Abstract
d-Arabinono-1,4-lactone oxidase, which catalyzes the terminal step in the biosynthesis of d-erythroascorbic acid in Saccharomyces cerevisiae, was functionally expressed in Escherichia coli inherently lacking the enzyme. The recombinant E. coli strain expressing the enzyme could overproduce d-erythroascorbic acid and l-ascorbic acid when supplied with d-arabinono-1,4-lactone and l-galactono-1,4-lactone, respectively.
Physiological and biochemical studies have revealed many functional roles of vitamin C, or l-ascorbic acid (ASC), which is biosynthesized by all higher plants and nearly all higher animals with the exceptions of humans, other primates, guinea pigs, some birds, and fish (4–6). Some known or proposed functions of ASC include its utilization as a free radical scavenger (3, 16), an oxidation-reduction system in electron transport (2), a cofactor for a number of enzymes (19, 20), and a controlling factor in plant cell development (1). However, the precise mechanisms of its function have not been established completely. In some lower eukaryotes, d-erythroascorbic acid (EASC) is present instead of ASC (10, 15). EASC is very similar to ASC in structure and physicochemical properties (21), suggesting that it may take the place of ASC in lower eukaryotes. We have previously reported that EASC is synthesized from d-arabinose through d-arabinono-1,4-lactone by a concerted reaction of d-arabinose dehydrogenase and d-arabinono-1,4-lactone oxidase (ALO) (10, 13, 14) and is an important antioxidant molecule in Saccharomyces cerevisiae (11). We also found that our proposed biosynthetic pathway for EASC is very similar to the biosynthetic pathway for ASC in plants, in which l-galactose and l-galactono-1,4-lactone are involved (23). It is worth noting that ALO can oxidize l-galactono-1,4-lactone as efficiently as d-arabinono-1,4-lactone (10). This means that both EASC and ASC can be produced by ALO, depending upon the substrate used. Here we report the success in the functional expression of ALO and the overproduction of EASC and ASC in Escherichia coli, which is inherently devoid of ALO and can normally make neither EASC nor ASC. This is the first description about functional expression of an enzyme catalyzing the biosynthesis of EASC and ASC in Escherichia coli. Because the organic synthesis of EASC is very difficult, we expect that these findings could open a facile route to an expedient and efficient method for the production of EASC and also of ASC that uses relatively simple biological tools.
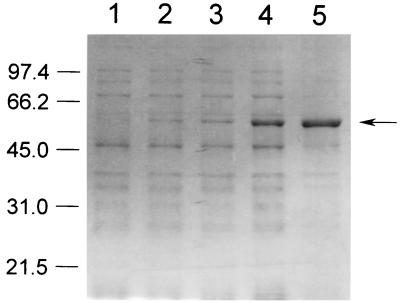
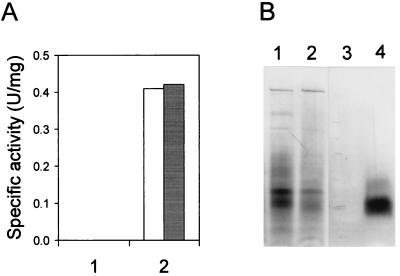
In order to express S. cerevisiae ALO in E. coli, oligonucleotide primers were synthesized on the basis of the nucleotide sequence of the ALO1 gene (11) with the sequences 5′-TTTCACCATATGTCTACTATCC-3′ (forward primer) and 5′-AAGGATCCTAGTCGGACAACTC-3′ (reverse primer). They were designed so that the amplified DNA could contain the entire open reading frame of the ALO1 gene with a NdeI site at the 5′ end and a BamHI site at the 3′ end. PCR was carried out with Pwo DNA polymerase (Roche Molecular Biochemicals) instead of Taq DNA polymerase to increase the fidelity of DNA synthesis. The template genomic DNA for PCR was prepared from S. cerevisiae ATCC 44774 according to the method of Wach et al. (22). The reaction mixture contained 0.5 μM each 5′ and 3′ primer, 0.2 mM deoxynucleoside triphosphate, 2.0 mM MgSO4, 1× PCR buffer (supplied by the manufacturer), and 0.5 μg of template genomic DNA and 2.5 U of Pwo