Abstract
Two strains of 1,1,1-trichloroethane (TCA)-degrading bacteria, TA5 and TA27, were isolated from soil and identified as Mycobacterium spp. Strains TA5 and TA27 could degrade 25 and 75 mg · liter of TCA−1 cometabolically in the presence of ethane as a carbon source, respectively. The compound 2,2,2-trichloroethanol was produced as a metabolite of the degradation process.
Volatile aliphatic chlorinated compounds such as 1,1,1-trichloroethane (TCA), trichloroethylene (TCE), and tetrachloroethylene have been detected in groundwater throughout Japan. These compounds are suspected carcinogens and hepatotoxins. Bioremediation is one of the most promising new technologies for cleaning up groundwater contamination because of its low cost and its potential for the complete destruction of pollutants. There are many reports on the degradation of TCE by aerobic bacteria (4, 8, 10, 12, 13, 16–18, 22–26) and some reports on anaerobic TCA degradation (1, 5, 9, 27), but there are few reports of aerobic TCA degradation. The methane-oxidizing bacterium Methylosinus trichosporium OB3b can degrade TCA (18), as can the ammonia-oxidizing bacterium Nitrosomonas europaea (7, 26), the propane-oxidizing bacteria Rhodococcus rhodochrous and Rhodococcus sp. strain Sm-1 (12), and Pseudomonas putida G786 containing cytochrome P450cam (11). However, TCA biodegradation has not been studied in detail. In this study, we isolated the TCA-degrading bacterial strains TA5 and TA27 and determined their biodegradation characteristics.
Isolation and identification of TCA-degrading bacteria.
We used an enrichment culture method to isolate TCA-degrading bacteria from various polluted soils under aerobic conditions. Small samples of soil (0.5 g each) were added to 69-ml serum bottles containing 15 ml of mineral salt medium each (22). The serum bottles were sealed with butyl rubber caps and crimped with aluminum rings. Ten milliliters of air were withdrawn from each bottle with a syringe, and 5 ml of methane, ethane, propane, or ethylene (as the carbon source) and 5 ml of oxygen were injected to replace the withdrawn air. TCA-saturated aqueous solution was added to each serum bottle by syringe to make a final concentration of 0.6 mg · liter−1. The cultures were incubated at 30°C while being shaken at 120 rpm. After each week of culture, 0.3 ml of culture broth was transferred to a new serum bottle containing 15 ml of fresh medium. After at least four transfers, the culture broth was spread onto an agar plate.
We isolated two strains of TCA-degrading bacteria, TA5 and TA27, from tetrachloroethylene-polluted soils. Both cell types were rod shaped, nonmotile, non-spore forming, gram positive, catalase positive, oxidase negative, and of the menaquinone-9 (H2) type. We found that both strains could utilize ethane, ethanol, and various other carbon compounds as their energy sources. Phylogenetic analysis was carried out with the genomic DNA of strains TA5 and TA27 being extracted by the method of Mizuguchi et al. and Sambrook et al., respectively (15, 20). The primers used for the PCR amplification of the 16S rRNA gene were 8FPL and 1492RPL (19). The PCR products were sequenced with a Big Dye Terminator cycle sequence kit (Applied Biosystems, Perkin-Elmer). The 16S rRNA gene sequences were compared with the 16S rRNA gene database from the DDBJ. Strains TA5 and TA27 had the highest levels of 16S rRNA gene similarity with Mycobacterium duvalii (level of similarity, 98.0%) (21) and Mycobacterium gilvum (level of similarity, 99.2%), respectively. From these physiological characteristics, it seemed that these strains were Mycobacterium spp. It has been reported that fast-growing mycobacteria can degrade various hazardous compounds. Mycobacterium chlorophenolicum can degrade pentachlorophenol (2, 14). Mycobacterium aurum can utilize vinyl chloride, the key enzyme for that degradation being alkene monooxygenase (6). Mycobacterium vaccae JOB-5 can degrade a number of the major compounds involved in groundwater pollution: benzene, toluene, propylbenzene, styrene, cis-1,2-dichloroethylene, and TCE (3, 24, 25). However, these strains could not degrade TCA. Ours is the first report of substantial TCA degradation by Mycobacterium sp. Isolates TA5 and TA27 could assimilate ethane and ethanol. Moreover, strain TA27 could assimilate propane and glucose. Both TA5 and TA27, grown on various carbon substrates, could degrade TCA. It seems that the TCA-degrading enzyme is constitutively produced.
