Abstract
The iap gene encodes the protein p60, which is common to all Listeria species. A previous comparison of the DNA sequences indicated conserved and species-specific gene portions. Based on these comparisons, a combination consisting of only five different primers that allows the specific detection and differentiation of Listeria species with a single multiplex PCR and subsequent gel analysis was selected. One primer was derived from the conserved 3′ end and is specific for all Listeria species; the other four primers are specific for Listeria monocytogenes, L. innocua, L. grayi, or the three grouped species L. ivanovii, L. seeligeri, and L. welshimeri, respectively. The PCR method, which also enables the simultaneous detection of L. monocytogenes and L. innocua, was evaluated against conventional biotyping with 200 food hygiene-relevant Listeria strains. The results indicated the superiority of this technique. Thus, this novel type of multiplex PCR may be useful for rapid Listeria species confirmation and for identification of Listeria species for strains isolated from different sources.
The genus Listeria comprises six characterized species: Listeria monocytogenes, L. innocua, L. ivanovii, L. seeligeri, L. welshimeri, and L. grayi (25). Among these gram-positive, nonsporulating and motile species, only L. monocytogenes is a human and animal pathogen, capable of causing severe infections like septicemia, encephalitis, and meningitis, especially in immunocompromised individuals, newborns, and pregnant women (26). L. monocytogenes belongs to the facultative intracellular bacteria that invades, replicates, and multiplies in a variety of mammalian cells (22). A number of genes and gene products necessary for the intracellular survival of this pathogen have been previously reviewed (22).
Several large outbreaks of listeriosis have been associated with contaminated commercial foodstuffs, such as vegetables, milk, and meat products, on which these bacteria can multiply even at low temperatures (26). Contamination not only is caused during food processing but also begins with the production of raw food materials in the environment. Some Listeria species like L. monocytogenes and L. innocua have been isolated from various environmental samples, e.g., soil, vegetation, and human and animal feces, indicating the widespread presence of the pathogen in nature (26). Due to its frequent occurrence in food, L. innocua can be considered an indicator bacterium for the presence of L. monocytogenes. However, little is known about the occurrence and distribution of other Listeria species. Species-specific identification with biochemical standard methods which include sugar fermentations or the CAMP phenomenon (27) are laborious and time-consuming and can require up to 7 days according to International Dairy Federation (IDF) standard 143:1995 (16). Moreover, isolates which demonstrated significant differences in main biochemical features were described previously (1, 4). Other, faster procedures like PCR and immunological or bacteriophage lysis techniques which might allow a more rapid monitoring of all Listeria species are limited for this purpose because they detect only the genus Listeria or only L. monocytogenes (7, 9, 12, 19, 23), thus lacking the ability to simultaneously characterize species other than L. monocytogenes. Especially would the coidentification of L. innocua be beneficial, as this species can be found associated with the occurrence of L. monocytogenes (11, 18), which association may lead to typing of only one of these two species.
To overcome some of these problems, we developed a novel multiplex PCR containing a minimum number of different primers. For this purpose, we used the previously characterized iap gene common to all members of the genus Listeria as the target because the comparison of all iap genes indicated there were conserved gene portions at the 5′ and 3′ ends, while the internal portions are species-specific (8). The iap gene of L. monocytogenes encodes the major extracellular protein p60 (20), which has been shown to be basically an essential murein hydrolase required for septum separation in a late step in cell division (3, 28). In addition, the L. monocytogenes p60 plays a role in the adherence of this organism to certain eukaryotic cells and confers immune protection to mice following infection with this pathogen (5, 13, 14, 21).
In several studies, iap-derived primers have been applied for the specific identification of L. monocytogenes (1, 4, 8, 10, 12). Recently, we developed a simple method for the simultaneous specific identification and differentiation of L. monocytogenes isolates by PCR amplification and size comparison of a hypervariable internal iap gene fragment. The encoded so-called Thr-Asn repeat domain, which is located on this fragment, also shows an extended-length polymorphism in strains of the same serotype (6). However, the new method described here enables the simultaneous detection of Listeria species and differentiation between L. monocytogenes, L. innocua, and two groups containing very rarely occurring species, the first containing L. seeligeri, L. welshimeri, and L. ivanovii and the second containing L. grayi and L. grayi subsp. murrayi, by a single amplification reaction.
