Abstract
The smudged eighty-eight butterfly Diaethria gabaza eupepla (Salvin & Godman, 1868) (Nymphalidae) is a vividly colored aposematic butterfly from Central and South America. A complete circular mitochondrial genome (mitogenome) of 15,156 bp from D. gabaza eupepla was assembled from a genome skimming Illumina sequence library. The AT-rich (80.5% AT) mitogenome consists of 13 protein-coding genes, 22 tRNAs, 2 rRNAs, and a control region in the typical butterfly gene order. Diaethria gabaza eupepla COX1 begins with an atypical CGA start codon and ATP6, COX1, COX2, CYTB, ND1, ND4, ND4L, and ND5 mRNAs contain incomplete stop codons completed by the addition of 3’ A residues. Phylogenetic reconstruction places Diaethria as the sister clade to Hamadryas within monophyletic nymphalid subfamily Biblidinae, consistent with previous phylogenetic hypotheses.
Keywords: Illumina sequencing, mitogenomics; Lepidoptera, Nymphalidae, Diaethria
The Living Prairie Mitogenomics Consortium is a structured inquiry exercise for undergraduates (Marcus et al. 2010) who assemble arthropod mitogenomes for improved DNA-based species identification and phylogenetics (Living Prairie Mitogenomics Consortium 2017, 2018, 2019, 2020, 2021). Participating students assembled, annotated, and analyzed sequence data (further curated by the instructor) and conducted a literature review for presentation here.
The butterflies of the genus Diaethria Billberg, 1820 (Nymphalidae: Biblidinae: Callicorini) are notable in that they possess color patterns on the ventral hind wings that resemble the numerals “88” or “89” and so are known by the common name “eighty-eight butterflies” (Dias et al. 2012). These distinctive patterns create a black, white, and red aposematic wing display intended to deter predators from eating these butterflies (Chai 1986; Pinheiro 1996). The genus includes a dozen species that occur between Texas, USA and Tierra del Fuego, Argentina (Dias et al. 2012). Male Diaethria are strongly attracted to urine-soaked sand and engage in mud-puddling behavior (DeVries 1987). The smudged eighty-eight butterfly Diaethria gabaza (Hewitson 1855) includes 3 subspecies, with subspecies D. gabaza eupepla (Salvin & Godman, 1868) occurring between Costa Rica and Colombia, where its larvae feed on climbing vines in the genus Serjania (Sapindaceae) (DeVries 1987).
Here we report the complete mitochondrial genome (mitogenome) sequence of D. gabaza eupepla from specimen Diae2019.1, collected by John R. MacDonald on 6 October 2019 at Finca Hartmann, Panama (GPS 8.844943 N, 82.760747 W) that has been pinned, spread, and deposited in the Wallis Roughley Museum of Entomology, University of Manitoba (http://www.wallisroughley.ca/, Jason Gibbs, Jason.Gibbs@umanitoba.ca) under the voucher number WRME0507740. This study was conducted with the approval of the University of Manitoba Office of Research Ethics & Compliance under permit number BF0155-1. Research was carried out in accordance with applicable national and international guidelines.
A leg was removed from the specimen and DNA was prepared using a DNEasy Blood and Tissue kit (Qiagen, Düsseldorf, Germany) with slight protocol modifications as described in McCullagh and Marcus (2015). DNA was sheared by sonication and a fragment library was prepared as previously described (Peters and Marcus 2017) using a NEBNext Ultra II DNA Library Prep Kit before sequencing by Illumina NovaSeq6000 (San Diego, CA) (Marcus 2018). Mitogenome assembly and annotation of the D. gabaza eupepla (GenBank accession MZ981736) was performed by mapping the resulting sequence library of 57,341,531 paired 150 bp reads (GenBank SRA PRJNA759138) to a Baeotus beotus reference mitogenome (Lepidoptera: Nymphalidae: Nympahlina: Coeini, MW566598 (Lalonde 2021)) using 5 iterations of the medium sensitivity settings of Geneious Prime 2021.1.1. Nuclear rRNA repeat sequences are increasingly recognized as being very useful for phylogenetic comparisons (Dodsworth 2015; Coissac et al. 2016; Marcus 2018; Krehenwinkel et al. 2019), so we also assembled the complete D. gabaza eupepla nuclear rRNA repeat (GenBank MZ981737) using a B. beotus (MW571038) (Lalonde 2021) reference sequence.
