Abstract
The T cell-mediated immune responses associated with asymptomatic infection (AS) of SARS-CoV-2 remain largely unknown. The diversity of T-cell receptor (TCR) repertoire is essential for generating effective immunity against viral infections in T cell response. Here, we performed the single-cell TCR sequencing of the PBMC samples from five AS subjects, 33 symptomatic COVID-19 patients and eleven healthy controls to investigate the size and the diversity of TCR repertoire. We subsequently analyzed the TCR repertoire diversity, the V and J gene segment deference, and the dominant combination of αβ VJ gene pairing among these three study groups. Notably, we revealed significant TCR preference in the AS group, including the skewed usage of TRAV1-2-J33-TRBV6-4-J2-2 and TRAV1-2-J33-TRBV6-1-J2-3. Our findings may shed new light on understanding the immunopathogenesis of COVID-19 and help identify optimal TCRs for development of novel therapeutic strategies against SARS-CoV-2 infection.
Keywords: SARS-CoV-2, COVID-19, Asymptomatic, TCR repertoires, scTCR-seq, Immunotherapy
1. Introduction
The ongoing pandemic of coronavirus disease 2019 (COVID-19) has posed a serious public health threat to the international communities. As of May 27, 2022, there have been 525,467,084 confirmed cases of COVID-19, including 6,285,171 deaths worldwide reported to the World Health Organization [1]. The clinical presentations of SARS-CoV-2 infections are highly variable, ranging from asymptomatic to fatal conditions [2], [3]. As the number of asymptomatic individuals continues to increase worldwide, the asymptomatic infection has raised concerns among global public health authorities [4]. It is conceivable that the host immune responses may play important roles in shaping the disease outcomes. Indeed, the asymptomatic subjects have been suggested to generate timely and well-coordinated protective immune responses against SARS-CoV-2 [5], [6]. Sekine et al. reported that SARS-CoV-2-specific T cells were detectable in antibody-seronegative exposed family members and convalescent individuals with a history of asymptomatic and mild COVID-19 [7]. Altmann et al. observed that T cell memory is potentially more durable across asymptomatic, mild, and severe COVID-19 than B cell memory [8]. Mazzoni et al. found that asymptomatic patients displayed a significantly lower magnitude of both cell-mediated and humoral immune responses to the virus, as compared to symptomatic individuals [9]. While Reynolds et al. reported that neutralizing antibody titers were maintained irrespective of symptoms [10]. However, the immune responses, especially the T cell-mediated responses and the underlying mechanisms in the asymptomatic COVID-19 patients remain to be further characterized.
T cells are important components of cellular immunity, killing virus-infected cells through T cell receptor (TCR)-mediated recognition of viral antigens [11], [12]. Approximately 95 % of the TCRs are heterodimers of alpha (α) and beta (β) subunits in humans, which are shaped through the recombination (rearrangement) of variable (V), diversity (D), and joining (J) gene segments for the specificity of given antigen [13]. T cells undergo clonal expansion to generate cells with identical TCRs after activation by pathogen/antigen [14]. Therefore, unraveling the size and the diversity of TCR repertoire in the asymptomatic subjects may help understand the T cell-mediated immune responses against SARS-CoV-2 infection. Previous studies reported the TCR bias in COVID-19 patients [15], [16], [17]. In addition, Minervina A. et al. revealed the dynamics of memory T cells in patients with mild COVID-19 infection by T-cell receptor (TCR) sequencing [18]. Snyder T. et al. developed a method for diagnosis of SARS-CoV-2 infection based on TCR sequencing of blood samples [19]. Moreover, Wang P. et al. designed a novel T cell-based vaccine for SARS-CoV-2 variants and sarbecoviruses [20]. However, the TCR repertoires in the asymptomatic subjects (AS) COVID-19 patients remain unknown.
In this study, we performed single-cell TCR sequencing (scTCR-seq) of PBMCs from five asymptomatic individuals (AS), 33 symptomatic COVID-19 patients (SM) and eleven healthy controls (HC), and revealed the TCR repertoire diversity, the V and J gene segment preference, and the dominant combination of αβ VJ gene pairing across different disease states. Our findings may help understand the T-cell mediated immunity and identify optimal TCRs for immunotherapy against SASR-CoV-2 infection.
2. Materials and methods
2.1. Study cohort and blood sample collection
This study was approved by the Ethics Committee of The First affiliated Hospital of Xi'an Jiaotong University (XJTU1AF2020LSK-015) and The Renmin Hospital of Wuhan University (WDRY2020-K130). All participants enrolled in this study offered the written informed consent by themselves or their surrogates. The case definition and classification of the study subjects followed the Guidelines of the World Health Organization (WHO) and the “Guidelines on the Diagnosis and Treatment of the Novel Coronavirus Infected Pneumonia” developed by the National Health Commission of People's Republic of China (Table S1) [21], [22], [23]. The study cohort included eleven healthy controls (HC, n = 11) and 38 SARS-CoV-2-infected individuals (COV, n = 38) consisting of five asymptomatic subjects (AS, n = 5) and 33 symptomatic patients (SM, n = 33) (Table 1, Table S1). The peripheral blood was collected into the standard EDTA-K2 Vacuette Blood Collection Tubes (Jiangsu Yuli Medical Equipment Co., Ltd., China; Cat.Y09012282) and the peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll density gradient centrifugation method as described [24]. PBMCs were stored in the liquid nitrogen storage tank (−196 °C) until being used for the studies. Of note, two of the patients (C19 and C26) had blood samples collected at two different time points during this study. All the experimental procedures were completed inside a biosafety level 2 (BSL-2) laboratory at the Department of Clinical Diagnostic Laboratories, Renmin Hospital of Wuhan University.
Table 1.