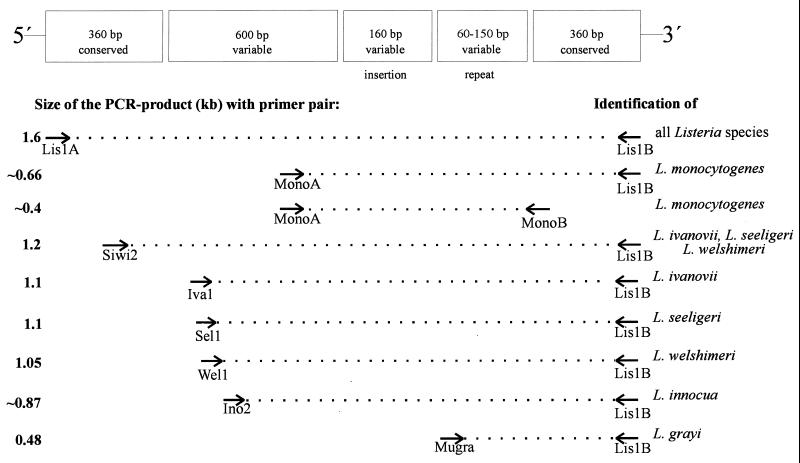
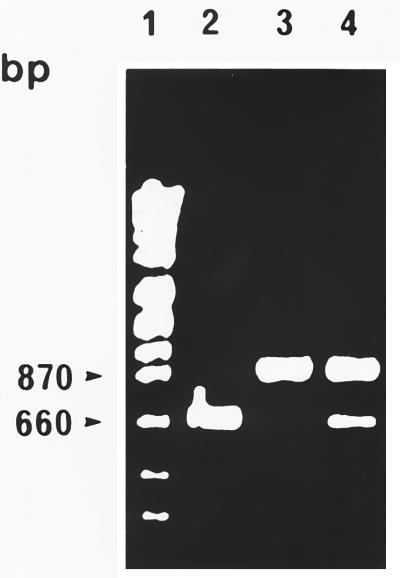
Based on the iap DNA sequence comparison, we previously selected primer combinations for the specific identification by PCR of all serotypes of L. monocytogenes (primers MonoA and MonoB) (Fig. 1) and all serotypes of L. innocua (primers Ino2 [5′-ACTAGCACTCCAGTTGTTAAAC-3′] and Lis1B [5′-TTATACGCGACCGAAGCCAAC-3′]) (8). PCR with the latter species leads to a specific product of 870 bp. In addition, the grouped species L. ivanovii, L. seeligeri, and L. welshimeri could be specifically identified by using primer pair Siwi2 (5′-TAACTGAGGTAGCGAGCGAA-3′) and Lis1B, which yields a PCR product comprising approximately 1.2 kb (8). Since primer Lis1B binds to the 3′ end of all listerial iap genes (8), this primer was selected to represent the fixed downstream primer along with four specific upstream primers in a multiplex PCR mix.
FIG. 1.
Binding regions of the primers within the iap genes selected for PCR identification of Listeria spp. The conserved and variable gene portions are schematically summarized according to Bubert et al. (5). Note that the third gene portion from the left site codes for a putative additional substrate binding domain which is not present in the iap genes of L. monocytogenes and L. innocua (28).
