Abstract
We compared and statistically evaluated the effectiveness of nine DNA extraction procedures by using frozen and dried samples of two silt loam soils and a silt loam wetland sediment with different organic matter contents. The effects of different chemical extractants (sodium dodecyl sulfate [SDS], chloroform, phenol, Chelex 100, and guanadinium isothiocyanate), different physical disruption methods (bead mill homogenization and freeze-thaw lysis), and lysozyme digestion were evaluated based on the yield and molecular size of the recovered DNA. Pairwise comparisons of the nine extraction procedures revealed that bead mill homogenization with SDS combined with either chloroform or phenol optimized both the amount of DNA extracted and the molecular size of the DNA (maximum size, 16 to 20 kb). Neither lysozyme digestion before SDS treatment nor guanidine isothiocyanate treatment nor addition of Chelex 100 resin improved the DNA yields. Bead mill homogenization in a lysis mixture containing chloroform, SDS, NaCl, and phosphate-Tris buffer (pH 8) was found to be the best physical lysis technique when DNA yield and cell lysis efficiency were used as criteria. The bead mill homogenization conditions were also optimized for speed and duration with two different homogenizers. Recovery of high-molecular-weight DNA was greatest when we used lower speeds and shorter times (30 to 120 s). We evaluated four different DNA purification methods (silica-based DNA binding, agarose gel electrophoresis, ammonium acetate precipitation, and Sephadex G-200 gel filtration) for DNA recovery and removal of PCR inhibitors from crude extracts. Sephadex G-200 spin column purification was found to be the best method for removing PCR-inhibiting substances while minimizing DNA loss during purification. Our results indicate that for these types of samples, optimum DNA recovery requires brief, low-speed bead mill homogenization in the presence of a phosphate-buffered SDS-chloroform mixture, followed by Sephadex G-200 column purification.
In the past decade, applications of molecular biological approaches have provided unique insights into the uncultured microbial communities of soils and waters because they avoid biases inherent in traditional culture-based microbiological methods (16). The validity of using molecular techniques in environmental studies depends on obtaining representative extracts of nucleic acids from entire microbial communities. Nucleic acid extraction methods, however, suffer from compounded inefficiencies in the individual component steps, including incomplete cell lysis, DNA sorption to soil surfaces, coextraction of enzymatic inhibitors from soil, and loss, degradation, or damage of DNA. Thus, studies of DNA extraction techniques (14, 23, 24, 27) have indicated that these techniques can introduce biases of their own.
In the initial efforts to extract DNA from sediments and soils workers used either cell extraction (recovery of cells from the soil matrix prior to cell lysis) or direct lysis within the soil matrix (13, 29, 35). Direct lysis techniques, however, have been used more because they yield more DNA and presumably a less biased sample of the microbial community diversity than cell extraction techniques yield (13, 21, 35). A major drawback of direct lysis methods is that more PCR-inhibitory substances are extracted along with the DNA (29, 36, 39). In addition, the number and diversity of the direct lysis DNA extraction protocols used for soils and sediments are daunting (11–13, 20, 27, 29, 30, 44), but each protocol usually includes from one to all three of the following basic elements: physical disruption, chemical lysis, and enzymatic lysis.
Four different physical disruption techniques, freeze-thawing (8, 16, 20, 31, 38), bead mill homogenization (4, 20, 22, 27, 35), ultrasonication (30), and grinding under liquid nitrogen (41, 44), have been described, and freeze-thaw disruption and bead mill homogenization are the most common. It is well established that bead mill homogenization yields more DNA than freeze-thaw disruption yields (20, 21, 27, 33). The drawbacks to bead mill homogenization include the fact that larger amounts of contaminating humic acids are recovered (21, 29, 33) and the fact that, in some instances, the DNA is sheared (21). The chemical lysis procedures used in the methods that have been described also vary, but they lysis mixtures can be categorized into mixtures that contain detergent (either sodium dodecyl sulfate [SDS] [4, 8, 12, 14, 15, 20, 25, 28, 36–38, 44] or Sarkosyl [14, 34, 37]), mixtures that contain NaCl, and mixtures that contain various buffers (usually Tris or phosphate, pH 7 to 8). The modifications of the basic chemical lysis techniques include high-temperature (60°C to boiling) incubation (4, 20, 34, 35), a phenol (8, 33, 38) or chloroform (12) extraction step, and incorporation of chelating agents (EDTA and Chelex 100) to inhibit nucleases and disperse soil particles (6, 18). The efficacy of diverse chemical lysis components remains largely unknown since only overall DNA recovery after cell lysis and subsequent purification is reported. Furthermore, the cellular lysis efficiency at each step of a protocol is reported only rarely (27). A final component of many DNA extraction techniques is enzymatic lysis. Lysozyme (4, 8, 11, 14, 31, 36, 38), proteinase K (25, 36, 44), achromopeptidase (7), and pronase E (14) have all been employed to promote cell lysis, and lysozyme digestion is the most widely used procedure. Because of a lack of comparative studies, it is unclear what effect the addition of an enzymatic lysis step has on DNA yield.
An added layer of complexity when the DNA yields obtained with different protocols are compared is the DNA purification method employed. For most soils that contain high concentrations of humic acids, agarose gel electrophoresis (16, 22, 25, 27, 28, 43, 44), Sephadex G-200 column chromatography (7, 8, 12, 20, 31, 39), and silica-based DNA binding (5, 27, 44) have been used individually or in combination to separate humic acids and other enzyme inhibitors from DNA. Purification efficiency is usually judged by the amount of DNA recovered and the success of the methods used to remove contaminants that inhibit PCR enzymes and other enzymes required for molecular analyses.
While the strengths and weaknesses of the various complete extraction and purification methods have been discussed previously (17, 20, 21, 27, 35, 39, 44), in general detailed comparisons of individual elements of the methods (e.g., the effectiveness of extraction with and without a chelating agent) have not been performed. Meaningful comparisons are also confounded by the variety of sample types examined and by the variable analytical procedures used to quantify DNA extraction efficiency and purity. Thus, selection of an appropriate DNA extraction and purification procedure from among the procedures that have been described to date remains a major problem in the application of molecular techniques to studies of soil and sediment microbial communities.
The objectives of this study were to compare the most common elements of DNA extraction and purification protocols and to use the information obtained to develop a comprehensive method for obtaining whole-community DNA from soil and sediment samples containing different quantities of organic matter. Nine different extraction procedures with contrasting physical, chemical, and enzymatic elements were evaluated on the basis of the following criteria: (i) efficiency of cell lysis, (ii) total yield of DNA, and (iii) molecular sizes of the DNA fragments. Four purification procedures were also evaluated by testing for the presence of PCR-inhibitory substances and the loss of DNA during the procedures. Bead mill homogenization conditions were then optimized with two different homogenization instruments in order to obtain the largest amount of DNA with the smallest amount of DNA shearing. The result was an optimized direct lysis DNA extraction and purification protocol for soils and sediments having diverse organic matter contents.
MATERIALS AND METHODS
Soil and sediment sampling and preservation.
Samples of a single type of soil (Collamer silt loam) from both an agricultural field and an adjacent forest and a wetland sediment that had the same silt loam texture but contained a different amount of organic matter (Table 1) were used in this study. The forest and agricultural soils were collected from McGowan Woods and an adjacent agricultural field (Cornell University, Ithaca, N.Y.), respectively. An ethanol-flamed spatula was used to collect approximately 400-g samples from the upper 2 to 5 cm of soil at each site. The soil samples were immediately transported to the laboratory in sterile, 200-ml Nalgene bottles. Approximately 400 ml of wetland sediment containing organic matter from decaying vegetation was collected at Sapsucker Woods (Ithaca, N.Y.) from the sediment-water interface by immersing and filling a previously sterilized 500-ml Nalgene bottle. The sample was immediately placed on ice and returned to the laboratory. Subsamples (approximately 100 g of soil or 200 ml of sediment) were set aside for use in chemical and particle size analyses. All of the samples used for DNA extraction were frozen at −20°C on the day of collection, lyophilized (Labconco Lyph-lock 4.5 lyophilizer), ground to a fine powder with a mortar and pestle, and stored at −20°C. All of the surfaces that contacted the soil were sterile or had previously been rinsed with a 10% (vol/vol) Clorox solution and then with sterile deionized water to remove any contaminating DNA.
