Summary
PIWI-interacting RNAs (piRNAs) guard germline genomes against the deleterious action of mobile genetic elements. PiRNAs use extensive base-pairing to recognize their targets and variable 3′ends could change the specificity and efficacy of piRNA silencing. Here, we identify conserved rules that ensure the generation of a single major piRNA 3′end in flies and mice. Our data suggest that the PIWI proteins initially define a short interval on pre-piRNAs that grants access to the ZUC-processor complex. Within this Goldilocks zone, the preference to cut in front of Uridine determines the ultimate processing site. We observe a mouse-specific roadblock that relocates the Goldilocks zone and generates an opportunity for consecutive trimming. Our data reveal a conserved hierarchy between length and sequence preferences that controls the piRNA sequence space. The unanticipated precision of 3′end formation bolsters the emerging understanding that the functional piRNA sequence space is tightly controlled to ensure effective defense.
Subject areas: Biological sciences, Cell biology, Molecular biology
Graphical abstract
Highlights
-
•
Every piRNA has a single major 3′end
-
•
Length preference trumps sequence preference during 3′end formation
-
•
A hierarchy of length and sequence preferences is conserved in flies and mice
-
•
Relocation of the Goldilocks zone generates opportunity for trimming in mice
Biological sciences; Cell biology; Molecular biology
Introduction
Target-recognition by complementary base-pairing places the sequences of the guide-RNAs at the center of all RNA silencing mechanisms (Bartel, 2018). In eukaryotes, short guide RNAs associate with Argonaute proteins to form RNA-induced silencing complexes (RISC) that defend against viruses, regulate gene expression, and protect genome integrity (Joshua-Tor and Hannon, 2011). Within RISC, the sequence of the guide RNA determines target specificity, whereas the Argonaute partner executes silencing mechanisms resulting in transcriptional and posttranscriptional restriction (Ghildiyal and Zamore, 2009; Stuwe et al., 2014). The base-pairing interactions of guide RNAs and their targets follow common themes (Sheu-Gruttadauria and MacRae, 2017). Universally, the most 5′ nucleotides of the guide RNAs are buried within the Argonaute protein and not available for base-pairing (Frank et al., 2010). The following nucleotides 2-7 are the first to engage with a target (Anzelon et al., 2021; Bartel, 2009). This ‘seed region’ dictates target specificity in microRNA (miRNA) silencing and provides a framework for studying other RNA silencing pathways (Bartel, 2018). Additional base-pairing across nucleotide 10 and 11 of the guide RNA is required for target-RNA cleavage by the intrinsic nuclease activity of Argonaute proteins (Rivas et al., 2005; Wang et al., 2008). In contrast to miRNA-guided silencing, target recognition by PIWI-interacting RNAs (piRNAs) requires extensive base-pairing along the 3′ half of the guide RNA and potentially generates more specific and stable piRNA:target pairs (Anzelon et al., 2021).
PiRNAs and their PIWI protein partners silence endogenous retroviruses and other mobile genetic elements (transposons) to protect the integrity of germline genomes (Czech et al., 2018; Iwasaki et al., 2015; Ozata et al., 2019). Mutants of key piRNA pathway genes universally result in the sterility of the animal (Lin, 2007; Siomi et al., 2011). PiRNAs comprise millions of non-conserved sequences. How these diverse small RNAs faithfully restrict all transposons while avoiding potentially deleterious off-target effects remains unknown. Thousands of different piRNAs can be parsed from a single long precursor transcript. Processing is initiated by an endonucleolytic cut. Either the ZUC-processor complex or piRNA-guided slicing generates a 5′ monophosphorylated fragment that can be loaded onto a PIWI protein to form a pre-piRNA complex (Iwasaki et al., 2015; Le Thomas et al., 2014). Consecutively, 3′end maturation is suggested to take place on the preassembled PIWI-pre-piRNA complex. The ZUC-processor complex generates a 3′ cut that releases the PIWI-piRNA complex from a potentially kilobases-long precursor (Han et al., 2015; Ipsaro et al., 2012; Mohn et al., 2015; Nishimasu et al., 2012). Following this second endonucleolytic cut, a 3′-5′ exonuclease trims the 3′end to its mature length (Figure 1A). This cut-n’-trim mechanism of piRNA 3′end formation has originally been suggested based on the observation that each PIWI protein associates with piRNAs of distinct lengths (Guzzardo et al., 2013). The footprint hypothesis suggests that 3′ends of piRNAs are determined by the physical impression of their PIWI-protein partner. Trimmers were identified in flies, silkworms, and mice (Ding et al., 2017; Feltzin et al., 2015; Hayashi et al., 2016; Izumi et al., 2016; Nishimura et al., 2018; Zhang et al., 2017). PNLDC1, a 3′-5′ exonuclease related to Poly(A)-Specific Ribonuclease (PARN) trims pre-piRNAs to their mature length and is required for fertility in mice (Anastasakis et al., 2016; Ding et al., 2017; Nishimura et al., 2018; Zhang et al., 2017). An unrelated exonuclease, Nibbler (Nbr) trims some piRNAs and selects miRNAs in flies but is dispensable for piRNA function (Feltzin et al., 2015; Hayashi et al., 2016). Fly piRNAs exhibit a preference for Uridine just after the mature 3′end (+1 position) corresponding to the signature of the ZUC-processor complex (Han et al., 2015; Mohn et al., 2015; Stein et al., 2019). A +1U preference has also been observed for untrimmed pre-piRNAs in mouse (Gainetdinov et al., 2018), and additional sequence preferences were found for ZUC-mediated 3′end formation in silkworm cell extracts (Izumi et al., 2020). Taken together, these studies suggest that 3′end processing is guided by sequence preferences rather than length constraints. Here, we systematically probe how length and sequence preferences integrate into the 3′end decision and identify a conserved hierarchy of events. Our study elucidates an unanticipated precision in 3′end formation that generates a single major 3′end according to hierarchical length and sequence decisions.
