Abstract
The relationship between structural and functional connectivity in the brain is a key question in systems neuroscience. Modern accounts assume a single global structure-function relationship that persists over time. Here we study structure-function coupling from a dynamic perspective, and show that it is regionally heterogeneous. We use a temporal unwrapping procedure to identify moment-to-moment co-fluctuations in neural activity, and reconstruct time-resolved structure-function coupling patterns. We find that patterns of dynamic structure-function coupling are region-specific. We observe stable coupling in unimodal and transmodal cortex, and dynamic coupling in intermediate regions, particularly in insular cortex (salience network) and frontal eye fields (dorsal attention network). Finally, we show that the variability of a region’s structure-function coupling is related to the distribution of its connection lengths. Collectively, our findings provide a way to study structure-function relationships from a dynamic perspective.
Subject terms: Network models, Dynamical systems
Temporal unwrapping analysis of diffusion weighted MRI connectivity and functional MRI scans reveals that the coupling between structure and function in the human brain is regionally heterogeneous and provides a framework to evaluate these relationships from a dynamic perspective.
Introduction
The brain is a network of anatomically connected neuronal populations. Inter-regional signaling via electrical impulses manifests as patterns of organized co-activations, termed “functional connectivity”. The coupling between structural connectivity (SC) and functional connectivity (FC) is a fundamental feature that reflects the integrity of neural signaling1. Historically, most studies have focused on static structure-function coupling over the course of a whole scanning session2.
However, over the past decade functional connectivity is increasingly conceptualized as a dynamic process3–5. Functional connectivity patterns display time-resolved fluctuations that are non-random6–10, highly organized11–14, individual-specific15, related to behavior16,17, and evolve over the lifespan18. As a result, structure-function coupling should fluctuate over multiple timescales. Indeed, multiple studies have reported evidence of dynamic structure-function relationships over the course of single recording sessions19,20, and over more protracted periods, including early childhood and young adult neurodevelopment21,22.
Importantly, previous studies on dynamic structure-function coupling worked under the assumption that structure-function relationships are uniform across the brain. Recent research suggests that structure-function coupling is regionally heterogeneous, such that structural and functional connectivity profiles are closely related in sensory (unimodal) cortex, but gradually decouple in transmodal cortex21,23–25. The systematic decoupling of structure and function along this unimodal-transmodal gradient is thought to reflect differentiation in micro-architectural properties2,26,27, including molecular, cellular, and laminar differentiation21,23,28,29. Indeed, computational models that implement regionally heterogeneous dynamics using micro-architectural properties make more accurate predictions of functional connectivity from structural connectivity30–33.
How do regional patterns of structure-function coupling fluctuate moment-to-moment? We considered two alternative possibilities. One possibility is that structure-function coupling is greater in transmodal cortex. Several recent studies have shown that static structure-function coupling is lower in transmodal cortex compared to unimodal cortex2,21,23,34. Given that transmodal cortex engages in multiple polysensory functions and functional relationships, a plausible explanation could be that greater variability in time-dependent structure-function coupling ultimately averages out and appears as lower static structure-function coupling. Another possibility is that structure-function coupling is greatest in regions that are intermediate in the putative unimodal-transmodal hierarchy. Numerous evidence points to diverse cytoarchitecture and connectional fingerprints in insular cortex35,36. By participating in a diverse set of connections with multiple brain regions, the insula is thought to dynamically engage in multiple cognitive systems37–39.
Here we derive time- and region-resolved patterns of structure-function coupling. We first estimate dynamic inter-regional co-fluctuation using a recently-developed temporal unwrapping method that does not require windowing9,14. We then reconstruct dynamic patterns of regional structure-function coupling and contextualize these patterns with respect to macroscale brain organization, including intrinsic networks, as well as functional and cellular hierarchies.
Results
The results are organized as follows. We first reconstruct frame-by-frame co-fluctuation matrices from regional BOLD time-series9,14. We then apply a multilinear model to estimate regional time-series of structure-function coupling23, before comparing regional fluctuations in structure-function coupling with large-scale intrinsic networks40, cortical hierarchies41, and cytoarchitectonic classes42. We also benchmark the extent to which dynamic fluctuations in structure-function coupling can be explained by topological and geometric embedding. Finally, we assess the correspondence between conventional (static) structure-function coupling and dynamic structure-function coupling. Data were derived from N = 327 healthy, unrelated participants from the Human Connectome Project (HCP43). Structural connectomes were reconstructed from diffusion MRI (dMRI). Static and dynamic functional connectivity were estimated from resting-state functional MRI (fMRI) (see Materials and Methods for detailed procedures). Analyses were performed using a network parcellation of 400 cortical nodes44.
Time-resolved structure-function coupling
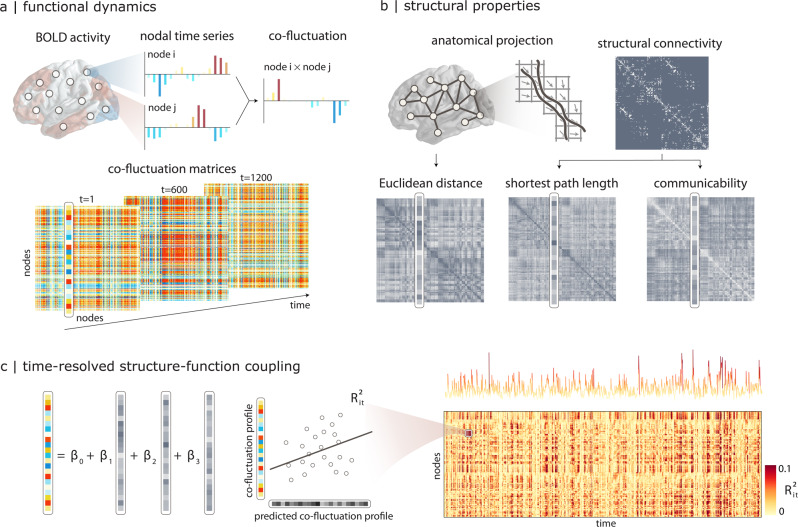
The temporal unwrapping procedure generates a node-by-node co-fluctuation matrix for each time point (Fig. 1a). We then use a multilinear regression model to predict the co-fluctuation profile of every node from its structural connectivity profile23,45. The model was fitted separately for each time point (Fig. 1c). The regression model incorporates multiple computational models of cortical communication1,46: (1) Euclidean distance, (2) shortest path length, and (3) communicability (Fig. 1b). Euclidean distance embodies the notion that proximal neurons may exchange information more easily, and is consistent with navigation-like communication47. Shortest path length is a statistic that embodies centralized routing-like communication48, while communicability is a statistic that embodies decentralized diffusion-like communication49,50. Note, however, that there exist multiple alternative statistics that measure the capacity of the network to transmit information, and the three chosen measures constitute a subset of that wider space51. All models were fitted independently for each individual participant.