TABLE 1.
Analysis of soils and sediment used for DNA extraction
| Soil or sedimenta | Soil horizon | Texture (%)
|
pHb | Organic carbon content (mg g [dry wt]−1)c | Bacterial content (109 cells g [dry wt]−1)d | ||
|---|---|---|---|---|---|---|---|
| Sand | Silt | Clay | |||||
| Collamer silt loam (agricultural) | A1 | 13 | 71 | 16 | 6.5 | 38 | 1.9 ± 0.5 |
| Collamer silt loam (forest) | 01 | 17 | 70 | 13 | 6.1 | 75 | 4.3 ± 1.4 |
| Wetland sediment (silt loam) | 17 | 68 | 15 | 5.7 | 442 | 11.0 ± 4.1 | |
Soil and sediment analysis.
The texture, organic carbon content, and pH of the soils and sediment were determined by standard methods at the Cornell Soil Analysis Facility in the Department of Soil, Crop, and Atmospheric Sciences (Table 1). The total bacterial cell count for each sample was estimated by using an acridine orange epifluorescence direct counting method (2, 10) and a Zeiss standard epifluorescence microscope equipped with a Zeiss 09 filter combination.
Experimental design for DNA extraction procedures.
A total of nine different procedures were evaluated by using each of the three sample types (Table 2). These procedures were designed to assess the effects of various combinations of SDS, organic solvents, chelation, enzymatic pretreatment with lysozyme, and bead mill homogenization or freeze-thaw lysis on DNA extraction and yield. Triplicate 50-mg samples of the wetland sediment and quadruplicate 100-mg samples of the agricultural and forest soils were added to 2-ml screw-cap plastic vials (Laboratory Products Sales, Rochester, N.Y.) containing 2 g of sterile, 0.1-mm-diameter zirconium-silica beads (BioSpec Products, Bartlesville, Okla.).
TABLE 2.
Enzymatic, chemical, and physical DNA extraction procedures tested with the three freeze-dried sediment and soil sample types shown in Table 1
| Procedure | Lysozyme pretreatmenta | Chemical treatmentb | Physical treatmentc | Reference |
|---|---|---|---|---|
| 1 | − | SDS | B | 27 |
| 2 | − | SDS-phenol | B | This paper |
| 3 | − | SDS-chloroform | B | 12 |
| 4 | − | SDS-Chelex 100 | B | 6 |
| 5 | + | SDS | FT | 11 |
| 6 | − | GTC | FT | 19 |
| 7 | − | SDS | FT | This paper |
| 8 | + | SDS | B | This paper |
| 9 | − | GTC | B | This paper |
For lysozyme pretreatment (procedures 5 and 8), 450-μl portions of a solution containing 150 mM NaCl, 100 mM EDTA (pH 8), and 15 mg of lysozyme per ml were added to vials, and the vials were incubated for 2 h at 37°C. After lysozyme pretreatment, 450-μl portions of an SDS lysis mixture (see below) were added to the vials.
For procedures 1 and 7, 450 μl of phosphate buffer (100 mM NaH2PO4, pH 8) and 450 μl of an SDS lysis mixture (100 mM NaCl, 500 mM Tris [pH 8], 10% [wt/vol] SDS) were added. For procedures 2 and 3, 300 μl of phosphate buffer, 300 μl of the SDS lysis mixture, and 300 μl of either buffered phenol (pH 8) or chloroform-isoamyl alcohol (24:1) were added. Procedure 4 was identical to procedure 1 except that 15% (wt/vol) Chelex 100 resin (Bio-Rad, Hercules, Calif.) was added to the SDS lysis mixture. The GTC treatments (procedures 6 and 9) consisted of 900 μl of 4 M GTC.
B, bead mill homogenization with the Mini Bead Beater at 1,600 rpm; FT, freeze-thaw treatment (three cycles consisting of 2 min in liquid N2 followed by 5 min in a 65°C water bath).
Enzymatic, chemical, and physical lysis treatments.
The enzymatic pretreatments, chemical treatments, and physical lysis conditions used for the nine DNA extraction procedures are shown in Table 2. Because the ratio of liquid volume to sediment volume influences lysis efficiency, the extract liquid volume used for all extractions was 900 μl. Bead mill homogenization was used in procedures 1 through 4, 8, and 9 after the chemical treatments were performed (Table 2). The tubes were shaken for two 5-min periods at 1,600 rpm in a Mini Bead Beater (BioSpec Products). Three freeze-thaw cycles were used in procedures 5 through 7. Each cycle consisted of immersion for 2 min in liquid nitrogen, followed by immersion for 5 min in a 65°C water bath.
Recovery and purification of DNA.
DNA released during each extraction procedure was recovered by the method of Herrick et al. (12). Briefly, beads and soil or sediment were separated from suspensions by centrifugation (10 s, 10,000 × g) with a microcentrifuge (model 235C; Fisher Scientific), and each liquid extract was transferred to a sterile 2-ml Eppendorf tube. The liquid remaining in the interstices of the bead bed was then collected by piercing the bottom of the tube with a hot 27-gauge needle and placing the pierced tube into a 15-ml, chloroform-safe, screw-cap polypropylene tube (Corning) containing a 1.5-ml Eppendorf tube with its top removed. The nested tubes were then centrifuged for 15 min (1,400 × g) with a tabletop centrifuge (model TJ6; Beckman). During centrifugation, liquid trapped in the bead bed drained into the lower 1.5-ml tube. This liquid was then pooled with the previously collected liquid from the sample.
The volume of each extract (except the extracts containing guanidinium isothiocyanate [GTC] [see below]) was reduced to 100 μl by removing water with butanol (32), and the concentrated extracts were purified on silica-based, SpinBind columns (FMC BioProducts) by using the manufacturer’s protocol. DNA was eluted from the SpinBind columns with 50 μl of warm (50°C) sterile water. Because butanol extraction did not effectively remove water from the GTC extracts, the DNA in each of these extracts was precipitated overnight at −20°C with an equal volume of isopropanol. The precipitate was pelleted by centrifugation, washed with 70% (vol/vol) ice-cold ethanol, dried, and dissolved in sterile, distilled H2O to a final volume of 100 μl (for the two types of soil extract) or 50 μl (for the sediment extract).
Estimating cellular lysis in extracted sediment samples.
The effects of the DNA extraction procedures on cellular lysis were estimated only with the wetland sediments, as follows. Triplicate 50-mg samples were subjected to each extraction procedure (Table 2) and the subsequent DNA recovery procedure as described above. Three formalin-preserved, freeze-dried samples were also processed from the DNA recovery step through the DAPI (4′,6-diamidino-2-phenylindole) staining step and served as “no-DNA-extraction” controls that accounted for cell loss due to sample manipulation. The sediment and beads remaining after the crude DNA extract was removed were transferred to a 10-ml screw-cap tube containing 4 ml of phosphate-buffered saline (145 mM NaCl, 500 mM NaH2PO4; pH 7.4) and 50 μl of a 37% (vol/vol) formalin solution. The tube was shaken for 15 min at 200 rpm on a rotary shaker to disperse the sediment particles and bacteria. The tube was then vortexed for 15 s, and the beads were allowed to settle for 10 s. One milliliter of the supernatant was centrifuged for 10 min at 10,000 × g to pellet the bacteria, and the pellet was then resuspended in 1.0 ml of phosphate-buffered saline by vortexing. The wash step was necessary to remove any remaining phenol or chloroform from the sample, which would have preferentially absorbed the fluorescent dyes. A 5-μl drop of the resuspended sample was mixed with a 5-μl drop of a DAPI solution (0.05 mg of DAPI/ml of deionized water) on an ethanol-washed slide and covered with a 22-mm2 coverslip. The total number of bacteria per milliliter of original suspension was determined by counting the number DAPI-stained blue fluorescent cells per microscopic field by using a Zeiss Standard 18 epifluorescence microscope equipped with a 100× Neofluar lens and an appropriate filter combination for DAPI. An area-to-volume calculation and appropriate dilution factors were used to convert the counting results to cells per milliliter and then to cells per gram (dry weight), as described previously (2). The efficiency of cell lysis for an extracted sample (expressed as a percentage) was calculated based on the difference between the bacterial cell count of the extracted preparation and the bacterial cell count of the no-DNA-extraction control.
