Abstract
Long non-coding RNAs (lncRNAs) play important roles in many biological processes. Knocking out or knocking down some lncRNAs will lead to lethality or infertility. These lncRNAs are called essential lncRNAs. Knowledges of essential lncRNAs are important in establishing minimal genomes of living cells, developing drug therapies and early diagnostic approaches for complex diseases. However, existing databases focus on collecting essential coding genes. Essential non-coding gene records are rare in existing databases. A comprehensive collection of essential non-coding genes, particularly essential lncRNA genes, is demanded. We manually curated 207 essential lncRNAs from literatures for establishing a database on essential lncRNAs, which is named as dbEssLnc (Database of essential lncRNAs). The dbEssLnc database has a web-based user-friendly interface for the users to browse, to search, to visualize and to blast search records in the database. The dbEssLnc database is freely accessible at https://esslnc.pufengdu.org. All data and source codes for mirroring the dbEssLnc database have been deposited in GitHub (https://github.com/yyZhang14/dbEssLnc).
Keywords: Essential lncRNA, Database, Knockout, Knockdown, dbEssLnc
1. Introduction
In human genome, only less than 3 % genes encode proteins. Most genes are non-coding genes [1]. These genes were once regarded as “junks” or “dark matters” in the genome [2]. Most of the non-coding genes can be transcribed into RNAs, which have little or no potential to be further translated into proteins. These RNAs are termed as non-coding RNAs. Over the past few years, functional analysis of non-coding RNAs, including long non-coding RNAs(lncRNAs), microRNAs(miRNAs), small interfering RNAs(siRNAs) and many others [3], receives overwhelming attentions.
LncRNAs are non-coding RNAs with more than 200nt in length [[3], [4]]. Several studies have revealed that lncRNAs play crucial roles in transcriptional regulations [5], epigenetic regulations [2], post-transcriptional regulations [6] and translational regulations [7]. LncRNAs are involved in various biological process through their interactions with proteins and other types of molecule [[8], [9]]. Many complex diseases, like cardiovascular disease [10], osteosarcoma [11] and neurological disease [4], are related to dysregulation of lncRNA genes [[12], [13]].
Some lncRNA genes are critical to normal cellular activities. Knocking out or knocking down such lncRNA genes would lead to lethality or infertility [[14], [15]]. These genes are regarded as essential lncRNA genes. According to the literature [16], knowledge of essential genes are critical in understanding genome organization and evolution, identifying and prioritizing drug targets, elucidating cellular networks and defining minimal genome in synthetic biology. Particularly, essential lncRNAs act as regulators for regulating stem cell pluripotency [[17], [18]]. In addition, lncRNA genes, which are essential in tumor cells but not in adjacent normal tissues, can be used as potential therapeutically targets to treat cancers [19]. Essential lncRNA genes also facilitate studies on the minimal requirement of a genome for living cells, which will eventually allow the artificial synthesis of complicated genomes [20]. They also regulates gene expressions governing hallmarks of cancer metastasis [21].
Several experimental efforts have been made in pinpointing essential lncRNA genes. For example, Sauvageau et al. find that knocking out of Fendrr, Peril, and Mdgt in mouse genome will result in perinatal and postnatal lethal phenotypes [22]. lncRNA Braveheart play critical roles in the establishment of the cardiovascular lineage during mammalian development [10]. Besides these experiments for identifying essential lncRNA genes following traditional definitions, cancer cell-line based experiments also provide insights in essential lncRNA genes. Since gene essentiality may be defined in different levels with different context [16], several oncogene and tumor suppressors can also be recognized as essential lncRNA genes. For example, knockdown of the oncogene MIR4435-1HG inhibits cell proliferation, and suppresses the migrating and invasive capacity of renal carcinoma cells [23]. In the context of cancer cellular level essentiality, it can be regarded as an essential lncRNA. Similar reports can also be found in gastric cancer for the oncogene LINC00461 [24]. For another example, the lncRNA PDCD4-AS1 is a tumor suppressor gene. Over-expression of which will effectively attenuate triple-negative breast cancer cell proliferation, migration and invasion, and will increase the apoptosis rate [7]. Therefore, this gene can be regarded as an essential lncRNA gene in the context of avoiding tumors.
With thousands of lncRNA being discovered in various species, great efforts have been made in establishing databases for depositing lncRNA gene records and annotations. For example, LNCipedia [25], which collected over 100 thousand human lncRNA transcript records with annotations from various sources. The NONCODE database v6 [26] is a comprehensive database of all types of non-coding RNAs. It recorded 173,112 human lncRNAs with annotations. The LncRNAWiki[27] is a knowledgebase of human lncRNAs with experimental evidences. Other examples include deepBase[28], lncRNAdb[29], LncBook[30], lncRNome[31], NRED[32], Cancer LncRNA Census[33], and lncRNAKB[34].