Fig. 1. Time-resolved structure-function coupling.
a The co-fluctuation of two brain regions i and j is calculated as the element-wise multiplication of the two z-scored fMRI BOLD activity time-series. The points of this time-series can be represented as one element in a co-fluctuation matrix. b Pairwise structural relationships are derived from structural connectivity networks reconstructed from diffusion MRI, including Euclidean distance between node centroids, shortest path length and communicability. c A multilinear regression model is used to predict a region’s co-fluctuation profile from its structural profile, using Euclidean distance, path length and communicability as predictors. The resulting coefficient of determination () indicates how well the structural connectivity profile predicts the functional connectivity profile for a particular brain region i at a particular time point t. The procedure generates a region × time matrix that captures the fluctuation of structure-function coupling for individual regions across time. The time-series shows time-resolved fluctuations in mean R2.
The multilinear model allows us to quantify regional structure-function coupling across time. For each brain region i and time point t, we measure the goodness of fit using the coefficient of determination between the predicted and the empirical functional profile (Fig. 1c). A value near 1 indicates strong coupling between the structural and functional profiles for the ith node at time t. These coefficients of determination are then assembled into a node × time structure-function coupling matrix. The procedure was carried out separately for each individual in the sample.
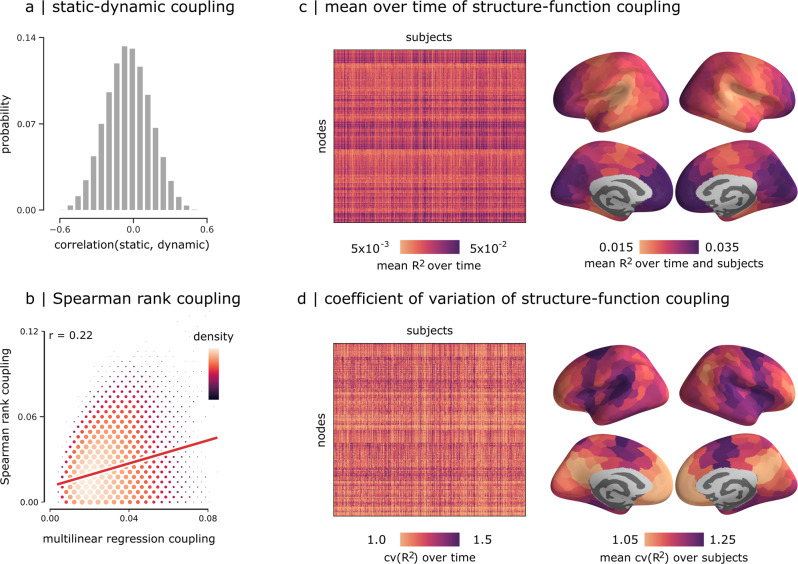
Figure 2a shows the Pearson correlations between dynamic structure-function coupling maps and the static structure-function coupling map reconstructed using the whole time-series. The coefficients span a wide distribution, encompassing both positive and negative values. A distribution of coefficients is mathematically expected given that the method is measuring the relationship between dynamic functional connectivity and static structural connectivity. In the present report, we further analyze how dynamic functional connectivity around a static structural connectivity reference yields fluctuations in structure-function correlations, and we map these fluctuations to the cortical hierarchy. Figure 2b shows the relationship between two alternative methods for estimating regional structure-function coupling. The abscissa shows structure-function coupling values estimated using the multilinear model described above, while the ordinate shows the same values estimated using the method described by Baum and colleagues21. The latter, which we term “Spearman rank coupling”, estimates structure-function coupling as the Spearman rank correlation between the structural and functional profiles of each node. The principal strength of the method is that it does not make arbitrary assumptions about which predictors to include; the principal weakness is that the correlation can only be computed between pairs of regions that have an underlying structural connection, potentially missing out on biologically-important dyadic relationships. Importantly, the two methods are positively correlated (r = 0.22), suggesting that, while the two methods offer qualitatively similar perspectives on structure-function coupling, they are not perfectly correlated, potentially because one is sensitive to direct monosynaptic relationships while the other also takes into account polysynaptic relationships.
Fig. 2. Dynamic structure-function coupling.
a Correlations between regional patterns of static and dynamic structure-function coupling. b Correlations between dynamic structure-function coupling estimated using a multilinear model23 versus coupling estimated using an alternative Spearman rank correlation method21. Scatter color and size represent the density. c Mean time-resolved structure-function coupling over time (left) and its mean over subjects (right). d Coefficient of variation of structure-function coupling across time (left), and its mean over subjects (right).
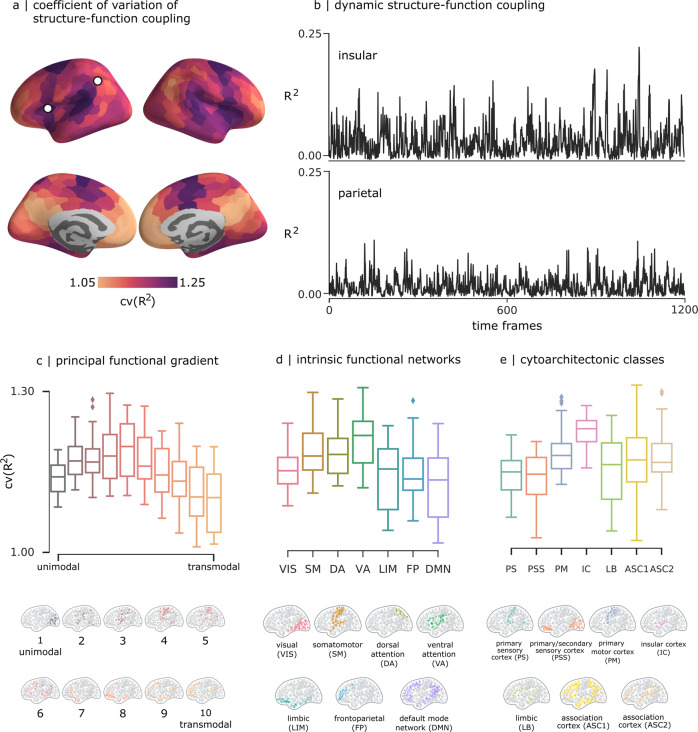
Figure 2c shows the mean structure-function coupling R2, while Fig. S1 shows the contribution of individual predictors. The relationship between R2 and co-fluctuation amplitude is shown in Fig. 3. To quantify the variability of structure-function coupling across time, we compute—separately for each participant—the coefficient of variation of R2 across time (cv(R2)). The coefficient of variation is the ratio of the standard deviation of R2 to the mean of R2. It is a standardized measure of dispersion of R2 values about the mean that captures the variability in structure-function coupling across time. In other words, cv(R2) allows us to compare the variability of structure-function coupling time-series that have different means. Figure 2d shows that cv(R2) is regionally heterogeneous and appears to be greatest in insular cortex, frontal eye fields, medial prefrontal, and medial occipital cortex. In the following section, we analyze this pattern in greater detail.
Fig. 3. Relationship with cortical hierarchies.
a Coefficient of variation of structure-function coupling, averaged over all participants. b Time-series of regional structure-function coupling shown for one region in parietal cortex (left) and one region in insular cortex (right) from one randomly selected participant. The mean coefficient of variation is displayed for three types of cortical annotations: c 10 equally-sized bins of the principal functional gradient41, d intrinsic functional networks40, and e von Economo cytoarchitectonic classes42.