Evaluation of DNA purification procedures.
DNA extraction procedure 4 (SDS-Chelex 100 treatment with bead mill homogenization) was found to produce the most PCR-inhibitory extracts of all of the DNA extraction procedures tested with the PCR inhibition assay (see below). Therefore, DNA extracted by this procedure was used to test DNA purification procedures, as follows. A 500-mg forest soil sample, a 500-mg agricultural soil sample, and a 200-mg sediment sample were extracted as described above. The final volumes of the crude extracts were adjusted to 1.5 ml with sterile, distilled water. The RNA in each crude extract was digested by adding 150 μl of an RNase solution (10 mg of RNase per ml, 100 mM Tris HCl, 10 mM EDTA [pH 8]) and incubating the preparation for 1 h at 37°C. Subsamples (25 to 100 μl) of the RNase-digested soil and sediment extracts were purified in quadruplicate by using the four methods (methods i to iv) described below.
(i) Method i.
Extract samples (100 μl) were added to SpinBind columns (FMC BioProducts) and were purified by using the manufacturer’s protocol. Briefly, DNA was bound to a silica support under chaotropic conditions, rinsed with Tris-buffered ethanol, and eluted with 100 μl of TE buffer.
(ii) Method ii.
Subsamples were subjected to agarose gel electrophoresis by loading 25 μl of crude extract onto a 1% (wt/vol) agarose gel and electrophoresing the preparation for 30 min at 5 V/cm. The ethidium bromide-stained DNA band was visualized under UV light and excised. DNA in the agarose slice was recovered by using SpinBind columns (FMC BioProducts) as recommended by the manufacturer.
(iii) Method iii.
One volume of sterile distilled water and 1 volume of 7.5 M ammonium acetate were added to 100 μl of crude extract, and the mixture was incubated on ice for 5 min to precipitate the protein. The sample was centrifuged for 10 min at 10,000 × g, and the supernatant was removed and mixed with 2 volumes of ice-cold ethanol and 20 μg of glycogen. The mixture was then allowed to stand overnight at −20°C. The precipitated DNA was pelleted by centrifugation for 15 min at 10,000 × g; then it was washed with 200 μl of ice-cold 70% (vol/vol) ethanol, air dried, and resuspended in 100 μl of sterile distilled water.
(iv) Method iv.
Sephadex G-200 spin columns (method of Erb and Wagner-Döbler [8], as modified by Boccuzzi et al. [3]) were used for DNA purification (12). Briefly, 1 g of dry Sephadex G-200 was hydrated, washed five times in high-salt TE buffer (10 mM Tris-HCl, 1 mM EDTA, 100 mM NaCl [pH 8]), and autoclaved. The suspension was packed aseptically into 1-ml syringes plugged at the bottom with sterile aquarium filter floss by repeated centrifugation (130 × g, 10 min) inside 15-ml sterile, screw-cap plastic tubes until the final compacted volume was 1.1 ml. The packed columns were washed with 100 μl of high-salt TE buffer, centrifuged (130 × g, 10 min), and transferred to new sterile 15-ml screw-cap plastic tubes. Subsamples (100 μl) of the crude extract were loaded into the syringes, and the DNA was eluted with three consecutive 100-μl portions of high-salt TE buffer, each followed by centrifugation (130 × g, 10 min). The final volume of eluted, purified DNA was reduced to 100 μl by butanol extraction (32).
PCR inhibition assay.
To determine whether PCR inhibitors were present in the purified DNA extracts, we designed an assay based on amplification of the 16S rRNA gene of Methylomonas albus BG8 (26). Equal volumes of purified DNA solutions from three or four replicate DNA purification runs were pooled and serially diluted in 10-fold increments with sterile, distilled water. A 10-μl portion of each dilution was added to a thin-walled PCR tube and amended with 0.5 ng of purified M. albus BG8 DNA in 5 μl of distilled water. The M. albus BG8 DNA was extracted and purified from cultures by using standard methods (1). Two drops of sterile, light mineral oil were then added to each tube. After the sample was heated to 80°C, 10 μl of a concentrated PCR buffer solution was added to each tube; each preparation contained (final concentrations) 50 mM KCl, 10 mM Tris buffer (pH 8.8), 1.5 mM MgCl2, 0.1 mg of bovine serum albumin per ml, 0.05% (vol/vol) Tween 20, 0.5 μM oligonucleotide PCR primer 10 γ (5), 0.5 μM oligonucleotide PCR primer P5 (11), each deoxynucleoside triphosphate at a concentration of 0.5 mM, and 0.025 U of Taq polymerase per μl. PCR amplification was carried out with a thermal cycler (model PTC-150; MJ Research, Watertown, Mass.) by using the following program: 95°C for 2 min, 30 cycles consisting of 95°C for 1 min, 60°C for 1 min, and 72°C for 2 min, and a final extension step consisting of 72°C for 5 min. After the amplification program was completed, 10 μl from each PCR tube was electrophoresed along with λ-HindIII DNA standards (Gibco/BRL) on a 1.5% (wt/vol) agarose gel at 4 V/cm for 1 to 2 h. The resulting 215-bp PCR product (M. albus BG8 amplicon) was visualized by staining with ethidium bromide. A PCR was considered positive if the expected 215-bp amplicon was detected. When the BG8 amplicon was detected at the same dilution of all three replicate soil or sediment samples, a score of 3 was assigned; likewise, scores of 1 and 2 were assigned to reflect successful amplification from one and two of the samples, respectively.
Influence of bead mill homogenization parameters on DNA extraction.
We examined the influence of bead mill homogenization speed on the DNA yield and fragment size by using the forest soil and two different bead mill homogenizers, the Mini Bead Beater (see above) and a Mini Bead Beater-8 (BioSpec Products). The homogenization speeds used for the Mini Bead Beater were calibrated between 1,600 and 5,000 rpm by using a stroboscope to measure the beating speed. For the trials, the homogenization time was constant (5 min) and extraction procedure 1 (Table 2) with SpinBind purification (as described above) was employed. The DNA yield was quantified as described below. The DNA fragment size distribution was determined by electrophoresis (3 V/cm for 1.5 h) on a 0.8% (wt/vol) agarose gel; the standards used were the λ-HindIII DNA standards described above.
The influence of the duration of bead mill homogenization (1, 2, 5, or 10 min) on DNA extraction efficiency was examined at two different speeds for each bead mill homogenizer (3,300 and 3,800 rpm for the Mini Bead Beater and 2,510 and 2,670 rpm for the Mini Bead Beater-8). In these experiments, extraction procedures 1 and 3 (see above; Table 2) were used in combination with SpinBind purification. The DNA yield was quantified as described below. The DNA fragment size was determined as described above.
DNA quantification.
The DNA in crude and purified extracts was quantified by comparing the fluorescence intensities of ethidium bromide-stained DNA sample bands to the fluorescence intensities of DNA standards on agarose gels by using the method of Zhou et al. (44). This method was modified by performing RNase digestion of the DNA extracts prior to electrophoresis and by including on the agarose gels negative control lanes containing only the loading dye in order to control for horizontal and vertical differences in the observed background densities of the stained gels (26). The DNA band densities on a gel (minus the background value) were measured by using an inverted, scanned, electronically printed image of the gel (42) and the public domain NIH Image program, which was developed at the U. S. National Institutes of Health (40a). A DNA standard curve was obtained for each agarose gel by using the densities (minus the background value) of the five or six smallest DNA fragments in four to six replicate lanes containing 100 to 250 ng of HindIII-digested λ DNA fragments (Gibco/BRL).
Replication and statistical analyses.