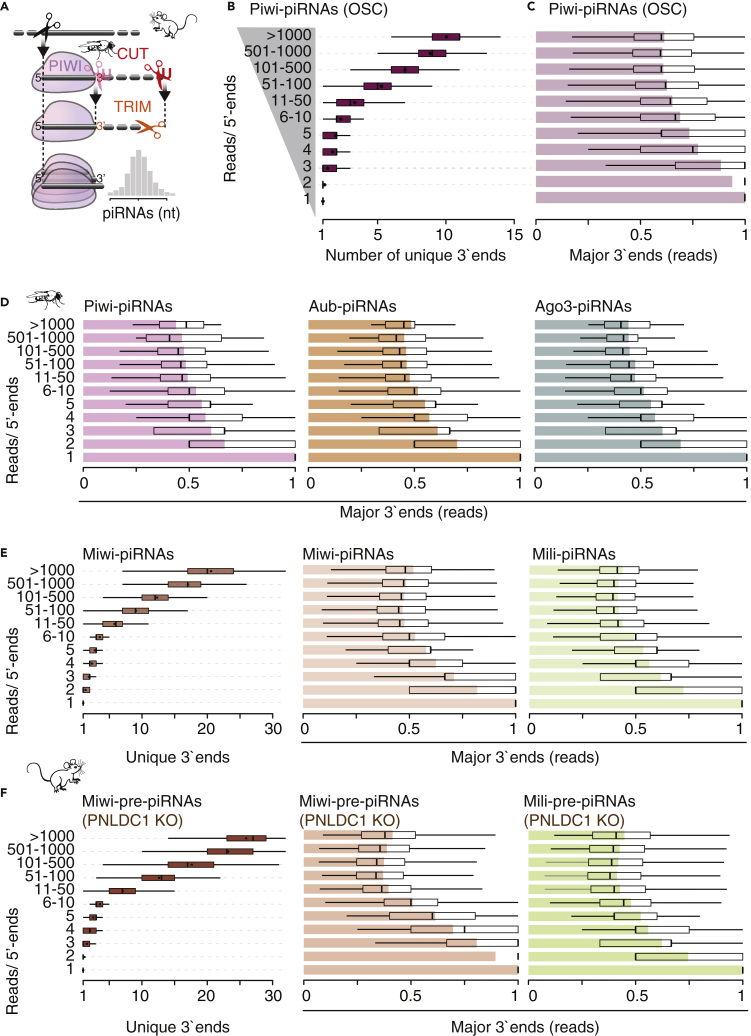
Figure 1.
PiRNAs have a single major 3′-end
(A) The cut-n’-trim model for piRNA 3′end formation proposes an initial endonucleolytic cut that generates a substrate for 3′-to-5′ exonucleolytic trimming. The resulting length distributions of piRNAs are suggested to resemble a footprint of their bound PIWI protein partner.
(B) Piwi-piRNAs that share the same 5′-end can have a number of different 3′-ends. Uniquely mapping Piwi-piRNAs from OSC (18–32 nt long) were grouped by the number of reads with a shared 5′-end. The number of different 3′-ends per 5′-end is depicted for each group.
(C and D) Most of the reads associated with the same 5′-end have a single preferred 3′-end. For each 5′-end, the most abundant 3′-end was identified (referred to as “major”), and its contribution to the abundance of the 5′-end was calculated. The mean contribution is shown as a bar plot, whereas quartiles are depicted as a boxplot. Only uniquely mapping piRNAs that are 18–32 nt long were used.
(E and F) In mice, both mature and untrimmed piRNAs can have many possible 3′-ends; however, only one contributes to approximately half of the abundance. Uniquely mapping reads were used for analyses (19–50nt). Datasets are publicly available (GSE156058; GSE83698; PRJNA421205).
Results
Every piRNA has a single major 3′end
The length of piRNAs varies from ∼23–29nt in flies and ∼23–35nt in mice (Czech et al., 2018; Iwasaki et al., 2015; Ozata et al., 2019). If piRNAs with the same 5′-end had a size distribution reflective of the total piRNA population, we would expect every 5′-end to be accompanied by multiple 3′-ends with the most prevalent represented by 20–30% of the small RNAs (Figure 1A). To test this hypothesis, we first counted the total number of diverse 3′ends for each piRNA 5′end. To account for sequencing depth, we grouped piRNA 5′ends by read count reflecting the representation in small RNA molecules. For the most abundant Piwi-piRNAs in ovarian somatic sheath cells (OSC), with more than 1000 reads originating from the same 5′position, we observed on average ten different 3′ends (Figure 1B). However, more than half of the reads were represented by a single major 3′end (Figure 1C). We observed similar results for Piwi-piRNAs in fly ovaries and Aubergine(Aub)-associated and Argonaute-3(Ago3)-associated piRNAs, where more than half of all reads associated with a specific 5′end shared the same 3′end (Figures 1D and S1) (ovary data (Hayashi et al., 2016)). Next, we analyzed available data for MIWI-bound and MILI-bound piRNAs in primary spermatocytes (Gainetdinov et al., 2018). Like for fly piRNAs, we observed many possible 3′ ends for mature mouse piRNAs, but one of them prevailed (Figure 1E). For highly represented MIWI-piRNAs (>1000 reads/5′end) we counted on average 21 different 3′ends. However, a single major end accounted for about half the reads, whereas the other half was represented by 20 different ends. Finally, we looked at murine pre-piRNAs that associated with MIWI or MILI in the absence of the trimming exonuclease (PNLDC1) (data (Gainetdinov et al., 2018)). Like mature piRNAs, pre-piRNAs had many possible 3′ends, but a single major end was represented by about half of all reads (Figure 1F). Taken together, our data reveal that each piRNA has a single major 3′end.