Hierarchical organization of dynamic structure-function coupling
We next consider how patterns of dynamic structure-function coupling reflect different features of cortical organization. Specifically, we focus on three widely studied cortical annotations, including the unimodal-transmodal principal functional gradient41, intrinsic functional networks44, and cytoarchitectonic classes42. In each case, we compute the mean coefficient of variation of structure-function coupling. Figure 3b shows exemplar time-series of structure-function coupling for nodes in insular and parietal cortex, exhibiting distinct variability patterns. Figure 3c–e shows that brain regions that occupy intermediate positions in the cortical hierarchy tend to display the most dynamic fluctuations in structure-function coupling. Specifically, we find the most variable fluctuations in the middle of the unimodal-transmodal hierarchy (classes 4-6), corresponding to the ventral attention/salience network and the insular cortex in the Yeo and Von Economo atlases, respectively, as well as the frontal eye fields, corresponding to the dorsal attention network. These observations are confirmed using spatial autocorrelation-preserving null models to test the null hypothesis that cv(R2) is uniform across the brain. The tests reveal significantly greater cv(R2) in intermediate positions of the unimodal-transmodal hierarchy (Fig. 2). Namely, the insular cortex and frontal eye fields, intermediate in the unimodal-transmodal hierarchy, have the most variable structure-function coupling, while unimodal and transmodal cortex have more stable structure-function coupling.
Relating static and dynamic structure-function coupling
In the previous section, we considered how structure-function coupling fluctuates around the mean. We next ask: how closely do dynamic patterns of structure-function coupling reflect static structure-function coupling? To address this question, we systematically compare the dynamic and static case. Taking into account all time points in the dynamic case, we compute (a) the proportion of time points for which dynamic coupling is greater than static coupling (“dynamic > static”), (b) how similar the dynamic patterns are to the static pattern (“bias”) and, (c) how tightly scattered the dynamic patterns are relative to the static pattern (“variance”) (Fig. 4a).
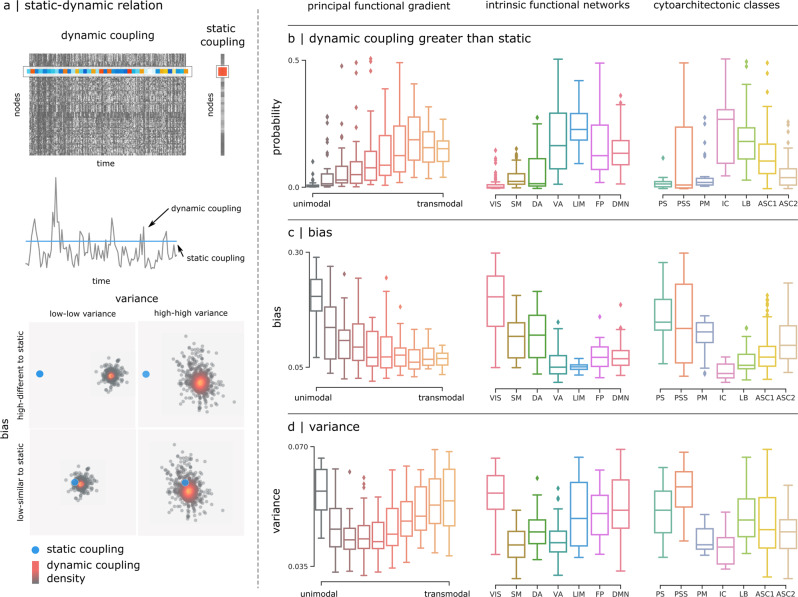
Fig. 4. Relating static and dynamic structure-function coupling.
a Top: static structure-function coupling is estimated using the functional connectivity matrix derived from the whole resting-state time-series23, and compared with dynamic coupling. The dynamic structure-function coupling of node i corresponds to the ith row of the dynamic coupling matrix, while the static coupling corresponds to the ith element of static coupling vector. Middle: dynamic values represented as a time-series (black line) that fluctuates around the single static coupling value (blue line). Bottom: dynamic coupling values are represented as a scattered distribution of points (black) around the static coupling value (blue point). The two are compared in different cortical annotations using three summary statistics: b the probability of having a larger dynamic coupling value compared to the static coupling, c the bias, and d the variance of the dynamic coupling to reproduce the static values.
We find that regions intermediate in the unimodal-transmodal hierarchy, corresponding to the insular cortex, tend to have relatively greater dynamic than static coupling compared with other groups in the hierarchy (up to 0.5, Fig. 4b). These regions also have the closest correspondence between dynamic and static coupling (Fig. 4c) and the lowest dynamical variance around the static case (Fig. 4d). Altogether, these results suggest that the relationship between dynamic and static coupling is not uniform across the brain, but strongly depends on the region’s position in the putative unimodal-transmodal hierarchy, with the closest correspondence between static and dynamic coupling observed in the middle of the hierarchy. Taken together with the results from the previous section, we reveal an interesting property about areas that are intermediate in the hierarchy, such as insular cortex and frontal eye fields. Namely, intermediate areas display the greatest overall fluctuations relative to the mean, but over time tend to follow and converge with static coupling.
Spatial and topological determinants of dynamic structure-function coupling
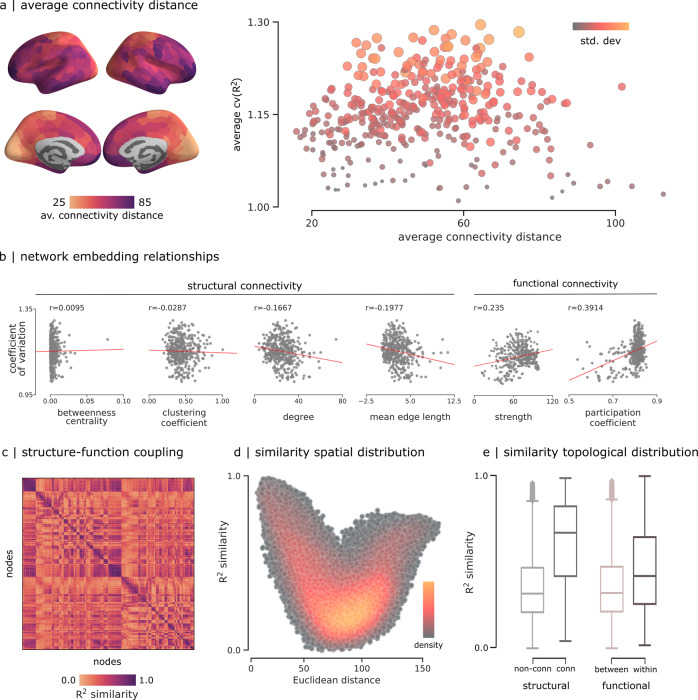
We finally seek to understand how dynamic local structure-function coupling depends on geometric, anatomical and functional embedding. Given that the unimodal-transmodal hierarchy possibly reflects a continuous gradient of connection lengths52–54, we ask whether dynamic structure-function coupling also reflects the distribution of connection lengths that a region participates in. Figure 5a shows the map of mean connectivity distance for each region53,54. We find that areas with very short and very long connection lengths tend to have more stable coupling, and areas with intermediate connection lengths tend to have more variable coupling. Figure 5b shows correlations between dynamic structure-function coupling and multiple measures of structural and functional network embedding, including betweenness, clustering and degree. Robust correlation analysis (biweight midcorrelation and percentage bend correlation55) suggests significant and stable correlations with structural degree (−0.1667; −0.1722; −0.1605), mean edge length (−0.1977; −0.1987; −0.1936), and functional strength (0.235; 0.1956; 0.1803). Altogether, these results suggest that the dynamic nature of structure-function coupling in “middle hierarchy” regions potentially originates from their connection length distribution.