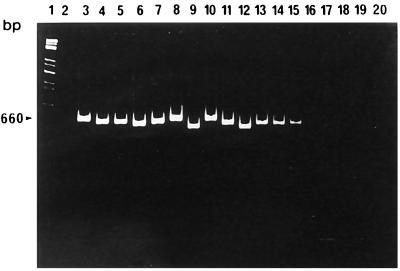
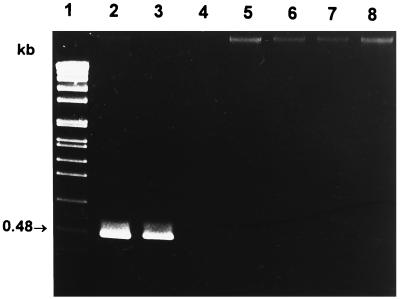
Initially, we ensured that primer combination MonoA (5′-CAAACTGCTAACACAGCTACT-3′) and Lis1B also identified all L. monocytogenes serotypes. Chromosomal DNA of bacteria used for amplification was prepared as described earlier (4). Reaction mixtures each contained 100 ng of each primer, 200 μM (each) deoxynucleoside triphosphate, 1.5 mM MgCl2, 1× PCR buffer, 50 to 100 ng of chromosomal DNA, and 1.5 U of Taq polymerase (Promega, Mannheim, Germany) according to standard protocols (15). PCR conditions are indicated in the figure legends. As shown in Fig. 2, L. monocytogenes strains belonging to all known serotypes could be specifically identified by a PCR product of approximately 660 bp. The slight size differences of the PCR products are due to the length polymorphism found in this amplified gene portion (see above). No cross-reactions with other listerial DNA were observed. We next selected a primer pair (MugraI and Lis1B) specific for the species L. grayi and L. grayi subsp. murrayi. As shown in Fig. 3, this primer pair yielded a specific PCR product of 480 bp in size only with these two species, while all other Listeria species gave no PCR product.
FIG. 2.
L. monocytogenes-specific PCR products with the primer pair MonoA and Lis1B. PCR conditions were as follows: 30 cycles, each at 95°C for 15 s, 58°C for 30 s, and 72°C for 45 s. Lanes: 1, molecular weight standard; 2, control reaction (all reagent ingredients except chromosomal DNA); 3, L. monocytogenes EGD serovar 1/2a (sv1/2a); 4, L. monocytogenes SLCC 2755 sv1/2b; 5, L. monocytogenes NCTC 5348 sv1/2c; 6, L. monocytogenes NCTC 5105 sv3a; 7, L. monocytogenes SLCC 5543 sv3b; 8, L. monocytogenes SLCC 2479 sv3c; 9, L. monocytogenes L 99 sv4a; 10, L. monocytogenes SLCC 4561 sv4ab; 11, L. monocytogenes SLCC 4013 sv4b; 12, L. monocytogenes ATCC 19116 sv4c; 13, L. monocytogenes ATCC 19117 sv4d; 14, L. monocytogenes ATCC 19118 sv4e; 15, L. monocytogenes SLCC 2482 sv7; 16, L. innocua sv6a; 17, L. welshimeri SLCC 5334; 18, L. seeligeri SLCC 3945; 19, L. ivanovii ATCC 19119; 20, L. grayi. PCR products were separated in a 4% polyacrylamide gel and stained with ethidium bromide.
FIG. 3.
L. grayi-specific PCR products with the primer combination MugraI and Lis1B. PCR conditions were as follows: 30 cycles, each at 95°C for 15 s, 58°C for 30 s, and 72°C for 45 s. Lanes: 1, molecular weight standard; 2, L. grayi (Institute for Milk Hygiene culture collection); 3, L. grayi subsp. murrayi (Institute for Milk Hygiene culture collection); 4, L. monocytogenes EGD; 5, L. innocua NCTC 11288 sv6a; 6, L. ivanovii ATCC 19119; 7, L. seeligeri SLCC 3945; 8, L. welshimeri SLCC 5334. PCR products were separated in a 1% agarose gel and stained with ethidium bromide.