All statistical analyses were performed by using Minitab statistical software (Minitab Inc., State College, Pa.). Three or four replicates per DNA extraction method were used for comparative statistical analyses of the nine DNA extraction procedures. The DNA yields were normalized to the yield obtained with extraction procedure 1 for each soil and sediment by dividing the DNA yield obtained with the procedure in question by the DNA yield obtained with procedure 1. For pairwise comparisons of the procedures, two-sample, one-sided t tests were used to determine if one procedure yielded significantly more DNA than another method. To determine the efficiency of DNA recovery after purification, four replicates were used for each purification method for each soil sample type. The percentage of DNA recovered for each sample and procedure was determined by comparing the DNA yield after purification to the DNA yield obtained with no purification.
Optimized DNA extraction-homogenization-purification protocol.
Quadruplicate 100-mg aliquots of freeze-dried agricultural soil, forest soil, or wetland sediment were added to a series of 2-ml screw-cap plastic vials containing 2 g of sterile, 0.1-mm-diameter zirconium-silica beads. Equal volumes (300 μl) of 100 mM NaH2PO4 (pH 8), an SDS lysis mixture (100 mM NaCl, 500 mM Tris [pH 8], 10% [wt/vol] SDS), and chloroform-isoamyl alcohol (24:1) were then added to each of the vials (DNA extraction procedure 3). Bead mill homogenization (2 min at 3,300 rpm with the Mini Bead Beater) was then used to physically disrupt the bacterial cells, and the crude DNA extract was recovered from the vials as described above. After the crude extract volume was reduced to 100 μl, the DNA was purified by using Sephadex G-200 columns and was quantified (see above).
RESULTS
Comparison of extraction procedures.
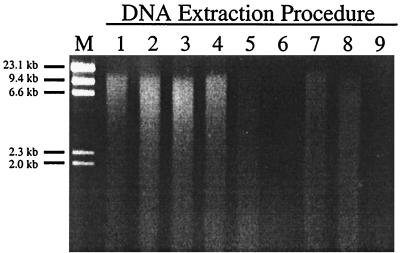
When the nine different DNA extraction procedures (Table 2) were used with freeze-dried samples, all of the procedures except procedures 6 and 9 yielded ethiduim bromide-stained bands in agarose gels (Fig. 1). Although GTC extraction is useful for RNA extraction from soils and sediments (19, 40), it is clearly not suitable for DNA extraction. The size distribution of the DNA fragments (16 to 3.5 kb) obtained with each of the remaining extraction procedures (procedures 1 through 5, 7, and 8) by gel electrophoresis revealed that some limited shearing took place during extraction regardless of the physical disruption method employed. The DNA yields varied considerably from sample to sample and with the extraction procedure used (Table 3). Generally, the DNA yields were greatest with the wetland sediment samples (range, 6 to 53 μg/g [dry weight]), intermediate with the forest soil (4 to 35 μg/g [dry weight]), and smallest with the agricultural soil (1.5 to 7.9 μg/g [dry weight]). For each sample, procedures that included bead mill homogenization and an organic solvent (either phenol or chloroform) along with SDS yielded larger amounts of DNA than procedures that included three freeze-thaw cycles with SDS, enzymatic pretreatment, and the chelator Chelex 100.
FIG. 1.
Comparison of DNA fragment size distributions and relative DNA yields obtained with the nine DNA extraction procedures described in Table 2. Five-microliter portions from three replicate extractions were pooled, digested with RNase, and electrophoresed on a 0.8% (wt/vol) agarose gel. The results obtained for wetland sediment are shown. DNA extracts obtained from forest and agricultural soils produced similar results. Lane M contained 500 ng of λ-HindIII marker.
TABLE 3.
Efficacy of DNA extraction procedures evaluated on the basis of DNA yield and cell lysis
| Procedure | Chemical treatmenta | Physical treatmentb | DNA yield (μg g [dry wt]−1)c
|
% Cell lysis with wetland sediment (mean ± SE)d | ||
|---|---|---|---|---|---|---|
| Agricultural soil | Forest soil | Wetland sediment | ||||
| 1 | SDS | B | 7.0 ± 0.5 | 26 ± 4 | 32 ± 1 | 68 ± 4 |
| 2 | SDS-phenol | B | 7.8 ± 0.2 | 31 ± 5 | 40 ± 4 | 77 ± 5 |
| 3 | SDS-chloroform | B | 7.9 ± 1.2 | 35 ± 7 | 53 ± 8 | 81 ± 4 |
| 4 | SDS-Chelex 100 | B | 6.2 ± 0.3 | 19 ± 5 | 32 ± 2 | 68 ± 7 |
| 5 | Lysozyme-SDS | FT | 1.5 ± 0.7 | 4 ± 3 | 6 ± 1 | 65 ± 4 |
| 6 | GTC | FT | <0.1 | <0.1 | 0.2 ± 0.3 | 64 ± 3 |
| 7 | SDS | FT | 2.7 ± 0.8 | 6 ± 3 | 6 ± 1 | 63 ± 6 |
| 8 | Lysozyme-SDS | B | 1.5 ± 1.2 | 9 ± 4 | 7 ± 4 | 69 ± 2 |
| 9 | GTC | B | <0.1 | <0.1 | <0.1 | 67 ± 4 |
See Table 2, footnotes a and b.
See Table 2, footnote c.
Calculated from the average density of ethidium bromide-stained DNA bands by using NIH Image software. The values are averages ± standard errors based on three or four independent extractions.
Estimated from the DAPI direct counts of bacterial cells per microscope field before and after DNA extraction. The values are average percentages of cells lysed ± standard errors based on three independent DNA extractions performed by using the same DNA extraction procedure.
Analysis of variance (ANOVA) showed that the sample type (soil or sediment) and the extraction procedure employed had significant effects on the DNA yield (P < 0.001 for both). There was also a significant soil-method interaction (P < 0.001), indicating that the effect of the DNA extraction method depended on the soil type tested. In order to compare DNA extraction procedures for the three sample types, it was necessary to normalize DNA yields on the basis of the average DNA yield obtained with one of the DNA extraction procedures (procedure 1 was selected). When this was done, ANOVA showed that the sample type did not have a significant effect on the DNA yield (P = 0.129), while the effect of the extraction method was very significant (P < 0.001).
Significant differences in DNA yields were detected when two-sample, one-sided t tests in which selected chemical and physical treatment parameters (e.g., bead mill homogenization versus three freeze-thaw cycles) were compared to the normalized data (Table 4). Bead mill homogenization was found to be superior to the freeze-thaw technique for obtaining DNA when otherwise identical procedures were compared. Similarly, procedures in which chloroform or phenol was included in the lysis mixture yielded more DNA than comparable procedures in which these organic solvents were not used. However, when a chelating agent (Chelex 100) or lysozyme pretreatment was included in the extraction procedure, the DNA yield decreased significantly (Tables 3 and 4). Procedure 3, which included bead mill homogenization, SDS, and chloroform in the extraction mixture, produced a greater normalized DNA yield than all of the other procedures, as determined by the two-sample t tests (Table 4). Therefore, procedure 3 was the optimum procedure for obtaining DNA in this study.
TABLE 4.
Pairwise statistical comparison of the principal parameters distinguishing the nine DNA extraction procedures
| Extraction variables or procedures | Procedures compareda | Normalized DNA yield (mean ± SE)b
|
Pc | |
|---|---|---|---|---|
| First procedure | Second procedure | |||
| Bead mill vs freeze-thaw lysis | 1 and 7 | 1.00 ± 0.09 | 0.22 ± 0.10 | <0.001 |
| Bead mill vs freeze-thaw lysis | 8 and 5 | 0.26 ± 0.15 | 0.18 ± 0.09 | 0.086 |
| Omission vs inclusion of Chelex 100 | 1 and 4 | 1.00 ± 0.09 | 0.86 ± 0.16 | 0.011 |
| Omission vs inclusion of lysozyme | 1 and 8 | 1.00 ± 0.09 | 0.26 ± 0.15 | <0.001 |
| Omission vs inclusion of lysozyme | 7 and 5 | 0.22 ± 0.10 | 0.18 ± 0.09 | 0.200 |
| Inclusion vs omission of phenol | 2 and 1 | 1.19 ± 0.13 | 1.00 ± 0.09 | <0.001 |
| Inclusion vs omission of chloroform | 3 and 1 | 1.35 ± 0.30 | 1.00 ± 0.09 | 0.001 |
| Procedure 3 vs procedure 2 | 3 and 2 | 1.35 ± 0.30 | 1.19 ± 0.13 | 0.066 |
| Procedure 3 vs procedure 4 | 3 and 4 | 1.35 ± 0.30 | 0.86 ± 0.16 | <0.001 |
| Procedure 3 vs procedure 5 | 3 and 5 | 1.35 ± 0.30 | 0.18 ± 0.09 | <0.001 |
| Procedure 3 vs procedure 7 | 3 and 7 | 1.35 ± 0.30 | 0.22 ± 0.10 | <0.001 |
| Procedure 3 vs procedure 8 | 3 and 8 | 1.35 ± 0.30 | 0.26 ± 0.15 | <0.001 |
See Materials and Methods and Table 2 for details concerning the extraction procedures.