However, none of the existing database focused on the essentiality of lncRNA genes. Existing essential gene databases, like DEG[35] and OGEE [36], focus on collecting essential protein-coding genes in bacteria, archaea, single cell eukaryotic organisms and cell-line experiments. Only a handful of essential non-coding gene records can be found in these databases. These records are still mainly in prokaryotic organisms. Most of the essential lncRNA records are still scattered in various literatures.
To this end, we developed a database system for essential lncRNA genes, which is named as dbEssLnc (https://esslnc.pufengdu.org). The dbEssLnc database has a web-based user-friendly interface, which allows users to browse, to search, to BLAST, to visualize and to download essential lncRNA gene information. We have deposited 201 essential lncRNA records in the dbEssLnc database. Since gene essentiality can be defined at different level with different context [16], we included reports not only from the direct gene knockout or knockdown experiments, but also from cell line-based tumor-related studies. Besides the general essential lncRNAs, the oncogenes and the tumor suppressors are also recorded as two additional categories. We believe that the dbEssLnc database will be a useful resource in computational biology, oncology and synthetic biology.
2. Materials and methods
2.1. Data curation
We collected the essential lncRNA data records manually from literatures. We searched PubMed database using keywords like “knockout or knockdown lncRNA”, “lncRNA lethality”, “long non-coding RNA”, and “essential lncRNA”. A total of 201 full texts of literatures were downloaded. We manually screened the literatures to find out details of essential lncRNAs, including the gene name and the source organism. We mapped the gene name to other databases, including NONCODE [26], HGNC [37], MGI [38] and Ensemble [39], to scratch related annotations, like NCBI gene id, NONCODE gene id, full gene name alias, lncRNA transcript sequences, and gene ontology annotations. These annotations are recorded along with the basic information of each essential lncRNA. The literature that report the essential lncRNA originally was linked using the PubMed ID.
Tissue specific expression profiles were collected from the NONCODE database for visualization purpose. The NONCODE database uses public RNA-seq data of human and mouse to estimate lncRNA expression levels. Human BodyMap 2.0 data from 16 human tissues (PRJEB2445) and mouse RNA-seq data set from six different tissues (PRJEB2476) were downloaded from the European Nucleotide Archive [40]. The expression level unit is FPKM/TPM. The expression profile of each lncRNA gene in various tissue types is presented by ECharts.
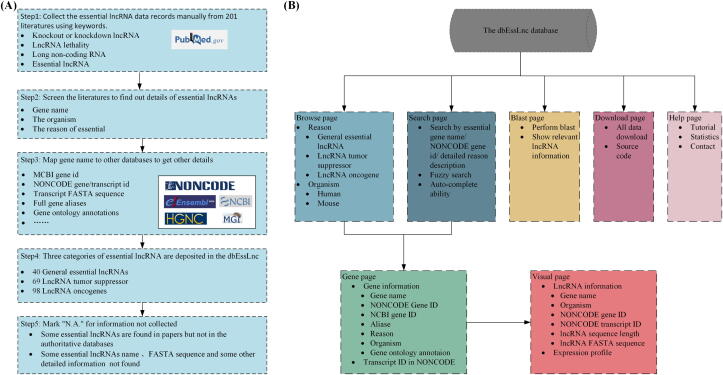
The gene name aliases vary in different reports. We manually mapped different aliases to standard names. However, in some cases, it is not possible to determine the exact mapping of records in related databases, such as the NCBI Genes and the NONCODE database. We keep this kind of reports as records in our database with annotations as N.A. The flowchart for the process of data curation and dbEssLnc website architecture are illustrated in Fig. 1.
Fig. 1.
The work flow of data curation process and the architecture of dbEssLnc. (A) The work flow is divided into 5 steps. Step1: use keywords to collect essential lncRNA data in PubMed. Step2: screen details of essential lncRNA from literatures. Step3: map gene names to other databases to get annotations. Step4: category essential lncRNAs, as general essential lncRNAs, lncRNA tumor suppressors and lncRNA oncogenes. Step5: mark “N.A.” for missing information. (B) Architecture of dbEssLnc website. dbEsslnc has four functional pages, including “Browse”, “Search”, “BLAST”, and “Download”. An additional “Help” page provides instructions on how to use the dbEssLnc database. Users may click the gene name in the “Browse” and the “Search” page to jump to the “Gene” page for annotations of the gene. By clicking gene transcript id on the “Gene” page, the “Visual” page would provide information about lncRNA transcripts.