TCA degradation characteristics.
The TCA degradation experiments were carried out as follows. Either strain TA5 or strain TA27 was added to a 69-ml serum bottle containing 15 ml of mineral salt medium (22), and the bottle was sealed with a butyl rubber cap and aluminum ring. Various concentrations of TCA and ethane were added to each serum bottle, and the cultures were incubated at 30°C while being shaken at 120 rpm. The TCA degradation was monitored by using gas chromatography to measure the concentration of gases in the headspace of the serum bottle. The water and gaseous concentrations of the TCA were calculated using Henry’s constant. A gas chromatograph (model GC-7AG; Shimadzu Co., Kyoto, Japan) equipped with a flame ionization detector using a glass column packed with Silicone DC550 (GL Science Co., Tokyo, Japan) was used for quantitative determination of the TCA and ethane. Oxygen in the gas phase was analyzed on a Molecular Sieve 5A packed column equipped with a thermal conductivity detector.
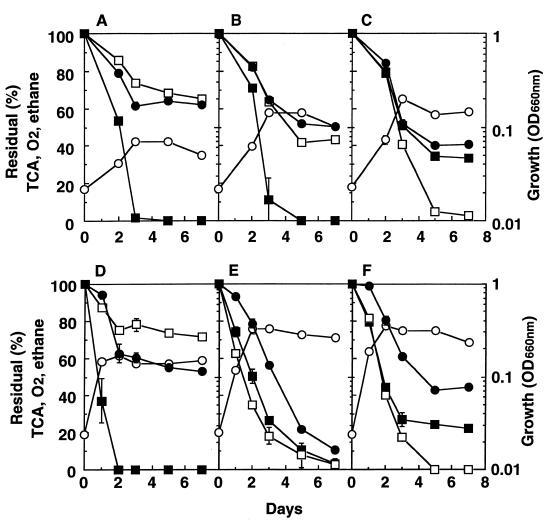
Figure 1 shows the effects of initial ethane concentrations of 3, 5, and 10% on the degradation of TCA at an initial concentration of 0.6 mg · liter−1. An initial ethane concentration of 3% yielded inadequate growth of both strains for effective TCA degradation (Fig. 1A and D). An initial ethane concentration of 10% yielded the highest growth rate of strain TA5, maximizing TCA degradation; 60% of the TCA was degraded within 5 days, after which the degradation almost stopped (Fig. 1C). It seemed that the cessation of TCA degradation was due to a lack of oxygen or ethane. Good growth of strain TA27 was observed at initial ethane concentrations of both 5 and 10% (Fig. 1E and F). At an initial ethane concentration of 5%, about 90% of the initial TCA was degraded within 7 days by strain TA27. However, TCA degradation was inhibited at an initial ethane concentration of 10% (Fig. 1F). A high concentration of ethane may competitively inhibit the degradation of TCA by strain TA27.
FIG. 1.
Effect of initial ethane concentration on TCA degradation by strains TA5 and TA27. (A to C) Strain TA5; (D to F) strain TA27. Initial ethane concentrations were 3% (A and D); 5% (B and E); and 10% (C and F). Initial pH: strain TA5, 6.8; strain TA27, 5.7. Symbols: ●, residual TCA; ○, bacterial growth (optical density at 660 nm [OD660nm]); ■, residual ethane concentration; □, residual O2 concentration. Error bars indicate the ranges for duplicate samples.
