Abstract
The mouse intestinal helminth Heligmosomoides polygyrus modulates host immune responses by secreting a transforming growth factor (TGF)-β mimic (TGM), to expand the population of Foxp3+ Tregs. TGM comprises five complement control protein (CCP)-like domains, designated D1-D5. Though lacking homology to TGF-β, TGM binds directly to the TGF-β receptors TβRI and TβRII and stimulates the differentiation of naïve T-cells into Tregs. However, the molecular determinants of binding are unclear. Here, we used surface plasmon resonance, isothermal calorimetry, NMR spectroscopy, and mutagenesis to investigate how TGM binds the TGF-β receptors. We demonstrate that binding is modular, with D1-D2 binding to TβRI and D3 binding to TβRII. D1-D2 and D3 were further shown to compete with TGF-β(TβRII)2 and TGF-β for binding to TβRI and TβRII, respectively. The solution structure of TGM-D3 revealed that TGM adopts a CCP-like fold but is also modified to allow the C-terminal strand to diverge, leading to an expansion of the domain and opening potential interaction surfaces. TGM-D3 also incorporates a long structurally ordered hypervariable loop, adding further potential interaction sites. Through NMR shift perturbations and binding studies of TGM-D3 and TβRII variants, TGM-D3 was shown to occupy the same site of TβRII as bound by TGF-β using both a novel interaction surface and the hypervariable loop. These results, together with the identification of other secreted CCP-like proteins with immunomodulatory activity in H. polygyrus, suggest that TGM is part of a larger family of evolutionarily plastic parasite effector molecules that mediate novel interactions with their host.
Keywords: TGF-β, TGF-β mimic, H.polygyrus, parasite, regulatory T cell, immune suppression, nuclear magnetic resonance, isothermal titration calorimetry, surface plasmon resonance, TGM
Abbreviations: CCP, complement control protein; CSP, chemical shift perturbation; HES, H. polygyrus excretory-secretory product; HpARI, H. polygyrus Alarmin Release Inhibitor; HpBARI, H. polygyrus Binds Alarmin Receptor and Inhibits; HSQC, 1H-15N shift correlation; HVL, hypervariable loop; ITC, isothermal titration calorimetry; SPR, surface plasmon resonance; TGF, transforming growth factor; TGM, transforming growth factor-β mimic
Helminth parasites are major human and animal health burdens in tropical regions of the world, with up to two billion infected humans worldwide (1, 2). The widespread association of helminths with mammals, together with the diversity of their lifecycles and niches, reflects an evolutionarily refined ability to manipulate the immune system using multiple molecular strategies (3, 4, 5). Helminth infections are often associated with an upregulation of regulatory T cells (Tregs), either through expansion of the host’s pre-existing Tregs or by inducing de novo differentiation of peripheral T cells into the Treg subset (6, 7, 8). Tregs potently promote immune tolerance by suppressing effector cell function (9), and in parasite-infected animals, they can restrict antiparasite immunity. Infection of mice with the intestinal helminth Heligmosomoides polygyrus increases the population of Tregs, and worm clearance can be induced by antibody-mediated depletion of Tregs (10). In a physiologic context, the pleiotropic cytokine transforming growth factor (TGF)-β can induce differentiation of naïve T cells into Tregs through the defining transcription factor Foxp3 (11, 12, 13). In accord with this, we demonstrated that H. polygyrus excretory-secretory products (HESs) stimulate the differentiation of naïve T cells into Tregs by signaling through the TGF-β receptors, TβRI and TβRII (14). In recent studies, the protein in HES responsible for stimulating the TGF-β pathway and inducing Tregs was identified as a secreted five-domain 420-amino acid protein, designated as TGF-β mimic, or transforming growth factor-β mimic (TGM) (15). TGM induces signaling in both murine (16) and human (17) T cells with an efficacy comparable to TGF-β itself and binds directly to the host TGF-β receptors, TβRI and TβRII, despite bearing no sequence similarity to TGF-β, or any other member of the TGF-β family (15).
TGF-β homodimers are comprised of two 112-amino acid cystine-knotted monomers tethered together by a single interchain disulfide bond. They signal by assembling a heterotetrameric complex with two pairs of two serine/threonine kinase receptors, known as the TGF-β type I and type II receptors, TβRI and TβRII (18, 19, 20). The three TGF-β isoforms, TGF-β1, TGF-β2, and TGF-β3, control a multitude of pathways in cellular differentiation (21, 22, 23) and immune homeostasis (12, 21, 24), and TGF-β-dependent differentiation of naïve CD4+ cells into CD4+ CD25+ Foxp3+ Tregs is essential for peripheral immune tolerance (11, 12). Mice lacking TGF-β1, which is expressed by most cells and tissues, exhibit perinatal mortality and develop multiorgan inflammatory disease and die after maternal TGF-β1 is depleted (21). The dysregulation of the TGF-β pathway has been implicated in the pathogenesis of several human diseases, including inflammatory bowel disease (25), renal and cardiac fibrosis (26, 27), and soft-tissue cancers (26, 28, 29). In the latter setting, TGF-β drives immune exclusion, which promotes cancer progression and can prevent effective checkpoint therapy (30, 31). Thus, TGF-β is a key therapeutic target in its own right (32, 33).
TGM, in contrast to the single-domain structure of TGF-β, is composed of five modular domains, designated D1 – D5, all with distant sequence homology to proteins of the complement control protein (CCP) family (15). CCP domains are approximately 60 to 65 amino acids in length with multiple short β-strands tethered together by two highly conserved disulfide bonds in a CysI-CysIII and CysII-CysIV topology (34). They are usually found in arrays and are present in numerous proteins, including the family of proteins that regulate complement, such as decay accelerating factor, factor H, and complement C3b/C4b receptor 1 (CR1) (34). In H. polygyrus, more than 30 CCP-containing proteins have been identified (35, 36), including in addition to TGM and nine TGM homologs (35), H. polygyrus Alarmin Release Inhibitor (HpARI) and H. polygyrus Binds Alarmin Receptor and Inhibits (HpBARI), which suppress innate and adaptive type II immune responses, by binding IL-33 and its receptor ST2, respectively (37, 38, 39). Similar to TGM, HpARI and HpBARI contain multiple CCP domains (three and two, respectively) and contain large insertions not present in canonical CCP domains (15, 37, 38).
Here, we characterized the individual domains of TGM and investigated the nature of the TGM:TβRI and TGM:TβRII binding interactions, using surface plasmon resonance (SPR), isothermal titration calorimetry (ITC), and NMR. Binding of TGM to the TGF-β receptors was found to be modular in nature, with D1-D2 and D3 binding TβRI and TβRII, respectively. TGM was additionally shown to bind to similar structural motifs on TβRI and TβRII as TGF-β, indicating that TGM truly mimics TGF-β, despite its lack of structural similarity. The solution structure of TGM-D3 was determined and showed that TGM-D3 assumes the overall fold of a CCP domain with two key differences: (1) a loop and a short helix replace two β-strands and (2) a long (23-amino acid) structurally ordered insertion within the hypervariable loop (HVL). These modifications lead to a significant lateral expansion of the domain and create potential interaction surfaces on opposite faces of the protein. Through NMR binding studies, as well as binding studies of TGM-D3 and TβRII variants, TGM-D3 is shown to engage TβRII through one of its two potential interaction surfaces, as well as through the HVL. These new structural data illuminate how H. polygyrus has adapted its own CCP domain–containing proteins for the purpose of protein mimicry and host immunomodulation.