Major 3′ends emphasize length preferences specific for each PIWI protein
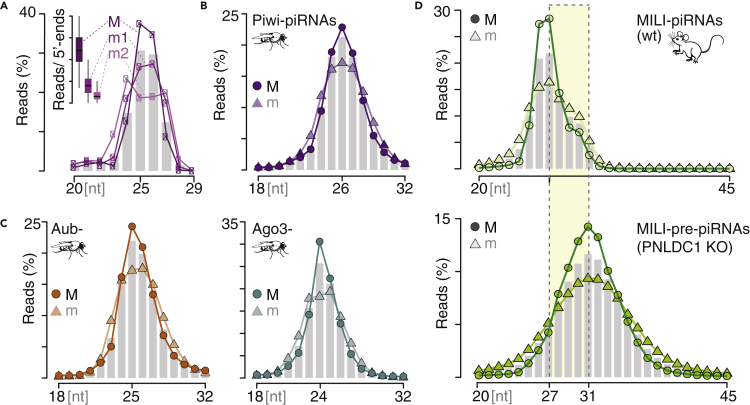
We next characterized the length preferences of major and minor 3′ends. The prevalence of a single major piRNA species associated with each 5′end eliminates 3′end variability as a reason for the broad length distribution of piRNA populations. It also suggests that individual major piRNAs (piRNAs that correspond to major 3′ends) are of different lengths, as no individual length accommodates more than half of all reads. To compare the length profiles of major (M) and minor (m) 3′ends, we normalized read counts within each group and plotted their distribution (Figure 2). Major Piwi-piRNAs are preferentially 25 or 26 nucleotides (nt) long in OSC (Figure 2A). In contrast, the second most abundant end (m1; ‘first minor end’) shows less preference for a particular length, and the next end (m2, ‘second minor end’) starts to show a bimodal length distribution. In fly ovaries, major Piwi-piRNAs are preferentially 26nt long (Figure 2B), whereas Aub-piRNAs and Ago3-piRNAs peak at lengths of 25 and 24nt, respectively (Figure 2C). Minor piRNAs universally show less length preference. Interestingly, both, trimmed piRNAs and untrimmed pre-piRNAs show an increased length preference associated with their major ends in mouse spermatocytes (Figures 2D and S2A). Major MILI-piRNAs peak at a length of 27nt, whereas MIWI-piRNAs peak at 30nt. Their major untrimmed counterparts are preferentially four nucleotides longer, resulting in 31nt long pre-piRNAs associated with MILI and 34nt long MIWI-pre-piRNAs. To test whether this four-nucleotide difference represents the preference of individual trimming events, we identified piRNA:pre-piRNA pairs and calculated the 3′distance of their major ends. Our data show that major pre-piRNAs associated with MIWI and MILI are preferentially four nucleotides longer than their mature counterparts (Figure S2B). The distinct difference of mature and pre-piRNAs in mice suggests a consistent roadblock that prevents access for the ZUC-processor on the pre-piRNA. Overall, the reinforced length preferences of major piRNAs bolster the hypothesis that 3′ends are defined through the footprint of their associated PIWI protein.
Figure 2.
Major 3′ends reinforce specific length preferences
(A) Piwi-piRNAs from OSC that correspond to major 3′-ends have an increased preference to be 25 or 26 nucleotides (nt) long. PiRNAs with the most abundant (major; M), the second most abundant (‘first minor end’; m1), and the third most abundant (‘second minor end’; m2) 3′-ends were grouped, and their length distribution was calculated. Reads were normalized to the total reads within each group. The bar plot depicts length distribution in nucleotides [nt]. The insert represents the contribution of Major (M) and minor (m1, m2) ends for each 5′-end.
(B and C) Fly Piwi-, Aubergine (Aub-), and Argonaute3 (Ago3-) piRNAs with major 3′-ends have a preference to be 26, 25, and 24 nt long, respectively. The length distribution of major (M) and all minor (m) 3′-ends is normalized to the respective group.
(D) Mature MILI-piRNAs that represent major 3′ends (M) have a stronger length preference than minor ends (m) in primary spermatocytes (SPI). MILI-pre-piRNAs are revealed in PNLDC1 knock-out (KO) spermatocytes (SPI) and exhibit a stronger length preference for major (M) compared to minor (m) sequences (data PRJNA421205).
Major 3′cuts bolster a preference for uridine following the cleavage site (+1U)
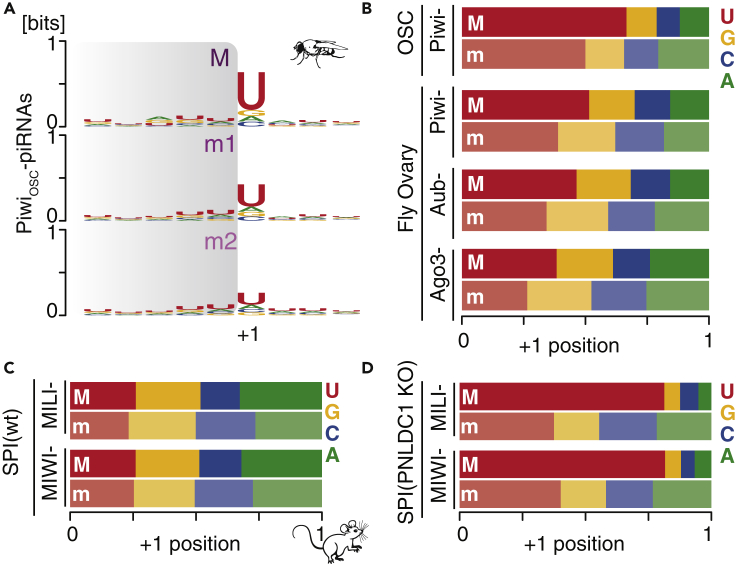
Next, we characterized sequence preferences at the major cut sites. The ZUC-processor complex establishes a preference for Uridine in the +1 position, and a +1U-bias has been observed for mature piRNAs in flies and untrimmed pre-piRNAs in mice (Han et al., 2015; Mohn et al., 2015). We observed a gradual decrease in +1U preference from the major (M) to the minor 3′ends (m1, 2) in Piwi-piRNAs (Figure 3A). Universally, we observed a stronger +1U preference for Major (M) compared to minor (m) piRNAs associated with all three PIWI proteins in flies (Figure 3B). Consistent with the idea that mature 3′ends in mouse are not guided by sequence preferences (Gainetdinov et al., 2018; Han et al., 2015), we did not observe a difference in +1U abundance between major and minor MILI-piRNAs and MIWI-piRNAs in mouse spermatocytes (Figure 3C). However, major pre-piRNAs showed a strong enrichment of +1U compared to minor 3′ends in PNLDC1 knockout mice (Figure 3D). Overall, we observed an increased preference for Uridine, one nucleotide downstream of major 3′ends when compared to minor ends, supporting the hypothesis that 3′ends are defined by sequence preference.