2.2. System design and implementation
All essential lncRNA gene data are deposited in a MySQL database. A user-friendly web-based interface was provided with dynamic web effects, which allows the user to browse, to search, to BLAST, to download and to visualize the data of essential lncRNAs. The web-based user interface of the dbEssLnc database is completely implemented using JavaScript, on both the frontend and the backend. The backend is driven by the Node.js engine with Express framework. The frontend is implemented using the Vue framework.
3. Results and discussions
3.1. Statistics of data records
We have a total of 207 essential lncRNAs records in the dbEssLnc database, including 40 general essential lncRNAs, 69 lncRNA tumor suppressor genes, and 98 lncRNA oncogenes. These statistics can be found in Tables 1 with details of organism and category distributions.
Table 1.
Statistics of essential lncRNA records.
| Organism | General essential lncRNA | Tumor suppressor | Oncogene | Total |
|---|---|---|---|---|
| Human | 8 | 67 | 98 | 173 |
| Mouse | 32 | 2 | 0 | 34 |
| Total | 40 | 69 | 98 | 207 |
3.2. Coverage analysis on human essential lncRNA genes
The number of the essential coding genes are usually recognized as about 10 % of all protein coding genes in human cells [[14], [16]]. Non-coding genes are usually thought to be less indispensable [41]. Therefore, we assume that the number of essential non-coding genes is less than 10 % of all non-coding genes. According to the most recent assemble of human genome [42], the number of genes in the human genome is about 60000, while almost 20,000 are coding genes. This makes the number of all non-coding genes about 40000. Previously, the number of lncRNA genes was estimated to be about 30000 [[43], [44]], which makes a majority of all non-coding genes. Combining all above numbers, we estimate the number of essential lncRNA genes in human genome should be roughly 3000 in various contexts. This number generally consists with a recent genome-wide computational estimation based on systems biology [45].
Based on the above estimations, dbEssLnc currently only covers a very small part of all essential lncRNA genes. This is due to the fact that gene essentiality was usually defined and discussed for protein-coding regions of the genome. Essentiality of non-coding regions was discussed mostly as computational prediction results based on datasets from coding regions [[41], [45], [46], [47]]. Experimentally characterized essential lncRNA genes are rarely reported. Although the CRISPR-Cas9 screening have provided very promising results on finding functional lncRNAs that affect cell growth in cell line experiments [[48], [49]], depositing these results in the dbEssLnc requires massive amount of manually curation and cross-reference verification works. These works are planned in the very near future.
3.3. Categories of essential lncRNA genes
In the dbEssLnc database, we divide all records into three categories: general essential lncRNAs, lncRNA oncogene and lncRNA tumor suppressor.
The general essential lncRNA are lncRNAs that are determined by direct gene knockout or knockdown experiments in vivo. They follow the traditional definition of essential genes. For example, the essentiality of Fendrr, Peril, and Mdgt in mouse [22] were determined in this way.
Since gene essentiality is a dynamic concept, which can be defined on both cellular level and organism level, and with different conditions and contexts [[14], [16]], cancer cell line experiments should be considered as gene essentiality evidences. Many literatures studied the functional roles of lncRNAs using cell line-based experiments in tumor cells [[50], [51]]. Screening essential lncRNA in human cancer cell line not only explain the differences between each cell line and cancer type, but also reflect the developmental origin, oncogenic drivers, paralogous gene expression pattern, and chromosomal structure [52].
Although cellular essential genes [16], which are determined in cancer cell lines, may be largely different to the set of organism essential genes in different conditions [14], it still provides us an opportunity to reveal essential genes in human, where direct gene inactivation experiments are obviously hampered. In addition, the OGEE database has already taken essential protein coding gene information from the CRISPR-Cas9 screening results on human cancer cell lines [36]. Therefore, we include the lncRNA oncogenes and lncRNA tumor suppressors as two additional categories in the dbEssLnc. Most information in these two categories are derived from cancer cell line experimental reports.
The lncRNA oncogenes, which have high level of expression and are required for cancer cell proliferation and survival, can be defined as essential lncRNA genes on the cellular level in the context of cancer cell line. Inactivation of these lncRNA genes will lead to a decrease in cancer cell proliferation, invasion and migration, and an increase in apoptosis [54]. Therefore, we include the lncRNA oncogene as a category of essential lncRNAs in the dbEssLnc database. For example, BRAF gene V600E mutation is a critical diagnostic, prognostic, and predictive biomarker in melanoma [55], primary brain tumors [56] and many kinds of cancers. The lncRNA BANCR, which is a BRAF-activated lncRNA, suppresses the proliferation, migration and invasion of cancer cells, and further induces apoptosis of hepatocellular carcinoma cells [57]. We include BANCR in the dbEssLnc database as an essential lncRNA oncogene. The curation of these records requires not only large-scale screening results, but also reports on functional characterizations.