DNA polymerase per 50 μl. The mixture was subjected to 30 cycles of the following treatment: denaturation for 1 min at 94°C, annealing for 1 min at 50°C, and extension for 2 min at 72°C. The amplified DNA fragment of 1.6 kb was cloned into pGEM-5Zf(+) (Promega) at the EcoRV site. From the resulting plasmid, a 1.6-kb NdeI-BamHI fragment containing the open reading frame of the ALO1 gene was isolated and ligated into the NdeI-BamHI sites of the vectors pET-3a and pET-15b (Novagen) to yield pET-ALO3 and pET-ALO15, respectively. The two final constructs, pET-ALO3 and pET-ALO15, were transformed into E. coli BL21(DE3), BL21(DE3)pLysS, and BL21(DE3)pLysE (Novagen). Among the six combinations, only BL21(DE3)pLysS harboring pET-ALO3 showed remarkable overexpression of a recombinant ALO on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) over the course of hours after induction with 1 mM isopropyl-β-d-thiogalactopyranoside (Fig. 1). The molecular mass of the recombinant enzyme nearly coincided with that of ALO purified from S. cerevisiae (around 60 kDa) (11). To investigate whether the recombinant ALO was functionally expressed or not, a spectrophotometric ALO assay was performed as described previously (10). We found substantial ALO activity in the cell lysate, which was obtained after sonication (5 cycles consisting of 15 s of sonication interspersed with 30 s of cooling on ice, with a sonication power of 40 W) followed by centrifugation at 12,000 × g for 1 h (Fig. 2A). The specific activity of the recombinant ALO was 0.41 U/mg. This value is approximately 12-fold higher than the specific activity of S. cerevisiae ALO measured after Triton X-100 solubilization (12). When the cell lysate of E. coli containing ALO was further centrifuged at 120,000 × g for 1 h, most of the ALO activity was detected in the precipitate, indicating that the enzyme is located in the membrane fraction. The activity staining for ALO, performed according to the method described previously (18), confirmed that the recombinant ALO was functionally expressed in E. coli (Fig. 2B). Interestingly, the recombinant ALO was active only when Triton X-100 was included in the gel. Considering these observations and the fact that ALO is believed to be a mitochondrial membrane protein in yeasts (10, 11, 18), the functional recombinant ALO seems to be targeted to the cell membrane of E. coli and combined with membrane fragments in the cell lysate. Although the activity of ALO was found mainly in the membrane fraction of the cell lysate, a large proportion of the expressed ALO was directed into the insoluble inclusion body with no enzymatic activity. Several approaches were also tried to alter nonfunctional expression to functional expression. However, lowering of the temperature, addition of sucrose to the culture medium, and induction with a low concentration of isopropyl-β-d-thiogalactopyranoside could not reduce the inclusion body formation effectively. In addition, some attempts to reconstitute the activity of non-functionally expressed ALO were also fruitless.
FIG. 1.
Time course expression pattern of ALO on SDS-PAGE. Lysates (20 μg of protein) of E. coli cells expressing ALO, which were obtained 0 min (lane 1), 15 min (lane 2), 30 min (lane 3), 1 h (lane 4), and 3 h (lane 5) after induction with 1 mM isopropyl-β-d-thiogalactopyranoside, were electrophoresed on a 10% polyacrylamide gel. Numbers on the left side refer to molecular masses of the standards (in kilodaltons). The arrow on the right side indicates ALO.
FIG. 2.