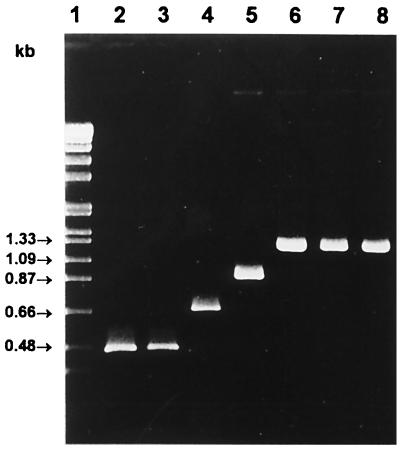
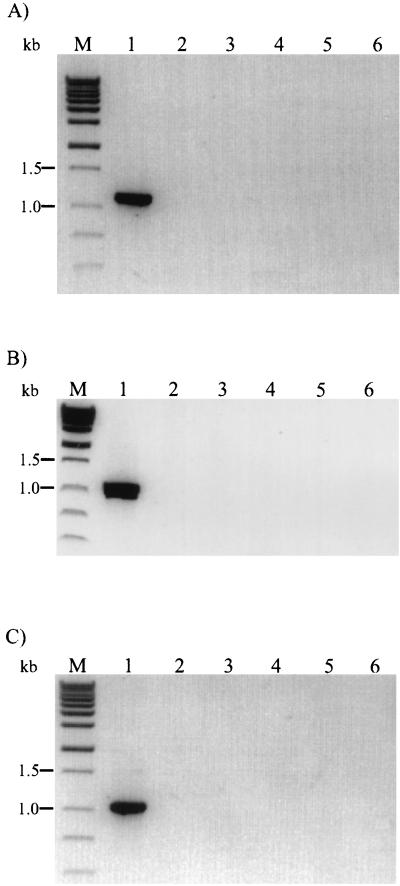
The fact that all specific PCR products can be easily distinguished by size comparisons in agarose gels enabled the generation of a multiplex PCR consisting of the four species-specific upstream primers MugraI, MonoA, Ino2, and Siwi2 and the conserved downstream primer Lis1B. The amplification procedures were the same as indicated above. As shown in Fig. 4, the presence of L. grayi or L. grayi subsp. murrayi led to the expected 480-bp product. When chromosomal DNA of L. monocytogenes was added to the reaction mix, the expected band of approximately 660 bp was observed. The species L. innocua was identified by the 870-bp DNA fragment, and the L. seeligeri-L. welshimeri-L. ivanovii group was identified by the occurrence of the 1.2-kb PCR product. No cross-reactions or additional bands were observed with other gram-positive or gram-negative bacterial species (data not shown). The control group included Bacillus subtilis, Brochothrix thermosphacta, Enterococcus faecalis, Staphylococcus aureus, Escherichia coli, and Salmonella typhimurium. The three rare species L. ivanovii, L. seeligeri, and L. welshimeri are not distinguishable by this method but can be easily differentiated on blood agar plates. However, to complete the list of specific identifications of all Listeria species by PCR, upstream primers in combination with Lis1B specific for each of these three species were selected. Primer Iva1 (5′-CTACTCAAGCGCAAGCGGCAC-3′) for L. ivanovii, primer Sel1 (5′-TACACAAGCGGCTCCTGCTCAAC-3′) for L. seeligeri, and primer Wel1 (5′-CCCTACTGCTCCAAAAGCAGCG-3′) for L. welshimeri were derived from species-specific internal iap gene portions. As shown in Fig. 5, all primer combinations led to the specific identification of the three species. In addition, specificity was confirmed with six different isolates of each species (data not shown).
FIG. 4.
Identification and differentiation of Listeria species by multiplex PCR containing the five primers MugraI, MonoA, Ino2, and Siwi2, and Lis1B (Lis-Mix). Reaction conditions were as follows: 30 cycles, each at 95°C for 15 s, 58°C for 30 s, and 72°C for 50 s. Lanes: 1, molecular weight standard; 2, L. grayi; 3, L. grayi subsp. murrayi; 4, L. monocytogenes EGD sv1/2a; 5, L. innocua sv6a; 6, L. ivanovii; 7, L. seeligeri; 8, L. welshimeri. PCR products were separated in a 1.2% agarose gel and stained with ethidium bromide.
FIG. 5.
Specific identification of L. ivanovii (A), L. seeligeri (B), and L. welshimeri (C) by PCR with primer pairs Iva1 and Lis1B, Sel1 and Lis1B, and Wel1 and Lis1B, respectively. PCR conditions were as follows: 35 cycles, each at 95°C for 15 s and 62°C for 30 s. Lanes in panel A: 1, L. ivanovii; 2, L. seeligeri; 3, L. welshimeri; 4, L. grayi; 5, L. monocytogenes EGD; 6, L. innocua. Lanes in panel B: 1, L. seeligeri; 2, L. ivanovii; 3, L. welshimeri; 4, L. grayi; 5, L. monocytogenes EGD; 6, L. innocua. Lanes in panel C: 1, L. welshimeri; 2, L. ivanovii; 3, L. seeligeri; 4, L. grayi; 5, L. monocytogenes EGD; 6, L. innocua. Lanes M, molecular weight markers. PCR products were separated in a 1.2% agarose gel and stained with ethidium bromide.