In the case of lncRNA tumor suppressors, it should be noted that suppressing expression of these genes may result in lethal phenotype of the organism due the progression of cancers. Tumor suppressor genes have negative regulatory effects on cell proliferation, including inhibiting cell proliferation, regulating cell cycle checkpoints, promoting apoptosis and participating in DNA damage repair [58]. These genes are essential for a normal organism in the context of avoiding tumors. For example, the importance of the TP53 gene as a tumor suppressor has been highlighted in human cancers for many years [59]. LncRNA MEG3 induces accumulation of p53 (TP53) protein, and selectively regulates p53 target gene expression [60]. Therefore, we define MEG3 as an essential lncRNA tumor suppressor.
3.4. Comparative analysis between human and mouse essential lncRNAs
Although dbEssLnc covers only very small part of all essential lncRNA genes, it still allows us to perform some simple comparative analysis between human and mouse essential lncRNAs.
8 homologous between human and mouse were found within the dbEssLnc database (Table 2). Although gene essentiality may not be conserved across species, orthologs sharing essentiality is a phenomenon that has been reported [[45], [60]]. However, whether this sharing of essentiality is a common phenomenon for lncRNA genes across species is still an open question.
Table 2.
Human and mouse homology lncRNA gene in the dbEssLnc database.
| Human essential lncRNA | Mouse essential lncRNA |
|---|---|
| FENDRR | Fendrr |
| MEG3 | Meg3 |
| MIAT | Miat |
| NEAT1 | Neat1 |
| LINC-PINT | Lncpint |
| TP53COR1 | Trp53cor1 |
| JPX | Jpx |
| GAS5 | Gas5 |
3.5. Case study
We take the Meg3 gene in mouse as an example to show how to use the dbEssLnc database to extract related information.
We provide four pages for users to access the dbEssLnc database functions, with an additional page for instructions and a home page. These four pages include “Browse”, “Search”, “BLAST”, and “Download”.
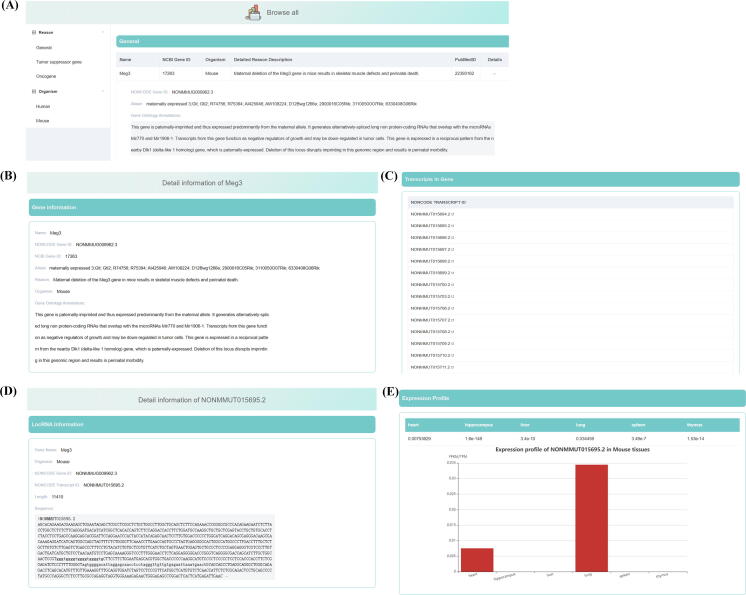
On the “Browse” page, all lncRNA records are categorized into groups, according to their reasons of essential and organism. Users can choose the category using the expandable sidebar on the left side of the page. Five different tables of records are displayed on the right side according to users' choices (Fig. 2A). These records are provided with links to external databases. In the category of general essential lncRNA, gene Meg3 is recorded to have NCBI gene ID 17,263 with detailed reason description: “Maternal deletion of the Meg3 gene in mice results in skeletal muscle defects and perinatal death”. The PubMed ID is linked to its original reports [60]. When the detail arrow is clicked to expand, gene name aliases, the NONCODE gene ID and gene ontology annotations are displayed. When the name of the gene is clicked, users are directed to a page called “gene”. On this page, all details of the gene will be displayed along with all gene transcript IDs and different splicing isoforms in NONCODE (Fig. 2B and 2C). Users can click NONCODE transcript ID to jump to the “visual” page. This page shows lncRNA information including lncRNA sequences in the FASTA format and the tissue specific expression levels (Fig. 2D and 2E). Clicking the three dots at the end of the sequence can expand or collapse the full sequence view.
Fig. 2.
The “Browse”, “Gene” and “Visual” page. (A) The “Browse” page have a sidebar for navigation and five tables; (B) The “Gene” page for gene annotations; (C) The “Gene” page for lncRNA transcript IDs; (D) The “Visual” page for lncRNA information; (E) The “Visual” page for lncRNA tissue specific expression profile.