Figure 3.
Major 3′-cuts show an increased preference for Uridine in the +1 position
(A) Piwi-piRNAs with major 3′-ends (M) have a stronger preference for Uridine in the +1 position (+1U) than minor 3′ends (m). Sequence preferences of the most abundant (major; M), the second most abundant (minor; m1), and the third most abundant (minor; m2) 3′-ends are depicted as sequence logos in bits. The observed part of the piRNAs is indicated in gray. The +1 position marks the nucleotide immediately following the piRNA 3′end.
(B) Major 3′-ends increase the +1U preference for piRNAs associated with all three PIWI proteins in Drosophila ovaries. The nucleotide composition in the +1 position was calculated for major (M) and all minor (m) piRNA 3′-ends.
(C) Neither major (M) nor minor (m) 3′ends of mature (trimmed) murine piRNAs exhibit a +1U preference.
(D) The fraction of +1U following a major 3′end is doubled compared to minor 3′ends of untrimmed pre-piRNAs in mice. (Uniquely mapping reads were used for all panels).
Length preference trumps sequence preference during 3′end formation
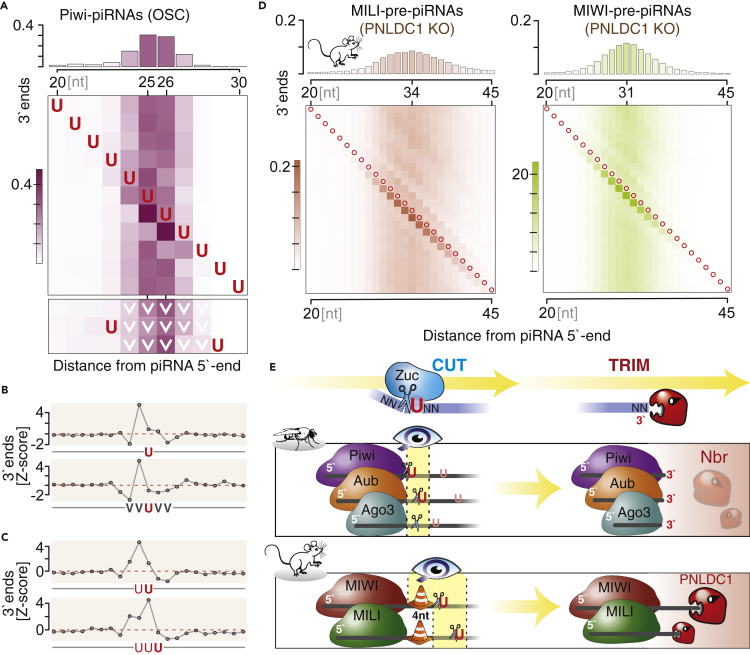
Our characterization of major piRNA 3′ends produced evidence for length and sequence preferences in flies and mice. Next, we aimed to identify how these preferences interact to define a single major 3′end. To this end, we probed the impact of Uridines at different positions relative to a preferred length (Figure 4). Piwi-piRNAs are preferentially 25 and 26 nt long. The presence of a Uridine one nucleotide downstream (position 26 and 27) results in a repositioning of 3′ends, and close to half of all piRNAs now end 5′ of this Uridine (Figure 4A). Uridines at positions outside of this preferred distance from the 5′end show no major effect on 3′end positioning. In the absence of any Uridine in a five-nucleotide window surrounding position 26, 3′ ends remain at positions 25 and 26 irrespectively. The effect of Uridine on the position of piRNA 3′ends can also be observed by calculating a Z-score for 3′ends across a local genomic interval, and when we restrict the nucleotide composition surrounding the Uridine to Adenine (A), Cytosine (C), or Guanosine (G) (Figure 4B). Finally, we ask what happens in the presence of Poly-Uridines. For Piwi-piRNAs, we observe a preference to position the 3′ end just before the last of two or three Uridines (Figure 4C). This preference might contribute to the observed 3’signature of ZUC-dependent piRNA processing in silkworm (Izumi et al., 2020) and to the enrichment of UVV triplets following the 3′end of murine piRNAs (Bornelöv et al., 2021). We observe similar positional effects for Uridines for Piwi-piRNAs, Aub-piRNAs, and Ago3-piRNAs in fly ovaries (Figure S3) and pre-piRNAs in mice (Figure 4D). Our data suggest that PIWI proteins establish a Goldilocks zone for Uridines to impact 3′end processing.
Figure 4.
Length preference is predominant over +1U preference
(A) For Piwi-piRNAs (OSC), the 3′-end is defined by the location of Uridine (U) within a narrow window located at a fixed distance from the 5′-end. The top panel depicts the length distribution of Piwi-piRNAs for orientation. Each row of the heatmap indicates fractions of piRNA 3′ends when the position of a single Uridine (U) is fixed across the interval. The heatmap at the bottom shows fractions of 3′ends in the absence of Uridines (U) across a continuous five nucleotide region (row 1), and upon requirement for a Uridine (U) immediately 5′ or 3′ of the fixed non-U (V) stretch. (V is C, G, or A).
(B) PiRNA 3′-ends are more likely to occur before Uridines. Z-score was calculated over a 51-nt window (central 21nt are depicted), with the Uridine (U) located in the center.
(C) In the presence of a poly-Uridine sequence (UU or UUU), piRNA 3′-ends are preferentially located before the last Uridine. (Z-score calculated across a 51-nt window; the central 21nt are depicted.)
(D) The presence of Uridines within an optimal length interval -the Goldilocks zone-shifts the preferred 3′ end of murine pre-piRNAs. Red circles indicate Uridines (U).