Results
TGM binds to TβRI and TβRII using D1-D2 and D3, respectively
Previous in vitro TGF-β bioassays demonstrated that only TGM domains 1 to 3 were required for induction of CD4+ CD25+ Foxp3+ Tregs from naïve murine T cells or activation of a TGF-β reporter in a mouse embryonic fibroblast cell line (35). Proteins lacking domains 4 and 5 (TGM-D123) retained ability to induce TGF-β signaling, albeit with reduced potency in T-cell assays, while removal of any or all of domains 1 to 3 completely abolished activity. TGM was furthermore shown to require both TβRI and TβRII to elicit TGF-β signaling (15), as TGM activity was inhibited by both SB431542, a TβRI kinase inhibitor (40), and ITD-1, which stimulates ubiquitin-dependent degradation of TβRII (41). Previous SPR measurements demonstrated that TGM binds TβRII with micromolar affinity, similar but weaker than TGF-β1 and -β3, but unlike TGF-β1 and TGF-β3, which only bind TβRI with low nanomolar affinity once bound to TβRII, TGM binds TβRI with low nanomolar affinity in the absence of TβRII (14).
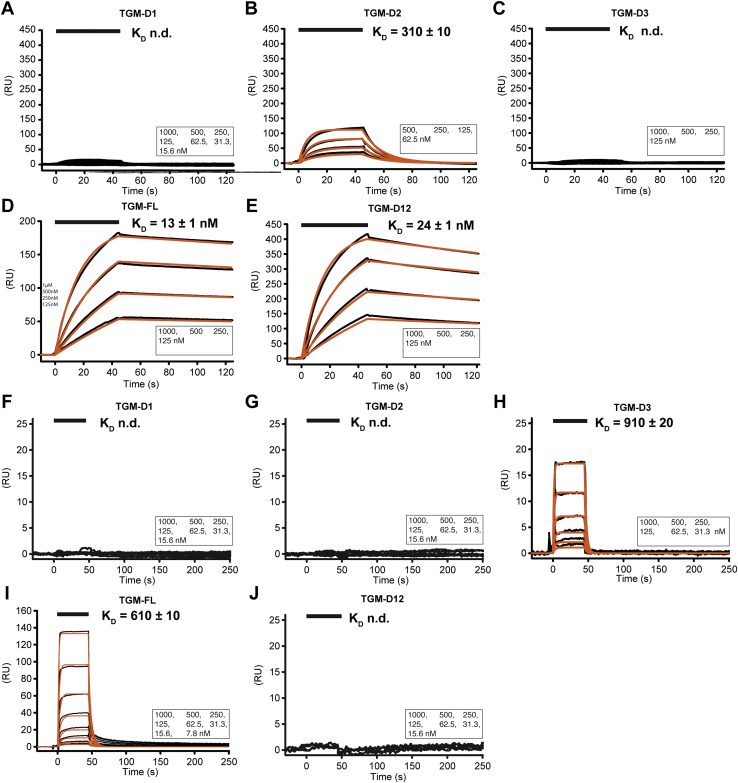
It is unknown which domains of TGM bind to TβRI and TβRII or if TβRI and TβRII directly contact one another, as in the TGF-β receptor complex. To investigate this, the individual domains TGM-D1, TGM-D2, and TGM-D3, along with full-length TGM (TGM-FL), were expressed and purified for SPR binding studies with the TGF-β receptors. The injection of these domains over biotinylated avi-tagged TβRI captured on a streptavidin-coated sensor chip yielded robust concentration-dependent responses when TGM-D2 or TGM-FL was injected, but not when TGM-D1 or TGM-D3 was injected (Fig. 1, A–D). The KD values derived by globally fitting the TGM-D2 and TGM-FL sensorgrams to a (1:1) kinetic model were 310 nM and 13 nM, respectively (Table 1). Thus, TGM-D2 is evidently the main binding partner for TβRI, but nonetheless lacks the full binding capacity of TGM. The same series of injections, performed over biotinylated avi-tagged TβRII captured on a streptavidin-coated sensor chip, yielded robust responses when TGM-D3 or TGM-FL was injected, but not when TGM-D1 or TGM-D2 was injected (Fig. 1,F–I). The KD values derived from the TGM-FL and TGM-D3 sensorgrams were 610 nM and 910 nM, respectively (Table 1). Thus, TGM-D3 accounts for most of the binding affinity of TGM-FL for TβRII.
Figure 1.
Binding of TβRI and TβRII by TGM-D1, TGM-D2, TGM-D3, TGM-D12, and TGM-FL as assessed by SPR.A–J, SPR sensorgrams obtained upon injection of TGM-D1 (A, F), TGM-D2 (B, G), TGM-D3 (C and H), TGM-FL (D and I), or TGM-D12 (E and J) over immobilized TβRI (A–E) or TβRII (F–J). Sensorgrams, obtained upon injections of a 2-fold dilution series of each TGM construct, are shown in black, with the fitted curves in orange (data for TGM-D1:TβRI, TGM-D3:TβRI, TGM-D1:TβRII, TGM-D2:TβRII, and TGM-D12:TβRII were not fit due to weak signal). Black bars shown above the sensorgrams specify the injection period. Injected concentrations are shown in the lower right. SPR, surface plasmon resonance.
Table 1.
TGM:TβRI and TGM:TβRII binding as assessed by SPR
| Surface | Analyte | Fitted parametersa |
|||
|---|---|---|---|---|---|
| kon (M−1 s−1) | koff (s−1) | KD (nM) | Rmax (RU) | ||
| TβRI | TGM-D1 | NDb | NDb | NDb | NDa |
| TβRI | TGM-D2 | (3.0 ± 0.1) × 105 | (9.1 ± 0.1) × 10−2 | 310 ± 10 | 89.6 ± 0.7 |
| TβRI | TGM-D3 | NDb | NDb | NDb | NDa |
| TβRI | TGM-D12 | (6.7 ± 0.1) × 104 | (1.6 ± 0.1) × 10−3 | 24 ± 1 | 429 ± 1 |
| TβRIc | TGM-FL | (5.9 ± 0.1) × 104 | (7.8 ± 0.2) × 10−4 | 13 ± 1 | 193 ± 1 |
| TβRII | TGM D1 | NDb | NDb | NDb | NDa |
| TβRII | TGM D2 | NDb | NDb | NDb | NDa |
| TβRII | TGM D1D2 | NDb | NDb | NDb | NDa |
| TβRII | TGM D3 | (6 ± 1) × 105 | 0.6 ± 0.1 | 910 ± 20 | 33.0 ± 0.4 |
| TβRII | TGM FL | (2 ± 6) × 107 | (1 ± 4) × 10−1 | 610 ± 10 | 215 ± 2 |
Fitted parameters were derived from kinetic analysis of a single injection series.
Not determined due to weak signal.
Measured on a lower density chip compared to that used for TβRI:TGM-D2 and TβRI:TGM-D12.
TGM-D3’s full and TGM-D2’s partial recapitulation of TGM binding affinity for TβRII and TβRI, respectively, suggested that TGM-D1 might contribute to binding of TβRI. Thus, we assessed binding of a construct containing both TGM-D1 and TGM-D2, designated TGM-D12, to TβRI and TβRII using SPR. This didomain construct bound robustly to TβRI, but did not bind at all to TβRII (Fig. 1, E and J). The KD derived from kinetic analysis of the TGM-D12:TβRI sensorgrams was 24 nM, which is within a factor of two of that of TGM-FL (Table 1). Thus, TGM-D1 also contributes to the binding to TβRI.