The D. gabaza eupepla circular 15,156 bp mitogenome assembly was composed of 22,600 paired reads with nucleotide composition: 39.8% A, 11.7% C, 7.8% G, and 40.7% T. The gene composition and order in D. gabaza eupepla is typical of the arrangement found in most butterfly mitogenomes (Park et al. 2016). The protein-coding genes start codons include: ATG (ATP6, COX2, COX3, CYTB, ND1, ND4), ATT (ND2, ND3, ND6), ATA (ND4L, ND5), and ATC (ATP8), while COX1 begins with an atypical CGA start codon as in many other insects (Liao et al. 2010). The mitogenome contains four protein-coding genes (COX1, COX2, ND4, ND5) with single-nucleotide (T) stop codons, and four protein-coding genes (ATP6, CYTB, ND1, ND4L) with two-nucleotide (TA) stop codons completed by post-transcriptional addition of 3′ A residues. The locations and structures of tRNAs were determined using ARWEN v.1.2 (Laslett and Canback 2008). All tRNAs exhibited cloverleaf secondary structure except that the dihydrouridine arm of trnS (AGN) was replaced by a loop. The size and structure of the mitochondrial rRNAs and control region are typical for Lepidoptera (McCullagh and Marcus 2015).
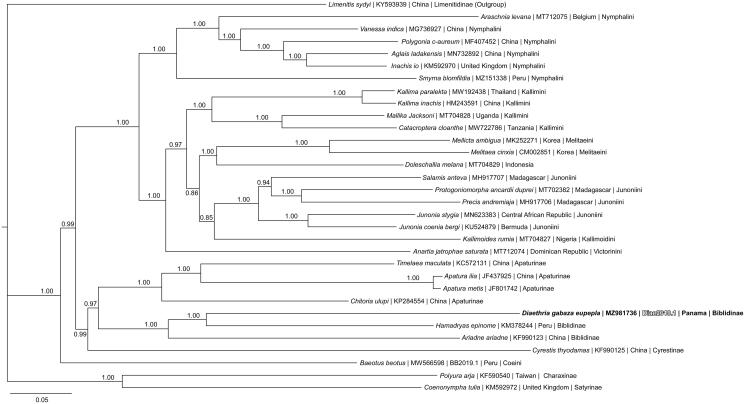
Phylogenetic reconstruction (Figure 1) was conducted using the complete mitogenome of D. gabaza eupepla and 31 other complete mitogenomes from the family Nymphalidae, including outgroup species Limenitis sydyi from nymphalid subfamily Limenitidinae and available from GenBank (Alexiuk et al. 2020; Hamilton et al. 2020; Lalonde and Marcus 2020; Payment et al. 2020; Alexiuk et al. 2021a, 2021b; Lalonde 2021). GenBank accession numbers are listed in Figure 1. Mitogenome sequences were aligned in CLUSTAL Omega (Sievers et al. 2011) and analyzed using Bayesian Inference with the GTR + I + G model (model selected using jModeltest 2.1.1 (Darriba et al. 2012)) in Mr. Bayes version 3.2.7 (Ronquist and Huelsenbeck 2003; Ronquist et al. 2012). As expected based on a previous phylogenetic hypothesis (Wahlberg et al. 2009), phylogenetic analysis placed D. gabaza eupepla as the sister taxon to Hamadryas epinome in a monophyletic clade with mitogenomes from nymphalid subfamily Biblidinae.
Figure 1.
The Bayesian phylogeny (GTR + I + G model, best state likelihood = −147,240.16, average deviation of split frequencies = 0.001131) of the Diaethria gabaza eupepla mitogenome, 31 additional mitogenomes from within family Nymphalidae, including outgroup species Limenitis sydyi (Nymphalidae: Limenitinae), produced by 10 million MCMC generations in MrBayes, with sampling every 1000 generations, and after discarding the first 250,000 generations as burn-in. The Bayesian posterior probability values determined by Mr Bayes are provided at each node. Each taxon in the analysis is labeled with species name, GenBank accession, the country of origin of the specimen with the sequenced mitogenome, and the nymphalid Tribe or Subfamily of the species.