Fig. 5. Spatial and topological determinants of structure-function coupling variability.
a Average connectivity distance calculated following53, and correlated with the average coefficient of variation of the structure-function coupling from Fig. 3a. Scatter color and size represent the standard deviation. b Coefficient of variation of structure-function coupling compared to network embedding metrics derived from structural and functional networks. To account for the possible effects of outliers, we also estimated these relationships using the biweight midcorrelation (r = 0.0031; −0.0349; −0.1722; −0.1987; 0.1956; 0.1647) and percentage bend correlation (r = − 0.0152; − 0.0359; − 0.1605; − 0.1936; 0.1803; 0.2980)55. c R2 similarity between pairs of nodes calculated as the Pearson correlation between pairs of regional structure-function coupling averaged across subjects (d) R2 similarity correlated with Euclidean distance. Colormap shows the density of the scatter plot. e R2 similarity values grouped by structural connectedness and functional intrinsic networks.
Interestingly, when we compute the group-average similarity of inter-regional structure-function time-courses (i.e., how similar are inter-regional fluctuations in structure-function coupling), we find a comparable relationship with Euclidean distance (Fig. 5d). Namely, regions that are physically close together and far apart tend to display similar fluctuations in structure-function coupling, and regions that are at intermediate distances from one another tend to display dissimilar fluctuations in coupling. Finally, we compare the similarity of structure-function coupling between regions with the structural and functional connectivity between those regions. We find that the mean similarity of structure-function coupling is greater for areas that are structurally connected than areas that are not (t(79798) = 80.95, p < 0.001) (Fig. 5e). Likewise, mean similarity of structure-function coupling is greater for areas that participate in the same intrinsic networks than those that are in different networks (t(79798) = 45.34, p < 0.001) (Fig. 5e). In other words, coordinated patterns of dynamic structure-function coupling are—as expected—driven by inter-regional structural and functional connectivity.
Discussion
Emerging theories emphasize dynamic functional interactions that unfold over structural brain networks3. Here, we study time- and region-resolved patterns of structure-function coupling. We find that dynamic coupling patterns reflect cortical hierarchies, with the most dynamic fluctuations in the insula and frontal eye fields. These graded patterns of dynamic coupling reflect the topological and geometric embedding of brain regions.
Our results build on recent work showing that structure-function coupling is not uniform across the brain, but highly region-specific21,23,24,32. These studies have consistently demonstrated that structure-function coupling is graded, with strong coupling in unimodal cortex and weak coupling in transmodal cortex. By applying a temporal unwrapping method to estimate functional co-fluctuation patterns from moment-to-moment, we show that structure-function coupling is not only regionally heterogeneous, but also highly dynamic19. Namely, we find that the extremes of the putative unimodal-transmodal hierarchy display more stable structure-function coupling, while regions intermediate in the hierarchy display more sizable fluctuations.
Interestingly, the most dynamic fluctuations were observed in insular cortex and frontal eye fields. In concert with the anterior cingulate and dorsolateral prefrontal cortices, the insula forms the ventral attention or salience network, which supports the orienting of attention to behaviorally-relevant stimuli, including sensory and autonomic signals related to the internal milieu35,36. By participating in a diverse set of interdigitated connections with multiple brain regions, the insula is thought to dynamically coordinate communication among multiple cognitive systems37–39. In particular, the posterior portion of the insula displays prominent functional connectivity with sensory regions, while the anterior portion is primarily connected with frontal areas involved in higher cognitive function39,56. In a similar vein, the frontal eye fields constitute a key node in the dorsal attention network, involved in biasing attention towards top-down goals and information foraging57,58.
Aligning these two findings, we observe a common functional theme of regions on the interface between higher-order heteromodal cognition and primary perceptual and internal states. We speculate that the greater variability in local structure-function coupling in the insula and frontal eye fields delineates a potential mechanism by which signals are flexibly routed through these unique cortical hubs across wide domains. These “middle hierarchy” regions must engage in particularly broad coordination patterns, integrating ongoing unimodal information processing with the more sustained and extended operations in heteromodal cortex. This information is likely weighted by salience and goal relevance, while also allowing novel ongoing sensory information to gain access to heteromodal cortex.
The graded nature of local structure-function coupling appears to be shaped by the geometric embedding of individual brain regions. Namely, we also find that regions with very short or very long connectivity distance tend to display stable coupling, while regions with intermediate connectivity distance, particularly insular cortex and frontal eye fields, display more variable structure-function coupling. These findings resonate with a growing appreciation for how geometric relationships shape topological relationships in the brain52,59–64. In particular, physical separation from sensorimotor cortex is thought to correspond to graded variation in connectivity distance, culminating in predominantly long-range functional connectivity in association cortex26,41,53,54,65. The particular distribution of connection lengths that “middle hierarchy” regions participate in—leaning neither toward overly short- or long-range connectivity—may support flexible reconfiguration and participation in multiple systems36–39,58, manifesting as variable structure-function coupling.
Our results build on a rapidly-developing literature on local structure-function relationships2. While traditional studies have focused on global structure-function relationships captured by a single forward model45,66–69, numerous recent reports point to region-specific structure-function coupling patterns21,23,24. These structure-function relationships undergo extensive maturation and lifespan trajectories21,70. Interestingly, regional differences in structure-function coupling are correlated with micro-architectural variations, including intracortical myelin and cellular composition21,23. This suggests that local circuit properties—invisible to macroscale connectivity reconstructions—may additionally drive structure-function coupling26. Consistent with this notion, multiple modeling studies have recently shown that biophysical models constrained by regionally heterogeneous micro-architectural information, such as myelination, gene expression and neurotransmitter receptor profiles, make more accurate predictions about functional connectivity compared to regionally homogeneous models31–33. How regional differences in micro-architecture shape moment-to-moment fluctuations in structure-function coupling remains an important question for future research.
The present results need to be interpreted with respect to multiple limitations. First, structural connectivity networks were reconstructed using diffusion weighted MRI, a method that is susceptible to systematic false positives and negatives71–74. Although the present findings are observed in individual participants and can be demonstrated using alternative methods, further development in computational tractometry is necessary. Second, the dynamics of the BOLD signal itself are influenced by multiple physiological confounds, including blood flow and respiration75,76. In the absence of concurrent measurements of cardiovascular and cerebrovascular factors, these results must be interpreted with caution. Likewise, it is important to note that there exist multiple alternative methods to quantify dynamic functional connectivity. We applied a recently-developed temporal unwrapping method that has been demonstrated to be robust to a wide range of methodological choices, including parcellation and global signal regression method, and are sensitive to individual differences9,14. The statistical properties of the underlying dynamic processes behind moment-to-moment functional dynamics of the human brain has been an area of active research for years77–82, and the applicability of these methods to studying structure-function relationships is increasingly recognized83,84.