Demographic and clinical features of the study cohort.
| Entire cohort (n = 49) | HC (n = 11) | AS (n = 5) | SM (n = 33) | P value | |
|---|---|---|---|---|---|
| Age, years (range 25 %–75 %) | 53.0 (45.0–66.0) | 40.0 (35.0–51.0) | 41.6 (23.5–61.0) | 59.9 (51.5–69.0) | <0.01 |
| Gender | 0.86 | ||||
| Female | 25 (51.0 %) | 6 (54.5 %) | 2 (40.0 %) | 17 (51.5 %) | / |
| Male | 24 (49.0 %) | 5 (45.5 %) | 3 (60.0 %) | 16 (48.5 %) | / |
| Mortality | 1 (2.0 %) | 0 | 0 | 1 (3.0 %) | 0.78 |
| Diagnostic tests | |||||
| Chest CT abnormal | 33 (67.3 %) | 0 | 0 | 33 (100 %) | <0.01 |
| Nucleic acid test (+) | 38 (77.6 %) | 0 | 5 (100 %) | 33 (100 %) | <0.01 |
| SARS-COV-2 IgM (+) | 22 (50 %) | 0 | 0 | 22 (66.7 %) | <0.01 |
| SARS-COV-2 IgG (+) | 36 (73.5 %) | 0 | 3 (60.0 %) | 33 (100 %) | <0.01 |
| Laboratory tests (SD) | |||||
| White blood cell count, ×109/L | 6.73 (2.84) | NA | 7.66 (2.93) | 6.58 (2.85) | 0.23 |
| Neutrophil count, ×109/L | 4.70 (2.72) | NA | 4.90 (2.19) | 4.67 (2.82) | 0.65 |
| Lymphocyte count, ×109/L | 1.30 (0.69) | NA | 1.79 (0.81) | 1.22 (0.66) | 0.12 |
| CRP, mg/L | 53.13 (36.10) | NA | 26.3 (n = 1) | 54.4 (36.48) | 0.58 |
| IL-2, pg/mL | 4.25 (1.90) | NA | 3.80 (0.46) | 4.33 (2.07) | 0.65 |
| IL-4, pg/mL | 3.07 (0.51) | NA | 2.99 (0.14) | 3.09 (0.56) | 0.77 |
| IL-6, pg/mL | 22.74 (52.43) | NA | 14.05 (16.69) | 24.12 (56.23) | 0.57 |
| IL-10, pg/mL | 8.02 (6.17) | NA | 10.26 (9.43) | 7.60 (5.72) | 0.82 |
| CD3, /μL | 838.94 (542.34) | NA | 1529.25 (264.62) | 749.87 (504.59) | 0.01 |
| CD4, /μL | 513.54 (325.23) | NA | 887.00 (225.54) | 465.35 (306.26) | 0.03 |
| CD8, /μL | 291.03 (222.41) | NA | 541.00 (143.77) | 258.77 (211.16) | 0.02 |
| CD19, /μL | 220.18 (155.09) | NA | 383.75 (116.52) | 199.07 (147.90) | 0.04 |
| IgM, g/L | 0.95 (0.35) | NA | 0.93 (0.54) | 0.95 (0.34) | 0.92 |
| IgG, g/L | 12.95 (4.86) | NA | 11.68 (2.43) | 13.04 (4.99) | 0.70 |
Notes: Data are n (%), age: median (ranging 25 %–75 %) or laboratory tests: mean with standard deviation (SD), unless otherwise stated. For statistical analyses, the Mann-Whitney U test was done for continuous variables which didn't conform to normal distribution and test for homogeneity of variance, and Pearson's χ2 test was done for categorical variables. Laboratory tests statistical analysis were conducted between with AS and SM group. P value <0.05 was considered as statistically significant. HC=healthy controls, AS = asymptomatic subjects, SM = symptomatic patients, NA = not available.
2.2. Clinical laboratory tests
All the clinical laboratory tests were conducted by the Clinical Laboratories at the Renmin Hospital of Wuhan University, including the routine blood tests, cytokines (IL-2, IL-4, IL-6, IL-10, AU5400, Beckman Coulter, Inc. USA), flow cytometry analysis of lymphocytes (CD3, CD4, CD8, CD19, CytoFLEX LX, Beckman Coulter, Inc. USA), and the SARS-CoV-2 specific IgM (Cat. 20203400769, YHLO Biotech) and SARS-CoV-2 specific IgG (Cat. 20203400770, YHLO Biotech) antibodies.
2.3. Preparation of single-cell suspensions
The frozen PBMC samples were retrieved from the liquid nitrogen storage tank, which were mixed with 10 mL of washing medium (90 % DMEM+10 % FBS) in a 15-mL polypropylene tube and centrifuged at 500g for 20 min. The supernatant was then removed (repeated this step twice). The cell pellets were resuspended with 500 μL of 1× PBS with 0.04 % BSA (A1933-25G, SIGMA), followed by adding 5 mL of 1× Red blood cell (RBC) lysis buffer (130-094-183, 10× stock solution, Miltenyi Biotech) and incubated at room temperature for 10 min to lyse the remaining red blood cells. After incubation, the cell suspension was centrifuged at 500g for 20 min at room temperature and then resuspended in 100 μL of Dead Cell Removal MicroBeads to remove dead cells using Miltenyi Dead Cell Removal Kit (130-090-101-1, Miltenyi Biotech). The live cells were collected and resuspended in 1× PBS with 0.04 % BSA and centrifuged at 300g for 3 min at 4 °C (repeated twice). The cell pellet was resuspended in 50 μL of 1× PBS with 0.04 % BSA. The cell viability was measured by the trypan blue exclusion method (15250-061, Invitrogen) and confirmed to be 85 % or higher. The cell number of the single cell suspension was counted using a Countess II Automated Cell Counter and the final concentration was adjusted to 700–1200 cells/μL.
2.4. Chromium 10× Genomics library construction and TCR sequencing
Cells were captured using the Chromium Single-Cell 5′ kit (V1) according to the manufacturer's instructions (10× Genomics), followed by cDNA amplification and library construction according to the standard protocols. The libraries were sequenced on an Illumina NovaSeq 6000 sequencing platform (paired-end multiplexing run, 150 bp) by LC-Bio Technology (Hangzhou, China) at a minimum depth of 5000 reads per cell. The sequencing results were demultiplexed and converted to FASTQ format using Illumina bcl2fastq software. To visualize the data, we further reduced the dimensionality of all cells by Seurat and used t-distributed stochastic neighbor embedding (t-SNE) to project the cells into 2D space. The steps included: (1) Using the LogNormalize method of the “Normalization” function of the Seurat software to calculate the expression level of genes; (2) the principal component analysis (PCA) was performed using the normalized expression level, within all the PCs, the top 10 PCs were used to conduct the clustering and t-SNE analysis; (3) using weighted shared nearest neighbor (SNN) graph-based clustering method to find clusters. The marker genes for each cluster were identified with the “bimod” (likelihood-ratio test) with default parameters via the FindAllMarkers function in Seurat. This selects marker genes that expressed in >10 % of the cells in a cluster and the average log (fold change) of >0.26. To further avoid interference of putative multiplets (where more than one cell was loaded into a given well on an array), cells in a defined cluster that had high expression of more than one cell type canonical marker gene were filtered to ensure the data quality. As a result, a total of 119,799 cells were used for the final analysis in this study. The nine cell types were integrated for further sub-clustering. After integration, genes were scaled to unit variance. Scaling, principal component analysis and clustering were performed as described above.