Functional expression of ALO in E. coli. (A) Spectrophotometric assay of ALO activity. The enzyme activities in the lysates obtained from the cells harboring the parental vector (lane 1) and from the cells expressing ALO (lane 2) were measured in the absence (□) or the presence (■) of 20 μM FAD. (B) Activity staining of ALO. The lysates (30 μg of protein) from the cells harboring the parental vector (lanes 1 and 3) and from the cells expressing ALO (lanes 2 and 4) were electrophoresed in a nondenaturing gel containing 0.1% Triton X-100, followed by Coomassie blue staining (lanes 1 and 2) and activity staining (lanes 3 and 4).
It has been reported that ALO contains a covalently bound flavin adenine dinucleotide (FAD) as a prosthetic group (10, 11, 18). In accordance with these reports, a putative binding site for covalently bound FAD of oxygen-dependent oxidoreductase (PROSITE: PS00862) has been found in the enzyme (11). To determine whether the recombinant ALO also maintains its covalently FAD-binding property during expression, the fluorescence detection assay for covalently bound FAD was performed as described by Nishikimi et al. (17). The lysate of the cells expressing ALO was subjected to SDS-PAGE and soaked in a solution containing 7% acetic acid. When illuminated with UV light for the detection of FAD fluorescence, the gel showed no fluorescent band at the expected position of ALO. Furthermore, the addition of FAD to the assay mixture did not enhance the activity of the recombinant ALO (Fig. 2A). Taken together, these observations suggested that a sufficient amount of FAD is noncovalently incorporated in the recombinant ALO.
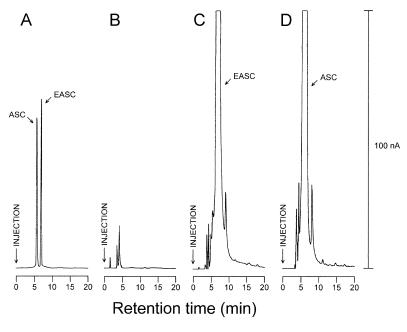
To measure the intracellular EASC, high-performance liquid chromatography (HPLC)-electrochemical detection was performed according to the method described previously (11). To induce the biosynthesis of EASC, E. coli cells were suspended in 0.2 M potassium phosphate (pH 6.1)–1 mM EDTA–10 mM d-arabinono-1,4-lactone and incubated for 2 h at 30°C. As expected for the cells harboring the empty vector, the electrochemical chromatogram did not show any distinct peak (Fig. 3B). In the case of the cells expressing ALO, however, a dominant peak was found at a retention time of 6.48 min, which was identical to that of EASC (Fig. 3C). The amount of EASC produced in the recombinant E. coli was calculated to be 178 μg · (g of wet cells)−1, which is approximately 31-fold higher than the amount present in S. cerevisiae (12). The incubation of the cells with 10 mM l-galactono-1,4-lactone swung the elution in favor of another, which had a retention time of 5.50 min and thereby corresponded to that of ASC (Fig. 3D). The amount of ASC produced in the cells was calculated to be 183 μg · (g of wet cells)−1. These results also mean that the useful dual expressions are possible, depending upon the substrates. Interestingly, EASC was also formed in small amounts when either d-glucose or d-arabinose was used instead of d-arabinono-1,4-lactone as a substrate (data not shown). However, ASC was never detected in either case. These observations suggested that d-arabinono-1,4-lactone is a natural constituent of E. coli but that l-galactono-1,4-lactone is not and that the presence of ALO is enough for E. coli to produce EASC.
FIG. 3.
HPLC-electrochemical detection of 10% trichloroacetic acid-soluble extracts of E. coli cells. (A) Standards, ASC and EASC, each at a concentration of 50 μM. (B) Extracts of the cells harboring the parental vector after incubation with 10 mM d-arabinono-1,4-lactone. (C) Extracts of the cells expressing ALO after incubation with 10 mM d-arabinono-1,4-lactone. (D) Extracts of the cells expressing ALO after incubation with 10 mM l-galactono-1,4-lactone. The maximum output current was adjusted to 100 nA, as indicated on the right side of the chromatograms.