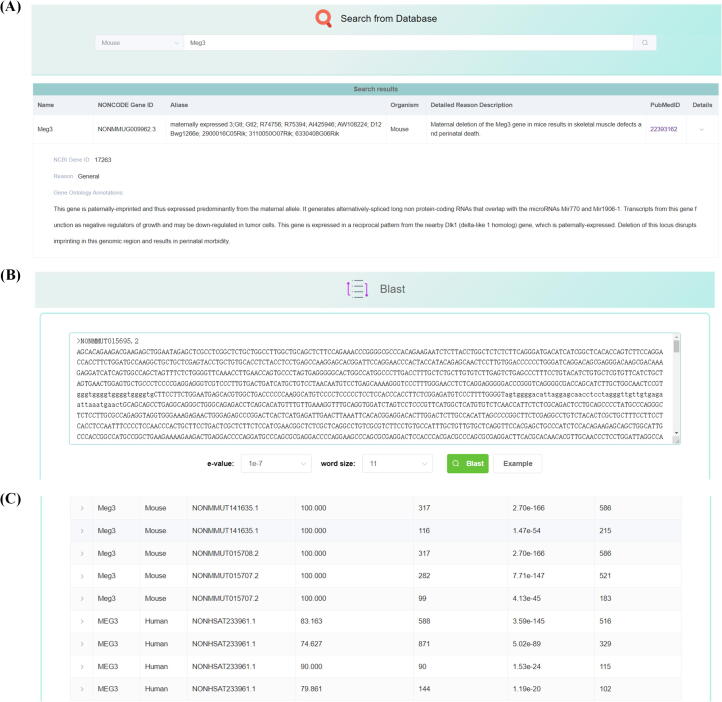
On the “Search” page, users can choose to search human or mouse essential lncRNAs, or a specific category of essential reasons. This is a full text search function. It offers a fuzzy search function that can automatically complete the user's input with the first few letters matching an existing name in the database. The query box supports wildcards queries with '_' and '%', where '_' matches any single character, and '%' any number of characters. After the gene name Meg3 is entered, clicking the search icon will return a record of Meg3 with the gene name, NONCODE ID, gene name aliases and detailed reason description fields. Clicking the details arrow will display the records details like in the “Browse” page (Fig. 3A). Copy the word “skeletal” and paste it in the query box, switch the search type to “General” and click the search icon again. Three genes, including “Dnm3os”, “Meg3” and ”RMRP“ are displayed, all with the word ”skeletal“ in the detailed reason description.
Fig. 3.
The “Search” page and the “Blast” page. (A) Select mouse from the selection box and search using the name Meg3 on the “Search” page; (B) users can copy a part of the mouse Meg3 sequence in FASTA format to the clipboard and paste it in the query box. After clicking the “Blast” button, dbEssLnc would find similar essential lncRNAs sequences in the database. (C) users can find that human MEG3 gene is in the list of similar essential lncRNA sequences.
On the “Blast” page, users can copy a part of the mouse Meg3 sequence in FASTA format and paste it in the query box (Fig. 3B). After properly adjusting the e-value and word size parameters, clicking the “Blast” button will start a BLAST search using the Meg3 sequence. The result will be shown several seconds later. The human MEG3 gene is in the list of similar essential lncRNA genes (Fig. 3C). Therefore, we find that the lncRNA gene MEG3 in human and Meg3 in mouse are both essential. Detail information of the alignment can be displayed by clicking the detail arrow on the left side.
In addition to the above pages, on the “Download” page, all essential lncRNA information and the source code of dbEssLnc can be downloaded. Users can click the corresponding file name to download the data tables. Source code can be obtained from the GitHub repository (https://github.com/yyZhang14/dbEssLnc), which allows the user to host their own mirror of dbEssLnc database on their own servers.
Finally, the “Help” page provides instructions on how to use the web interfaces of dbEssLnc. Step-by-step instructions were provided in the “Tutorial” section. The “Statistics” section gives the data distribution in the dbEssLnc. The “Contact” provides the approach to contact the authors of the database.
We have tested the website of dbEssLnc on both Windows and MacOS platform with mainstream web browsers, including IE, Firefox, Google Chrome and Safari. All these browsers can view and operate the website normally.