Sample demultiplexing, barcode processing and single-cell 5′ gene counting were completed by using the Cell Ranger pipeline (https://support.10xgenomics.com/single-cell geneexpression/software/pipelines/latest/what-is-cell-ranger, version 3.1.0). The scRNA-seq data were aligned to Ensembl genome GRCh38 reference genome. The Cell Ranger Seurat (version 3.1.1) was used for dimensional reduction, clustering, and analysis of scRNA-seq data. Approximately 200,000 cells passed the quality control threshold: genes that expressed in less than one cell were removed, number of genes expressed per cell >500 as low cut-off, UMI counts <500, and the percentage of mitochondrial-DNA derived gene-expression < 25 %.
In detail, the TCR Barcoded, full-length TCR V(D)J segments were enriched from the amplified 5′ cDNA libraries using the Chromium Single-Cell V(D)J Enrichment kit according to the manufacturer's protocol (10× Genomics). The TCR transcripts were enriched in separate reactions from the same amplified cDNA material if both T cells were expected to be present in the partitioned cell population.
Cell Ranger pipeline (https://support.10xgenomics.com/single-cell-vdj) that processes Chromium single cell 5′ RNA-seq output for V(D)J was employed to assemble, quantify, and annotate paired V(D)J transcript sequences. The workflow of Cell Ranger started by demultiplexing the Illumina sequencer's base call files (BCLs) for each flowcell directory into FASTQ files. Cell calling was performed independently of V(D)J read filtering and assembly. Clonal expansion cells shared the same paired receptor sequences (presumably derived from the same progenitor cell). Cell barcodes were grouped into clonotypes if they shared the same set of productive CDR3 nucleotide sequences. The repertoire information of all samples was exhibited based on the following features: CDR3 abundance, CDR3 base length, variable (V) and joining (J) segment usage (the frequency of associated reads for each V/J gene presented in the samples), the length of V/J gene in CDR3 region, and V-J gene paired frequency in CDR3 junctions. We employed the hierarchical cluster (https://stat.ethz.ch/R-manual/Rdevel/library/stats/html/hclust.html) and Kruskal's Non-metric Multidimensional Scaling (isoMDS, https://cran.r-project.org/web/packages/MASS/MASS.pdf) to estimate the Immune Repertoires Similarity. To avoid batch effects, we followed the standard operational procedures (SOP) recommended by the manufacturer (10× Genomics). In particular, all the datasets in this study were generated by the same operators at the same laboratories using the same experimental protocols and sequencing platforms.
2.5. Statistical analysis
All the statistical analyses were performed using SPSS (Statistical Package for the Social Sciences) (version 18.0 software, SPSS Inc.) unless otherwise stated. The clonal diversity of each sample was calculated using Shannon's entropy, which took into consideration of both abundance and the frequency distribution of each CDR3 sequence [25]. Entropy was calculated by summing the product of the frequency of each clone with the log2 of that frequency via CalcDiversityStats function of VDJTools, which was calculated by several re-sampling steps (down-sampling to the size of the smallest dataset) [26]. The normalized entropy of clonotype frequency distribution was calculated by dividing by log [number of clonotypes]. The higher Shannon's entropy, the more diverse the distribution of CDR3 clones. The categorical variables were described as frequency rates and percentages, whereas continuous variables were described using mean or median with interquartile range (IQR) values. For analysis of two groups, independent group t-test was used when the data were normally distributed; otherwise, the Mann-Whitney test was used. For analysis of three and more groups, One-way Anova test was performed when the data were normally distributed; otherwise, Kruskal-Wallis test was used. In addition, Bonferroni correction was used for the multiple comparisons. P < 0.05 was considered as statistically significant.
2.6. Data availability
The raw sequence data reported in this paper have been deposited in GEO, under accession code GSE165080 and GSE169441 are publicly accessible at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE165080 [27] and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE169441. Other supporting raw data are available from the corresponding author upon reasonable request. Source data are provided with this paper.
3. Results
3.1. Identification of eleven T cell subtypes by single-cell RNA sequencing (scRNA-seq)
We recruited eleven healthy controls (HC), five asymptomatic subjects (AS) and 33 symptomatic patients (SM) in this stud and summarized the demographic and clinical features of the study subject (Fig. 1, Tables 1, S1). Of note, the asymptomatic subjects were tested positive for the nucleic acids of SASR-CoV-2 by RT-PCR but were lack of clinical symptoms and abnormal chest CT images. On the other hand, all 33 symptomatic patients were positive for the viral nucleic acids and presented clinical symptoms and abnormal chest CT images. In addition, we revealed that the CD3+, CD4+, CD8+, and CD19+ cell counts were significantly higher in AS than SM patients (1529.25 ± 264.62 cells/μL vs 749.87 ± 504.59 cells/μL; 887.00 ± 225.54 cells/μL vs 465.35 ± 306.26 cells/μL; 541.00 ± 143.77/μL vs 258.77 ± 211.16 cells/μL; 383.75 ± 116.52 vs 199.07 ± 147.90 cells/μL, P < 0.05) (Tables 1, S1).
Fig. 1.
Identification of eleven T cell subtypes by single-cell RNA sequencing (scRNA-seq).
