.L., Rainer, J., Haschka, M.D., Geley, S. and Villunger, A. (2015) Beclin 1 is dispensable for chromosome congression and proper outer kinetochore assembly. EMBO Rep. 16, 1233–1236 10.15252/embr.201540731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsabar, M., Mock, C.S., Venkatachalam, V., Reyes, J., Karhohs, K.W., Oliver, T.G.et al. (2020) A switch in p53 dynamics marks cells that escape from DSB-induced cell cycle arrest. Cell Rep. 32, 107995 10.1016/j.celrep.2020.107995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holland, A.J., Fachinetti, D., Zhu, Q., Bauer, M., Verma, I.M., Nigg, E.A.et al. (2012) The autoregulated instability of Polo-like kinase 4 limits centrosome duplication to once per cell cycle. Genes Dev. 26, 2684–2689 10.1101/gad.207027.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Adams, S.D., Csere, J., D'angelo, G., Carter, E.P., Romao, M., Arnandis, T.et al. (2021) Centrosome amplification mediates small extracellular vesicle secretion via lysosome disruption. Curr. Biol. 31, 1403–1416.e7 10.1016/j.cub.2021.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arnandis, T., Monteiro, P., Adams, S.D., Bridgeman, V.L., Rajeeve, V., Gadaleta, E.et al. (2018) Oxidative stress in cells with extra centrosomes drives non-cell-autonomous invasion. Dev. Cell 47, 409–424.e9 10.1016/j.devcel.2018.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Santaguida, S. and Amon, A. (2015) Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol. 16, 473–485 10.1038/nrm4025 [DOI] [PubMed] [Google Scholar]
- 72.Nigg, E.A. and Holland, A.J. (2018) Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 19, 297–312 10.1038/nrm.2017.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sladky, V.C., Knapp, K., Szabo, T.G., Braun, V.Z., Bongiovanni, L., van den Bos, H.et al. (2020) PIDDosome-induced p53-dependent ploidy restriction facilitates hepatocarcinogenesis. EMBO Rep. 21, e50893 10.15252/embr.202050893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sladky, V.C., Eichin, F., Reiberger, T. and Villunger, A. (2021) Polyploidy control in hepatic health and disease. J. Hepatol. 75, 1177–1191 10.1016/j.jhep.2021.06.030 [DOI] [PubMed] [Google Scholar]
- 75.Celton-Morizur, S., Merlen, G., Couton, D., Margall-Ducos, G. and Desdouets, C. (2009) The insulin/Akt pathway controls a specific cell division program that leads to generation of binucleated tetraploid liver cells in rodents. J. Clin. Invest. 119, 1880–1887 10.1172/jci38677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Margall-Ducos, G., Celton-Morizur, S., Couton, D., Brégerie, O. and Desdouets, C. (2007) Liver tetraploidization is controlled by a new process of incomplete cytokinesis. J. Cell Sci. 120, 3633–3639 10.1242/jcs.016907 [DOI] [PubMed] [Google Scholar]
- 77.Sladky, V.C. and Villunger, A. (2020) Uncovering the PIDDosome and Caspase-2 as regulators of organogenesis and cellular differentiation. Cell Death Differ. 27, 2037–2047 10.1038/s41418-020-0556-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Di Donato, N., Jean, Y.Y., Maga, A.M., Krewson, B.D., Shupp, A.B., Avrutsky, M.I.et al. (2016) Mutations in CRADD result in reduced caspase-2-mediated neuronal apoptosis and cause megalencephaly with a rare lissencephaly variant. Am. J. Hum. Genet. 99, 1117–1129 10.1016/j.ajhg.2016.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ha, H.J. and Park, H.H. (2018) RAIDD mutations underlie the pathogenesis of thin lissencephaly (TLIS). PLoS ONE 13, e0205042 10.1371/journal.pone.0205042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Harel, T., Hacohen, N., Shaag, A., Gomori, M., Singer, A., Elpeleg, O.et al. (2017) Homozygous null variant in CRADD, encoding an adaptor protein that mediates apoptosis, is associated with lissencephaly. Am. J. Med. Genet. A 173, 2539–2544 10.1002/ajmg.a.38347 [DOI] [PubMed] [Google Scholar]
- 81.Koprulu, M., Shabbir, R.M.K., Zaman, Q., Nalbant, G., Malik, S. and Tolun, A. (2021) CRADD and USP44 mutations in intellectual disability, mild lissencephaly, brain atrophy, developmental delay, strabismus, behavioural problems and skeletal anomalies. Eur. J. Med. Genet. 64, 104181 10.1016/j.ejmg.2021.104181 [DOI] [PubMed] [Google Scholar]
- 82.Polla, D.L., Rahikkala, E., Bode, M.K., Määttä, T., Varilo, T., Loman, T.et al. (2019) Phenotypic spectrum associated with a CRADD founder variant underlying frontotemporal predominant pachygyria in the Finnish population. Eur. J. Hum. Genet. 27, 1235–1243 10.1038/s41431-019-0383-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zaki, M.S., Accogli, A., Mirzaa, G., Rahman, F., Mohammed, H., Porras-Hurtado, G.L.et al. (2021) Pathogenic variants in PIDD1 lead to an autosomal recessive neurodevelopmental disorder with pachygyria and psychiatric features. Eur. J. Hum. Genet. 29, 1226–1234 10.1038/s41431-021-00910-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sheikh, T.I., Vasli, N., Pastore, S., Kharizi, K., Harripaul, R., Fattahi, Z.et al. (2021) Biallelic mutations in the death domain of PIDD1 impair Caspase-2 activation and are associated with intellectual disability. Transl. Psychiatry 11, 1 10.1038/s41398-020-01158-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Harripaul, R., Vasli, N., Mikhailov, A., Rafiq, M.A., Mittal, K., Windpassinger, C.et al. (2018) Mapping autosomal recessive intellectual disability: combined microarray and exome sequencing identifies 26 novel candidate genes in 192 consanguineous families. Mol. Psychiatry 23, 973–984 10.1038/mp.2017.60 [DOI] [PubMed] [Google Scholar]
- 86.Kurki, M.I., Saarentaus, E., Pietiläinen, O., Gormley, P., Lal, D., Kerminen, S.et al. (2019) Contribution of rare and common variants to intellectual disability in a sub-isolate of northern Finland. Nat. Commun. 10, 410 10.1038/s41467-018-08262-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Niizuma, K., Endo, H., Nito, C., Myer, D.J., Kim, G.S. and Chan, P.H. (2008) The PIDDosome mediates delayed death of hippocampal CA1 neurons after transient global cerebral ischemia in rats. Proc. Natl Acad. Sci. US.A. 105, 16368–16373 10.1073/pnas.0806222105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Troy, C.M., Rabacchi, S.A., Friedman, W.J., Frappier, T.F., Brown, K. and Shelanski, M.L. (2000) Caspase-2 mediates neuronal cell death induced by beta-amyloid. J. Neurosci. 20, 1386–1392 10.1523/JNEUROSCI.20-04-01386.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ribe, E.M., Jean, Y.Y., Goldstein, R.L., Manzl, C., Stefanis, L., Villunger, A.et al. (2012) Neuronal caspase 2 activity and function requires RAIDD, but not PIDD. Biochem. J. 444, 591–599 10.1042/BJ20111588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Helming, L. and Gordon, S. (2009) Molecular mediators of macrophage fusion. Trends Cell Biol. 19, 514–522 10.1016/j.tcb.2009.07.005 [DOI] [PubMed] [Google Scholar]