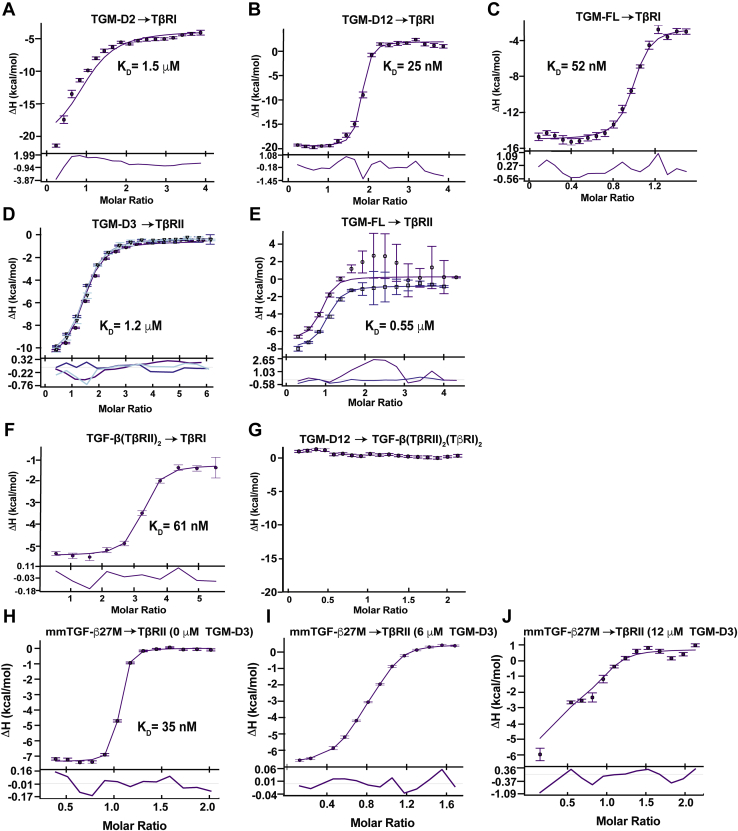
ITC experiments, which in contrast to SPR are carried out entirely in solution and do not require any tagging, were also performed to assess binding of the individual domains of TGM to TβRI and TβRII. In accord with the SPR results, titration of TGM-D2, TGM-D12, and TGM-FL into TβRI and TGM-D3 and TGM-FL into TβRII yielded readily measurable binding isotherms with large negative enthalpies (Figs. 2, A–E and S1, A–E). In contrast, titration of TGM-D1 and TGM-D3 over a similar range of concentrations into TβRI and TGM-D1 and TGM-D2 into TβRII did not (Fig. S1, F–M). In further accord with the SPR results, the fitted KD values for binding of TGM-D12 to TβRI and TGM-D3 to TβRII were comparable to those of TGM-FL and were generally consistent with those measured by SPR (Table S2). In contrast, and as expected based on the SPR results, the KD for binding of TGM-D2 to TβRI was significantly increased (ca. 50-fold) relative to TGM-FL (Table S2). ITC, in addition to providing KD values, also provides values for the stoichiometry, and as shown, TGM-FL binds both TβRI and TβRII with near 1:1 stoichiometry (Table S2). The near 1:1 stoichiometry is also observed for the TGM subdomains shown to bind TβRI and TβRII, TGM-D12 and TGM-D3, respectively, but for TGM-D2 binding to TβRI, the stoichiometry was closer to 0.5. The differing stoichiometry for binding of TGM-D2 and TGM-D12 to TβRI is likely due the weaker affinity of the former interaction, which makes accurate data fitting difficult. Thus, as discussed in a following section, an alternative method was used and this established 1:1 stoichiometry for the TGM-D2 to TβRI interaction.
Figure 2.
TGM binding to TβRI and TβRII and competition with TGF-β by ITC.A–E, integrated heats for the injection of TGM-D2 (A), TGM-D12 (B), or TGM-FL (C) into TβRI, and TGM-D3 (D) or TGM-FL (E) into TβRII, together with the fit (smooth line) and residuals (below) to a 1:1 binding model. Error bars indicate bias in the NITPIC estimation of the integrated heats. F and G, TGM-D12 and TGF-β(TβRII)2 competitive binding to TβRI. Integrated heats obtained upon injection of TGF-β(TβRII)2 (F) into TβRI or TGM-D12 into TβRI with a saturating concentration of TGF-β(TβRII)2 binary complex (G). H–J, TGM-D3 and mmTGF-β27Μ competitive binding to TβRII. Integrated heat obtained upon injection of 150 μM mmTGF-β27M into 15 μM TβRII in the sample cell with 0 μM (H), 6.0 μM (I), or 12.0 μM (J) TGM-D3. The data points correspond to the integrated heats and the smooth lines a global fit over the three datasets to a 1:1 binding model with competition. ITC, isothermal titration calorimetry.
TGM competes with TGF-β for binding to TβRI and TβRII
To assess potential shared binding sites on TβRI and TβRII, ITC competition experiments were performed in which KDs and enthalpies for TβRI and TβRII binding to their partners were measured under noncompetitive and competitive conditions. In the case of TβRI, titration of the TGF-β3(TβRII)2 complex into TβRI yielded a fitted KD of 61 nM (Fig. 2F and Table S3), which is similar to the KD of 25 nM when TGM-D12 was titrated into TβRI (Fig. 2B and Table S2). However, unlike TGM-D12:TβRI binding which had a large negative enthalpy, −19 kcal mol−1 (Table S2), binding of TGF-β3(TβRII)2 to TβRI had a much smaller negative enthalpy, −4.2 kcal mol−1, even at an increased temperature (Table S3). In light of similar KDs, but significantly different enthalpies, the competition experiment with TβRI was performed by titrating TGM-D12 into the cell loaded with the TGF-β3(TβRII)2(TβRI)2 ternary complex (Fig. 2G). This yielded no heat, indicating that TGM-D12 and TGF-β3(TβRII)2 compete for binding to TβRI.
TGF-β3, and TGF-β homodimers in general, is well known to be practically insoluble in the unbound form, except under either very acidic (pH 4.0) or basic (pH 11.0) conditions (42). Therefore, competition experiments in which TβRII is titrated into TGM (or TGM-D3), together with TGF-β3 as a competitor, are not feasible. To circumvent this, mmTGF-β27M, an engineered TGF-β monomer, which is soluble at neutral pH and binds TβRII in a manner indistinguishable from TGF-β3, was used (43). In the absence of competitor, titration of mmTGF-β27M into TβRII yielded a large negative enthalpy and a sharp binding transition, consistent with a low nanomolar binding affinity as previously reported (Fig. 2H) (43). In the presence of increasing concentrations of competitor, either 6 or 12 μM TGM-D3, there was a readily detectable increase in the curvature, consistent with competitive binding (Fig. 2,I and J). In order to analyze the data, the integrated heats from the three experiments, together with fitted KD and enthalpy for the TGM-D3:TβRII interaction (Table S2), were globally fit to a simple competitive binding model to derive the KD for high affinity mmTGF-β27M:TβRII binding (Fig. 2, H–J and Table S3). The KD was found to be 35 nM, in accord with previous SPR measurements for the TβRII:TGF-β interaction with immobilized TGF-β1 or TGF-β3 (43). This demonstrates that TGM-D3 and mmTGF-β27M, and by logical extension TGM and TGF-β homodimers, compete for binding to TβRII.