Acknowledgments
Thanks to Melanie Lalonde and Josephine Payment for assistance with DNA extraction and for bioinformatics pipeline development. The authors thank Genome Quebec for assistance with library preparation and sequencing.
Funding Statement
This work was supported by Natural Sciences and Engineering Research Council of Canada under Grant RGPIN-2016-06012 and by the University of Manitoba under the University Research Grants Program.
Contributor Information
Collaborators: Jan-Glynnis C. Alex, Mackenzie R. Alexiuk, Katrina J. Audet, Somtochukwu D. Azubuike, Amber S. Bezte, Madison B. Boychuk, Natalie L. Cale, Lara A. Carroll, Gabriela Y. Castro, Joshua Cheng, Janam Chopra, Gregory A. Corkal, Carla Louise M. Dizon, Moeez Farooq, Cecilia C. Flores, Ruzzell C. Flores, Tadbeer Grewal, Maria Haguisan, Kousha Kamal, Haziqa B. Kassim, Spencer J. Kauenhofen, Anushka Kothari, Suseong Lee, Yanju Li, Raju Majumdar, Ana Markovic, Hope K. Mcauley, Ashley M. McKay, Deesha Nayar, Gurasis Osahan, Jordan A. Paul, Dhruvrajsinh Raolji, Alexandria Reimer, Talia R. Shafai, Simarjit Singh, Shloke Srivastava, Jackie Wang, Tristan B. Wolfe, Shirly J. Xie, and Jeffrey M. Marcus
Disclosure statement
No potential conflict of interest was reported by the authors.
Author contributions
Jeffrey Marcus was responsible for the conception and design of the study as well as the initial drafting of the paper. All of the authors were involved in the analysis and interpretation of the data, revising the manuscript critically for intellectual content, and gave the final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ981736–MZ981737. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA759138, SRX11982685, and SAMN21156917 respectively.
References
- Alexiuk MR, Marcus JM, Lalonde M.. 2020. The complete mitochondrial genome of the Jackson’s Leaf butterfly Mallika jacksoni (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA B Resour. 5:3316–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexiuk MR, Lalonde MM, Marcus JM.. 2021a. Phylogenetic analysis of the complete mitochondrial genome of the Japanese peacock butterfly Aglais io geisha (Stichel 1907) (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA B Resour. 6(10):3082–3084. ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexiuk MR, Lalonde MML, Marcus JM.. 2021b. Phylogenetic analysis of the complete mitochondrial genome of the Blomfild's Beauty butterfly Smyrna blomfildia (Fabricius 1781) (Insecta: Lepidoptera: Nymphalidae: Nymphalini). Mitochondrial DNA B Resour. 6(11):3199–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai P. 1986. Field observations and feeding experiments on the responses of rufous-tailed jacamars (Galbula ruficauda) to free-flying butterflies in a tropical rainforest. Biol J Linn Soc Lond. 29(3):161–189. [Google Scholar]
- Coissac E, Hollingsworth PM, Lavergne S, Taberlet P.. 2016. From barcodes to genomes: extending the concept of DNA barcoding. Mol Ecol. 25(7):1423–1428. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries PJ. 1987. The butterflies of Costa Rica and their natural history. Princeton, NJ: Princeton University Press. [Google Scholar]
- Dias FMS, Carneiro E, Casagrande MM, Mielke O.. 2012. Biology and external morphology of immature stages of the butterfly, Diaethria candrena candrena. J Insect Sci. 12:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodsworth S. 2015. Genome skimming for next-generation biodiversity analysis. Trends Plant Sci. 20(9):525–527. [DOI] [PubMed] [Google Scholar]
- Hamilton RV, Marcus JM, Lalonde M.. 2020. The complete mitochondrial genome of the black dead leaf butterfly Doleschallia melana (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA B Resour. 5(3):3306–3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krehenwinkel H, Pomerantz A, Henderson JB, Kennedy SR, Lim JY, Swamy V, Shoobridge JD, Graham N, Patel NH, Gillespie RG, et al.. 2019. Nanopore sequencing of long ribosomal DNA amplicons enables portable and simple biodiversity assessments with high phylogenetic resolution across broad taxonomic scale. GigaScience. 8(5):giz006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde M. 2021. Phylogenetic analysis of the complete mitochondrial genome of the graphic beauty butterfly Baeotus beotus (Doubleday 1849) (Lepidoptera: Nymphalidae: Nymphalinae: Coeini). Mitochondrial DNA B: Resour. 6(4):1516–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde MML, Marcus JM.. 2020. The complete mitochondrial genome of the Malagasy clouded mother-of-pearl butterfly Protogoniomorpha ancardii duprei (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA B Resour. 5:3261–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D, Canback B.. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175. [DOI] [PubMed] [Google Scholar]