(A) A schematic diagram of the study design for single-cell RNA sequencing (scRNA-seq) and T cell receptor sequencing (TCR-seq). Peripheral blood mononuclear cells (PBMCs) were collected from eleven healthy controls (HC), five asymptomatic subjects (AS) and 33 symptomatic COVID-19 patients (SM), followed by scRNA-seq and TCR-seq using 10xGenomics platform. (B) The T-distributed stochastic neighbor embedding (t-SNE) plots showing the marker genes used for identification of the following eleven T cell subtypes: activated effector T (GZMH), DNT (DDX17), effector memory T (GZMK), γδT (TRDV2), MAIT (TRAV1-2), memory T cells (DDIT4), naive T (CCR7), NKT (KLRD1), pro-NKT (MK167), Th17 (CRIP2), and Treg (FOXP3). The plots are color-labeled based on the expression level of the respective marker gene in log scale, which was calculated via LogNormalize method of the “NormalizeData” function of the Seurat software.
To profile the immune cell landscape of COVID-19 patients with different clinical presentations, we performed deep single-cell RNA sequencing on PBMCs from the healthy subjects and COVID-19 patients [27]. We detected eleven T cell subclusters, including activated effector T, double negative T (DNT), effector memory T, γδT, mucosal associated invariant T (MAIT), memory T, naive T, NKT, pro-NKT, T helper cell 17 (Th17), and regulatory T (Treg) cells (Fig. 1A). We also showed the representative marker genes for the corresponding T cell subsets, such as activated effector T (GZMH), DNT (DDX17), effector memory T (GZMK), γδT (TRDV2), MAIT (TRAV1-2), memory T cells (DDIT4), naive T (CCR7), NKT (KLRD1), pro-NKT (MK167), Th17 (CRIP2), and Treg (FOXP3) (Fig. 1B, Table S2). The ridge plot shows the distribution identity of these marker genes in the eleven T cell clusters (Fig. S1B). We further investigated TCR repertoire on T cells from the PBMCs among AS, SM, and HC by single-cell TCR sequencing (scTCR-seq). Of note, except some of the CD4−CD8− T cells, >90 % of cells in all subsets exhibited TCR (Fig. S1A).
3.2. Distinct T cell clonal expansions associated with AS and SM patients
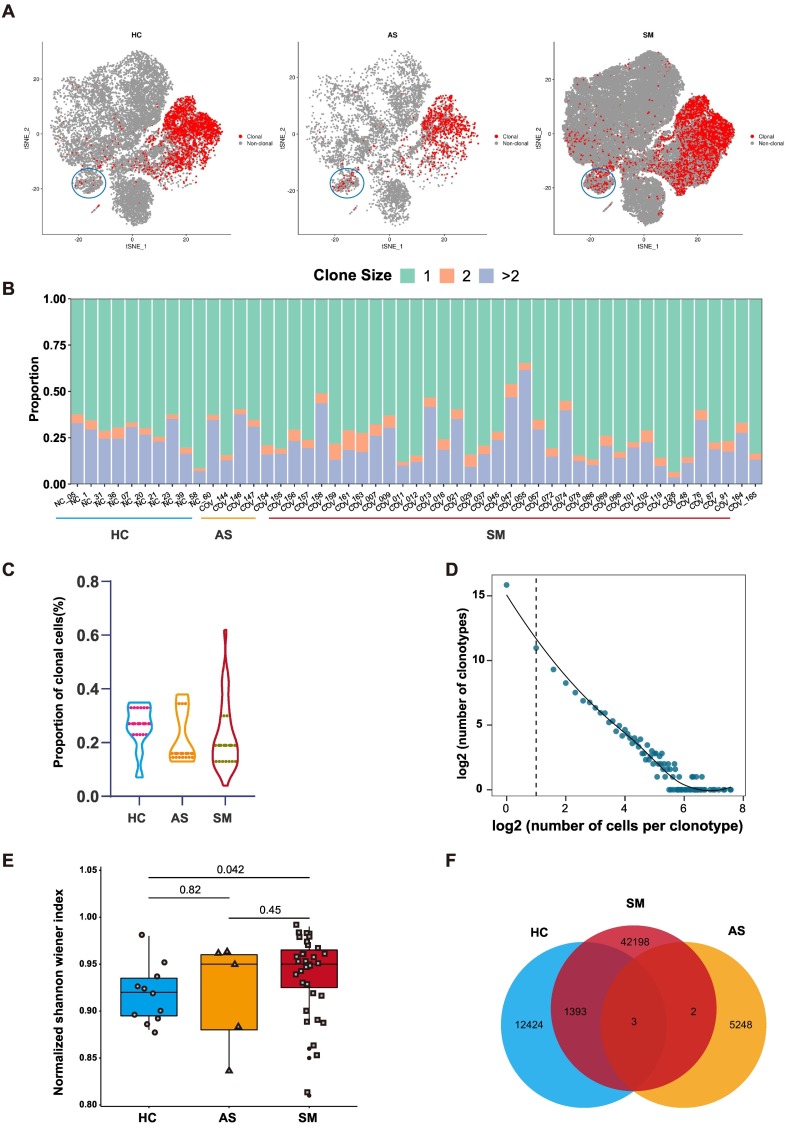
In the cell-mediated immune responses, activated T cells specifically recognized the amino acid fragments of antigen presented by antigen presenting cells (APC) through TCRs, by which the immune clearance was achieved and distinctive TCR repertoire was greatly enriched. Therefore, it is important to understand the difference in TCR repertoires between asymptomatic and symptomatic patients of COVID-19 for the prognosis and optimizing the treatments. To illustrate the difference between clonal expansion and TCR bias when T cells were activated by SARS-CoV-2, we performed scTCR-seq with complete TCR α and β chains. We characterized the T cell clonal expansion status in the HC, AS and SM, respectively. We found that more clonal expansion occurred in MAIT cell cluster of AS and SM compared to HC (Figs. 2A, S1C). As expected, most T cell clones were unique (Fig. 2B, Table S4). We then calculated the relative proportion of the clonal T cells in HC, AS and SM groups, respectively. Our data suggested that there was no significant difference in the relative proportion of the clonal T cells among the three groups (Fig. 2C). However, the number of the clonotypes in SM (ranged from 3 to 191) and the clonal diversity were significantly different from the HC group. Clonal diversity in each sample was calculated using Shannon's entropy and considered both the CDR3 abundance and the frequency distribution of CDR3 sequence (Fig. 2D, E, Table S3). To clarify whether different TCR repertoire in each group was due to the different T cell subsets, we calculated the proportion of each T cell subsets in the mean level of HC, AS, SM, respectively (Fig. S1D). Overall, there was no significant difference of T cell subsets in all groups and the differences of TCR repertoire in each group was not due to the different T cell subsets. Finally, the Venn diagram showed that there were 42,198 TCRs in SM patients, 5248 in AS and 12,424 in HC, respectively. Interestingly, there were 1393 shared TCRs between SM and HC, but only two shared TCRs between SM and AS and three shared TCRs among SM, AS, and HC (Fig. 2F).