To test the suitability of the multiplex PCR for coidentification of different Listeria species with a single reaction, we applied PCRs containing the chromosomal DNAs of L. innocua and L. monocytogenes. As shown in Fig. 6, the 660- and 870-bp bands which lead to the identification of both species could be obtained. The multiplex PCR also led to two clear products of the expected sizes when other bacterial combinations, such as L. innocua and L. grayi, L. monocytogenes and L. grayi, and L. monocytogenes and L. ivanovii, were used. However, when PCR was performed with the chromosomal DNA of a third Listeria species, unclear bands occasionally appeared; thus, the method might not be advisable for the simultaneous identification of more than two Listeria species from the four different groups. These additional amplification products might occur by in vitro recombination of PCR products, which can be generated by the Taq DNA polymerase jumping between templates during amplification (24).
FIG. 6.
Parallel identification and differentiation of L. monocytogenes and L. innocua by multiplex PCR containing the Lis-Mix primers. Reaction conditions were the same as indicated in the legend of Fig. 4. Lanes: 1, molecular weight marker; 2, L. monocytogenes EGD; 3, L. innocua sv6b; 4, L. monocytogenes EGD and L. innocua sv6b. PCR products were separated in a 1.2% agarose gel and stained with ethidium bromide.
Next, the multiplex PCR was evaluated with pure cultures derived from the type culture collection of the Institute for Milk Hygiene, Vienna, Austria. All isolates were characterized by using either IDF method 143:1995 (16) or International Organization for Standardization (ISO) method 11290-1 (17) as the standard method to demonstrate the species-specific biochemical features of relevance. A total of 199 pure cultures, which had been isolated mostly from foodstuffs but also from clinical samples, were screened by the iap-specific multiplex technique. The numbers of isolates were 100 and 49 for L. monocytogenes and L. innocua, respectively, and a total of 50 for the L. seeligeri-L. welshimeri-L. ivanovii and L. grayi-L. grayi subsp. murrayi groups. All strains from the type culture collection were grown in tryptone soy broth at 37°C overnight and subjected to standard detection procedures according to IDF or to multiplex PCR analyses.
Overall conformity of microbiological versus multiplex PCR results was found in 100% of the analyzed L. monocytogenes and L. innocua strains. In a single case the multiplex PCR detected L. innocua in addition to L. monocytogenes. Microbiological reinvestigation revealed that the L. monocytogenes culture was indeed contaminated with L. innocua. With regard to conformity between results from both methods, the L. seeligeri-L. welshimeri-L. ivanovii group results proved to be slightly divergent. Out of 50 isolates tested, 35 strains showed concordant results after a first PCR trial. One isolate was shown to be L. monocytogenes whereas four isolates proved to be L. innocua by PCR. However, the results obtained by PCR were confirmed by repeated microbiological investigation by plating the cultures onto Rapid Lis-Agar and confirmation of suspect colonies was obtained by CAMP reaction. Triplicate PCR trials did not lead to a positive amplification of 10 isolates although microbiological investigation by IDF standard 143:1995 identified the presence of five L. seeligeri strains, four L. welshimeri strains, and one L. ivanovii strain. Repeated microbiological investigation proved these cultures contained nonlisterial cells, thus confirming results obtained by PCR. PCR identified conclusively all 50 strains for this group, whereas only 35 strains were correctly identified by standard biotyping.
In conclusion, we have developed a novel multiplex PCR for the specific identification and differentiation of L. monocytogenes, L. innocua, L. grayi, and the three grouped species L. ivanovii, L. seeligeri, and L. welshimeri. The latter three can be differentiated by subsequent PCRs; thus, a complete PCR identification protocol specific for each Listeria species is now available. In contrast to what is required by other typing methods, e.g., API Listeria (2), no pure cultures are required for accurate typing of Listeria spp. However, this method reaches its limit when more than two Listeria species from the four different groups are present. Moreover, the comparison with conventional biotyping indicates that identification of Listeria species by PCR with iap as a stable chromosomal target gene is more reliable, less time-consuming, and less laborious, and thus very cost-effective. In addition, this multiplex PCR may also allow a more systematic study of the occurrence of Listeria spp. in food or in the environment. The application for the direct detection of Listeria spp. in food samples by this method is in progress.
