Figure 1.
PiRNAs have a single major 3′-end
(A) The cut-n’-trim model for piRNA 3′end formation proposes an initial endonucleolytic cut that generates a substrate for 3′-to-5′ exonucleolytic trimming. The resulting length distributions of piRNAs are suggested to resemble a footprint of their bound PIWI protein partner.
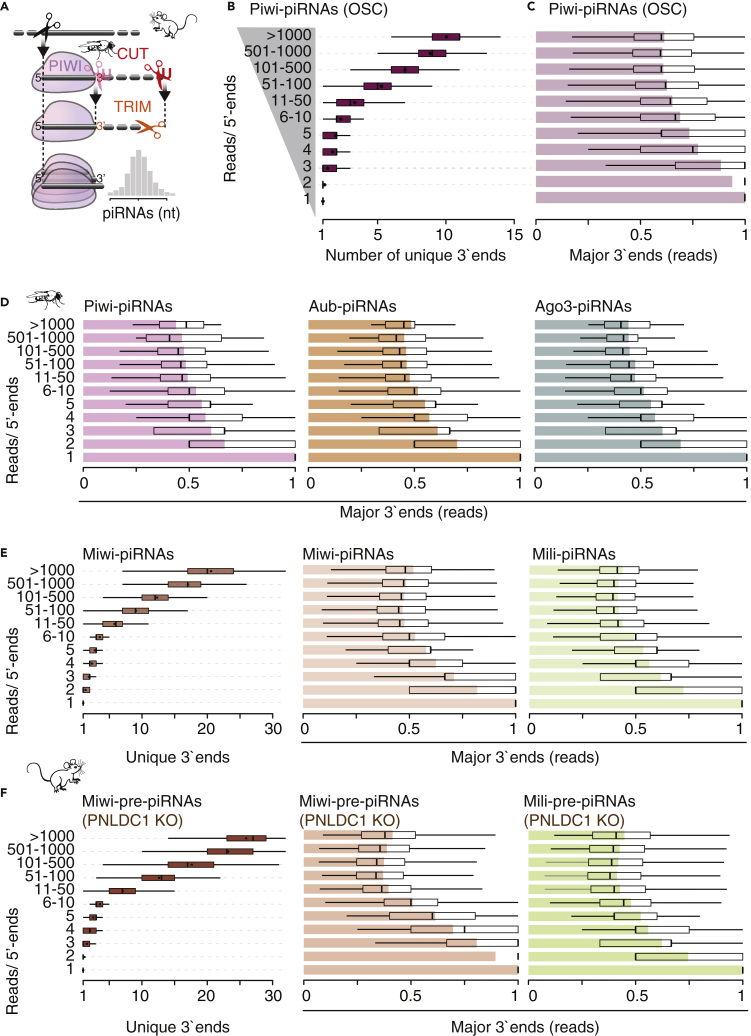
(B) Piwi-piRNAs that share the same 5′-end can have a number of different 3′-ends. Uniquely mapping Piwi-piRNAs from OSC (18–32 nt long) were grouped by the number of reads with a shared 5′-end. The number of different 3′-ends per 5′-end is depicted for each group.
(C and D) Most of the reads associated with the same 5′-end have a single preferred 3′-end. For each 5′-end, the most abundant 3′-end was identified (referred to as “major”), and its contribution to the abundance of the 5′-end was calculated. The mean contribution is shown as a bar plot, whereas quartiles are depicted as a boxplot. Only uniquely mapping piRNAs that are 18–32 nt long were used.
(E and F) In mice, both mature and untrimmed piRNAs can have many possible 3′-ends; however, only one contributes to approximately half of the abundance. Uniquely mapping reads were used for analyses (19–50nt). Datasets are publicly available (GSE156058; GSE83698; PRJNA421205).