All DNA yields were normalized relative to the DNA yield obtained with procedure 1 (SDS treatment and bead mill homogenization with the Mini Bead Beater at 1,600 rpm).
Calculated probability that the DNA yield from the first procedure in the pairwise comparison is less than or equal to the DNA yield from the second procedure.
Cellular lysis.
The effects of the DNA extraction procedures on cell lysis were examined only with the wetland sediment. The extent of lysis varied between 63 and 81% (Table 3). All of the procedures, even the GTC treatments from which no DNA was recovered, resulted in considerable cellular lysis. ANOVA indicated that the DNA extraction procedure also had a significant effect on cellular lysis (P = 0.003). Two-sample, one-sided t tests confirmed that extraction with an organic solvent (phenol or chloroform) produced significantly greater lysis than extraction without an organic solvent (P < 0.001). Although bead mill homogenization resulted in greater lysis than three freeze-thaw cycles (68% ± 4% versus 63% ± 6%), the effect was not statistically significant (P = 0.17) and did not account for the significantly greater DNA yield. Rather, the vigorous shaking during homogenization probably resulted in the liberation of more DNA from lysed organisms into the extraction mixture instead of the DNA remaining associated with soil particles and cell debris. The results of cellular lysis in the presence of Chelex 100 (procedure 4) were also not statistically different from the results of extraction with no Chelex 100 (procedure 1) (P = 0.48). As expected, there was a strong positive correlation between DNA yield and extent of cellular lysis (r = 0.71) in for the procedures that yielded significant amounts of DNA (procedures 1 through 5, 7, and 8).
Comparison of DNA purification methods.
DNA extraction procedure 4 (SDS-Chelex 100 chemical treatment combined with bead mill homogenization) resulted in the darkest DNA-containing extracts. These extracts were also found to be the most inhibited extracts in the PCR inhibition assay. Indeed, compared to all other DNA extraction procedures, procedure 4 required at least one additional 10-fold dilution to successfully PCR amplify DNA (data not shown). Thus, procedure 4 was used for subsequent tests involving four different DNA purification procedures (Table 5). In these tests the level of DNA recovery was expressed as a percentage of the DNA in the crude extract. The values obtained with the four purification methods were very different (Table 5). ANOVA demonstrated that both sample type and purification method had a significant effect on DNA recovery (P ≤ 0.001 each). When levels of DNA recovery were compared across sample types, the Sephadex G-200 column purification method resulted in the highest levels of DNA recovery (85% ± 8%), followed by the ammonium acetate precipitation method (76% ± 8%), the SpinBind purification method (68% ± 23%), and the gel electrophoresis purification method (29% ± 17%). As determined by one-sided, two-sample t tests, the level of DNA recovery after Sephadex G-200 column purification was found to be significantly greater than the level of recovery after gel electrophoresis purification (P = 0.018). However, the statistical evidence that the Sephadex G-200 column purification method yielded more DNA than the SpinBind and ammonium acetate precipitation methods was not as strong (P = 0.18 and P = 0.13, respectively).
TABLE 5.
Efficacy of DNA purification procedures as evaluated by DNA recovery and inhibition of the PCR
| Purification method | % DNA recovery (mean ± SE)a
|
No. of successful PCR amplifications with the following dilutions of extractb:
|
|||||
|---|---|---|---|---|---|---|---|
| Agricultural soil | Forest soil | Wetland sediment | 100 | 10−1 | 10−2 | 10−3 | |
| No purification | 100 ± 7 | 100 ± 9 | 100 ± 15 | 0 | 0 | 0 | 2f |
| SpinBind column | 83 ± 3 | 80 ± 5 | 41 ± 8 | 0 | 1c | 3 | 3 |
| Gel electrophoresis | 40 ± 12 | 38 ± 12 | 10 ± 1 | 0 | 2d | 3 | 3 |
| Ammonium acetate precipitation | 85 ± 4 | 76 ± 9 | 69 ± 6 | 0 | 0 | 0 | 3 |
| Sephadex G-200 column | 80 ± 7 | 95 ± 6 | 80 ± 8 | 0 | 2e | 3 | 3 |
DNA yield was calculated by using the average density of ethidium bromide-stained DNA bands analyzed with NIH Image software.
DNA from each soil or sediment was diluted, spiked with purified DNA, PCR amplified, and scored on the basis of the presence of a 215-bp BG8 amplicon as described in Materials and Methods.
PCR amplification was successful only from forest soil DNA.
PCR amplification was successful only from forest soil and wetland sediment DNA.
PCR amplification was successful only from agricultural soil and wetland sediment DNA.
PCR amplification was successful only from agricultural soil and forest soil DNA.
For molecular analyses, one of the most important attributes of extracted DNA is its purity. To ensure that a test was sensitive, purity was estimated by using a PCR inhibition assay in which M. albus BG8 genomic DNA (0.5 ng) was added to DNA extracts and used as the template for PCR amplification of the 16S rRNA gene (Table 5). The total DNA concentrations (native DNA plus added DNA) in the PCR mixtures ranged from 0.8 to 1.9 ng/μl for the agricultural soil, from 1.2 to 3.1 ng/μl for the forest soil, and from 0.5 to 5.0 ng/μl for the wetland sediment. The 16S rRNA gene of M. albus BG8 was successfully amplified from unpurified DNA only after 1,000- to 10,000-fold dilution (Table 5). In contrast, the Sephadex G-200 column and gel electrophoresis purification methods allowed PCR amplification to occur at the lowest sample dilutions (10-fold) in two of the three samples and in all of the samples diluted 100-fold (Table 5). Thus, the Sephadex G-200 and electrophoretic purification methods were superior to the other methods for removing PCR-inhibiting substances. However, when both DNA yield and purity were considered, the Sephadex G-200 column procedure was deemed superior on the basis of a higher DNA yield (Table 5).
Optimization of bead mill homogenization parameters.
The effects of two key bead mill homogenization parameters (speed and duration) on DNA extraction were examined by using the forest soil samples. The homogenization devices tested (Mini Bead Beater and Mini Bead Beater-8) were from the same manufacturer (BioSpec Products), but they differed in sample capacity, design, and mechanical specifications. The Mini Bead Beater holds a single 2-ml tube, moves with wrist action through a 1.6-cm shake distance with a soft reversal, and operates at relatively high speeds (up to 5,000 rpm). In contrast, the Mini Bead Beater-8 holds eight 2-ml tubes and moves with piston action through a 3.2-cm shake distance with a sharp reversal; it operates at relatively low speeds (up to 2,700 rpm).