Collectively, the present work identifies patterns of local structure-function coupling that are systematically organized across the cortex and highly dynamic. The temporal coupling of structure and function points towards a rich and under-explored feature of the brain that may potentially help to understand how functions and cognitive processes are flexibly implemented and deployed.
Methods
Data acquisition
Structural and functional data were obtained from the Human Connectome Project (s900 release43). Scans from 327 healthy young participants (age range 22–35 years) with no familial relationships were used, including individual measures of diffusion MRI and four resting-state functional MRI time-series (two scans on day 1 and two scans on day 2, each of 15 min long). Data were processed following the procedure described in ref. 28,85.
Structural network reconstruction
Gray matter was parcellated into 400 cortical regions according to the Schaefer functional atlas44. Structural connectivity between regions was estimated for each participant using deterministic streamline tractography. First, the distribution of fiber orientation for each region was generated using the multi-shell multi-tissue constrained spherical deconvolution algorithm from the MRtrix3 package86,87 (https://www.mrtrix.org/). After that, the structural connectivity weight between any two regions was given by the number of streamlines normalized by the mean length of streamlines and the mean surface area of the two regions. This normalization reduces bias towards long fibers during streamline reconstruction, as well as the bias from differences in region sizes.
Functional time-series reconstruction
Functional MRI data were corrected for gradient nonlinearity, head motion (using a rigid body transformation), and geometric distortions (using scan pairs with opposite phase encoding directions (R/L, L/R)88). BOLD time-series were then subjected to a high-pass filter (>2000s FWHM) to correct for scanner drifts, and to the ICA-FIX process to remove additional noise89. The data was parcellated in the same atlas used for structural networks.
Time-resolved structure-function coupling
To estimate region- and time-resolved structure-function coupling, we first constructed temporal co-fluctuation matrices. We started by calculating the element-wise product of the z-scored BOLD time-series between pairs of brain regions14. Region pairs with an activity on the same side of the baseline will have a positive co-fluctuation value, whereas two regions that fluctuate in opposite directions at the same time will have a negative co-fluctuation value (Fig. 1a). The average across time of these co-fluctuation matrices recovers the Pearson correlation coefficient that is often used to define functional connectivity.
To define region-specific structure-function coupling, we constructed a multilinear regression model to predict the co-fluctuation profile of a node i from its geometric and structural connectivity profile to all other nodes j ≠ i23. Predictors included Euclidean distance, shortest path length, and communicability. Euclidean distance was calculated between node centroids. Shortest path length refers to the shortest contiguous sequence of edges between 2 nodes. Communicability (Cij) between two nodes i and j is defined as the weighted sum of all paths and walks between those nodes23,49. For a weighted adjacency matrix A, communicability is calculated as , where is the diagonal matrix of the generalized node degree matrix50. Shortest path length was implemented using Brainconn (https://github.com/fiuneuro/brainconn), a Python version of the Brain Connectivity Toolbox. Weighted communicability was implemented in netneurotools (https://github.com/netneurolab/netneurotools), an open-source Python package for network neuroscience. We used the minmax-normalized weighted structural connectivity matrix for each individual, and a negative log transformation was applied to the structural connectivity weights before calculating the shortest path length51.
Concretely, for region i, subject s, time point t, we have,
| 1 |
where coflucs,t,i is the co-fluctuation profile, predicted by Euclidean distance disti, shortest path length spls,i, and weighted communicability cmcs,i. The regression coefficients {β0, β1, β2, β3} were estimated by ordinary least squares. Coupling was measured using adjusted R-squared , a metric for goodness of fit. The regression was applied for individual profiles of brain regions, generating a cortical map of coupling values at each time point for each subject. We therefore define structure-function relationships as the goodness of fit for the linear regression model and, in keeping with previous literature, we refer to model fit as “coupling”.
Static and dynamic structure-function coupling
The multilinear regression model, when applied without temporal expansion, generates one R2 value per brain region, which we refer to here as static coupling23. By incorporating temporal co-fluctuation patterns, we obtained structure-function coupling measure per region as a frame-by-frame time-series, which we call dynamic coupling. To assess how dynamic coupling differs from static coupling, we frame the question as comparing a single observation (static) with a distribution (dynamic). We defined three summary statistics: (1) the probability of having a larger dynamic coupling value compared to the static coupling, (2) the bias, and (3) the variance of the dynamic coupling to reproduce the static values. Bias was used to evaluate how dynamic values deviate from the static value. It was calculated as the median of the difference between the dynamic coupling values and the static coupling. Small values of bias indicate that dynamic coupling values are close to the static coupling values, while large values indicate deviation. Variance was used to evaluate the extent of scattering of the dynamic values. It was calculated as the standard deviation of the distribution formed by the dynamic values. More specifically, we used the difference between the 84th percentile and the 16th percentile (±1 standard deviation from the mean under normality) to provide a more robust estimation of variability in case of possible outliers, extreme values, or skewed distributions. Thus a low variance value means that the distribution had low variability, and high variance value indicates the opposite.
Cortical annotations
Patterns of dynamic local structure-function coupling were contextualized relative to three common annotations: (1) 7 intrinsic functional networks as defined in ref. 40, 7 cytoarchitectonic classes described in ref. 42,90, and 10 functional hierarchy groups as defined in ref. 23, based on the principal functional gradient reported in ref. 41. Collectively, these three partitions of the brain are thought to reflect multimodal hierarchies91.
Null models
To assess correspondence between coupling maps and cortical annotations, we applied spatial autocorrelation-preserving permutation tests, termed “spin tests”23,92,93. In this model, the cortical surface is projected to a sphere using the coordinates of the vertex closest to the center of mass of each parcel. The sphere is then randomly rotated, generating surface maps with randomized topography, where each parcel has a reassigned value. The parcels corresponding to the medial wall were assigned to the closest rotated parcel23,29,94. The rotation was applied to one hemisphere and then mirrored to the other hemisphere. This corresponds to “Vázquez-Rodríguez” method described in ref. 93. The method was chosen based on the benchmarking in ref. 93 because (a) it is was consistently most conservative method in the simulation and empirical analyses, (b) it was designed for parcellated data and did not have to discard permutations when parcels were rotated into the medial wall. We generated 10,000 spin permutations using netneurotools (https://github.com/netneurolab/netneurotools). Details of spatially-constrained null models in neuroimaging (https://github.com/netneurolab/markello_spatialnulls) were described in ref. 93.
Predictor contributions
Predictor contributions in the supplementary figure were calculated using dominance analysis95,96. The analysis estimates the relative importance of predictors by constructing all possible subsets of the predictor variables and re-fitting the regression model for each combination. The “total dominance” statistic is adopted as a summary measure quantifying the contribution of each predictor to the overall goodness of fit. This method, among other procedures for interpreting multilinear regression models, can account for multicollinearity and is sensitive to potential patterns in the model97. This paper used a re-implementation of the Dominance-Analysis (https://github.com/dominance-analysis/dominance-analysis) package in netneurotools (https://github.com/netneurolab/netneurotools).
Supplementary information
Acknowledgements
We thank Golia Shafiei, Justine Hansen, Laura Suárez, Ross Markello, Vincent Bazinet, Louis-Philippe Robichaud and Filip Milisav for comments and suggestions on the manuscript. This research was undertaken thanks in part to funding from the Canada First Research Excellence Fund, awarded to McGill University for the Healthy Brains for Healthy Lives initiative. BM acknowledges support from the Natural Sciences and Engineering Research Council of Canada (NSERC Discovery Grant RGPIN #017-04265) and from the Canada Research Chairs Program.