Fig. 2.
TCR clonal expansion and clonotype of AS and SM.
(A) The t-SNE plot shows the B cells expansions in the HC, AS and SM, clonal was marked with red color. (B) The TCR distribution of asymptomatic AS, SM, and HC. Unique (n = 1), duplicated (n = 2), and clonal (n > 2) TCRs are labeled with different colors. (C) The violin plots showing the proportion of clonal cells of HC, AS and SM. (D) The association between the number of T cell clones and the number of cells per clonotype. The dashed line separates nonclonal and clonal cells. LOESS fitting is labeled as the solid line showing negative correlation between the two axes. (E) Box plot showing the clonal diversity of HC, AS and SM. Each dot represents the clonal diversity of each sample. P-value was calculated using Wilcoxon test. (F) Venn diagram showing the common and specific V-J genes of HC, AS and SM. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. A number of significant variable and joining genes of the TCR αβ chain were correlated with disease symptoms
CDR3 is the focus of the immune repertoire because it is encoded by three genes: variable (V), diversity (D), and joining (J) genes, which leads to the highest degree of diversity in CDR3 [28]. Each immune protein in the immune repertoire has a wide variety of subtypes and plays a vital role in health. Therefore, the more subtypes of immune proteins, the more effective they are against pathogens. Here, we investigated the bias of V and J gene segments in AS and SM, and discovered that the frequency of several significant V and J genes in the TCR αβ chain were increased in AS, including TRAJ16 and TRBJ2-1, whereas the frequency of TRAV2, TRAJ8, TRAJ40, TRBV3-1 and TRBV5-1 were the highest in SM, and TRBJ2-5 was highest in HC (Fig. 3A, Table S5). Moreover, TRAV2, TRAJ8, TRAJ40, TRBV3-1 and TRBV5-1 were found to be correlated with the disease symptoms (Fig. 3B). Taken together, these results revealed the TCR V J bias in AS and SM, suggesting that there was a possible association between TCR V and J gene segments with the symptoms of COVID-19.
Fig. 3.
TCR variable (V) and joining (J) gene usage comparison among AS, SM, and HC.
(A) The frequency and differential analysis of V and J gene segments of α and β chains between each group (P < 0.05). (B) Spearman rank order correlation analysis showing the V and J gene segments of α and β chains are significantly associated with the symptoms.
3.4. Differences in TRA/B rearrangements among AS, SM, and HC
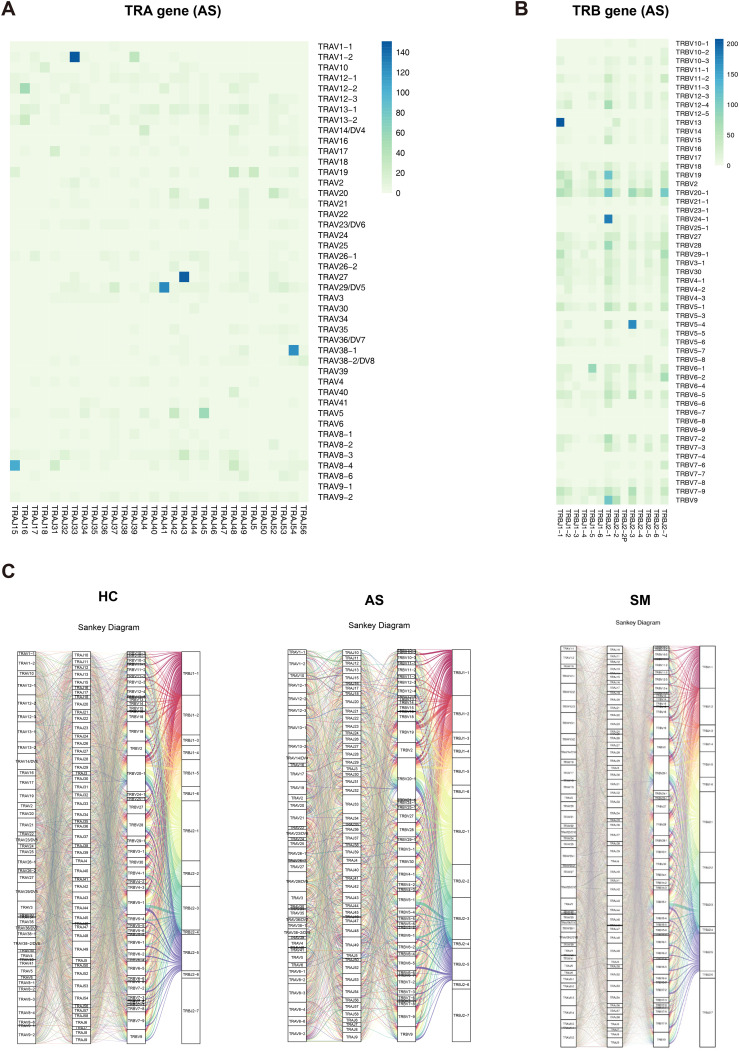
We then compared the V-J pairing of α and β chains separately. We found seven V-J pairs (TRAV27-TRAJ43, TRAV29/DV5-TRAJ41, TRAV38-1-TRAJ54, TRAV8-4-TRAJ15, TRBV13-TRBJ-1, TRBV24-1-TRBJ2-1 and TRBV5-4-TRBJ2-4) in AS, which were the most frequent pairs among three groups (Figs. 4A, B, S2A and Table S6). There were five unique TRAV-TRAJ pairs (i.e., TRAV26-1-TRAJ7, TRAV3-TRAJ50, TRAV34-TRAJ56, TRAV39-TRAJ24 and TRAV8-2-TRA J46) in AS, 179 unique TRAV-TRAJ pairs (e.g., TRAV1-1-TRAJ43, TRAV10-TRAJ11, TRAV10-TRAJ3 and TRAV10-TRAJ47) in SM, and 27 unique TRAV-TRAJ pairs (e.g., TRAV2-TRAJ53, TRAV20-TRAJ46, TRAV25-TRAJ46, and TRAV26-TRAJ10) in HC. In addition, there were 62 TRAV-TRAJ pairs shared by AS and SM whereas only two TRAV-TRAJ pairs shared by AS and HC. Approximately 95.18 %, 68.12 %, and 76.19 % of the T cells used these common VJ pairs in AS, SM and HC, respectively (Fig. S2B, Table S6). For TRBV-TRBJ pairs, there were two unique gene pairs (TRBV21-1-TRBJ2-6 and TRBV7-4-TRBJ2-4) in AS. Moreover, nine pairs were shared between AS and SM. There were 28 and four unique gene pairs in SM and HC, respectively. There were 504 TRBV-TRBJ pairs that were shared by all groups (Fig. S2C, and Table S4). For the sample-specific αβ VJ pairs, the Sankey diagrams displayed the COVID-19 specific αβ VJ pairs that occurred more than once. There were 1642, 13,910, and 4373 αβ VJ pairs in AS, SM and HC, respectively. The most frequent αβ VJ pairs in AS included TRAV1-2-J33-TRBV6-4-J2-2, TRAV1-2-J33-TRBV6-4-J2-3, TRAV1-2-J33-TRBV6-1-J2-3, whereas TRAV1-2-J33-TRBV6-4-J2-3 and TRAV1-2-J33-TRBV6-4-J2-1 were the most frequent pairs in SM and HC, respectively (Fig. 4C, Table S6).