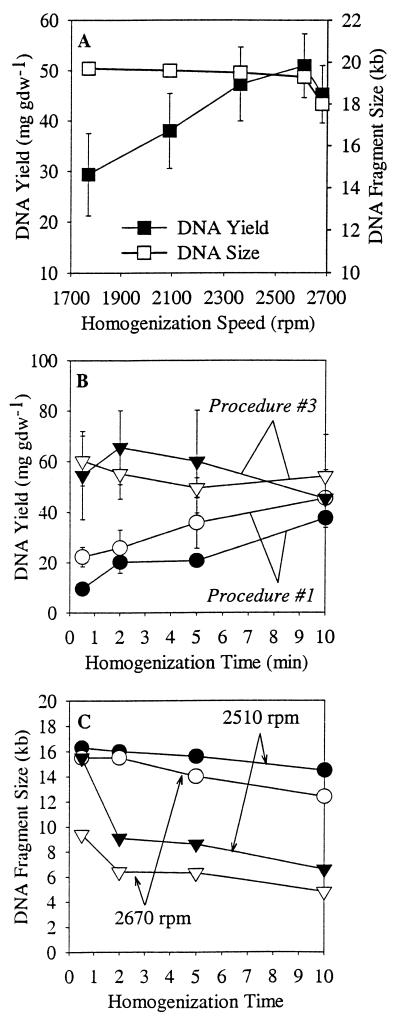
When extraction procedure 1 (Table 2) was used with the Mini Bead Beater, the DNA size decreased dramatically at speeds greater than 3,300 rpm (Fig. 2A). Addition of chloroform (procedure 3) did not influence DNA shearing at the higher speeds (data not shown). Similar DNA shearing was not observed when samples were processed with extraction procedure 1 by using the Mini Bead Beater-8 operated at the maximum speed (2,700 rpm) (Fig. 3A). The size of the largest DNA fragment was generally greater with the Mini Bead Beater-8 over the entire performance range (18 to 20 kb) than with the Mini Bead Beater (8 to 18 kb). With the Mini Bead Beater, the DNA yield was greatest at a medium speed, 3,300 rpm (Fig. 2A), while with the Mini Bead Beater-8, the maximum DNA yield was obtained at speeds ranging from 2,500 to 2,600 rpm (Fig. 3A).
FIG. 2.
Effects of Mini Bead Beater homogenization speed and duration on DNA yield and maximum DNA fragment size. (A) Influence of homogenization speed on DNA yield and fragment size when DNA extraction procedure 1 was used. (B and C) Influence of homogenization time on DNA yield (B) and maximum fragment size (C) at two homogenization speeds (3,300 and 3,800 rpm) when DNA extraction procedure 3 was used. The error bars indicate standard errors (n = 4). gdw, gram (dry weight).
FIG. 3.
Effects of Mini Bead Beater-8 homogenization speed and duration and extraction protocol on DNA yield and maximum DNA fragment size. (A) Influence of homogenization speed on DNA yield and fragment size when DNA extraction procedure 1 was used. (B and C) Influence of homogenization time on DNA yield (B) and maximum fragment size (C) at two homogenization speeds (2,510 rpm [open symbols] and 2,670 rpm [solid symbols]) when DNA extraction procedure 1 (circles) and procedure 3 (triangles) were used. The error bars indicate standard errors (n = 4). gdw, gram (dry weight).
When the duration of homogenization with the Mini Bead Beater during procedure 1 was changed at two critical speeds (3,300 and 3,800 rpm), the size and yield of DNA changed dramatically (Fig. 2B and C). The DNA yield and size at 3,300 rpm were significantly greater than the DNA yield and size at 3,800 rpm, and longer homogenization times resulted in smaller DNA fragments. Again, the presence of chloroform (procedure 3) during homogenization had little effect on DNA size when the Mini Bead Beater was used. When both DNA yield and size were examined, the optimal speed and duration for the Mini Bead Beater were determined to be 3,300 rpm and 120 s, respectively.
Varying the Mini Bead Beater-8 homogenization speed during procedure 3 produced a DNA yield and fragment size pattern that was very different from that obtained with the Mini Bead Beater under the same conditions (Fig. 3B and C). When the homogenization time was varied with procedure 1, the DNA yield increased with longer homogenization times, but the DNA fragment size decreased slightly (from 16 to 13 kb). When chloroform was included in the extraction preparation (procedure 3), the DNA yield was substantially greater (Fig. 3B), but the resulting average DNA fragment size was much smaller (Fig. 3C). With procedure 3 and the Mini Bead Beater-8, there were two optimal settings depending on the needs of the experiment. To recover the greatest amount of DNA, 2,510 rpm for 120 s was used. When high-molecular-weight DNA was more important, the correct settings for speed and duration were 2,510 rpm and 30 s, respectively.
DNA recovery with the optimized extraction-homogenization-purification protocol.
The DNA yields obtained with the agricultural soil, the forest soil, and the wetland sediment when the optimized protocol (corrected for purification losses) was used were 14.7 ± 2.8, 75.6 ± 9.2, and 107.9 ± 2.1 μg/g (dry weight), respectively (Table 6). These yields corresponded to extraction efficiencies of 7.7, 17.6, and 9.8 fg of DNA per bacterial cell, respectively, based on the acridine orange fluorescence microscopic counts shown in Table 1. The levels of DNA recovery from the agricultural soil, forest soil, and wetland sediment, based on an assumed average value of 9 fg of DNA per cell (38), were 86, 195, and 109%, respectively.
TABLE 6.
DNA extraction efficiency of the optimized extraction-homogenization-purification protocol based on DNA yield and acridine orange fluorescent direct counts
| Soil or sedimenta | DNA yield (μg g [dry wt]−1)b | Cellular DNA content (fg cell−1) | Recovery efficiency (%) |
|---|---|---|---|
| Agricultural soil | 14.7 ± 2.8 | 7.7 | 86 |
| Forest soil | 75.6 ± 9.2 | 17.6 | 195 |
| Wetland sediment | 107.9 ± 2.1 | 9.8 | 109 |
See Table 1 for a description of soils and fluorescent bacterial direct counts.
DNA yield based on 85% DNA recovery during Sephadex G-200 purification. Mean ± standard error.
DISCUSSION
Selection of optimal DNA extraction and purification procedures.
Data in Tables 3 through 5 and Fig. 1 through 3 provide criteria for choosing procedures and conditions in order to achieve specific DNA extraction goals. The experimental setup which was used allowed us to statistically evaluate the efficacy of individual elements of DNA extraction and purification procedures, including incorporation of chelators and organic solvents into the lysis mixtures. Our results support the results of previous studies (20, 21, 27, 33) in which higher DNA yields were obtained with bead mill homogenization than with freeze-thaw physical lysis. However, we found that including lysozyme pretreatment, Chelex 100, or GTC had detrimental effects and either decreased the DNA yield (lysozyme and GTC) or increased humic acid recovery (Chelex 100). Including organic solvents (phenol and chloroform) substantially increased the DNA yield (19 and 35%, respectively).
Although all four purification procedures tested had some beneficial effect, the methods differed with respect to both the quantity and the quality of the DNA recovered. Sephadex G-200 column and silica-based SpinBind column purification resulted in the greatest DNA yield and removed more contaminants than either gel electrophoresis or ammonium acetate precipitation. Sephadex G-200 columns were superior to silica-based columns based on both yield and purity, but they required more preparation time and low-speed centrifugation (130 × g) during packing to ensure that the Sephadex G-200 beads remained intact (3). Although not tested in this study, Sepharose 4B spin columns have recently been reported to rival Sephadex G-200 spin columns in terms of both DNA recovery and purification (17). Initial tests in our laboratory showed that humic acids in our extracts were retained slightly longer on the Sepharose 4B columns. A careful comparison of the effectiveness of Sephadex G-200 and Sepharose 4B columns packed at low speeds should be made with each soil or sediment type to determine which purification system is better for each type of sample.
None of the four purification methods tested completely removed all of the contaminants, and additional purification steps involving hexadecyltrimethylammonium bromide (CTAB) and polyvinylpolypyrrolidone either during or after extraction may be required to reduce polysaccharide contamination (25, 28, 44) and humic acid contamination (7, 13, 15, 20, 25, 35, 43, 44), respectively. In a direct comparison, these two compounds yielded equivalent purification efficiencies, but the levels of DNA recovery were much greater with CTAB (44). Initial experiments performed in our laboratory with polyvinylpolypyrrolidone extraction after cell lysis also revealed that there was a significant loss of DNA during purification (unpublished data). Initial extraction of crude extracts with CTAB followed by Sephadex G-200 column purification may result in even greater purification of the extracts than Sephadex G-200 column purification alone. DNA losses and purification efficiencies, however, should be quantified.
Selection of optimal bead mill homogenization conditions.