TGM binds TβRI with high affinity due to direct binding of both TGM-D1 and TGM-D2
The SPR and ITC experiments have shown that TGM-D12 recapitulates nearly the full-binding affinity of TGM-FL to TβRI, while TGM-D2 alone is 20 to 50 fold weaker. In spite of the apparent contribution of TGM-D1, its direct binding to TβRI was not detected using either SPR or ITC. In order to investigate the possibility that TGM-D1 does directly bind TβRI, but too weakly to be detected within the range of affinities possible by SPR or ITC, we prepared 15N-labeled TGM-D1, as well as 15N-TGM-D2 and 15N-TGM-D3, and examined binding to TβRI using NMR spectroscopy.
The two-dimensional (2D) 1H-15N shift correlation (HSQC) spectra of both TGM-D2 and TGM-D3 were both well dispersed, with numerous peaks outside of the random coil limit (7.8–8.5 ppm in the 1H dimension), demonstrating that these proteins are natively folded (Fig. S2, A and C). The number of backbone amide signals for TGM-D3 was close to the number expected (77 observed, 81 expected), while for TGM-D2, the total number of signals exceeded that expected (106 observed, 76 expected). To determine if the additional signals in TGM-D2 were due to sample heterogeneity, for example, as a result of slow conformational dynamics, HSQC ZZ-exchange spectra with mixing times ranging between 0 to 250 ms were recorded (44). These experiments identified at least 12 pairs of peaks undergoing exchange on this timescale, indicating that the protein is undergoing a slow conformational transition that leads to two forms in solution (Fig. S2, A and B). The process responsible was not investigated but might be proline cis:trans isomerization, as this is known to occur on slow timescales (45) and TGM-D2 has four additional proline residues relative to TGM-D3 (Table S1).
TGM-D1, in contrast to TGM-D2 and TGM-D3, had poor signal dispersion, with most peaks clustered in the random coil region of the spectrum (Fig. S3A). To investigate the possibility that TGM-D1 was natively folded, but aggregated, CHAPS in increasing concentrations was added to the buffer and the protein concentration was decreased. This led to the appearance of a large number of peaks outside of the random coil region (Fig. S3, B–D). The spectrum with 20 μM TGM-D1 and 10 mM CHAPS in the buffer had roughly the expected number of peaks (46) but also a few intense peaks in the random coil region of the spectrum. Thus, TGM-D1 appears to be natively folded, but perhaps still partially aggregated under these conditions.
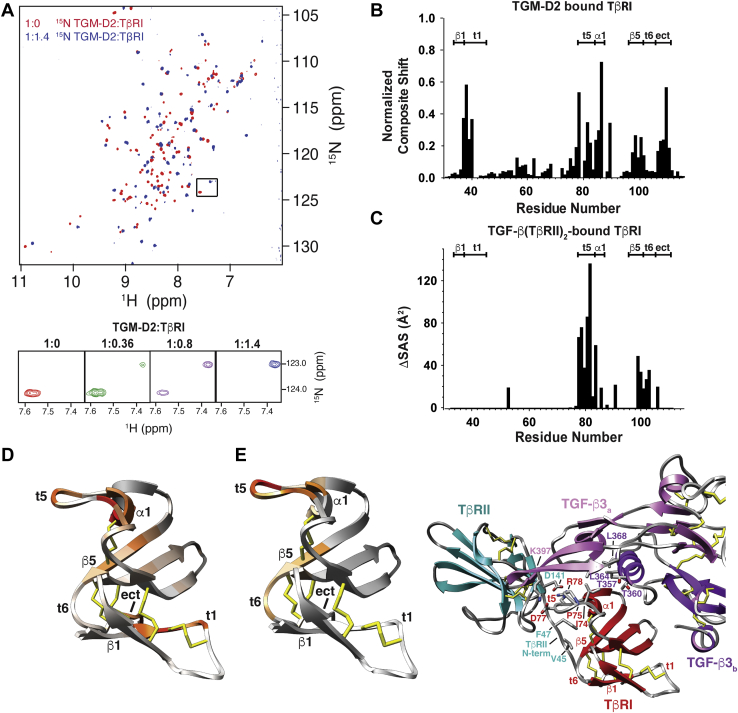
To assess binding, 15N-labeled TGM-D1, TGM-D2, or TGM-D3 was combined with increasing amounts of unlabeled TβRI, ranging from 0 to 1.4 equivalents. This resulted in significant perturbations in the backbone amide signals of TGM-D2 (Fig. 3A), but not those of either TGM-D1 or TGM-D3 (Fig. S4, A and B), consistent with the SPR and ITC results. The signals of 15N-TGM-D2 underwent slow-exchange conversion from the free to the bound form as increasing amounts of TβRI were added, but were not fully converted to the bound form until more than 0.8 equivalents of TβRI were added (Fig. 3A). Thus, TGM-D2 appears to bind TβRI with 1:1 stoichiometry, not 0.5:1 as suggested by the ITC titration. The binding of TβRI was further shown to resolve the conformational doubling apparent in TGM-D2 (Fig. S4, C and D), indicating that binding stabilizes TGM-D2 in one of its two forms.
Figure 3.
Binding of TGM-D2 to TβRI.A, 1H-15N HSQC spectra of 0.2 mM 15N TGM-D2 alone (red) overlaid with the spectrum of the same sample, but with 1.2 M equivalents of unlabeled TβRI added (blue). Spectra were recorded in 25 mM sodium phosphate, 50 mM sodium chloride, and 5% 2H2O, pH 7.0, at 310 K. Expansion of the boxed region with intermediate titration points is shown below. B and D, plot of the composite shift perturbations of TβRI upon binding to TGM-D2 (B) and a depiction of these on the structure of TβRI from PDB 2PJY (D). Structure is colored using a scale where white indicates minimal composite shift perturbation and red indicates maximal. C and E, plot of the difference in solvent-accessible surface area for individual residues of TβRI between the free and bound form (PDB 2PJY) (C) and a depiction of these on the structure of TβRI from PDB 2PJY (E, left). Structure is colored using a scale where gray indicates minimal SAS and red indicates maximal SAS. Shown also in (E) (right) is the structure of one side of the TGF-β3(TβRII)2(TβRI)2 complex, with the two monomers of TGF-β3 depicted in pink and magenta, TβRII in cyan, and TβRI in red. Key residues at the interface between TGF-β3:TβRII and TβRI are shown. HSQC, 1H-15N shift correlation.
The spectrum of 15N TGM-D1 with 1.4 equivalents of unlabeled TβRI added was recorded with 10 mM CHAPS in the buffer. This might impede binding, and thus, a role of TGM-D1 in binding TβRI cannot be excluded. Thus, the converse experiment was performed, with 15N-labeled TβRI combined with 1.2 M equivalents of unlabeled TGM-D1, TGM-D2, or TGM-D3, all in buffers lacking CHAPS. The addition of TGM-D2 caused large perturbations in most of the signals of TβRI, whereas addition of TGM-D3 led to no perturbations, consistent with the inverse experiments (Fig. S5, A and B). The addition of TGM-D1 in the absence of CHAPS resulted in the weakening or full disappearance of most of the TβRI backbone signals, along with small chemical shift perturbations of other signals (Fig. S5C). The disappearance of these signals is likely due to 15N-TβRI binding TGM-D1 and being incorporated into a TGM-D1 aggregate. Thus, TGM-D1 does appear to bind TβRI and the high affinity of TGM-FL for TβRI is likely a result of multivalent binding, in which TGM-D1 and TGM-D2 both directly bind TβRI.