Author contributions
Conception of project: ZQL, BVR, BM. Analysis: ZQL, BVR. Writing – original draft: ZQL, BVR, BM. Writing – editing: RNS, BCB, RFB.
Peer review
Peer review information
Communications Biology thanks Daniele Marinazzo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Enzo T Tagliazucchi and Luke Grinham. Peer reviewer reports are available.
Data availability
The MRI data that support the findings of this study are available from the Human Connectome Project, https://www.humanconnectome.org/study/hcp-young-adult.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhen-Qi Liu, Bertha Vázquez-Rodríguez.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-022-03466-x.
References
- 1.Avena-Koenigsberger A, Misic B, Sporns O. Communication dynamics in complex brain networks. Nat. Rev. Neurosci. 2018;19:17. doi: 10.1038/nrn.2017.149. [DOI] [PubMed] [Google Scholar]
- 2.Suárez, L. E., Markello, R. D., Betzel, R. F. & Misic, B. Linking structure and function in macroscale brain networks. Trends Cogn. Sci. 24, 302–315 (2020). [DOI] [PubMed]
- 3.Lurie DJ, et al. Questions and controversies in the study of time-varying functional connectivity in resting fMRI. Netw. Neurosci. 2020;4:30–69. doi: 10.1162/netn_a_00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Preti MG, Bolton TA, Van De Ville D. The dynamic functional connectome: State-of-the-art and perspectives. NeuroImage. 2017;160:41–54. doi: 10.1016/j.neuroimage.2016.12.061. [DOI] [PubMed] [Google Scholar]
- 5.Shine JM, Poldrack RA. Principles of dynamic network reconfiguration across diverse brain states. NeuroImage. 2018;180:396–405. doi: 10.1016/j.neuroimage.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Zalesky A, Fornito A, Cocchi L, Gollo LL, Breakspear M. Time-resolved resting-state brain networks. Proc. Natl. Acad. Sci. USA. 2014;111:10341–10346. doi: 10.1073/pnas.1400181111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang C, Glover GH. Time–frequency dynamics of resting-state brain connectivity measured with fMRI. NeuroImage. 2010;50:81–98. doi: 10.1016/j.neuroimage.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Betzel RF, Fukushima M, He Y, Zuo X-N, Sporns O. Dynamic fluctuations coincide with periods of high and low modularity in resting-state functional brain networks. NeuroImage. 2016;127:287–297. doi: 10.1016/j.neuroimage.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esfahlani FZ, et al. High-amplitude cofluctuations in cortical activity drive functional connectivity. Proc. Natl. Acad. Sci. 2020;117:28393–28401. doi: 10.1073/pnas.2005531117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liégeois R, Laumann TO, Snyder AZ, Zhou J, Yeo BTT. Interpreting temporal fluctuations in resting-state functional connectivity MRI. NeuroImage. 2017;163:437–455. doi: 10.1016/j.neuroimage.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Allen EA, et al. Tracking whole-brain connectivity dynamics in the resting state. Cereb. Cortex. 2014;24:663–676. doi: 10.1093/cercor/bhs352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shine JM, et al. The dynamics of functional brain networks: integrated network states during cognitive task performance. Neuron. 2016;92:544–554. doi: 10.1016/j.neuron.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vidaurre D, Smith SM, Woolrich MW. Brain network dynamics are hierarchically organized in time. Proc. Natl. Acad. Sci. 2017;114:12827–12832. doi: 10.1073/pnas.1705120114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faskowitz J, Esfahlani FZ, Jo Y, Sporns O, Betzel RF. Edge-centric functional network representations of human cerebral cortex reveal overlapping system-level architecture. Natw. Neurosci. 2020;23:1644–1654. doi: 10.1038/s41593-020-00719-y. [DOI] [PubMed] [Google Scholar]
- 15.Jo, Y., Faskowitz, J., Esfahlani, F. Z., Sporns, O. & Betzel, R. F. Subject identification using edge-centric functional connectivity. NeuroImage238, 118204 (2021). [DOI] [PubMed]
- 16.Eichenbaum A, Pappas I, Lurie D, Cohen JR, D’Esposito M. Differential contributions of static and time-varying functional connectivity to human behavior. Netw. Neurosci. 2021;5:145–165. doi: 10.1162/netn_a_00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez-Castillo J, et al. Tracking ongoing cognition in individuals using brief, whole-brain functional connectivity patterns. Proc. Natl. Acad. Sci. 2015;112:8762–8767. doi: 10.1073/pnas.1501242112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Battaglia D, et al. Dynamic functional connectivity between order and randomness and its evolution across the human adult lifespan. NeuroImage. 2020;222:117156. doi: 10.1016/j.neuroimage.2020.117156. [DOI] [PubMed] [Google Scholar]
- 19.Fukushima M, et al. Structure–function relationships during segregated and integrated network states of human brain functional connectivity. Brain Struct. Funct. 2018;223:1091–1106. doi: 10.1007/s00429-017-1539-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukushima M, Sporns O. Structural determinants of dynamic fluctuations between segregation and integration on the human connectome. Commun. Biol. 2020;3:1–11. doi: 10.1038/s42003-020-01331-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baum GL, et al. Development of structure–function coupling in human brain networks during youth. Proc. Natl. Acad. Sci. 2020;117:771–778. doi: 10.1073/pnas.1912034117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu, Z., Jamison, K. W., Sabuncu, M. R. & Kuceyeski, A. Heritability and interindividual variability of regional structure-function coupling. Nat. Commun.12, 4894 (2021). [DOI] [PMC free article] [PubMed]
- 23.Vázquez-Rodríguez B, et al. Gradients of structure–function tethering across neocortex. Proc. Natl. Acad. Sci. 2019;116:21219–21227. doi: 10.1073/pnas.1903403116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Preti MG, Van De Ville D. Decoupling of brain function from structure reveals regional behavioral specialization in humans. Nat. Commun. 2019;10:1–7. doi: 10.1038/s41467-019-12765-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paquola C, et al. Microstructural and functional gradients are increasingly dissociated in transmodal cortices. PLoS Biol. 2019;17:e3000284. doi: 10.1371/journal.pbio.3000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buckner RL, Krienen FM. The evolution of distributed association networks in the human brain. Trends Cogn. Sci. 2013;17:648–665. doi: 10.1016/j.tics.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 27.Mesulam M-M. From sensation to cognition. Brain J. Neurol. 1998;121:1013–1052. doi: 10.1093/brain/121.6.1013. [DOI] [PubMed] [Google Scholar]
- 28.Shafiei G, et al. Topographic gradients of intrinsic dynamics across neocortex. Elife. 2020;9:e62116. doi: 10.7554/eLife.62116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen, J. Y. et al. Mapping gene transcription and neurocognition across human neocortex. Nat Hum Behav5, 1240–1250 (2021). [DOI] [PubMed]
- 30.Kong, X. et al. Anatomical and functional gradients shape dynamic functional connectivity in the human brain. 2021.03.15.435361, https://www.biorxiv.org/content/10.1101/2021.03.15.435361v1 (2021).