Fig. 4.
Differences in TRA/B rearrangement among AS, SM, and HC.
(A, B) The Heatmaps showing the VJ gene pairs of TRA (A) and TRB (B) in AS. The color range indicating the usage counts of specific V-J gene pairs. (C) Sankey diagram showing COVID-19 specific αβ VJ pairs that occurred more than once.
4. Discussion
In this study, we first reviewed and analyzed the clinical laboratory data and found that the CD3+, CD4+, and CD8+ T cell counts were significantly lower in SM compared with AS and normal reference values, suggesting that T cell exhaustion might be associated with the disease symptoms [29]. We then conducted the scTCR-seq and revealed >90 % of cells in all subsets with TCR information, demonstrating that direct sequence-based immune profiling is a powerful approach for understanding the dynamics of the immune responses [30].
We subsequently examined the T cell clonal expansion status in HC, AS and SM, respectively. We found that more clonal expansion occurred in the MAIT cluster from AS and SM compared to HC. Previous studies have shown that MAIT cells were associated with the mucosa immunity to a variety of microbial infections [31], [32], [33], suggesting that these clonal MAIT cells may play an important role in the immune responses to SARS-CoV-2 infection in AS and SM patients. The findings that clone patterns of TCRs were reused and the clonotype diversity was significantly different in AS and SM may help us to better understand the TCR repertoires in AS subjects and SM patients. Next, we also investigated the bias of the V and J gene segments in AS and SM and found the frequency of TRAJ16 increased in AS compared with HC, which was consistent with the previous report in the graft-versus-host disease (GVHD) [34]. Interestingly, GVHD and COVID-19 shared similar pathological consequences [35]. In addition, TRBJ2-1 was highly expressed in AS compared with SM. Since SARS-CoV-2 infection was shown to affect the cardiovascular system, and previous studies have demonstrated the importance of TRBJ2-1 in HLA-I restricted tumor-associated peptide and acute myocardial infarction [36], [37], suggesting that TRBJ2-1 might be associated with the cardiovascular disorders in COVID-19 patients [38], [39]. TRBJ2-5 was the highest in HC subjects whereas TRAV2, TRAJ8, TRAJ40, TRBV3-1 and TRBV5-1 were the highest in SM. These skewed V and J gene segments might be associated with the symptomatic COVID-19. Finally, we uncovered the common and unique the V-J pairing of α and β chain rearrangements in AS, SM and HC. In particular, we identified the most frequent αβ VJ pairs in AS subjects, including TRAV1-2-J33-TRBV6-4-J2-2, TRAV1-2-J33-TRBV6-4-J2-3, TRAV1-2-J33-TRBV6-1-J2-3, which may contribute to the antiviral immunity and symptom-free state in the AS group. Of note, McGregor et al. reported the TRAV1-2-TRAJ33 TCR-α rearrangement in peripheral T-cell lymphomas derived from innate-like T-cells [40]. The preference of αβ VJ pairs in AS may have important implications for development of novel therapeutic strategies and vaccine candidates against SARS-CoV-2 infection. Due to the frequent mutations of SARS-CoV-2, T cell-based vaccines will receive more and more attention. Indeed, several TCR-based vaccine candidates have been developed, highlighting the significance of the TCR-related studies [20], [41].
Some of the limitations of this study include relatively small sample size, variations in the time-points for the sample collections and relevant statistical weakness. Another limitation was lack of experimental validations on some of the important skewed TCR genes and its TRA/B rearrangements in the peripheral blood samples to rule out the random differences in pairings due to diversity in genes in the population or influences from other individual differences such as viral infections, chronic illness, differences in HLA alleles.
The complexity and dynamics of the T-cell repertoires may render huge challenges for study of the immunopathogenesis of COVID-19. However, the scTCR-seq technology allowed us to conduct unbiased research to understand the contributions of T cell-mediated immune responses to.
SARS-CoV-2 infection. Taken together, this study revealed the basic features of TCR repertoires in response to SARS-CoV-2 infection and provided novel insights into the mechanisms of T-cell immunity, which may help to identify optimal TCRs for development of novel therapeutic strategies and vaccine candidates against SARS-CoV-2 infection.
Abbreviations
- COVID-19
Coronavirus disease 2019
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus-2
- TCR
T cell receptor
- scTCR-seq
Single-cell TCR sequencing
- HC
Healthy controls
- AS
Asymptomatic infections
- SM
Symptomatic COVID-19 patients
- PBMCs
Peripheral blood mono-nuclear cells
The following are the supplementary data related to this article.
Cohort characteristics.
T cell cluster identification and differentially expressed genes.
scTCR-seq depth.
Clonotype proportion of each COVID-19 patients and healthy controls.
TRAB VJ genes.
TRAB VJ pairs.