A major effort was made in this study to optimize bead mill homogenization parameters (speed and duration) in order to obtain large quantities of high-molecular-weight DNA. Previous work has shown that bead mill homogenization may yield either highly sheared DNA or high-molecular-weight DNA (21, 22), and we were interested in reconciling these contradictory findings. We found that shearing occurred during longer homogenization times and at higher speeds. Furthermore, including chloroform in the extraction mixture, particularly when the Mini Bead Beater-8 was employed, also enhanced shearing through an unknown mechanism. Ample quantities of high-molecular-weight DNA, however, could be obtained by using either bead mill homogenizer with or without chloroform by closely monitoring bead mill homogenization speed and duration.
When bead mill homogenization conditions are selected, it is critical to first identify the intended use of the purified DNA. If the DNA is to be cloned, large fragments are required in order to minimize the number of clones that need to be screened. On the other hand, if the DNA is to be used in PCR, DNA fragment size may not be as important as DNA yield (although chimera formation can be a problem). In the case of the Mini Bead Beater, there appeared to be one set of conditions (120 s at 3,300 rpm) that maximized both DNA yield and DNA fragment size when DNA extraction procedure 3 was used (Table 7). In the case of the Mini Bead Beater-8, however, there were two sets of homogenization conditions that either maximized DNA yield or maximized DNA fragment size (Table 7). To maximize the DNA fragment size for cloning, we used bead mill homogenization with the Mini Bead Beater-8 at 2,510 rpm for 30 s and DNA extraction procedure 3. This method produced ample quantities of high-molecular-weight DNA suitable for cloning. If a high DNA yield was the primary concern and smaller DNA fragment sizes (6 to 9 kb) were not detrimental to the subsequent experimental objectives, then bead mill homogenization with the Mini Bead Beater-8 at 2,510 rpm for 120 s combined with DNA extraction procedure 3 was the method of choice. Regardless of the bead mill homogenization method or extraction procedure employed, Sephadex G-200 column purification and SpinBind column (silica-based) purification should be used to remove PCR-inhibitory substances based on a need for high levels of DNA recovery and short purification times, respectively.
TABLE 7.
Optimal bead mill homogenization conditions for obtaining maximum DNA yield and/or maximum DNA fragment size
| Homogenizer | Extraction method | Homogenization conditions
|
DNA characteristicsa
|
Purification method | ||
|---|---|---|---|---|---|---|
| Speed (rpm) | Time (s) | Yield | DNA fragment size | |||
| Mini Bead Beater | Procedure 3 | 3,300 | 120 | Best | Best | Sephadex G-200 |
| Mini Bead Beater-8 | Procedure 3 | 2,510 | 30 | OK | Best | Sephadex G-200 |
| Mini Bead Beater-8 | Procedure 3 | 2,510 | 120 | Best | OK | Sephadex G-200 |
DNA was characterized on the basis of total yield and size of the largest DNA fragments. Best indicates either maximum yield or maximum DNA fragment size compared to all other conditions tested. OK indicates adequate but not maximal DNA yield or fragment size.
Evaluation of the optimized extraction-homogenization-purification protocol.
An estimate of the overall efficiency of obtaining whole-community DNA is one of the most valuable criteria for assessing an extraction technique. Two methods, recovery of DNA from microorganisms added to soil samples and recovery of DNA from a known quantity of indigenous microorganisms, have been used to assess extraction efficiency. We avoided the former method for assessing recovery of DNA from soil microbes for two reasons. Added organisms do not represent the full diversity of indigenous microorganisms (large and small organisms with different capacities for lysis), and the use of such organisms does not account for soil matrix effects (partial to full protection of bacteria associated with the surfaces or interiors of soil particles) that limit cell lysis. We felt that the latter method, comparing DNA recovery to direct counts of indigenous microorganisms by using a DNA-binding fluorochrome, such as DAPI, provided a better comparison of DNA methods. DNA yields and fluorescent direct counts of bacterial cells are only occasionally reported, and the range of reported values is 0.5 to 5.36 fg of DNA per bacterial cell (8, 9, 27, 35, 44). This is equivalent to 6 to 60% DNA recovery based on an assumed average value of 9 fg of DNA per cell (38). When our optimized extraction, homogenization, and purification protocol with the Mini Bead Beater was applied to the agricultural soil, the forest soil, and the wetland sediment, the DNA recovery efficiencies were 86, 195, and 109%, respectively. These high values underscore the effectiveness of the optimized DNA extraction procedure used for freeze-dried samples. These results also show that it is necessary to assess nonbacterial DNA pools. There are several possible explanations for extraction efficiencies greater than 100%. A large number of eukaryotic microorganisms or substantial amounts of high-molecular-weight plant and animal detrital DNA could account for DNA extraction efficiencies greater than 100%. In addition, DNA extraction efficiencies are based on fluorescent direct counts of bacteria in freeze-dried soils and sediments. Initial cell lysis during the freeze-drying process could lead to an incomplete census of microorganisms and falsely inflate DNA extraction efficiencies. We found that DNA yields decreased rapidly for refrigerated samples and decreased slowly over several weeks for frozen samples and that freeze-drying a sample minimized DNA loss. Future work should explore the benefits of the freeze-drying process, which include sample preservation, initial cell lysis, and increased lysis efficiency due to cell wall damage during the freeze-drying process.
Two factors, DNA extraction efficiency and cell lysis, were used to gauge the effectiveness of our optimized DNA extraction and purification procedure. Although the cell lysis and DNA extraction efficiencies were quite high, complete cell lysis during DNA extraction still remains elusive. Like the results of a previous study done in our laboratory (27), fluorescent direct counts showed that very small bacteria persisted inside soil particles after the optimized DNA extraction protocol was used. On the basis of this observation, we concluded that there was still some extraction bias towards larger, more exposed bacterial cells with the optimized DNA extraction and purification protocol. A primary assumption in this work was that greater DNA recovery reflected a more representative (diverse) sample of DNA from the microbial community. While DNA yield is not the best way to estimate diversity, the use of quantitative DNA diversity measures in this study would have been very time-consuming. New tools to rapidly compare the DNA diversities of extracts are needed to better estimate the effectiveness of DNA extraction protocols.
It is important to reiterate the remarkable complexity of soil and sediment types and the fact that there are multiple factors that may affect the performance of a DNA extraction method. Generalizations from the present study may be limited to silt loam soils and sediments having different organic matter contents; however, our results also provide useful guidelines that may be applied to developing protocols for other types of samples as well.
ACKNOWLEDGMENTS
This research was supported by grant ES 05950-03 from the NIEHS/Superfund Basic Research and Education Program.
We are very grateful to David Hinman and Barbara Eaglesham for technical support. We thank BioSpec Products for use of the Mini Bead Beater-8. We especially thank Jim Herrick for his careful review of and insightful comments on an earlier version of the manuscript.