TGM-D2 and TGF-β:TβRII bind a similar set of residues on TβRI
The ITC competition experiments clearly demonstrated that TGM-D12 and TGF-β3(TβRII)2 complex compete with one another for binding TβRI, suggesting that TGM-D12 recognizes and binds a set of residues that partially or fully overlap with that bound by TGF-β3(TβRII)2. To further investigate, we prepared a sample of 13C,15N TβRI bound to a slight excess of unlabeled TGM-D2 (as the complex with TGM-D12 proved to be intractable) and assigned the backbone HN, N, C, CO, and Cβ resonances for all nonproline residues, except for Cys41-Thr42, Ser69-Cys71, Ala87, and Ser90-Thr92 (Fig. S6B). To identify potential interface residues, the assigned chemical shifts for TGM-D2-bound TβRI were compared to those previously reported for unbound TβRI under similar buffer conditions (Figs. 3B and S6A) (18). The largest chemical shift perturbations (CSPs) fell within three regions. The first is the C-terminal end of β1 and the turn that follows (t1), amino acids 32 to 40 (Fig. 3, B and D). This region of TβRI does not interact at all with TGF-β(TβRII)2 (Fig. 3, C and E). The second, turn 5 (t5), also known as the Pro-Arg-Asp-Arg-Pro (PRDRP) prehelix extension, and the short 1 turn helix that follows (α1) (Fig. 3, B and D), is the contact between TGF-β and TβRII and residues 78 to 87 of TβRI (Fig. 3, C and E), while the third is β-strand 5 and the following extended C-terminus (Fig. 3, B and D) which is the interface between the structurally ordered N-terminal tail of TβRII and residues 97 to 110 of TβRI (Fig. 3, C and E). Thus, one domain of TGM, D2, has evolved not only to replicate the binding properties of two host proteins (TGFβ and TβRII) but also to form a third novel site that may confer its overall higher affinity for the receptor.
TGM-D3 and TGF-β bind a similar set of residues on TβRII
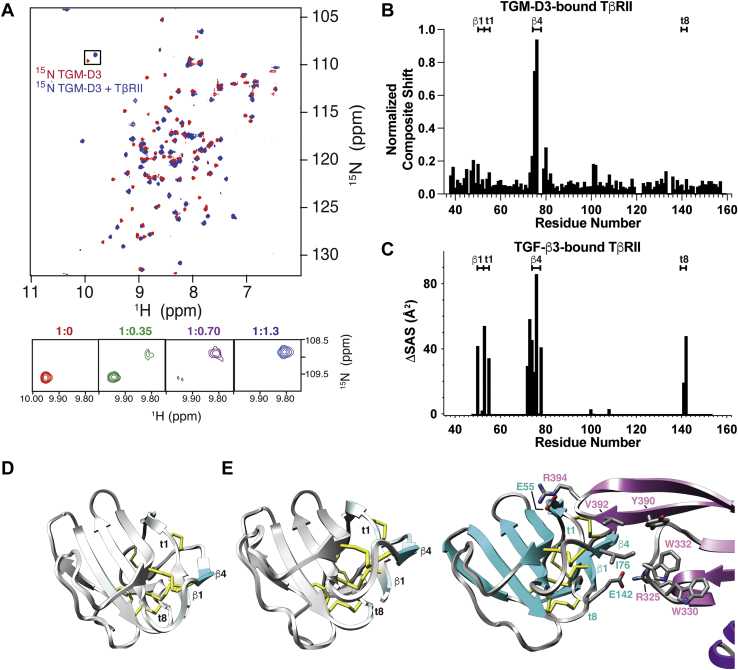
The binding of 15N TGM-D1, TGM-D2, and TGM-D3 by unlabeled TβRII and 15N TβRII by unlabeled TGM-D1, TGM-D2, and TGM-D3 was also investigated using NMR. This revealed multiple perturbations in 15N TGM-D3 signals, but none with 15N TGM-D1 or 15N TGM-D2 when unlabeled TβRII was added (Figs. 4A and S7, A and B); similarly, many of the signals of 15N TβRII were perturbed by TGM-D3, but not by TGM-D1 or TGM-D2 (Fig. S7, C–E). These results, in addition to being internally consistent, also conformed to the overall conclusions derived from the earlier analyses by SPR and ITC.
Figure 4.
Binding of TGM-D3 to TβRII.A, 1H-15N HSQC spectra of 0.2 mM 15N TGM-D3 alone (red) overlaid with the spectrum of the same sample, but with 1.2 M equivalents of unlabeled TβRII added (blue). Spectra were recorded in 25 mM sodium phosphate, 50 mM sodium chloride, and 5% 2H2O, pH 6.0, at 310 K. Expansion of the boxed region with intermediate titration points is shown below. B and D, plot of the composite shift perturbations of TβRII upon binding to TGM-D3 (B) and a depiction of these on the structure of TβRII from PDB 1KTZ (D). Structure is colored using a scale where white indicates minimal composite shift perturbation and cyan indicates maximal. C and E, plot of the difference in solvent accessible surface area for individual residues of TβRII between the free and bound form (PDB 1KTZ) (C) and a depiction of these on the structure of TβRI from PDB 2PJY (E, left). Structure is colored using a scale where gray indicates minimal SAS and cyan indicates maximal SAS. Shown also in panel E (right) is the structure of one side of the TGF-β3(TβRII)2 complex, with the two monomers of TGF-β3 depicted in pink and magenta and TβRII in cyan. Key residues at the interface between TGF-β3 and TβRII are shown. HSQC, 1H-15N shift correlation.
To identify specific residues of TβRII that are recognized and bound by TGM-D3, the backbone signals of 15N,13C TβRII bound to unlabeled TGM-D3 were assigned (Fig. S8B) and compared to those previously reported for the unbound form (Fig. S8A) (47). The largest chemical shift perturbations, as deduced from a composite of the HN, N, C, Cβ, and CO resonances, fell within a narrow region from residues 75 to 77 (Fig. 4, B and D). This region corresponds closely with the primary region of TβRII that binds TGF-β (Fig. 4, C and E left), demonstrating that the same motif of TβRII, the β4 edge strand that binds deeply in the cleft between the fingers 1 to 2 and 3 to 4 (47, 48) of TGF-β, is also engaged by TGM-D3. The binding of TGM-D3 leads to only minor shift perturbations outside of TβRII β4 (Fig. 4B), whereas TGF-β3 also directly engages residues 50 to 55 and 141 to 142 (Fig. 4, C and E right). Thus, while these might still be contacted by TGM-D3, as suggested by small composite shift perturbations in these regions (Fig. 4B), it appears that these contacts may not be as intimate as those with TGF-β.