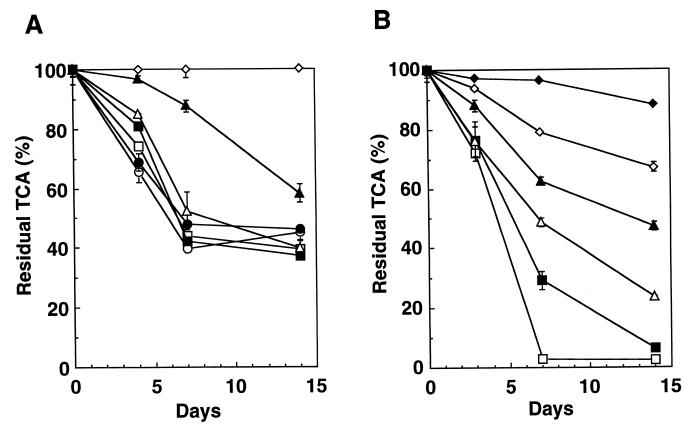
The relative degradation of TCA by strains TA5 and TA27 was affected by the initial concentration of TCA. The rate of TCA degradation by strain TA5 was high at low initial TCA concentrations: at an initial TCA concentration of between 0.05 and 5 mg · liter−1, more than 50% of the TCA was degraded within 7 days (Fig. 2A), after which the cell growth stopped. However, at an initial TCA concentration of 25 mg · liter−1, the growth rate decreased, and only 40% of the TCA was degraded within 14 days. The degradation discontinued when cell growth stopped due to the lack of oxygen. It seemed that the degradation of TCA by strain TA5 was associated with cell growth. Strain TA27 degraded more than 95 and 40% of the TCA within 7 days when the initial TCA concentrations were 0.5 and 25 mg · liter−1, respectively, while 10% of the TCA was degraded within 14 days when the initial TCA concentration was 75 mg · liter−1 (Fig. 2B). The percentage of degradation decreased with increasing initial TCA concentrations. This may be due to the production of toxic metabolites.
FIG. 2.
Effect of TCA concentration on TCA degradation by strains TA5 and TA27. (A) Strain TA5; (B) strain TA27. Initial pH: strain TA5, 6.8; strain TA27, 5.7. Initial ethane concentrations: strain TA5, 10%; strain TA27, 5%. Initial TCA concentrations (in mg · liter−1): ⧫, 75; ◊, 50; ▴, 25; ▵, 5; ■, 2.5; □, 0.5; ●, 0.25; ○, 0.05. Error bars indicate the ranges for duplicate samples.
Temporal pattern of TCA degradation products.
To isolate and identify water-soluble TCA metabolites, after 7 days the culture broth was acidified to pH 2 and extracted three times with the same volume of diethyl ether (16). The organic phase was concentrated to approximately 2 ml at reduced pressure and was methylated with diazomethane; this was followed by gas-chromatographic mass spectrometry. Two peaks (peaks A and B) were detected in both strains. Peak B was identified as 2,2,2-trichloroethanol. We were unable to identity the compound represented by peak A, but we determined that it was not a chlorinated compound.
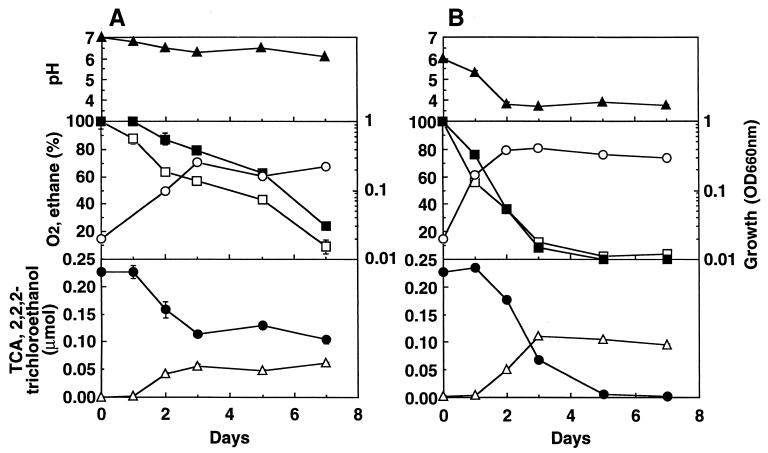
Figure 3 shows the temporal pattern of residual TCA, 2,2,2-trichloroethanol production, residual oxygen and ethane concentrations, cell growth, and the change of pH at an initial TCA amount of 0.23 μmol. This experiment was carried out at the favorable initial pH levels of 6.8 and 5.7 for TCA degradation of strains TA5 and TA27, respectively. The concentrations of TCA, oxygen, and ethane decreased with the increasing growth of strain TA5, while cell growth reached a maximum after 3 days and when 55% of the initial TCA had been degraded.
FIG. 3.
Temporal pattern of TCA degradation by strains TA5 and TA27. (A) Strain TA5; (B) strain TA27. Initial ethane concentrations: strain TA5, 10%; strain TA27, 5%. Initial pH: strain TA5, 6.8; strain TA27, 5.7. Symbols: ●, residual TCA amount; ▵, 2,2,2-trichloroethanol production; ■, residual ethane concentration; □, residual O2 concentration; ○, bacterial growth (optical density at 660 nm [OD660nm]); ▴, pH. Error bars indicate the ranges for duplicate samples.