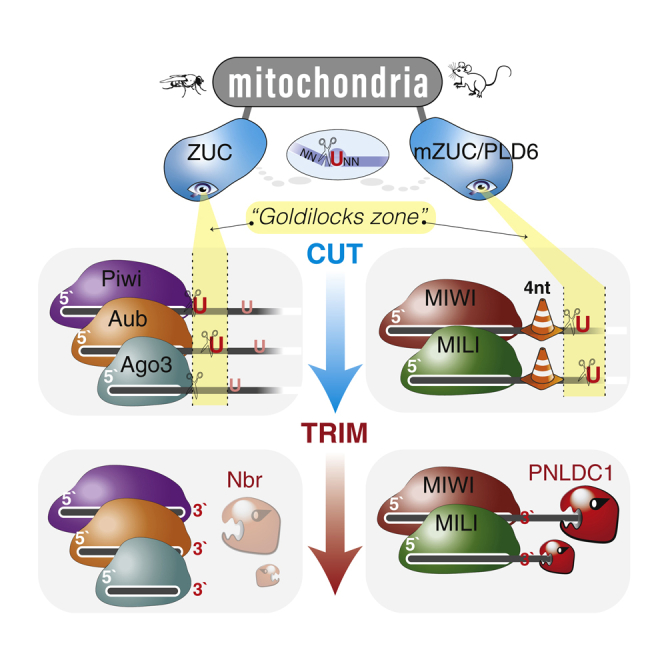
(E) The revised model for piRNA 3′end formation shows a hierarchical integration of length and sequence preferences to generate a single major piRNA species. First PIWI determines a specific interval for access to the ZUC-processor complex on the pre-piRNA. Within this Goldilocks zone, the ZUC-processor preferentially cuts 5′ of a Uridine (U). A mouse-specific roadblock changes the position of the Goldilocks zone and leaves substrate for consecutive trimming by PNLDC1. The final major piRNA likely reflects a footprint of its associated PIWI protein.
Overall, a model emerges that integrates length and sequence preferences to establish a single major 3′end for piRNAs in flies and mice (Figure 4E). Universally, length preferences trump sequence preferences in a hierarchical decision. In flies, the ZUC-processor complex seems to have access to the pre-piRNA immediately downstream of PIWI’s footprint, which does not leave much room for trimming after the initial cut. In contrast, ZUC’s visibility is shifted by a consistent distance on murine pre-piRNAs. After the initial cut, the roadblock seems to be lifted and PNLDC1 faithfully trims four nucleotides to establish the mature 3′end with major sequences that are determined by length preferences.
Discussion
Each PIWI protein establishes a distinct Goldilocks zone for 3′end processing on pre-piRNAs. Identifying whether these specific footprints indeed reflect the geometry of different PIWI proteins requires further structural studies (Anzelon et al., 2021; Matsumoto et al., 2016; Yamaguchi et al., 2020). Length preferences could reflect the most stable conformation of PIWI proteins in complexity with their guide RNAs or optimal (in)stability for effective turnover. Transcriptional silencing might benefit from increased stability of piRISC:target interactions and a longer residence time on chromatin. However, there is no simple relationship between length and silencing mode. In Drosophila, the nuclear PIWI protein—Piwi—associates with the longest piRNAs, whereas in mouse and human the longest piRNAs are found in the cytoplasmic PIWIL1 (MIWI or HIWI) (Aravin et al., 2006; Girard et al., 2006; Klenov et al., 2007; Lau et al., 2006; Vagin et al., 2006; Yin and Lin, 2007). Cytoplasmic slicing of target transcripts by piRISC bears similarity to target cleavage by Argonaute complexes during RNA interference (RNAi), and additional base-pairing interactions are likely to impact turnover kinetics (Becker et al., 2019; Joshua-Tor and Hannon, 2011; Tolia and Joshua-Tor, 2007).
The positioning of the initial 3′ CUT in mice suggests a mouse specific roadblock that could be a component of the ZUC-processor complex. Neither ZUC/PLD6 nor the conserved Tudor protein Papi/TDRKH are significantly different in size in mice compared to flies (Izumi et al., 2016; Liu et al., 2011; Nishida et al., 2018; Saxe et al., 2013). However, two piRNA pathway genes, the RNA binding protein MAELSTROM (MAEL) and the Zinc-finger protein GTSF1, act in piRNA biogenesis in mice, whereas their function is shifted to the nuclear effector phase of piRNA silencing in flies (Aravin et al., 2009; Castaneda et al., 2014; Chen et al., 2015; Ipsaro et al., 2021; Muerdter et al., 2013; Ohtani et al., 2013; Sato and Siomi, 2015; Sienski et al., 2012; Yoshimura et al., 2018). The observed inaccessibility could be a shadow cast by one of these two ominous piRNA pathway genes or another unidentified component of the murine ZUC-processor complex.
PiRNA length determines the sequence space available for target interaction. In contrast to the well-characterized seed-recognition of miRNAs, seed-interactions of piRNAs with a target are weaker and compensated by extensive base-pairing across the piRNA length (Anzelon et al., 2021; Bartel, 2018). Longer piRNAs have the potential for stronger and more specific interactions, and precision of a major 3′end might be required to control specificity. The notion that every piRNA has a single major 3′end contributes to our emerging understanding that the functional piRNA sequence space is regulated to ensure effective genome defense (Genzor et al., 2021).
Limitations of the study
Like most models for piRNA processing and function, our conclusions are based on signatures in piRNA datasets. Future studies are required to identify the molecular players and directly probe our working models. It would be particularly interesting to elucidate the identity of the observed roadblock in 3′ end processing of pachytene piRNAs that generates substrate for 3′ trimming.
STAR★Methods
Key resources table
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Astrid D. Haase (astrid.haase@nih.gov).
Material availability
This work does not report original experimental data.
Method details
Data access and initial processing of sequencing data
A total of 12 datasets were analyzed in this study. OSC data were obtained from [GEO accession number: GSE156058]. FH-Piwi-IP dataset was generated by combining three replicates with the following SRA accession numbers: SRR12430857, SRR12430858, SRR12430859. Drosophila ovary data were obtained from [GSE83698]. SRA accession numbers are as follows: fly-Piwi-IP = SRR3715419, fly-Aub-IP = SRR3715420, fly-Ago3-IP = SRR3715421. Mouse data were obtained from [NCBI BioProject accession number PRJNA421205]. Datasets were initially processed as previously described (Genzor et al., 2021). The packages utilized include: data.table (Matt Dowle, 2021), dplyr (Hadley Wickham et al., 2022), ggpubr (Kassambara, 2020), GenomicRanges (Lawrence et al., 2013), stringr (Wickham, 2019), parallel (Team, 2021a). All computational code is available in ‘Supplemental Code’.