Supplementary materials
Funding
This study is supported in part by the Science and Technology Department, Shaanxi Province (Grant No. 2020ZDXM2-SF-02) (CZ and BS) and the operational funds from The First Affiliated Hospital of Xi'an Jiaotong University (CZ and BS).
CRediT authorship contribution statement
Conceptualization: CZ and BS; Methodology: CZ, HB, JM, WM, TJ, XW, HQ, QZ, FH, XC; Resources: CZ, BS, DL, BZ; Writing: CZ, JM, HB; Review and editing: CZ, HB, JM, BS; Supervision: CZ, BS; Funding: CZ and BS; Project administration: CZ.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Author Weikang Mao is employed by the LC-BIO TECHNOLOGIES (HANGZHOU) CO., LTD., China. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are extremely grateful to all the patients and their families for participation of this study and providing the valuable information. We also thank our colleagues at The First Affiliated Hospital of Xi'an Jiaotong University and The Renmin Hospital of Wuhan University for their kind help and strong support throughout the course of this study.
Editor: Mohandas Narla
References
- 1.World Health Organization WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int/
- 2.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 3.Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oran D.P., Topol E.J. Prevalence of asymptomatic SARS-CoV-2 infection : a narrative review. Ann. Intern. Med. 2020;173:362–367. doi: 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hosoki K., Chakraborty A., Sur S. Molecular mechanisms and epidemiology of COVID-19 from an allergist's perspective. J. Allergy Clin. Immunol. 2020;146:285–299. doi: 10.1016/j.jaci.2020.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohn M.K., Hall A., Sepiashvili L., Jung B., Steele S., Adeli K. Pathophysiology of COVID-19: mechanisms underlying disease severity and progression. Physiology (Bethesda) 2020;35:288–301. doi: 10.1152/physiol.00019.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekine T., Perez-Potti A., Rivera-Ballesteros O., Stralin K., Gorin J.B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168. doi: 10.1016/j.cell.2020.08.017. e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altmann D.M., Boyton R.J. SARS-CoV-2 T cell immunity: specificity, function, durability, and role in protection. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abd6160. [DOI] [PubMed] [Google Scholar]
- 9.Mazzoni A., Maggi L., Capone M., Spinicci M., Salvati L., Colao M.G., Vanni A., Kiros S.T., Mencarini J., Zammarchi L., et al. Cell-mediated and humoral adaptive immune responses to SARS-CoV-2 are lower in asymptomatic than symptomatic COVID-19 patients. Eur. J. Immunol. 2020;50:2013–2024. doi: 10.1002/eji.202048915. [DOI] [PubMed] [Google Scholar]
- 10.Reynolds C.J., Swadling L., Gibbons J.M., Pade C., Jensen M.P., Diniz M.O., Schmidt N.M., Butler D.K., Amin O.E., Bailey S.N.L., et al. Discordant neutralizing antibody and T cell responses in asymptomatic and mild SARS-CoV-2 infection. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abf3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schamel W.W., Alarcon B., Minguet S. The TCR is an allosterically regulated macromolecular machinery changing its conformation while working. Immunol. Rev. 2019;291:8–25. doi: 10.1111/imr.12788. [DOI] [PubMed] [Google Scholar]
- 12.Halle S., Halle O., Forster R. Mechanisms and dynamics of T cell-mediated cytotoxicity in vivo. Trends Immunol. 2017;38:432–443. doi: 10.1016/j.it.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Dash P., Fiore-Gartland A.J., Hertz T., Wang G.C., Sharma S., Souquette A., Crawford J.C., Clemens E.B., Nguyen T.H.O., Kedzierska K., et al. Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature. 2017;547:89–93. doi: 10.1038/nature22383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang H., Sikora M.J., Islam S., Chowdhury R.R., Chien Y.H., Scriba T.J., Davis M.M., Steinmetz L.M. Select sequencing of clonally expanded CD8(+) T cells reveals limits to clonal expansion. Proc. Natl. Acad. Sci. U. S. A. 2019;116:8995–9001. doi: 10.1073/pnas.1902649116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni L., Cheng M.L., Feng Y., Zhao H., Liu J., Ye F., Ye Q., Zhu G., Li X., Wang P., et al. Impaired cellular immunity to SARS-CoV-2 in severe COVID-19 patients. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.603563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang F., Gan R., Zhen Z., Hu X., Li X., Zhou F., Liu Y., Chen C., Xie S., Zhang B., et al. Adaptive immune responses to SARS-CoV-2 infection in severe versus mild individuals. Signal Transduct Target Ther. 2020;5:156. doi: 10.1038/s41392-020-00263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarjour N.N., Masopust D., Jameson S.C. T cell memory: understanding COVID-19. Immunity. 2021;54:14–18. doi: 10.1016/j.immuni.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minervina A.A., Komech E.A., Titov A., Bensouda Koraichi M., Rosati E., Mamedov I.Z., Franke A., Efimov G.A., Chudakov D.M., Mora T., et al. Longitudinal high-throughput TCR repertoire profiling reveals the dynamics of T-cell memory formation after mild COVID-19 infection. elife. 2021;10 doi: 10.7554/eLife.63502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snyder T.M., Gittelman R.M., Klinger M., May D.H., Osborne E.J., Taniguchi R., Zahid H.J., Kaplan I.M., Dines J.N., et al. Noakes M.T. Magnitude and dynamics of the T-cell response to SARS-CoV-2 infection at both individual and population levels. medRxiv. 2020 doi: 10.1101/2020.07.31.20165647. [DOI] [Google Scholar]
- 20.Wang P., Xu Z., Zhou W., Jin X., Xu C., Luo M., Ma K., Cao H., Huang Y., Lin X., et al. Identification of potential vaccine targets for COVID-19 by combining single-cell and bulk TCR sequencing. Clin. Transl. Med. 2021;11 doi: 10.1002/ctm2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Government of China . 8nd ed. 2021. New Coronavirus Pneumonia Prevention and Control Program.http://www.nhc.gov.cn/jkj/s3577/202105/6f1e8ec6c4a540d99fafef52fc86d0f8/files/4a860a7e85d14d55a22fbab0bbe77cd9.pdf [Google Scholar]