ASC is one of the most well-known biological antioxidants. It functions as a free radical scavenger and acts as a primary defense. However, there are few definite answers to its precise mechanism of action and other roles. According to our previous study (11), EASC should be considered an antioxidant of high standing no less than ASC should be. Besides its role as an antioxidant, EASC is expected to carry out various functions similar to those of ASC. Uncovering many veiled functions and mechanisms of EASC entails the isolation of larger quantities for more detailed works. However, the methods for organic synthesis of EASC so far developed (9, 21) are very complicated, laborious, and costly. And even though some organic chemical approaches were tried to synthesize ASC starting from l-galactono-1,4-lactone (7, 8), a byproduct of the beet sugar industry, there has not been any suggestion about using bacterial methods, which are unquestionably the most familiar to us. In E. coli, this is the first demonstration that an EASC- or ASC-producing enzyme is overexpressed in a functional form. It will be very interesting to extend our study so as to make E. coli cells capable of overproducing EASC or ASC by adding just some sugars. There are two enzymes reported to be involved in the biosynthetic pathway of EASC in yeasts; one of them is ALO, and the other is d-arabinose dehydrogenase (13, 14). Therefore, coexpression of ALO and another sugar dehydrogenase may give us the most useful system for EASC or ASC overproduction.
Acknowledgments
This work was supported by a research grant to the SRC (Research Center for Molecular Microbiology, Seoul National University) from the Korea Science and Engineering Foundation (KOSEF).
REFERENCES
- 1.Arrigoni O. Ascorbate system in plant development. J Bioenerg Biomembr. 1994;26:407–419. doi: 10.1007/BF00762782. [DOI] [PubMed] [Google Scholar]
- 2.Asard H, Horemans N, Caubergs R J. Transmembrane electron transport in ascorbate-loaded plasma membrane vesicles from higher plants involves a b-type cytochrome. FEBS Lett. 1992;306:143–146. doi: 10.1016/0014-5793(92)80986-q. [DOI] [PubMed] [Google Scholar]
- 3.Bendich A, Machlin L J, Scandurra O, Burton G W, Wayner D D M. The antioxidant role of vitamin C. Adv Free Radical Biol Med. 1986;2:419–444. [Google Scholar]
- 4.Burns J J. Missing step in man, monkey and guinea pig required for the biosynthesis of l-ascorbic acid. Nature. 1957;180:533. doi: 10.1038/180553a0. [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee I B. Evolution and the biosynthesis of ascorbic acid. Science. 1973;182:1271–1272. doi: 10.1126/science.182.4118.1271. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhuri C R, Chatterjee I B. l-Ascorbic acid synthesis in birds: phylogenetic trend. Science. 1969;164:435–436. doi: 10.1126/science.164.3878.435. [DOI] [PubMed] [Google Scholar]
- 7.Csiba M, Cleophax J, Petit S, Gero S D. An efficient three step synthesis of vitamin C from l-galactono-1,4-lactone, a by-product of the sugar industry. Tetrahedron Lett. 1992;33:5059–5060. [Google Scholar]
- 8.Csiba M, Cleophax J, Petit S, Gero S D. An expedient and practical three-step synthesis of vitamin C from a byproduct of the sugar industry: the l-galactono-1,4-lactone pathway. J Org Chem. 1993;58:7281–7282. [Google Scholar]