Counting numbers of unique 3′-ends
Uniquely mapping piRNAs (NH = 1) were collapsed by sequence while retaining their abundance information (# of reads per unique sequence). The following sequences were collapsed again by the coordinate of their 5′-end while retaining the total read abundance and the number of unique sequences that share the 5′-end coordinates. These 5′-ends were subsequently grouped into discrete bins based on their total read abundance (bins shown on y axis). Amounts of unique sequences sharing 5′-ends were depicted for each bin using geom_boxplot() function from the ggplot (Wickham, 2016) package for R (Team, 2021a). See (Figures 1B, 1E, 1F, and S1A).
Defining the “major 3′-end” and determining its contribution
Uniquely mapping piRNA sequences (NH = 1) were grouped by their 5′-end coordinates. For each 5′-end, the most abundant sequence was identified as “major end”. Ties for the title of “major end” were solved in a random fashion to avoid biases. The 5′-ends were subsequently grouped into discrete bins based on their total read abundance (bins shown on y axis). For each bin, the contribution of the “major end” was determined by dividing the abundance of the “major end” by the total abundance of all sequences sharing the respective 5′-end. The mean value for each bin was depicted as bar plot using geom_barplot() while the distribution of contribution values within each bin was depicted as a boxplot using geom_boxplot() from the ggplot package (Wickham, 2016). See (Figures 1C, 1D, and 1E).
Ranking 3′-ends
Uniquely mapping piRNA sequences (NH = 1) were once again grouped by their 5′-end coordinates. For each 5′-end, the most abundant sequence (with major 3′-end) was identified as “M”, the second most abundant sequence was identified as “m1” and the third as “m2”. The contribution of these sequences to the abundance of the 5′-end was calculated in the same fashion as previously. The distribution of these contribution values was plotted as a boxplot in the insert of Figure 2A. The size distributions of “M”, “m1”, and “m2” were calculated by counting the number of reads at different sizes and normalizing by the total number of reads in “M”, “m1”, or “m2” bins. See (Figure 2A).
Size distribution of the “major 3′-end”
For each 5′-end, a “major end” was identified in the same manner as described previously. Size distribution of all “major ends” was determined by counting the number of “major end” reads at different lengths and normalizing by the total number of “major end” reads. Same analysis was performed on “minor ends” whereby the “minor ends” are any sequences that were not identified as “major ends”. The size distribution of the entire library was depicted as bar plot using the ggplot package for R (Wickham, 2016). See (Figures 2B–2D, and S2A).
Distance between major 3′-ends in trimmed vs mature piRNAs
The major 3′-end was defined for uniquely mapping piRNA sequences (NH = 1) in WT and PNLDC KO samples. Only major 3′-end containing sequences whose 5′-end was present in both datasets were kept. The resulting sequences in the WT sample were subsequently paired with the corresponding sequence (of the same 5′-end) in PNLDC-KO sample. Only pairs where the PNLDC-KO sequence was longer than the WT sequence were kept. The nucleotide length difference between these two sequences was determined and depicted as a bar plot using the ggplot package (Wickham, 2016). See (Figure S2B).
Determining the composition of the +1 nucleotide
Uniquely mapping piRNA sequences (NH = 1) were categorized into “major ends” and “minor ends” in a manner described previously. The identity of the nucleotide immediately after the 3′-end of piRNAs (referred to as the +1 nucleotide) was obtained from the corresponding genome (dm6 (Team, 2014) for fly and mm10 (Team, 2021b) for mouse). The composition of this nucleotide was depicted as a stacked bar plot using the ggplot package. Sequences logos were generated using the ggseqlogo (Wagih, 2017) package for R. See (Figure 3).
Sequence context specific 3′-end distribution
Uniquely mapping piRNA sequences (NH = 1) were grouped into non-mutually exclusive bins based on the sequence context downstream of the piRNA 5′-end. The distribution of 3′-ends for each sequence context was determined by counting the number of reads at different sizes and normalizing by the total amount of reads within the sequence context. This distribution was plotted as a heatmap using the geom_tile() function within the ggplot package (Wickham, 2016). See (Figures 4A and 4D).
Z score for 3′-ends
Genomic sequence immediately downstream and upstream of all piRNA (uniquely mapping only) 3′-ends was isolated. A sequence context of interest (ex: “UUU”) was searched for within a 51nt window surrounding the 3′-end. If found, information regarding the position of this sequence context in respect to the 3′-end was recorded. Data from all successfully located sequence contexts were collated to determine total numbers of piRNA 3′-ends at given distances from the sequence context of interest. The total number of piRNA 3′-ends for each position was converted to a Z score by determining the number of standard deviations from the mean (within the full 51nt window). See (Figures 4B and 4C).
Quantification and statistical anlaysis
All the calculations and statistical analyses were carried out using previously published R packages listed in the Key resources table and described in Method details.
Additional resources
URL for github: https://github.com/HaaseLab/piRNA_ThreePrimeEnds.
Acknowledgments
We thank M Hafner for critical comments on the manuscript, all members of the Haase and Hafner labs for helpful discussions, and E He (NIH Medical Arts) for help with the model. This work would not have been possible without the support of the NIH high-performance computing group and funding by the intramural research program of the NIDDK (DK075111).
Author contributions
D.S. designed and performed all experiments with help from P.G. A.D.H. and D.S. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Published: June 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104427.