(Parts of this work were presented at the XIIIth International Symposium on Problems of Listeriosis Congress in 1998 in Halifax, Nova Scotia, Canada.)
Acknowledgments
We thank D. Cunningham (Merck KGaA, Darmstadt, Germany) for critically reading the manuscript.
This work was supported by grant BMFT KI88059 from the Bundesministerium für Forschung und Technologie, by the Universitätsbund Würzburg, and by Merck KGaA. B.Y. received a grant (KMAF-SGRP-198043-3) from the Korean Ministery of Agriculture and Forestry.
REFERENCES
- 1.Allerberger F, Dierich M, Petranyi G, Lalic M, Bubert A. Nonhemolytic strains of Listeria monocytogenes detected in milk products using VIDAS immunoassay kit. Zentbl Hyg. 1997;200:189–195. [PubMed] [Google Scholar]
- 2.Bille J, Catimel B, Bannerman E, Jaquet C, Yersin M-N, Caniaux I, Monget D, Rocourt J. API Listeria, a new and promising one-day system to identify Listeria isolates. Appl Environ Microbiol. 1992;58:1857–1860. doi: 10.1128/aem.58.6.1857-1860.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bubert A, Kestler H, Götz M, Böckmann R, Goebel W. The Listeria monocytogenes iap gene as an indicator gene for the study of PrfA-dependent regulation. Mol Gen Genet. 1997;256:54–62. doi: 10.1007/s004380050545. [DOI] [PubMed] [Google Scholar]
- 4.Bubert A, Riebe J, Schnitzler N, Schönberg A, Goebel W, Schubert P. Isolation of catalase-negative Listeria monocytogenes strains from listeriosis patients and their rapid identification by anti-p60 antibodies and/or PCR. J Clin Microbiol. 1997;35:179–183. doi: 10.1128/jcm.35.1.179-183.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bubert A, Kuhn M, Goebel W, Köhler S. Structural and functional properties of the p60 proteins from different Listeria species. J Bacteriol. 1992;174:8166–8171. doi: 10.1128/jb.174.24.8166-8171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bubert, A., M. Rauch, W. Goebel, D. Mack, and H.-A. Elsner. Submitted for publication.
- 7.Bubert A, Schubert P, Köhler S, Frank R, Goebel W. Synthetic peptides derived from the Listeria monocytogenes p60 protein as antigens for the generation of polyclonal antibodies specific for secreted cell-free L. monocytogenes p60 proteins. Appl Environ Microbiol. 1994;60:3120–3127. doi: 10.1128/aem.60.9.3120-3127.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bubert A, Köhler S, Goebel W. The homologous and heterologous regions within the iap gene allow genus- and species-specific identification of Listeria spp. by polymerase chain reaction. Appl Environ Microbiol. 1992;58:2625–2632. doi: 10.1128/aem.58.8.2625-2632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deneer H, Boychuk I. Species-specific identification of Listeria monocytogenes by DNA amplification. Appl Environ Microbiol. 1991;57:606–609. doi: 10.1128/aem.57.2.606-609.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elsner H-A, Sobottka I, Bubert A, Albrecht H, Laufs R, Mack D. Catalase-negative Listeria monocytogenes causing lethal sepsis and meningitis in an adult hematologic patient. Eur J Clin Microbiol Infect Dis. 1996;15:965–967. doi: 10.1007/BF01690521. [DOI] [PubMed] [Google Scholar]
- 11.Ferron P, Michard J. Distribution of Listeria spp. in confectioners’ pastries from western France: comparison of enrichment methods. Int J Food Microbiol. 1993;18:289–303. doi: 10.1016/0168-1605(93)90152-7. [DOI] [PubMed] [Google Scholar]
- 12.Furrer B, Candrian U, Hoefelein C, Luethy J. Detection and identification of Listeria monocytogenes in cooked sausage products and in milk by in vitro amplification of haemolysin gene fragments. J Appl Bacteriol. 1991;70:372–379. doi: 10.1111/j.1365-2672.1991.tb02951.x. [DOI] [PubMed] [Google Scholar]