- 9.Gan L X, Seib P A. Synthesis of d-erythroascorbic acid from d-glucose. Carbohydr Res. 1991;220:117–125. [Google Scholar]
- 10.Huh W-K, Kim S-T, Yang K-S, Seok Y-J, Hah Y C, Kang S-O. Characterisation of d-arabinono-1,4-lactone oxidase from Candida albicans ATCC 10231. Eur J Biochem. 1994;225:1073–1079. doi: 10.1111/j.1432-1033.1994.1073b.x. [DOI] [PubMed] [Google Scholar]
- 11.Huh W-K, Lee B-H, Kim S-T, Kim Y-R, Rhie G-E, Baek Y-W, Hwang C-S, Lee J-S, Kang S-O. d-Erythroascorbic acid is an important antioxidant molecule in Saccharomyces cerevisiae. Mol Microbiol. 1998;30:895–903. doi: 10.1046/j.1365-2958.1998.01133.x. [DOI] [PubMed] [Google Scholar]
- 12.Huh W-K. Ph.D. thesis. Seoul, Korea: Seoul National University; 1998. [Google Scholar]
- 13.Kim S-T, Huh W-K, Kim J-Y, Hwang S-W, Kang S-O. d-Arabinose dehydrogenase and biosynthesis of erythroascorbic acid in Candida albicans. Biochim Biophys Acta. 1996;1297:1–8. doi: 10.1016/0167-4838(96)00077-5. [DOI] [PubMed] [Google Scholar]
- 14.Kim S-T, Huh W-K, Lee B-H, Kang S-O. d-Arabinose dehydrogenase and its gene from Saccharomyces cerevisiae. Biochim Biophys Acta. 1998;1429:29–39. doi: 10.1016/s0167-4838(98)00217-9. [DOI] [PubMed] [Google Scholar]
- 15.Nick J A, Leung C T, Loewus F A. Isolation and identification of erythroascorbic acid in Saccharomyces cerevisiae and Lypomyces starkeyi. Plant Sci. 1986;46:181–187. [Google Scholar]
- 16.Niki E. Vitamin C as an antioxidant. World Rev Nutr Diet. 1991;64:1–30. doi: 10.1159/000418567. [DOI] [PubMed] [Google Scholar]
- 17.Nishikimi M, Kiuchi K, Yagi K. Detection of l-gulono-γ-lactone oxidase on SDS-polyacrylamide gels by the fluorescence of its covalently bound flavin. FEBS Lett. 1977;81:323–325. doi: 10.1016/0014-5793(77)80545-0. [DOI] [PubMed] [Google Scholar]
- 18.Nishikimi M, Noguchi E, Yagi K. Occurrence in yeast of l-galactonolactone oxidase which is similar to a key enzyme for ascorbic acid biosynthesis in animals, l-gulonolactone oxidase. Arch Biochem Biophys. 1978;191:479–486. doi: 10.1016/0003-9861(78)90386-7. [DOI] [PubMed] [Google Scholar]
- 19.Peterkofsky B. Ascorbate requirement for hydroxylation and secretion of procollagen: relationship to inhibition of collagen synthesis in scurvy. Am J Clin Nutr. 1991;54:1135S–1140S. doi: 10.1093/ajcn/54.6.1135s. [DOI] [PubMed] [Google Scholar]
- 20.Rebouche C J. Ascorbic acid and carnitine biosynthesis. Am J Clin Nutr. 1991;54:1147S–1152S. doi: 10.1093/ajcn/54.6.1147s. [DOI] [PubMed] [Google Scholar]
- 21.Shao Y-Y, Seib P A, Kramer K J, Van Galen D A. Synthesis and properties of d-erythroascorbic acid and its vitamin C activity in the tobacco hornworm (Manduca sexta) J Agric Food Chem. 1993;41:1391–1396. [Google Scholar]
- 22.Wach A, Pick H, Philippsen P. Procedures for isolating yeast DNA for different purposes. In: Johnston J R, editor. Molecular genetics of yeasts. Oxford, United Kingdom: IRL Press; 1994. pp. 1–16. [Google Scholar]
- 23.Wheeler G L, Jones M A, Smirnoff N. The biosynthetic pathway of vitamin C in higher plants. Nature. 1998;393:365–369. doi: 10.1038/30728. [DOI] [PubMed] [Google Scholar]