4. Conclusion
Distinguishing essential genes from non-essential genes has been a long-standing question in genetics. With the advancement of technology, a big progress has been made in performing genome-wide essentiality screening among diverse species. Most existing studies focused on the essentiality of protein-coding genes. The essentiality of non-coding genes are much less discussed due to various technical reasons [14]. The essentiality of lncRNA genes can be defined on both cellular and organism levels. Particularly, lncRNA gene essentiality is useful in cancer related studies. However, existing lncRNA databases do not highlight essentiality information. This is the primary reason that we develop the dbEssLnc database. Although large scale CRISPR-Cas9 screening have provided very promising information in cancer cell lines [62], computational prediction approaches, like GIC [46] and SGII [45] cannot efficiently utilize these results. A database like dbEssLnc will facilitate the development of these computational prediction approaches.
As far as we can tell, dbEssLnc database is the first data resource focusing on collecting essential lncRNA information. We provide a relatively comprehensive and accurate collection of essential lncRNAs with a flexible web-based user interface. The establishment of dbEssLnc will promote the study of the minimal genome for living cells. These essential lncRNAs will also provide insights on developing therapeutic and early diagnostic approaches for tumors and other complex disorders. It is also expected that computational prediction methods will be developed for identifying essential lncRNAs. More records will be deposited in the dbEssLnc database in the very near future. We will keep tracking newly discovered essential lncRNA reports and update the dbEssLnc database in time.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by National Natural Science Foundation of China [NSFC 61872268] and National Key R&D Program of China [2018YFC0910405].
References
- 1.Aliperti V., Skonieczna J., Cerase A. Long non-coding RNA (lncRNA) roles in cell biology, neurodevelopment and neurological disorders. Noncoding RNA. 2021;7:36. doi: 10.3390/ncrna7020036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schaukowitch K., Kim T.-K. Emerging epigenetic mechanisms of long non-coding RNAs. Neuroscience. 2014;264:25–38. doi: 10.1016/j.neuroscience.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J., Zhu S., Meng N., He Y., Lu R., Yan G.-R. ncRNA-Encoded Peptides or Proteins and Cancer. Mol Ther. 2019;27:1718–1725. doi: 10.1016/j.ymthe.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts T.C., Morris K.V., Wood M.J.A. The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130507. doi: 10.1098/rstb.2013.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long Y., Wang X., Youmans D.T., Cech T.R. How do lncRNAs regulate transcription? Sci Adv. 2017;3:eaao2110. doi: 10.1126/sciadv.aao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dykes I.M., Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genom Proteom Bioinform. 2017;15:177–186. doi: 10.1016/j.gpb.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D., Wang Z., Zhang L., Sun S. LncRNA PDCD4-AS1 alleviates triple negative breast cancer by increasing expression of IQGAP2 via miR-10b-5p. Transl Oncol. 2021;14 doi: 10.1016/j.tranon.2020.100958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrè F., Colantoni A., Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17:106–116. doi: 10.1093/bib/bbv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L., Zhou Y., Li H. LncRNA, miRNA and lncRNA-miRNA interaction in viral infection. Virus Res. 2018;257:25–32. doi: 10.1016/j.virusres.2018.08.018. [DOI] [PubMed] [Google Scholar]
- 10.Klattenhoff C.A., Scheuermann J.C., Surface L.E., Bradley R.K., Fields P.A., Steinhauser M.L., et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng B., Pan R., Ou X., Wang T., Wang W., Nie Y., et al. LncRNA RBM5-AS1 promotes osteosarcoma cell proliferation, migration, and invasion. Biomed Res Int. 2021;2021:5271291. doi: 10.1155/2021/5271291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases - PubMed n.d. https://pubmed.ncbi.nlm.nih.gov/30285109/ (accessed September 15, 2021). [DOI] [PMC free article] [PubMed]
- 13.Gao Y., Shang S., Guo S., Li X., Zhou H., Liu H., et al. Lnc2Cancer 3.0: an updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data. Nucleic Acids Res. 2021;49:D1251–D1258. doi: 10.1093/nar/gkaa1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartha I., di Iulio J., Venter J.C., Telenti A. Human gene essentiality. Nat Rev Genet. 2018;19:51–62. doi: 10.1038/nrg.2017.75. [DOI] [PubMed] [Google Scholar]