- 22.World Health Organization . 2020. Laboratory Testing for 2019 Novel Coronavirus (2019-nCoV) in Suspected Human Cases.https://www.who.int/health-topics/coronavirus/laboratory-diagnostics-for-novel-coronavirus [Google Scholar]
- 23.World Health Organization . 2020. COVID-19: Surveillance, Case Investigation and Epidemiological Protocols.https://www.who.int/internal-publications-detail/considerations-in-the-investigation-of-cases-and-clusters-of-covid-19 [Google Scholar]
- 24.Fuss I.J., Kanof M.E., Smith P.D., Zola H. Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr Protoc Immunol. 2009 doi: 10.1002/0471142735.im0701s85. Chapter 7, Unit7 1. [DOI] [PubMed] [Google Scholar]
- 25.Cui J.H., Lin K.R., Yuan S.H., Jin Y.B., Chen X.P., Su X.K., Jiang J., Pan Y.M., Mao S.L., Mao X.F., et al. TCR repertoire as a novel indicator for immune monitoring and prognosis assessment of patients with cervical cancer. Front. Immunol. 2018;9:2729. doi: 10.3389/fimmu.2018.02729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider-Hohendorf T., Mohan H., Bien C.G., Breuer J., Becker A., Gorlich D., Kuhlmann T., Widman G., Herich S., Elpers C., et al. CD8(+) T-cell pathogenicity in rasmussen encephalitis elucidated by large-scale T-cell receptor sequencing. Nat. Commun. 2016;7:11153. doi: 10.1038/ncomms11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X., Bai H., Ma J., Qin H., Zeng Q., Hu F., Jiang T., Mao W., Zhao Y., Chen X., et al. Identification of distinct immune cell subsets associated with asymptomatic infection, disease severity, and viral persistence in COVID-19 patients. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.812514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu P., Liu D., Yang X., Gao J., Chen Y., Xiao X., Liu F., Zou J., Wu J., Ma J., et al. Characterization of human alphabetaTCR repertoire and discovery of D-D fusion in TCRbeta chains. Protein Cell. 2014;5:603–615. doi: 10.1007/s13238-014-0060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louati N., Rekik T., Menif H., Gargouri J. Blood lymphocyte T subsets reference values in blood donors by flow cytometry. Tunis Med. 2019;97:327–334. [PubMed] [Google Scholar]
- 30.Freeman J.D., Warren R.L., Webb J.R., Nelson B.H., Holt R.A. Profiling the T-cell receptor beta-chain repertoire by massively parallel sequencing. Genome Res. 2009;19:1817–1824. doi: 10.1101/gr.092924.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K.D., Hodeib S., Korol C., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Napier R.J., Adams E.J., Gold M.C., Lewinsohn D.M. The role of mucosal associated invariant T cells in antimicrobial immunity. Front. Immunol. 2015;6:344. doi: 10.3389/fimmu.2015.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dias J., Sobkowiak M.J., Sandberg J.K., Leeansyah E. Human MAIT-cell responses to Escherichia coli: activation, cytokine production, proliferation, and cytotoxicity. J. Leukoc. Biol. 2016;100:233–240. doi: 10.1189/jlb.4TA0815-391RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiRienzo C.G., Murphy G.F., Friedman T.M., Korngold R. T-cell receptor V(alpha) usage by effector CD4+Vbeta11+ T cells mediating graft-versus-host disease directed to minor histocompatibility antigens. Biol. Blood Marrow Transplant. 2007;13:265–276. doi: 10.1016/j.bbmt.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy G.F. COVID-19 and graft-versus-host disease: a tale of two diseases (and why age matters) Lab. Investig. 2021;101:274–279. doi: 10.1038/s41374-020-00520-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan K.F., Gully B.S., Gras S., Beringer D.X., Kjer-Nielsen L., Cebon J., McCluskey J., Chen W., Rossjohn J. Divergent T-cell receptor recognition modes of a HLA-I restricted extended tumour-associated peptide. Nat. Commun. 2018;9:1026. doi: 10.1038/s41467-018-03321-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhong Z., Wu H., Zhang Q., Zhong W., Zhao P. Characteristics of T cell receptor repertoires of patients with acute myocardial infarction through high-throughput sequencing. J. Transl. Med. 2019;17:21. doi: 10.1186/s12967-019-1768-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guzik T.J., Mohiddin S.A., Dimarco A., Patel V., Savvatis K., Marelli-Berg F.M., Madhur M.S., Tomaszewski M., Maffia P., D'Acquisto F., et al. COVID-19 and the cardiovascular system: implications for risk assessment, diagnosis, and treatment options. Cardiovasc. Res. 2020;116:1666–1687. doi: 10.1093/cvr/cvaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.AlShahrani I., Hosmani J., Shankar V.G., AlShahrani A., Togoo R.A., Yassin S.M., Khan S., Patil S. COVID-19 and cardiovascular system-a comprehensive review. Rev. Cardiovasc. Med. 2021;22:343–351. doi: 10.31083/j.rcm2202041. [DOI] [PubMed] [Google Scholar]
- 40.McGregor S., Shah A., Raca G., Mirza M.K., Smith S.M., Anastasi J., Vardiman J.W., Hyjek E., Gurbuxani S. PLZF staining identifies peripheral T-cell lymphomas derived from innate-like T-cells with TRAV1-2-TRAJ33 TCR-alpha rearrangement. Blood. 2014;123:2742–2743. doi: 10.1182/blood-2014-02-555482. [DOI] [PubMed] [Google Scholar]
- 41.Mysore V., Cullere X., Settles M.L., Ji X., Kattan M.W., Desjardins M., Durbin-Johnson B., Gilboa T., Baden L.R., et al. Walt D.R. Protective heterologous T cell immunity in COVID-19 induced by the trivalent MMR and Tdap vaccine antigens. Med (N Y) 2021;2:1050–1071. doi: 10.1016/j.medj.2021.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cohort characteristics.
T cell cluster identification and differentially expressed genes.
scTCR-seq depth.
Clonotype proportion of each COVID-19 patients and healthy controls.
TRAB VJ genes.
TRAB VJ pairs.
Supplementary materials
Data Availability Statement
The raw sequence data reported in this paper have been deposited in GEO, under accession code GSE165080 and GSE169441 are publicly accessible at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE165080 [27] and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE169441. Other supporting raw data are available from the corresponding author upon reasonable request. Source data are provided with this paper.