The growth of strain TA27 reached a maximum after 2 days and when 22% of the initial TCA had been degraded; by 5 days, 99% of the TCA had been degraded and the pH had decreased. A decrease of more than 2 U of pH revealed the production of acidic metabolites and the capability to degrade TCA at low pH levels. The major product of TCA degradation by both strains was 2,2,2-trichloroethanol. The 0.13 and 0.23 μmol of TCA consumed within 7 days was converted to 0.06 and 0.09 μmol of 2,2,2-trichloroethanol by strains TA5 and TA27, respectively; the rates of TCA conversion to 2,2,2-trichloroethanol by strains TA5 and TA27 were 46 and 39%, respectively. These low percentages may have resulted from other conversion products being made, or they may be due to strains TA5 and TA27 degrading the 2,2,2-trichloroethanol. The degradation products of 2,2,2-trichloroethanol need to be identified.
In summary, we isolated ethane-utilizing Mycobacterium spp. that could degrade TCA at an initial concentration as high as 75 mg · liter−1. 2,2,2-Trichloroethanol was a TCA metabolite. We believe that these data will be useful in the development of bioremediation techniques for TCA-contaminated sites.
Nucleotide sequence accession number.
The nucleotide sequences of strains TA5 and TA27 have been deposited in the DDBJ, EMBL, and GenBank databases under accession no. AB028483 and AB028482, respectively.
REFERENCES
- 1.Bouwer E J, McCarty P L. Transformations of 1- and 2-carbon halogenated aliphatic organic compounds under methanogenic conditions. Appl Environ Microbiol. 1983;45:1286–1294. doi: 10.1128/aem.45.4.1286-1294.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briglia M, Eggen R I L, van Elsas D J, de Vos W M. Phylogenetic evidence for transfer of pentachlorophenol-mineralizing Rhodococcus chlorophenolicus PCP-I to the genus Mycobacterium. Int J Syst Bacteriol. 1994;44:494–498. doi: 10.1099/00207713-44-3-494. [DOI] [PubMed] [Google Scholar]
- 3.Burback B L, Perry J J. Biodegradation and biotransformation of groundwater pollutant mixtures by Mycobacterium vaccae. Appl Environ Microbiol. 1993;59:1025–1029. doi: 10.1128/aem.59.4.1025-1029.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Folsom B R, Chapman P J, Pritchard P H. Phenol and trichloroethylene degradation by Pseudomonas cepacia G4: kinetics and interactions between substrates. Appl Environ Microbiol. 1990;56:1279–1285. doi: 10.1128/aem.56.5.1279-1285.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gälli R, McCarty P L. Biotransformation of 1,1,1-trichloroethane, trichloromethane, and tetrachloromethane by a Clostridium sp. Appl Environ Microbiol. 1989;55:837–844. doi: 10.1128/aem.55.4.837-844.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartmans S, de Bont J A M. Aerobic vinyl chloride metabolism in Mycobacterium aurum L1. Appl Environ Microbiol. 1992;58:1220–1226. doi: 10.1128/aem.58.4.1220-1226.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hommes N G, Russell S A, Bottomley P J, Arp D J. Effects of soil on ammonia, ethylene, chloroethane, and 1,1,1-trichloroethane oxidation by Nitrosomonas europaea. Appl Environ Microbiol. 1998;64:1372–1378. doi: 10.1128/aem.64.4.1372-1378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jahng D, Wood T K. Trichloroethylene and chloroform degradation by a recombinant pseudomonad expressing soluble methane monooxygenase from Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1994;60:2473–2482. doi: 10.1128/aem.60.7.2473-2482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klecka G M, Gonsior S J, Markham D A. Biological transformations of 1,1,1-trichloroethane in subsurface soils and ground water. Environ Toxicol Chem. 1990;9:1437–1451. [Google Scholar]