TGM-D3 structure and dynamics
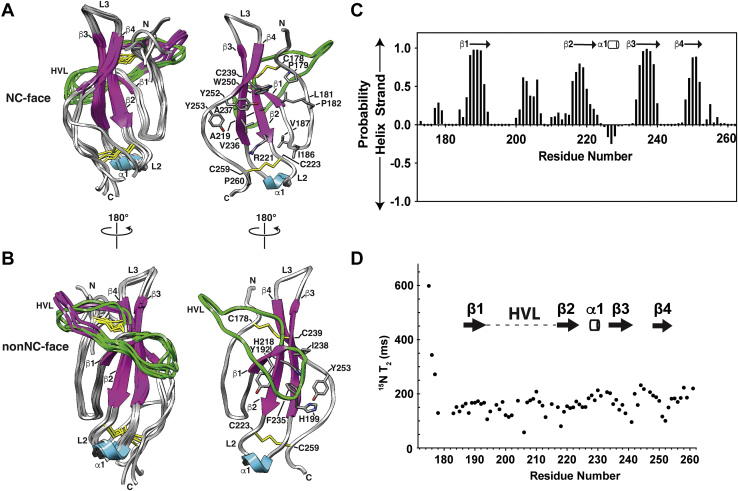
The structure of TGM-D3 was determined based on near-complete chemical shift assignments for both the backbone and side chains, 1H-1H NOE distance restraints, 1H-15N, 13Cα-1Hα, and 13CO-15N RDCs, and 3JHN-Hα J-couplings, with relevant statistics presented in Table S4. TGM-D3 is comprised of four β-strands (Tyr189-Gly193, Thr217-Arg221, Glu234-Lys241, and Ser248-Tyr252) arranged into a highly twisted antiparallel β-sheet with a β1:β2:β3:β4 topology (Fig. 5, A and B). The first β-strand is present in some but not all of the lowest-energy structures. There is also a 310 helix (Gln228-Ala230) connecting β2 and β3 in some, but not all of the lowest-energy structures (Fig. 5, A and B). The structures are consistent with a PECAN analysis of secondary shifts (49), with four high probability extended regions predicted between residues 184 to 191, 216 to 222, 234 to 241, and 248 to 252, and a low probability helical region from residues 226 to 228 (Fig. 5C). The secondary shifts also predict, with lower probability, extended regions between residues 177 to 179 and 201 to 206. The former corresponds to the N-terminal region (Fig. 5A), while the latter corresponds to the middle section of the 23-residue HVL loop that connects β1 and β2 (Fig. 5B). This section of the HVL extends perpendicularly across the C-terminal end of β1 and packs on its N-terminal end against several bulky hydrophobic residues that emanate from the surface of the twisted sheet, including Tyr192, His218, Ile238, and Phe235 (Fig. 5B). The HVL is mostly converged among the ten lowest energy structures, with an average backbone pairwise RMSD of 1.74 Å. The segments from residues 177 to 179 and 201 to 206, although highly extended, do not form hydrogen bonds that define a β-strand and thus are not classified as such in the calculated structures.
Figure 5.
Structure and backbone dynamics of TGM-D3.A and B, shown on the left are an ensemble of the five lowest-energy NMR structures of the unbound form of TGM-D3: β-strands, magenta; loops, gray; 310 helix, cyan; disulfide bonds, yellow, two conformations of HVL highlighted in green and pink. Key structural features are indicated. Orientations shown differ by a 180-degree rotation around the y-axis, with orientation shown in (A) highlighting the face of the protein that includes N- and C-terminus (NC-face) and the orientation shown in (B) highlighting the opposite face (non-NC-face). Shown on the right are single representative structures, with the four cysteines that form the two disulfide bonds and the side chains of key residues highlighted. C, PECAN-based prediction of TGM-D3 secondary structure. Positive values indicate β-strand probability; negative values indicate helical probability. Spectra recorded in 25 mM sodium phosphate, 50 mM sodium chloride, and 5% 2H2O, pH 6.0, at 310 K. Secondary structure elements shown above correspond to those deduced from the calculated TGM-D3 solution structure. D, backbone 15N T2 relaxation times for TGM-D3 plotted per individual residue with structural features mapped. HVL, hypervariable loop.
The Cys178-Cys239 disulfide pins the N-terminus to one end of the β-sheet, while the C-terminus is pinned to the other end by the Cys223-Cys259 disulfide (Fig. 5, A and B). The core of the protein is localized in the region circumscribed by the extended N-terminal segment on one side and β4 and the extended segment that follows on the other side (Fig. 5A). The hydrophobic residues in the core include Leu181 and Pro182 from the extended N-terminal segment, Ile186, Val187, and Tyr189 from β1, Ala219 and the hydrophobic portion of the side chain of Arg221 from β2, Val236 and Ala237 from β3, and Trp250 and Tyr252 from β4 (Fig. 5A).
The backbone 15N T2 relaxation times, which are sensitive to fast (ns-ps) timescale motions that result from low-amplitude fluctuations of the backbone, are significantly increased in the N-terminal tail and modestly increased near the C-terminal end of the HVL and in the shorter loops connecting β2-β3 and β3-β4 (Fig. 5D). The increases in 15N T2 indicate increased flexibility in these regions, especially the N-terminal tail which does not converge in the final ensemble of structures. The other loop regions converge reasonably well, consistent with their more modest increases in 15N T2 (Fig. 5D), although one exception is the HVL, which adopts two conformations, in which the C-terminal portion of the HVL either ascends or descends as it contacts the extended N-terminus (Fig. 5B, green and pink, respectively). There is a low percentage of Ramachandran outliers in the structure, but these are present in regions that are completely or partially unstructured, including the N- and C-termini and the C-terminal portion of the HVL.
TGM-D3 is a remodeled CCP domain with a lateral expansion to expose hydrophobic sites
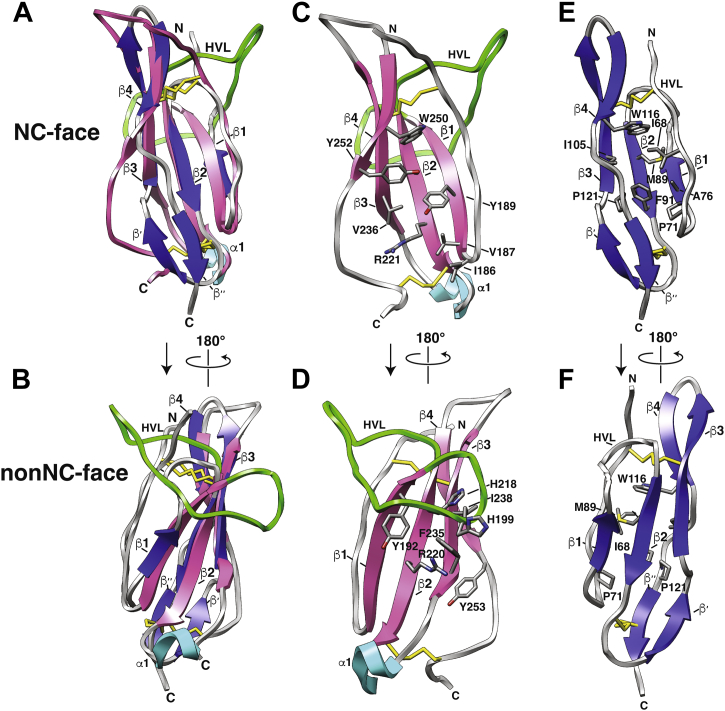
Structures with the closest similarity to TGM-D3, as identified by a DALI (50, 51) search of the Protein Data Bank, were all CCP-containing proteins, as anticipated based on previous bioinformatic analyses (15). Structural overlays show that the top hit, 1CKL (human CD46), as well as other top hits 2PSM (IL-15Rα), 1H2P (CD55), 5FO9 (CR1), and 5FOA (decay accelerating factor), have close correspondence of the four β-strands and the CysI -CysIII and CysII -CysIV disulfides that form the core of the TGM-D3 fold (Fig. 6, A and B). However, in spite of the considerably longer length of TGM-D3 than that of the top-scoring CCP domains (90 and 65–75 residues, respectively), TGM-D3 lacks two short β-strands, one in the loop connecting β2 and β3, designated β′, and another at the C-terminus, designated β’’, present in all of the top-scoring CCP domains (Fig. 6, C–F, respectively). In conventional CCP domains, the pairing of the β′ and β’’ strands, together with the II-IV disulfide that bridges the β’’ strand to the C-terminal end of β2, serves to draw the C-terminal segment toward the loop connecting β2-β3, thereby creating a closed cavity that is packed with hydrophobic residues (Fig. 6, E and F). In TGM-D3, there is a significant lateral expansion of the domain due to the absence of the β′ and β’’ strands, which leads to a pronounced divergence of the extended segment that follows β4 away from the structurally ordered N-terminal segment (Fig. 6D). This lateral expansion is evident whether the protein is viewed from the face that includes the N- and C-terminus, designated as the NC-face (Fig. 6C), or the opposite face, designated as the non-NC-face, that includes the extended HVL (Fig. 6D). The expansion on the NC-face of the protein leaves several hydrophobic residues partly exposed to solvent, including Ile186, Val187, and Tyr252 (Fig. 6C). The expansion on the non-NC-face also leads to the partial exposure of several hydrophobic residues, including Tyr253 in the extended segment following β4 and Tyr192 and Phe235 which form part of the surface against which the HVL packs (Fig. 6D). Overall, the remodeling of TGM-D3 leads to a considerable lateral expansion of the domain and creates potential interaction surfaces on both the NC- and non-NC-faces for binding to TβRII.