- 31.Demirtaş M, et al. Hierarchical heterogeneity across human cortex shapes large-scale neural dynamics. Neuron. 2019;101:1181–1194. doi: 10.1016/j.neuron.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P, et al. Inversion of a large-scale circuit model reveals a cortical hierarchy in the dynamic resting human brain. Sc. Adv. 2019;5:eaat7854. doi: 10.1126/sciadv.aat7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deco, G. et al. Dynamical consequences of regional heterogeneity in the brain’s transcriptional landscape. Sci. Adv.7, eabf4752 (2021). [DOI] [PMC free article] [PubMed]
- 34.Valk, S. L. et al. Genetic and phylogenetic uncoupling of structure and function in human transmodal cortex. Nat Commun13, 2341 (2022). [DOI] [PMC free article] [PubMed]
- 35.Allen M. Unravelling the neurobiology of interoceptive inference. Trends Cogn. Sci. 2020;24:265–266. doi: 10.1016/j.tics.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Uddin LQ. Salience processing and insular cortical function and dysfunction. Nat. Rev. Neurosci. 2015;16:55–61. doi: 10.1038/nrn3857. [DOI] [PubMed] [Google Scholar]
- 37.Ghaziri J, et al. The corticocortical structural connectivity of the human insula. Cereb. Cortex. 2017;27:1216–1228. doi: 10.1093/cercor/bhv308. [DOI] [PubMed] [Google Scholar]
- 38.Kurth F, Zilles K, Fox PT, Laird AR, Eickhoff SB. A link between the systems: functional differentiation and integration within the human insula revealed by meta-analysis. Brain Struct. Funct. 2010;214:519–534. doi: 10.1007/s00429-010-0255-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tian Y, Zalesky A. Characterizing the functional connectivity diversity of the insula cortex: Subregions, diversity curves and behavior. NeuroImage. 2018;183:716–733. doi: 10.1016/j.neuroimage.2018.08.055. [DOI] [PubMed] [Google Scholar]
- 40.Yeo, B. T. et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol.106, 1125–1165 (2011). [DOI] [PMC free article] [PubMed]
- 41.Margulies DS, et al. Situating the default-mode network along a principal gradient of macroscale cortical organization. Proc. Natl. Acad. Sci. 2016;113:12574–12579. doi: 10.1073/pnas.1608282113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Economo, C. F. & Koskinas, G. N. Die cytoarchitektonik der hirnrinde des erwachsenen menschen (J. Springer, 1925).
- 43.Van Essen DC, et al. The human connectome project: a data acquisition perspective. Neuroimage. 2012;62:2222–2231. doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaefer A, et al. Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity mri. Cereb. Cortex. 2018;28:3095–3114. doi: 10.1093/cercor/bhx179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goñi J, et al. Resting-brain functional connectivity predicted by analytic measures of network communication. Proc. Natl. Acad. Sci. 2014;111:833–838. doi: 10.1073/pnas.1315529111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seguin C, Tian Y, Zalesky A. Network communication models improve the behavioral and functional predictive utility of the human structural connectome. Netw. Neurosci. 2020;4:980–1006. doi: 10.1162/netn_a_00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seguin C, Van Den Heuvel MP, Zalesky A. Navigation of brain networks. Proc. Natl. Acad. Sci. 2018;115:6297–6302. doi: 10.1073/pnas.1801351115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fornito, A., Zalesky, A. & Bullmore, E. Fundamentals of Brain Network Analysis (Academic Press, 2016).
- 49.Estrada E, Hatano N. Communicability in complex networks. Phys. Rev. E. 2008;77:036111. doi: 10.1103/PhysRevE.77.036111. [DOI] [PubMed] [Google Scholar]
- 50.Crofts JJ, Higham DJ. A weighted communicability measure applied to complex brain networks. J. R. Soc. Interface. 2009;6:411–414. doi: 10.1098/rsif.2008.0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Avena-Koenigsberger A, et al. Path ensembles and a tradeoff between communication efficiency and resilience in the human connectome. Brain Struct. Funct. 2017;222:603–618. doi: 10.1007/s00429-016-1238-5. [DOI] [PubMed] [Google Scholar]
- 52.Sepulcre J, et al. The organization of local and distant functional connectivity in the human brain. PLoS Comput. Biol. 2010;6:e1000808. doi: 10.1371/journal.pcbi.1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oligschläger S, et al. Gradients of connectivity distance are anchored in primary cortex. Brain Struct. Funct. 2017;222:2173–2182. doi: 10.1007/s00429-016-1333-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oligschläger S, et al. Gradients of connectivity distance in the cerebral cortex of the macaque monkey. Brain Struct. Funct. 2019;224:925–935. doi: 10.1007/s00429-018-1811-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vallat R. Pingouin: statistics in python. J. Open Source Softw. 2018;3:1026. doi: 10.21105/joss.01026. [DOI] [Google Scholar]
- 56.Royer J, et al. Myeloarchitecture gradients in the human insula: Histological underpinnings and association to intrinsic functional connectivity. NeuroImage. 2020;216:116859. doi: 10.1016/j.neuroimage.2020.116859. [DOI] [PubMed] [Google Scholar]
- 57.Corbetta M, Shulman GL. Control of goal-directed and stimulus-driven attention in the brain. Nat. Rev. Neurosci. 2002;3:201–215. doi: 10.1038/nrn755. [DOI] [PubMed] [Google Scholar]
- 58.Corbetta M, Patel G, Shulman GL. The reorienting system of the human brain: from environment to theory of mind. Neuron. 2008;58:306–324. doi: 10.1016/j.neuron.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stiso J, Bassett DS. Spatial embedding imposes constraints on neuronal network architectures. Trends Cogn. Sci. 2018;22:1127–1142. doi: 10.1016/j.tics.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 60.Roberts JA, et al. The contribution of geometry to the human connectome. NeuroImage. 2016;124:379–393. doi: 10.1016/j.neuroimage.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 61.Ercsey-Ravasz M, et al. A predictive network model of cerebral cortical connectivity based on a distance rule. Neuron. 2013;80:184–197. doi: 10.1016/j.neuron.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Betzel RF, et al. Generative models of the human connectome. NeuroImage. 2016;124:1054–1064. doi: 10.1016/j.neuroimage.2015.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mišić B, et al. The functional connectivity landscape of the human brain. PLoS One. 2014;9:e111007. doi: 10.1371/journal.pone.0111007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Betzel RF, Griffa A, Hagmann P, Mišić B. Distance-dependent consensus thresholds for generating group-representative structural brain networks. Net. Neurosci. 2019;3:475–496. doi: 10.1162/netn_a_00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Larivière S, et al. Functional connectome contractions in temporal lobe epilepsy: Microstructural underpinnings and predictors of surgical outcome. Epilepsia. 2020;61:1221–1233. doi: 10.1111/epi.16540. [DOI] [PubMed] [Google Scholar]