Supplemental information
Data and code availability
All sequencing data are publicly available: GSE156058 (Genzor et al., 2021), GSE83698 (Hayashi et al., 2016), PRJNA421205 (Gainetdinov et al., 2018). Original code is available in ‘Supplemental Code’ and on github: https://github.com/HaaseLab/piRNA_ThreePrimeEnds. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Anastasakis D., Skeparnias I., Shaukat A.N., Grafanaki K., Kanellou A., Taraviras S., Papachristou D.J., Papakyriakou A., Stathopoulos C. Mammalian PNLDC1 is a novel poly(A) specific exonuclease with discrete expression during early development. Nucleic Acids Res. 2016;44:8908–8920. doi: 10.1093/nar/gkw709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzelon T.A., Chowdhury S., Hughes S.M., Xiao Y., Lander G.C., MacRae I.J. Structural basis for piRNA targeting. Nature. 2021;597:285–289. doi: 10.1038/s41586-021-03856-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., van der Heijden G.W., Castaneda J., Vagin V.V., Hannon G.J., Bortvin A. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet. 2009;5:e1000764. doi: 10.1371/journal.pgen.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. Metazoan MicroRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker W.R., Ober-Reynolds B., Jouravleva K., Jolly S.M., Zamore P.D., Greenleaf W.J. High-throughput analysis reveals rules for target RNA binding and cleavage by AGO2. Mol. Cell. 2019;75:741–755.e11. doi: 10.1016/j.molcel.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornelöv S., Czech B., Hannon G.J. An evolutionarily conserved stop codon enrichment at the 5′ ends of mammalian piRNAs. bioRxiv. 2021 doi: 10.1101/2021.10.27.464999. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaneda J., Genzor P., van der Heijden G.W., Sarkeshik A., Yates J.R., 3rd, Ingolia N.T., Bortvin A. Reduced pachytene pi RNA s and translation underlie spermiogenic arrest in M aelstrom mutant mice. EMBO J. 2014;33:1999–2019. doi: 10.15252/embj.201386855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.M., Campbell E., Pandey R.R., Yang Z., McCarthy A.A., Pillai R.S. Metazoan Maelstrom is an RNA-binding protein that has evolved from an ancient nuclease active in protists. RNA. 2015;21:833–839. doi: 10.1261/rna.049437.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B., Munafo M., Ciabrelli F., Eastwood E.L., Fabry M.H., Kneuss E., Hannon G.J. piRNA-guided genome defense: from biogenesis to silencing. Annu. Rev. Genet. 2018;52:131–157. doi: 10.1146/annurev-genet-120417-031441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D., Liu J., Dong K., Midic U., Hess R.A., Xie H., Demireva E.Y., Chen C. PNLDC1 is essential for piRNA 3′ end trimming and transposon silencing during spermatogenesis in mice. Nat. Commun. 2017;8:819. doi: 10.1038/s41467-017-00854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltzin V.L., Khaladkar M., Abe M., Parisi M., Hendriks G.J., Kim J., Bonini N.M. The exonuclease Nibbler regulates age-associated traits and modulates piRNA length in Drosophila. Aging Cell. 2015;14:443–452. doi: 10.1111/acel.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F., Sonenberg N., Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- Gainetdinov I., Colpan C., Arif A., Cecchini K., Zamore P.D. A single mechanism of biogenesis, initiated and directed by PIWI proteins, explains piRNA production in most animals. Mol. Cell. 2018;71:775–790.e5. doi: 10.1016/j.molcel.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genzor P., Konstantinidou P., Stoyko D., Manzourolajdad A., Marlin Andrews C., Elchert A.R., Stathopoulos C., Haase A.D. Cellular abundance shapes function in piRNA-guided genome defense. Genome Res. 2021;31:2058–2068. doi: 10.1101/gr.275478.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M., Zamore P.D. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A., Sachidanandam R., Hannon G.J., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Guzzardo P.M., Muerdter F., Hannon G.J. The piRNA pathway in flies: highlights and future directions. Curr. Opin. Genet. Dev. 2013;23:44–52. doi: 10.1016/j.gde.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadley Wickham R.F., Henry L., Müller K. 2022. Dplyr: A Grammar of Data Manipulation. [Google Scholar]
- Han B.W., Wang W., Li C., Weng Z., Zamore P.D. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science. 2015;348:817–821. doi: 10.1126/science.aaa1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi R., Schnabl J., Handler D., Mohn F., Ameres S.L., Brennecke J. Genetic and mechanistic diversity of piRNA 3′-end formation. Nature. 2016;539:588–592. doi: 10.1038/nature20162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsaro J.J., Haase A.D., Knott S.R., Joshua-Tor L., Hannon G.J. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature. 2012;491:279–283. doi: 10.1038/nature11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipsaro J.J., O'Brien P.A., Bhattacharya S., Palmer A.G., 3rd, Joshua-Tor L. Asterix/Gtsf1 links tRNAs and piRNA silencing of retrotransposons. Cell Rep. 2021;34:108914. doi: 10.1016/j.celrep.2021.108914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki Y.W., Siomi M.C., Siomi H. PIWI-interacting RNA: its biogenesis and functions. Annu. Rev. Biochem. 2015;84:405–433. doi: 10.1146/annurev-biochem-060614-034258. [DOI] [PubMed] [Google Scholar]
- Izumi N., Shoji K., Sakaguchi Y., Honda S., Kirino Y., Suzuki T., Katsuma S., Tomari Y. Identification and functional analysis of the pre-piRNA 3′ trimmer in silkworms. Cell. 2016;164:962–973. doi: 10.1016/j.cell.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi N., Shoji K., Suzuki Y., Katsuma S., Tomari Y. Zucchini consensus motifs determine the mechanism of pre-piRNA production. Nature. 2020;578:311–316. doi: 10.1038/s41586-020-1966-9. [DOI] [PubMed] [Google Scholar]
- Joshua-Tor L., Hannon G.J. Ancestral roles of small RNAs: an Ago-centric perspective. Cold Spring Harb. Perspect. Biol. 2011;3:a003772. doi: 10.1101/cshperspect.a003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A. 2020. Ggpubr: ‘ggplot2’ Based Publication Ready Plots. [Google Scholar]