Figure 6.
TGM-D3 comparison to CCP domains.A and B, alignment of TGM-D3 to representative CCP domain, PDB 1CKL (blue), as viewed from the NC-face (A) or non-NC-face (B). TGM-D3: β-strands, magenta; loops, gray; 310 helix, cyan, HVL, green; PDB 1CKL: β-strands, blue; loops, including HVL, gray. Key structural features are indicated. C and D, TGM-D3 in the same orientation as shown in A and B, respectively. Side chains of key residues on both the NC- and non-NC-faces are highlighted. E and F, PDB 1CKL in the same orientation as shown in A and B, respectively. Side chains of residues in the hydrophobic core are highlighted. CCP, complement control protein; HVL, hypervariable loop.
TGM-D3 engages its partner, TβRII, using structural motifs unique to TGM-D3
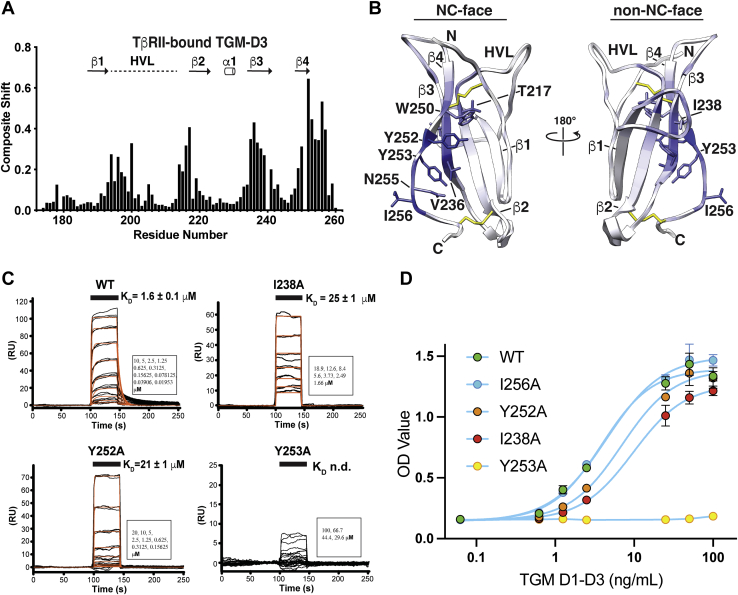
To identify the binding interface on TGM-D3 for TβRII, the backbone of 15N,13C TGM-D3 was fully assigned as bound to unlabeled TβRII (Fig. S9B). This enabled differences in the assigned chemical shifts to be computed relative to the free form (Figs. 7A and S9A). These differences showed that the regions of TGM-D3 most strongly perturbed upon binding TβRII ranged from residues 234 to 243 and 249 to 257, which correspond to most of β3 and β4, as well as a few residues that extend beyond the end of β4 (Fig. 7, A and B). The regions perturbed to a lesser extent include residues 214 to 219 and 193 to 200, which correspond to the N-terminal end of β2 and the N-terminal end of the HVL. The residues maximally perturbed on β3 and β4 are present on the NC-face and non-NC-face of TGM-D3 and include Tyr252 and Val236 and Ile238, Tyr253, and Ile256, respectively (Fig. 7B).
Figure 7.
Binding of TβRII to TGM-D3.A and B, composite shift perturbations of TGM-D3 upon binding to TβRII (A) and a depiction of these on the structure of TβRII (B). Secondary structure shown above the composite shifts in A corresponds to the secondary structure as deduced from the TGM-D3 solution structure. Structure in B is colored using a scale where white indicates minimal composite shift perturbation and dark blue indicates maximal shift perturbation. C, binding of TβRII by TGM-D3 variants as assessed by SPR. SPR sensorgrams obtained upon injection of WT, I238A, Y252A, or Y253A TGM-D3 over immobilized TβRII. Sensorgrams, obtained upon injection of a 2-fold dilution series of each TGM construct, are shown in black, with the fitted curves in orange (data for Y253A were not fit due to weak signal). Black bars shown above the sensorgrams specify the injection period. Injected concentrations are shown in the lower right. D, impact of TGM-D3 mutations on TGF-β signaling as measured through the MFB-F11 TGF-β responsive bioassay. I256A (blue), Y252A (orange), I238A (red), and Y253A (yellow) were assessed for TGF-β signaling and compared to WT TGM1-D13 (green). SPR, surface plasmon resonance.
To determine whether TβRII binds to the NC- or non-NC-face and to determine which residues contribute greatest to binding, we substituted residues of TGM-D3 within the NC- and non-NC-faces that could potentially interact with TβRII with alanine and assessed TβRII binding using SPR. The specific residues chosen for substitution included Val236, Tyr252, and Asn255 on the NC-face and Arg198, His199, Phe235, Ile238, Tyr253, and Ile256 on the non-NC-face. Lys254 and Lys258 in the extended region following β-strand 4 were also substituted. The variants were screened for native folding by recording the one-dimensional (1D) 1H NMR spectrum. Though some had small shifts in some of the resolved methyl and amide signals, all were found to be natively folded and none presented any evidence of gross folding abnormalities (Fig. S10).
The SPR response amplitudes were attenuated for several of the variants, including R198A, F235A, V236A, I238A, Y252A, and Y253A, indicating that the binding affinity was diminished (Figs. 7C and S11, A–H). The responses were nonetheless sufficient to obtain satisfactory fits for all of the variants, except Y253A, which was severely diminished (Figs. 7C and S11, A–H). The KD values for the R198A, I238A, Y252A, and K254A variants were increased by more than 20-fold, while for the F235A and V236A variants, more modest increases of about 4-fold were observed (Table S5). The side chains of Arg198, Lys254, Tyr253, and Ile238 are all on the non-NC-face of the protein, while that of Tyr252, immediately adjacent to Tyr253, is located on the NC-face. The other residues located nearby Tyr253 that underwent large backbone CSPs, Asn255, and Ile256 resulted in little to no attenuation of binding upon substitution with alanine. Tentatively, this suggests the binding site for TβRII resides on the non-NC-face of the protein in the region that is formed by residues protruding from β3, the C-terminal end of β4, and the N-terminal end of the HVL. The large CSPs for residues with side chains on the NC-face of the protein is likely because their backbone atoms are contacted by TβRII, for example, Val236, Tyr252, and Asn255, or because of indirect transmission of binding-induced perturbations through the disulfide-stabilized structure, for example, Thr217, Trp250, and Ile256.