- Liao F, Wang L, Wu S, Li Y-P, Zhao L, Huang G-M, Niu C-J, Liu Y-Q, Li M-G.. 2010. The complete mitochondrial genome of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae). Int J Biol Sci. 6(2):172–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Living Prairie Mitogenomics Consortium. 2017. The complete mitochondrial genome of the lesser aspen webworm moth Meroptera pravella (Insecta: Lepidoptera: Pyralidae). Mitochondrial DNA B Resour. 2:344–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Living Prairie Mitogenomics Consortium. 2018. The complete mitochondrial genome of the giant casemaker caddisfly Phryganea cinerea (Insecta: Trichoptera: Phryganeidae). Mitochondrial DNA B Resour. 3:375–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Living Prairie Mitogenomics Consortium. 2019. The complete mitochondrial genome of the North American pale summer sedge caddisfly Limnephilus hyalinus (Insecta: Trichoptera: Limnephilidae). Mitochondrial DNA B Resour. 4:413–415. [Google Scholar]
- Living Prairie Mitogenomics Consortium. 2020. The complete mitochondrial genome of the brown pansy butterfly, Junonia stygia (Aurivillius, 1894), (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA B Resour. 5:41–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Living Prairie Mitogenomics Consortium. 2021. The complete mitochondrial genome of the Indian leafwing butterfly Kallima paralekta (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA B Resour. 6:274–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus JM. 2018. Our love-hate relationship with DNA barcodes, the Y2K problem, and the search for next generation barcodes. AIMS Genet. 5(1):1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus JM, Hughes TM, McElroy DM, Wyatt RE.. 2010. Engaging First year undergraduates in hands-on research experiences: the upper green river barcode of life project. J Coll Sci Teach. 39:39–45. [Google Scholar]
- McCullagh BS, Marcus JM.. 2015. The complete mitochondrial genome of Lemon Pansy, Junonia lemonias (Lepidoptera: Nymphalidae: Nymphalinae). J Asia-Pacific Ent. 18(4):749–755. [Google Scholar]
- Park JS, Kim MJ, Jeong SY, Kim SS, Kim I.. 2016. Complete mitochondrial genomes of two gelechioids, Mesophleps albilinella and Dichomeris ustalella (Lepidoptera: Gelechiidae), with a description of gene rearrangement in Lepidoptera. Curr Genet. 62(4):809–826. [DOI] [PubMed] [Google Scholar]
- Payment JE, Marcus JM, Lalonde M.. 2020. The complete mitochondrial genome of the African leaf butterfly Kallimoides rumia (Insecta: Lepidoptera: Nymphalidae). Mitochondrial DNA B Resour. 5:3415–3417.33458190 [Google Scholar]
- Peters MJ, Marcus JM.. 2017. Taxonomy as a hypothesis: testing the status of the Bermuda buckeye butterfly Junonia coenia bergi (Lepidoptera: Nymphalidae). Syst Entomol. 42(1):288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro C. 1996. Palatability and escaping ability in Neotropical butterflies: tests with wild kingbirds (Tyrannus melancholicus, Tyrannidae). Biol J Linn Soc Lond. 59(4):351–365. [Google Scholar]
- Ronquist F, Huelsenbeck JP.. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP.. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al.. 2011. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7(539):539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg N, Leneveu J, Kodandaramaiah U, Peña C, Nylin S, Freitas AVL, Brower A.. 2009. Nymphalid butterflies diversify following near demise at the Cretaceous/Tertiary boundary. Proc Biol Sci. 276(1677):4295–4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. MZ981736–MZ981737. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA759138, SRX11982685, and SAMN21156917 respectively.