- Klenov M.S., Lavrov S.A., Stolyarenko A.D., Ryazansky S.S., Aravin A.A., Tuschl T., Gvozdev V.A. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A., Toth K.F., Aravin A.A. To be or not to be a piRNA: genomic origin and processing of piRNAs. Genome Biol. 2014;15:204. doi: 10.1186/gb4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. piRNAs in the germ line. Science. 2007;316:397. doi: 10.1126/science.1137543. [DOI] [PubMed] [Google Scholar]
- Liu L., Qi H., Wang J., Lin H. PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development. 2011;138:1863–1873. doi: 10.1242/dev.059287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto N., Nishimasu H., Sakakibara K., Nishida K.M., Hirano T., Ishitani R., Siomi H., Siomi M.C., Nureki O. Crystal structure of silkworm PIWI-clade argonaute siwi bound to piRNA. Cell. 2016;167:484–497.e9. doi: 10.1016/j.cell.2016.09.002. [DOI] [PubMed] [Google Scholar]
- Matt Dowle A.S. 2021. data.table: Extension of ‘data.Frame’. [Google Scholar]
- Mohn F., Handler D., Brennecke J. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science. 2015;348:812–817. doi: 10.1126/science.aaa1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F., Guzzardo P.M., Gillis J., Luo Y., Yu Y., Chen C., Fekete R., Hannon G.J. A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila. Mol. Cell. 2013;50:736–748. doi: 10.1016/j.molcel.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K.M., Sakakibara K., Iwasaki Y.W., Yamada H., Murakami R., Murota Y., Kawamura T., Kodama T., Siomi H., Siomi M.C. Hierarchical roles of mitochondrial Papi and Zucchini in Bombyx germline piRNA biogenesis. Nature. 2018;555:260–264. doi: 10.1038/nature25788. [DOI] [PubMed] [Google Scholar]
- Nishimasu H., Ishizu H., Saito K., Fukuhara S., Kamatani M.K., Bonnefond L., Matsumoto N., Nishizawa T., Nakanaga K., Aoki J., et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature. 2012;491:284–287. doi: 10.1038/nature11509. [DOI] [PubMed] [Google Scholar]
- Nishimura T., Nagamori I., Nakatani T., Izumi N., Tomari Y., Kuramochi-Miyagawa S., Nakano T. PNLDC1, mouse pre-piRNA Trimmer, is required for meiotic and post-meiotic male germ cell development. EMBO Rep. 2018;19:e44957. doi: 10.15252/embr.201744957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani H., Iwasaki Y.W., Shibuya A., Siomi H., Siomi M.C., Saito K. DmGTSF1 is necessary for Piwi-piRISC-mediated transcriptional transposon silencing in the Drosophila ovary. Genes Dev. 2013;27:1656–1661. doi: 10.1101/gad.221515.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozata D.M., Gainetdinov I., Zoch A., O'Carroll D., Zamore P.D. PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet. 2019;20:89–108. doi: 10.1038/s41576-018-0073-3. [DOI] [PubMed] [Google Scholar]
- Rivas F.V., Tolia N.H., Song J.J., Aragon J.P., Liu J., Hannon G.J., Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- Sato K., Siomi M.C. Functional and structural insights into the piRNA factor Maelstrom. FEBS Lett. 2015;589:1688–1693. doi: 10.1016/j.febslet.2015.03.023. [DOI] [PubMed] [Google Scholar]
- Saxe J.P., Chen M., Zhao H., Lin H. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 2013;32:1869–1885. doi: 10.1038/emboj.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu-Gruttadauria J., MacRae I.J. Structural foundations of RNA silencing by argonaute. J. Mol. Biol. 2017;429:2619–2639. doi: 10.1016/j.jmb.2017.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G., Donertas D., Brennecke J. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi M.C., Sato K., Pezic D., Aravin A.A. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Stein C.B., Genzor P., Mitra S., Elchert A.R., Ipsaro J.J., Benner L., Sobti S., Su Y., Hammell M., Joshua-Tor L., Haase A.D. Decoding the 5′ nucleotide bias of PIWI-interacting RNAs. Nat. Commun. 2019;10:828. doi: 10.1038/s41467-019-08803-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuwe E., Toth K.F., Aravin A.A. Small but sturdy: small RNAs in cellular memory and epigenetics. Genes Dev. 2014;28:423–431. doi: 10.1101/gad.236414.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team T.B.D. 2014. BSgenome.Dmelanogaster.UCSC.dm6: Full Genome Sequences for Drosophila Melanogaster (UCSC version dm6) [Google Scholar]
- Team R.C. 2021. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Team T.B.D. 2021. BSgenome.Mmusculus.UCSC.mm10: Full Genome Sequences for Mus musculus (UCSC Version Mm10, Based on GRCm38.p6) [Google Scholar]
- Tolia N.H., Joshua-Tor L. Slicer and the argonautes. Nat. Chem. Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- Vagin V.V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Wagih O. 2017. Ggseqlogo: A‘ggplot2’ Extension for Drawing Publication-Ready Sequence Logos. [Google Scholar]
- Wang Y., Sheng G., Juranek S., Tuschl T., Patel D.J. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Springer-Verlag; 2016. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Wickham H. 2019. Stringr: Simple, Consistent Wrappers for Common String Operations. [Google Scholar]
- Yamaguchi S., Oe A., Nishida K.M., Yamashita K., Kajiya A., Hirano S., Matsumoto N., Dohmae N., Ishitani R., Saito K., et al. Crystal structure of Drosophila Piwi. Nat. Commun. 2020;11:858. doi: 10.1038/s41467-020-14687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- Yoshimura T., Watanabe T., Kuramochi-Miyagawa S., Takemoto N., Shiromoto Y., Kudo A., Kanai-Azuma M., Tashiro F., Miyazaki S., Katanaya A., et al. Mouse GTSF 1 is an essential factor for secondary pi RNA biogenesis. EMBO Rep. 2018;19:e42054. doi: 10.15252/embr.201642054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Guo R., Cui Y., Zhu Z., Zhang Y., Wu H., Zheng B., Yue Q., Bai S., Zeng W., et al. An essential role for PNLDC1 in piRNA 3′ end trimming and male fertility in mice. Cell Res. 2017;27:1392–1396. doi: 10.1038/cr.2017.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data are publicly available: GSE156058 (Genzor et al., 2021), GSE83698 (Hayashi et al., 2016), PRJNA421205 (Gainetdinov et al., 2018). Original code is available in ‘Supplemental Code’ and on github: https://github.com/HaaseLab/piRNA_ThreePrimeEnds. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.