- 66.Honey CJ, Kötter R, Breakspear M, Sporns O. Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proc. Natl. Acad. Sci. USA. 2007;104:10240–10245. doi: 10.1073/pnas.0701519104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Honey CJ, et al. Predicting human resting-state functional connectivity from structural connectivity. Proc. Natl. Acad. Sci. USA. 2009;106:2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mišić B, et al. Network-level structure-function relationships in human neocortex. Cereb. Cortex. 2016;26:3285–3296. doi: 10.1093/cercor/bhw089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mišić B, et al. Cooperative and competitive spreading dynamics on the human connectome. Neuron. 2015;86:1518–1529. doi: 10.1016/j.neuron.2015.05.035. [DOI] [PubMed] [Google Scholar]
- 70.Zamani Esfahlani, F., Faskowitz, J., Slack, J., Mišić, B. & Betzel, R. F. Local structure-function relationships in human brain networks across the lifespan. Nat. Commun.13, 2053 (2022). [DOI] [PMC free article] [PubMed]
- 71.Jones DK, Knösche TR, Turner R. White matter integrity, fiber count, and other fallacies: the do’s and don’ts of diffusion mri. NeuroImage. 2013;73:239–254. doi: 10.1016/j.neuroimage.2012.06.081. [DOI] [PubMed] [Google Scholar]
- 72.de Reus MA, van den Heuvel MP. Estimating false positives and negatives in brain networks. NeuroImage. 2013;70:402–409. doi: 10.1016/j.neuroimage.2012.12.066. [DOI] [PubMed] [Google Scholar]
- 73.Thomas C, et al. Anatomical accuracy of brain connections derived from diffusion mri tractography is inherently limited. Proc. Natl. Acad. Sci. USA. 2014;111:16574–16579. doi: 10.1073/pnas.1405672111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maier-Hein KH, et al. The challenge of mapping the human connectome based on diffusion tractography. Nat. Commun. 2017;8:1–13. doi: 10.1038/s41467-017-01285-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Colenbier N, et al. Disambiguating the role of blood flow and global signal with partial information decomposition. NeuroImage. 2020;213:116699. doi: 10.1016/j.neuroimage.2020.116699. [DOI] [PubMed] [Google Scholar]
- 76.Tsvetanov KA, et al. The effects of age on resting-state BOLD signal variability is explained by cardiovascular and cerebrovascular factors. Psychophysiology. 2021;58:e13714. doi: 10.1111/psyp.13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tagliazucchi E, Balenzuela P, Fraiman D, Chialvo D. Criticality in large-scale brain fMRI dynamics unveiled by a novel point process analysis. Front. Physiol. 2012;3:15. doi: 10.3389/fphys.2012.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tagliazucchi E, Siniatchkin M, Laufs H, Chialvo DR. The voxel-wise functional connectome can be efficiently derived from co-activations in a sparse spatio-temporal point-process. Front. Neurosci. 2016;10:381. doi: 10.3389/fnins.2016.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Petridou N, Gaudes CC, Dryden IL, Francis ST, Gowland PA. Periods of rest in fMRI contain individual spontaneous events which are related to slowly fluctuating spontaneous activity. Hum. Brain Mapp. 2013;34:1319–1329. doi: 10.1002/hbm.21513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu G-R, et al. A blind deconvolution approach to recover effective connectivity brain networks from resting state fMRI data. Med. Image Anal. 2013;17:365–374. doi: 10.1016/j.media.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 81.Liu X, Duyn JH. Time-varying functional network information extracted from brief instances of spontaneous brain activity. Proc. Natl. Acad. Sci. 2013;110:4392–4397. doi: 10.1073/pnas.1216856110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Karahanoğlu FI, Van De Ville D. Transient brain activity disentangles fMRI resting-state dynamics in terms of spatially and temporally overlapping networks. Nat. Commun. 2015;6:7751. doi: 10.1038/ncomms8751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Novelli, L. & Razi, A. A mathematical perspective on edge-centric brain functional connectivity. Nat. Commun.13, 2693 (2022). [DOI] [PMC free article] [PubMed]
- 84.Pope, M., Fukushima, M., Betzel, R. F. & Sporns, O. Modular origins of high-amplitude co-fluctuations in fine-scale functional connectivity dynamics. Proc. Natl. Acad. Sci. U.S.A.118, e2109380118 (2021). [DOI] [PMC free article] [PubMed]
- 85.Park B-y, et al. Signal diffusion along connectome gradients and inter-hub routing differentially contribute to dynamic human brain function. NeuroImage. 2021;224:117429. doi: 10.1016/j.neuroimage.2020.117429. [DOI] [PubMed] [Google Scholar]
- 86.Dhollander, T., Raffelt, D. & Connelly, A. Unsupervised 3-tissue response function estimation from single-shell or multi-shell diffusion mr data without a co-registered t1 image. In ISMRM Workshop on Breaking the Barriers of Diffusion MRI, vol. 5, 5 (2016).
- 87.Tournier J-D, et al. Mrtrix3: A fast, flexible and open software framework for medical image processing and visualisation. NeuroImage. 2019;202:116137. doi: 10.1016/j.neuroimage.2019.116137. [DOI] [PubMed] [Google Scholar]
- 88.de Wael RV, et al. Anatomical and microstructural determinants of hippocampal subfield functional connectome embedding. Proc. Natl. Acad. Sci. 2018;115:10154–10159. doi: 10.1073/pnas.1803667115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Salimi-Khorshidi G, et al. Automatic denoising of functional mri data: combining independent component analysis and hierarchical fusion of classifiers. Neuroimage. 2014;90:449–468. doi: 10.1016/j.neuroimage.2013.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Scholtens LH, de Reus MA, de Lange SC, Schmidt R, van den Heuvel MP. An mri von economo–koskinas atlas. Neuroimage. 2018;170:249–256. doi: 10.1016/j.neuroimage.2016.12.069. [DOI] [PubMed] [Google Scholar]
- 91.Huntenburg JM, Bazin P-L, Margulies DS. Large-scale gradients in human cortical organization. Trends Cogn. Sci. 2018;22:21–31. doi: 10.1016/j.tics.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 92.Alexander-Bloch AF, et al. On testing for spatial correspondence between maps of human brain structure and function. Neuroimage. 2018;178:540–551. doi: 10.1016/j.neuroimage.2018.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Markello, R. D. & Misic, B. Comparing spatial null models for brain maps. NeuroImage236, 118052 (2021). [DOI] [PubMed]
- 94.Shafiei G, et al. Spatial patterning of tissue volume loss in schizophrenia reflects brain network architecture. Biol. Psychiat. 2020;87:727–735. doi: 10.1016/j.biopsych.2019.09.031. [DOI] [PubMed] [Google Scholar]
- 95.Budescu DV. Dominance analysis: A new approach to the problem of relative importance of predictors in multiple regression. Psychol. Bull. 1993;114:542–551. doi: 10.1037/0033-2909.114.3.542. [DOI] [Google Scholar]
- 96.Azen R, Budescu DV. The dominance analysis approach for comparing predictors in multiple regression. Psychol. Methods. 2003;8:129–148. doi: 10.1037/1082-989X.8.2.129. [DOI] [PubMed] [Google Scholar]
- 97.Kraha, A., Turner, H., Nimon, K., Zientek, L. & Henson, R. Tools to support interpreting multiple regression in the face of multicollinearity. Front. Psychol. 3, 44 (2012). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MRI data that support the findings of this study are available from the Human Connectome Project, https://www.humanconnectome.org/study/hcp-young-adult.