- 10.Koh S-C, Bowman J P, Sayler G S. Soluble methane monooxygenase production and trichloroethylene degradation by a type I methanotroph, Methylomonas methanica 68-1. Appl Environ Microbiol. 1993;59:960–967. doi: 10.1128/aem.59.4.960-967.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lefever M R, Wackett L P. Oxidation of low molecular weight chloroalkanes by cytochrome P450cam. Biochem Biophys Res Commun. 1994;201:373–378. doi: 10.1006/bbrc.1994.1711. [DOI] [PubMed] [Google Scholar]
- 12.Malachowsky K J, Phelps T J, Teboli A B, Minnikin D E, White D C. Aerobic mineralization of trichloroethylene, vinyl chloride, and aromatic compounds by Rhodococcus species. Appl Environ Microbiol. 1994;60:542–548. doi: 10.1128/aem.60.2.542-548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonald I R, Uchiyama H, Kambe S, Yagi O, Murrell J C. The soluble methane monooxygenase gene cluster of the trichloroethylene-degrading methanotroph Methylocystis sp. strain M. Appl Environ Microbiol. 1997;63:1898–1904. doi: 10.1128/aem.63.5.1898-1904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Middeldorp P J M, Briglia M, Salkinoja-Salonen M S. Biodegradation of pentachlorophenol in natural soil by inoculated Rhodococcus chlorophenolicus. Microb Ecol. 1990;20:123–139. doi: 10.1007/BF02543872. [DOI] [PubMed] [Google Scholar]
- 15.Mizuguchi Y, Fukunaga M, Taniguchi H. Plasmid deoxyribonucleic acid and translucent-to-opaque variation in Mycobacterium intracellulare 103. J Bacteriol. 1981;146:656–659. doi: 10.1128/jb.146.2.656-659.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakajima T, Uchiyama H, Yagi O, Nakahara T. Novel metabolite of trichloroethylene in a methanotrophic bacterium, Methylocystis sp. M, and hypothetical degradation pathway. Biosci Biotech Biochem. 1992;56:486–489. doi: 10.1271/bbb.56.486. [DOI] [PubMed] [Google Scholar]
- 17.Nelson M J K, Montgomery S O, Mahaffey W R, Pritchard P H. Biodegradation of trichloroethylene and involvement of an aromatic biodegradative pathway. Appl Environ Microbiol. 1987;53:949–954. doi: 10.1128/aem.53.5.949-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oldenhuis R, Vink R L J M, Janssen D B, Witholt B. Degradation of chlorinated aliphatic hydrocarbons by Methylosinus trichosporium OB3b expressing soluble methane monooxygenase. Appl Environ Microbiol. 1989;55:2819–2826. doi: 10.1128/aem.55.11.2819-2826.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reysenbach A-L, Wickham G S, Pace N R. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1994;60:2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 21.Stanford J L, Gunthorpe W J. A study of some fast-growing scotochromogenic mycobacteria including species descriptions of Mycobacterium gilvum (new species) and Mycobacterium duvalii (new species) Br J Exp Pathol. 1971;52:627–637. [PMC free article] [PubMed] [Google Scholar]
- 22.Uchiyama H, Nakajima T, Yagi O, Tabuchi T. Aerobic degradation of trichloroethylene by a new type II methane-utilizing bacterium, strain M. Agric Biol Chem. 1989;53:2903–2907. [Google Scholar]
- 23.Uchiyama H, Nakajima T, Yagi O, Nakahara T. Role of heterotrophic bacteria in complete mineralization of trichloroethylene by Methylocystis sp. strain M. Appl Environ Microbiol. 1992;58:3067–3071. doi: 10.1128/aem.58.9.3067-3071.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanderberg L A, Burback B L, Perry J J. Biodegradation of trichloroethylene by Mycobacterium vaccae. Can J Microbiol. 1995;41:298–301. doi: 10.1139/m95-041. [DOI] [PubMed] [Google Scholar]
- 25.Vanderberg L A, Perry J J. Dehalogenation by Mycobacterium vaccae JOB-5: role of the propane monooxygenase. Can J Microbiol. 1994;40:169–172. doi: 10.1139/m94-029. [DOI] [PubMed] [Google Scholar]
- 26.Vannelli T, Logan M, Arciero D M, Hooper A B. Degradation of halogenated aliphatic compounds by the ammonia-oxidizing bacterium Nitrosomonas europaea. Appl Environ Microbiol. 1990;56:1169–1171. doi: 10.1128/aem.56.4.1169-1171.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogel T M, McCarty P L. Abiotic and biotic transformations of 1,1,1-trichloroethane under methanogenic conditions. Environ Sci Technol. 1987;21:1208–1213. [Google Scholar]