REFERENCES
- 1.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1997. [Google Scholar]
- 2.Beloin R M, Sinclair J L, Ghiorse W C. Distribution and activity of microorganisms in subsurface sediments of a pristine study site in Oklahoma. Microb Ecol. 1988;16:85–97. doi: 10.1007/BF02097407. [DOI] [PubMed] [Google Scholar]
- 3.Boccuzzi V M, Straube W L, Ravel J, Colwell R R, Hill R T. Abstracts of the 95th General Meeting of the American Society for Microbiology 1995. Washington, D.C.: American Society for Microbiology; 1995. Removal of contaminating substances from environmental samples prior to PCR by Sephadex G-200 Spun columns, abstr. N-168; p. 361. [Google Scholar]
- 4.Bruce K D, Horns W D, Hobman J L, Osborn A M, Strike P, Ritchie D A. Amplification of DNA from native populations of soil bacteria by using the polymerase chain reaction. Appl Environ Microbiol. 1992;58:3413–3416. doi: 10.1128/aem.58.10.3413-3416.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brusseau G A, Bulygina E S, Hanson R S. Phylogenetic analysis and development of probes for differentiating methylotrophic bacteria. Appl Environ Microbiol. 1994;60:626–636. doi: 10.1128/aem.60.2.626-636.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryant J E. Detection of Cryptosporidium parvum oocysts in soil using the polymerase chain reaction. M.S. thesis. Ithaca, N.Y: Cornell University; 1995. [Google Scholar]
- 7.DeGrange V, Bardin R. Detection and counting of Nitrobacter populations in soil by PCR. Appl Environ Microbiol. 1995;61:2093–2098. doi: 10.1128/aem.61.6.2093-2098.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erb R W, Wagner-Döbler I. Detection of polychlorinated biphenyl genes in polluted sediments by direct DNA extraction and polymerase chain reaction. Appl Environ Microbiol. 1993;59:4065–4073. doi: 10.1128/aem.59.12.4065-4073.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuhrman J A, Lee S H, Masuchi Y, Davis A A, Wilcox R M. Characterization of marine prokaryotic communities via DNA and RNA. Microb Ecol. 1994;28:133–145. doi: 10.1007/BF00166801. [DOI] [PubMed] [Google Scholar]
- 10.Haldeman D L, Amy P S, Ringelberg D, White D C, Garen R E, Ghiorse W C. Microbial growth and resuscitation alter community structure after perturbation. FEMS Microbiol Lett. 1995;17:27–38. [Google Scholar]
- 11.Herrick J B, Madsen E L, Batt C A, Ghiorse W C. Polymerase chain reaction amplification of naphthalene catabolic and 16S rRNA gene sequences from indigenous sediment bacteria. Appl Environ Microbiol. 1993;59:687–694. doi: 10.1128/aem.59.3.687-694.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrick J B, Miller D N, Madsen E L, Ghiorse W C. Extraction, purification, and amplification of microbial DNA from sediments and soils. In: Burke J F, editor. PCR: essential techniques. New York, N.Y: John Wiley & Sons; 1996. pp. 130–133. [Google Scholar]
- 13.Holben W E, Jansson J K, Chelm B K, Tiedje J M. DNA probe method for the detection of specific microorganisms in the soil bacterial community. Appl Environ Microbiol. 1988;54:703–711. doi: 10.1128/aem.54.3.703-711.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holben W E. Isolation and purification of bacterial DNA from soil. In: Weaver R W, Angle S, Bottomley P, Bezdicek D, Smith S, Tabatabai A, Wollum A, editors. Methods of soil analysis, part 2. Microbiological and biochemical properties. Madison, Wis: Soil Science Society of America, Inc.; 1994. pp. 727–751. [Google Scholar]
- 15.Holben W E. Isolation and purification of bacterial community DNA from environmental samples. In: Hurst C J, Knudsen G R, McInerney M J, Stetzenbach L D, Walter M V, editors. Manual of methods in environmental microbiology. Washington, D.C.: American Society for Microbiology; 1997. pp. 431–436. [Google Scholar]
- 16.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson C R, Harper J P, Willoughby D, Rosen E E, Churchill P F. A simple, efficient method for separation of humic substances and DNA from environmental samples. Appl Environ Microbiol. 1997;63:4993–4995. doi: 10.1128/aem.63.12.4993-4995.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobsen C S, Rasmussen O F. Development and application of a new method to extract bacterial DNA from soil based on separation of bacteria from soil with cation-exchange resin. Appl Environ Microbiol. 1992;58:2458–2462. doi: 10.1128/aem.58.8.2458-2462.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King G M. Associations of methanotrophs with the roots and rhizomes of aquatic vegetation. Appl Environ Microbiol. 1994;60:3220–3227. doi: 10.1128/aem.60.9.3220-3227.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuske C R, Banton K L, Adorada D L, Stark P C, Hill K K, Jackson P J. Small-scale DNA sample preparation method for field PCR detection of microbial cells and spores in soil. Appl Environ Microbiol. 1998;64:2463–2472. doi: 10.1128/aem.64.7.2463-2472.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leff L G, Dana J R, McArthur J V, Shimkets L J. Comparison of methods of DNA extraction from stream sediments. Appl Environ Microbiol. 1995;61:1141–1143. doi: 10.1128/aem.61.3.1141-1143.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liesack W, Stackebrandt E. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J Bacteriol. 1992;174:5072–5078. doi: 10.1128/jb.174.15.5072-5078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madsen E L. Epistemology of environmental microbiology. Environ Sci Technol. 1998;32:429–539. [Google Scholar]
- 24.Madsen E L. A critical analysis of methods for determining the composition and biogeochemical activities of soil microbial communities in situ. Soil Biochem. 1996;9:287–370. [Google Scholar]
- 25.Malik M, Kain J, Pettigrew C, Ogram A. Purification and molecular analysis of microbial DNA from compost. J Microbiol Methods. 1994;20:183–196. [Google Scholar]
- 26.Miller D N. Biogeochemical, microbiological, and molecular insights into the controls of methane emissions from a forested wetland. Ph.D. thesis. Ithaca, N.Y: Cornell University; 1996. [Google Scholar]
- 27.Moré M I, Herrick J B, Silva M C, Ghiorse W C, Madsen E L. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl Environ Microbiol. 1994;60:1572–1580. doi: 10.1128/aem.60.5.1572-1580.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogram A. Isolation of nucleic acids from environmental samples. In: Burlage R S, Atlas R, Stahl D, Geesey G, Sayler G, editors. Techniques in microbial ecology. New York, N.Y: Oxford University Press; 1998. pp. 273–288. [Google Scholar]
- 29.Ogram A, Sayler G S, Barkay T. The extraction and purification of microbial DNA from sediments. J Microbiol Methods. 1987;7:57–66. [Google Scholar]
- 30.Picard C, Ponsonnet C, Paget E, Nesme X, Simonet P. Detection and enumeration of bacteria in soil by direct DNA extraction and polymerase chain reaction. Appl Environ Microbiol. 1992;58:2717–2722. doi: 10.1128/aem.58.9.2717-2722.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rochelle P A, Fry J C, Parkes R J, Weightman A J. DNA extraction for 16S rRNA gene analysis to determine genetic diversity in deep sediment communities. FEMS Microbiol Lett. 1992;100:59–66. doi: 10.1111/j.1574-6968.1992.tb14019.x. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Smalla K, Cresswell N, Mendonca-Hagler L C, Wolters A, van Elsas J D. Rapid DNA extraction protocol from soil for polymerase chain reaction-mediated amplification. J Appl Bacteriol. 1993;74:78–85. [Google Scholar]
- 34.Smith G B, Tiedje J M. Isolation and characterization of a nitrite reductase gene and its use as a probe for denitrifying bacteria. Appl Environ Microbiol. 1992;58:376–384. doi: 10.1128/aem.58.1.376-384.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steffan R J, Goksoyr J, Bej A K, Atlas R M. Recovery of DNA from soils and sediments. Appl Environ Microbiol. 1988;54:2908–2915. doi: 10.1128/aem.54.12.2908-2915.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tebbe C C, Vahjen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl Environ Microbiol. 1993;59:2657–2665. doi: 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trevors J T, Lee H, Cook S. Direct extraction of DNA from soil. Microb Releases. 1992;1:111–115. [Google Scholar]
- 38.Tsai Y, Olson B H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai Y, Olson B H. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl Environ Microbiol. 1992;58:2292–2295. doi: 10.1128/aem.58.7.2292-2295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai Y, Park M J, Olson B H. Rapid method for direct extraction of mRNA from seeded soils. Appl Environ Microbiol. 1991;57:765–768. doi: 10.1128/aem.57.3.765-768.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a.U. S. National Institutes of Health. 1999. [Online.] NIH Image program. National Institutes of Health, Washington, D.C. http://rsb.info.nih.gov/nih-image. [7 September 1999, last date accessed.]
- 41.Volossiouk T, Robb E J, Nazar R N. Direct DNA extraction for PCR-mediated assays of soil organisms. Appl Environ Microbiol. 1995;61:3972–3976. doi: 10.1128/aem.61.11.3972-3976.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winans S C, Rooks M J. Sensitive, economic laboratory photodocumentation using standard video camera and thermal printer. BioTechniques. 1993;14:902. [PubMed] [Google Scholar]
- 43.Young C C, Burghoff R L, Keim L G, Minak-Bernero V, Lute J R, Hinton S M. Polyvinylpyrrolidone-agarose gel electrophoresis purification of polymerase chain reaction-amplifiable DNA soils. Appl Environ Microbiol. 1993;59:1972–1974. doi: 10.1128/aem.59.6.1972-1974.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou J, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]