- 13.Geginat G, Lalic M, Kretschmar M, Goebel W, Hof H, Palm D, Bubert A. Th1 cells specific for a secreted protein of Listeria monocytogenes are protective in vivo. J Immunol. 1998;160:6046–6056. [PubMed] [Google Scholar]
- 14.Harty J, Pamer E G. CD8 T lymphocytes specific for the secreted p60 antigen protect against Listeria monocytogenes. J Immunol. 1995;154:4642–4650. [PubMed] [Google Scholar]
- 15.Innis M A, Gelfand D H, Sninsky J J. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. [Google Scholar]
- 16.International Dairy Federation. Milk and milk products—detection of Listeria monocytogenes. IDF standard 143A:1995. Brussels, Belgium: International Dairy Federation; 1995. [Google Scholar]
- 17.International Organization of Standardization. Microbiology of food and animal feeding stuffs: horizontal method for the detection and enumeration of Listeria monocytogenes. Part 1: detection method. ISO standard 11290-1. Geneva, Switzerland: International Organization of Standardization; 1996. [Google Scholar]
- 18.Jeyasekaran G, Karunasagar I, Karunasagar I. Incidence of Listeria spp. in tropical fish. Int J Food Microbiol. 1996;31:333–340. doi: 10.1016/0168-1605(96)00980-4. [DOI] [PubMed] [Google Scholar]
- 19.Johnson W M, Tyler S D, Ewan E P, Ashton F E, Wang G, Rozee K R. Detection of genes coding for listeriolysin and Listeria monocytogenes antigen A (lmaA) in Listeria spp. by the polymerase chain reaction. Microb Pathog. 1992;12:79–86. doi: 10.1016/0882-4010(92)90068-y. [DOI] [PubMed] [Google Scholar]
- 20.Köhler S, Leimeister-Wächter M, Chakraborty T, Lottspeich F, Goebel W. The gene coding for protein p60 of Listeria monocytogenes and its use as a specific probe for Listeria monocytogenes. Infect Immun. 1990;58:1943–1950. doi: 10.1128/iai.58.6.1943-1950.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn M, Goebel W. Identification of an extracellular protein of Listeria monocytogenes possibly involved in intracellular uptake by mammalian cells. Infect Immun. 1989;57:55–61. doi: 10.1128/iai.57.1.55-61.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn M, Goebel W. Molecular studies on the virulence of Listeria monocytogenes. Genet Eng. 1995;17:31–51. [PubMed] [Google Scholar]
- 23.Loessner M J, Rees C E D, Stewart G S A B, Scherer S. Construction of luciferase reporter bacteriophage A511::luxAB for rapid and sensitive detection of viable Listeria cells. Appl Environ Microbiol. 1996;62:1133–1140. doi: 10.1128/aem.62.4.1133-1140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pääbo S, Irwin D M, Wilson A C. DNA damage promotes jumping between templates during enzymatic amplification. J Biol Chem. 1990;265:4718–4721. [PubMed] [Google Scholar]
- 25.Rocourt J, Cossart P. Listeria monocytogenes. In: Doyle M P, Beuchat L R, Montville T J, editors. Food microbiology fundamentals and frontiers. Washington, D.C.: ASM Press; 1997. pp. 337–352. [Google Scholar]
- 26.Schuchat A, Swaminathan B, Broome C V. Epidemiology of human listeriosis. Clin Microbiol Rev. 1991;4:169–183. doi: 10.1128/cmr.4.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seeliger H P R, Jones D. Genus Listeria Pirie 1940, 383AL. In: Sneath P H A, Mair N S, Sharpe M E, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 2. Baltimore, Md: Williams & Wilkins; 1986. pp. 1235–1245. [Google Scholar]
- 28.Wuenscher M, Köhler S, Bubert A, Gerike U, Goebel W. The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J Bacteriol. 1993;175:3491–3501. doi: 10.1128/jb.175.11.3491-3501.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]