- 15.Joshi M., Rajender S. Long non-coding RNAs (lncRNAs) in spermatogenesis and male infertility. Reprod Biol Endocrinol. 2020;18:103. doi: 10.1186/s12958-020-00660-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rancati G., Moffat J., Typas A., Pavelka N. Emerging and evolving concepts in gene essentiality. Nat Rev Genet. 2018;19:34–49. doi: 10.1038/nrg.2017.74. [DOI] [PubMed] [Google Scholar]
- 17.Rosa A., Ballarino M. Long Noncoding RNA Regulation of Pluripotency. Stem Cells Int. 2016;2016:1797692. doi: 10.1155/2016/1797692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tu J., Tian G., Cheung H.-H., Wei W., Lee T.-L. Gas5 is an essential lncRNA regulator for self-renewal and pluripotency of mouse embryonic stem cells and induced pluripotent stem cells. Stem Cell Res Ther. 2018;9:71. doi: 10.1186/s13287-018-0813-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartwell L.H., Szankasi P., Roberts C.J., Murray A.W., Friend S.H. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- 20.Koonin E.V. How many genes can make a cell: the minimal-gene-set concept. Annu Rev Genomics Hum Genet. 2000;1:99–116. doi: 10.1146/annurev.genom.1.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song X., Wang H., Wu J., Sun Y. Long noncoding RNA SOX2-OT knockdown inhibits proliferation and metastasis of prostate cancer cells through modulating the miR-452-5p/HMGB3 axis and inactivating Wnt/β-catenin pathway. Cancer Biother Radiopharm. 2020;35:682–695. doi: 10.1089/cbr.2019.3479. [DOI] [PubMed] [Google Scholar]
- 22.Sauvageau M., Goff L.A., Lodato S., Bonev B., Groff A.F., Gerhardinger C., et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. ELife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu K., Hu L., Lv X., Chen J., Yan Z., Jiang J., et al. Long non-coding RNA MIR4435-1HG promotes cancer growth in clear cell renal cell carcinoma. Cancer Biomark. 2020;29:39–50. doi: 10.3233/CBM-201451. [DOI] [PubMed] [Google Scholar]
- 24.Shi X., You X., Zeng W.-C., Deng Y.-J., Hong H.-L., Huang O.-X., et al. Knockdown of LINC00461 inhibits cell proliferation and induces apoptosis in gastric cancer by targeting LSD1. Eur Rev Med Pharmacol Sci. 2019;23(10769–75) doi: 10.26355/eurrev_201912_19779. [DOI] [PubMed] [Google Scholar]
- 25.Volders P.-J., Anckaert J., Verheggen K., Nuytens J., Martens L., Mestdagh P., et al. LNCipedia 5: towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019;47:D135–D139. doi: 10.1093/nar/gky1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao L., Wang J., Li Y., Song T., Wu Y., Fang S., et al. NONCODEV6: an updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic Acids Res. 2021;49:D165–D171. doi: 10.1093/nar/gkaa1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.L L, Z L, C L, D Z, Q L, C F, et al. LncRNAWiki 2.0: a knowledgebase of human long non-coding RNAs with enhanced curation model and database system. Nucleic Acids Res. 2022;50 doi: 10.1093/nar/gkab998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie F., Liu S., Wang J., Xuan J., Zhang X., Qu L., et al. deepBase v3.0: expression atlas and interactive analysis of ncRNAs from thousands of deep-sequencing data. Nucleic Acids Res. 2021;49:D877–D883. doi: 10.1093/nar/gkaa1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quek X.C., Thomson D.W., Maag J.L.V., Bartonicek N., Signal B., Clark M.B., et al. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43:D168–D173. doi: 10.1093/nar/gku988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma L., Cao J., Liu L., Du Q., Li Z., Zou D., et al. LncBook: a curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res. 2019;47:D128–D134. doi: 10.1093/nar/gky960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhartiya D, Pal K, Ghosh S, Kapoor S, Jalali S, Panwar B, et al. lncRNome: a comprehensive knowledgebase of human long noncoding RNAs. Database (Oxford) 2013;2013:bat034. 10.1093/database/bat034. [DOI] [PMC free article] [PubMed]
- 32.Dinger M.E., Pang K.C., Mercer T.R., Crowe M.L., Grimmond S.M., Mattick J.S. NRED: a database of long noncoding RNA expression. Nucleic Acids Res. 2009;37:D122–D126. doi: 10.1093/nar/gkn617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vancura A., Lanzós A., Bosch-Guiteras N., Esteban M.T., Gutierrez A.H., Haefliger S., et al. Cancer LncRNA Census 2 (CLC2): an enhanced resource reveals clinical features of cancer lncRNAs. NAR. Cancer. 2021;3 doi: 10.1093/narcan/zcab013. zcab013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seifuddin F., Singh K., Suresh A., Judy J.T., Chen Y.-C., Chaitankar V., et al. lncRNAKB, a knowledgebase of tissue-specific functional annotation and trait association of long noncoding RNA. Sci Data. 2020;7:326. doi: 10.1038/s41597-020-00659-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo H., Lin Y., Liu T., Lai F.-L., Zhang C.-T., Gao F., et al. DEG 15, an update of the Database of Essential Genes that includes built-in analysis tools. Nucleic Acids Res. 2021;49:D677–D686. doi: 10.1093/nar/gkaa917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gurumayum S., Jiang P., Hao X., Campos T.L., Young N.D., Korhonen P.K., et al. OGEE v3: Online GEne Essentiality database with increased coverage of organisms and human cell lines. Nucleic Acids Res. 2021;49:D998–D. doi: 10.1093/nar/gkaa884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gray K.A., Yates B., Seal R.L., Wright M.W., Bruford E.A. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res. 2015;43:D1079–D1085. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ringwald M., Richardson J.E., Baldarelli R.M., Blake J.A., Kadin J.A., Smith C., et al. Mouse genome informatics (MGI): latest news from MGD and GXD. Mamm Genome. 2021 doi: 10.1007/s00335-021-09921-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howe K.L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., et al. Ensembl 2021. Nucleic Acids Res. 2021;49:D884–D891. doi: 10.1093/nar/gkaa942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cummins C., Ahamed A., Aslam R., Burgin J., Devraj R., Edbali O., et al. The European nucleotide archive in 2021. Nucleic Acids Res. 2022;50:D106–D110. doi: 10.1093/nar/gkab1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wells A., Heckerman D., Torkamani A., Yin L., Sebat J., Ren B., et al. Ranking of non-coding pathogenic variants and putative essential regions of the human genome. Nat Commun. 2019;10:5241. doi: 10.1038/s41467-019-13212-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nurk S., Koren S., Rhie A., Rautiainen M., Bzikadze A.V., Mikheenko A., et al. The complete sequence of a human genome. Science. 2022;376:44–53. doi: 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hon C.-C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J.L., Gough J., et al. An atlas of human long non-coding RNAs with accurate 5’ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xin X.-H., Zhang Y.-Y., Gao C.-Q., Min H., Wang L., Du P.-F. SGII: systematic identification of essential lncRNAs in mouse and human genome with lncRNA-protein-protein heterogeneous interaction network. Front Genet. 2022;13 doi: 10.3389/fgene.2022.864564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng P., Chen J., Meng Y., Zhou Y., Yang J., Cui Q. Defining essentiality score of protein-coding genes and long noncoding RNAs. Front Genet. 2018;9:380. doi: 10.3389/fgene.2018.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian D, Wenlock S, Kabir M, Tzotzos G, Doig AJ, Hentges KE. Identifying mouse developmental essential genes using machine learning. Dis Model Mech 2018;11:dmm034546. 10.1242/dmm.034546. [DOI] [PMC free article] [PubMed]
- 48.Liu S.J., Horlbeck M.A., Cho S.W., Birk H.S., Malatesta M., He D., et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355 doi: 10.1126/science.aah7111. aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu S., Li W., Liu J., Chen C.-H., Liao Q., Xu P., et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat Biotechnol. 2016;34:1279–1286. doi: 10.1038/nbt.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J., Su Z., Lu S., Fu W., Liu Z., Jiang X., et al. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin Chim Acta. 2018;485:229–233. doi: 10.1016/j.cca.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 51.Hao T., Huang S., Han F. LINC-PINT suppresses tumour cell proliferation, migration and invasion through targeting miR-374a-5p in ovarian cancer. Cell Biochem Funct. 2020;38:1089–1099. doi: 10.1002/cbf.3565. [DOI] [PubMed] [Google Scholar]
- 52.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Torry D.S., Cooper G.M. Proto-oncogenes in development and cancer. Am J Reprod Immunol. 1991;25:129–132. doi: 10.1111/j.1600-0897.1991.tb01080.x. [DOI] [PubMed] [Google Scholar]
- 54.Ito T., Tanaka Y., Murata M., Kaku-Ito Y., Furue K., Furue M. BRAF heterogeneity in melanoma. Curr Treat Options Oncol. 2021;22:20. doi: 10.1007/s11864-021-00818-3. [DOI] [PubMed] [Google Scholar]
- 55.Maraka S., Janku F. BRAF alterations in primary brain tumors. Discov Med. 2018;26:51–60. [PubMed] [Google Scholar]
- 56.Li J., Wang J., Zhou W., Zhang S., Le Y., He R. Downregulation of BRAF-activated non-coding RNA suppresses the proliferation, migration and invasion, and induces apoptosis of hepatocellular carcinoma cells. Oncol Lett. 2017;14:4751–4757. doi: 10.3892/ol.2017.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joyce C., Rayi A., Kasi A. StatPearls Publishing; StatPearls, Treasure Island (FL): 2021. Tumor-Suppressor Genes. [PubMed] [Google Scholar]
- 58.Aubrey B.J., Strasser A., Kelly G.L. Tumor-suppressor functions of the TP53 Pathway. Cold Spring Harb Perspect Med. 2016;6 doi: 10.1101/cshperspect.a026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou Y., Zhang X., Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48:R45–R53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gu Z., Steinmetz L.M., Gu X., Scharfe C., Davis R.W., Li W.-H. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- 61.Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]