To assess the contribution of TβRII residues to binding, we performed a similar analysis in which we substituted Ile73, Ser75, and Ile76 in β4 with either alanine (Ile73 or Ile76) or leucine (Ser75). To investigate the possibility that residues of TβRII that formed hydrogen-bonded ion pairs with the fingertip regions of TGF-β, Asp55, and Glu142, also interact with TGM-D3, we also substituted these with asparagine and glutamine, respectively. The SPR measurements showed that all variants within β4 perturbed binding. The S75L variant in the center of the strand increased the KD by nearly 200-fold, whereas the I76A and I73A variants increased the KD by 26- and 7-fold, respectively (Figs S11, E–H and Table S5). The variants at flanking positions also significantly increased the KD, with D55N and E142Q variants increasing the KD by 63- and 17-fold, respectively (Figs S11, I–J and Table S5). Thus, in spite of the modest binding-induced CSPs at these positions, these residues nonetheless contribute significantly to binding. These residues may interact with TGM-D3 Arg198 and Lys254, which when mutated to alanine increased the KD for binding TβRII by over 30-fold (Figs. S11 and Table S5). Thus, TGM-D3 appears to closely mimic TGF-β by engaging TβRII not only through β4 but also by interacting with Asp55 and Glu142 which flank β4 in the structure of TβRII (Fig. 4E, right).
To ascertain if the residues in TGM-D3 important for TβRII binding were also functionally important, we evaluated the effect of four representative substitutions on signaling activity using the highly sensitive MFB-F11 TGF-β reporter bioassay (52). The substitutions chosen for study ranged from Y253A, which dramatically decreased TβRII binding, to I238A and Y252A which led to significant, but not as severe, reductions and I256A which led to no reduction. The substitutions were studied in the context of TGM constructs lacking domains 4 and 5 and were produced using mammalian cell expression, as done previously for full-length TGM and truncated forms, including TGM D1-D3 (15, 35). The results were overall consistent with those from the SPR binding studies, with the Y253A substitution blunting the signaling, except at the highest doses tested, and the I238A and Y252A substitutions, diminishing the signaling potency (EC50 9.7 and 7.0 ng/ml, respectively, vs. 3.9 ng/ml for WT), but to a lesser degree than the Y253A substitution. The I256A substitution, which led to no reduction in TβRII binding affinity, was essentially equipotent with respect to WT (EC50 4.5 ng/ml vs. 3.9 ng/ml for WT).
Discussion
The genome of the mouse helminth H. polygyrus encodes a highly expanded family of CCP-containing proteins, several of which have been identified in its secretome to regulate host immune responses (15, 37, 39, 53). TGM, together with its five adult (TGM-2 to -6) and four larval (TGM-7 to -10) homologs, are among the proteins in this family, and at least two of these, TGM and TGM-2, have been shown to regulate immunosuppressive signaling through the Treg pathway (35). Though potency of signaling through TGM is similar to that of TGF-β (15, 16), protein–protein binding kinetics and the amplitude and kinetics of signaling in murine reporter cell lines and primary murine T cells is distinct, with increased Treg potency and decreased fibrotic gene response (15, 16).
The results presented here demonstrate that TGM binds the TGF-β receptors in a modular manner, with TGM-D2 and TGM-D3 as the main partners for TβRI and TβRII, respectively. The binding of TβRI is potentiated by TGM-D1, and this is likely mediated by a composite interface formed by both TGM-D1 and TGM-D2, not allostery, as the NMR titration data presented in Fig. S5C show that TGM-D1 directly, albeit weakly, binds TβRI. It is common for CCP-containing proteins to bind partners through arrays of CCPs, with avidity playing an important role (34). In addition, in multidomain CCP-containing proteins, the domains tend to be connected by short linkers and assume a relatively defined orientation to one another. In TGM, the linker connecting D1-D2 and D2-D3 is limited to just a few residues (Fig. S1). In addition, if one excludes the first four residues of TGM-D3, which are an artifact of the way the domain was produced, the entire domain, including the N- and C-termini, is overall quite rigid (Fig. 5C). The CCP domains that comprise TGM may therefore form a relatively extended structure with the domain orientations restricted to one another. In the case of TGM domains 1 and 2, this may be important for forming the shared interface that recognizes and binds TβRI. In the case of TGM domains 2 and 3, this may be important in positioning the type I and type II receptors with an appropriate spacing, and possibly also orientation, to enable efficient transphosphorylation and signaling.
The assembly of TβRI:TβRII signaling heterodimers by TGM is distinct compared to TGF-β homodimers, which assemble a (TβRI:TβRII)2 heterotetramer, first by binding TβRII with moderate to high affinity (KD ca. 50 nM) and in turn by recruiting and binding TβRI through a composite TGF-β:TβRII interface (KD ca. 30 nM) (18, 54, 55, 56). Though further studies are required, differences in kinetics of assembly, as well as the stoichiometry of the TGM vs. TGF-β signaling complexes, might account for at least some of the differences in the amplitude and kinetics of signaling that have been observed. These differences might also contribute to TGM’s gene expression profile, which is skewed away from extracellular matrix accumulation toward immunosuppression. Though domains 4 and 5 of TGM do not appear to be involved in ligating the TGF-β receptors, they might have other roles, such as targeting TGM to T cells or other cell types to enable effective immunosuppression in vivo.
The ITC competition experiments and NMR assignments of the free and bound forms of TβRI and TβRII demonstrate that TGM-D2 and TGM-D3 mimic TGF-β by engaging the same primary motifs of the receptors: the -PRDRP- prehelix extension, β5, and the extended C-terminus in TβRI and the β4 edge strand, as well as flanking acidic residues, Asp55 and Glu142, in TβRII. The fact that TGM-D2 engages not only the same regions of TβRI as both TGF-β and TβRII, but also an additional region, namely the C-terminal end of β1 and the turn that follows, suggests that this domain alone has extensively adapted to enable TGM’s high affinity for TβRI. This affinity is notably further augmented by TGM-D1, which evidently must recognize and bind TβRI at sites other than those bound by TGM-D2.
TGM-D3 is distinct from almost all other reported CCP domains in that its HVL is significantly extended (15). The structure of TGM-D3 and 15N T2 measurements show that the HVL extends laterally around the domain and is structurally ordered. The N-terminal end of the HVL packs against a triad of aromatic residues that protrude from the non-NC-face of the protein, including Tyr192, His218, and Phe235. This may serve to rigidify this portion of the HVL and position it to engage TβRII. The structure of TGM-D3 further shows that it is expanded laterally compared to canonical CCP domains. This is due to the elimination of the β′ and β’’ strands, which allows the C-terminal end of β4 and the extended segment that follows to diverge away from β1 and the extended N-terminus, against which it packs in canonical CCP domains. The expansion of the domain leads to partial exposure of several hydrophobic residues on both the NC- and non-NC-faces. These structural modifications of TGM-D3, together with the tentative identification of the binding site for TβRII on the non-NC-face of the protein created by these modifications, suggest that TGM-D3 accommodates TβRII by engaging its edge β-strand, β4, through hydrophobic residues on non-NC-face and that it stabilizes TβRII by interacting with Asp55 and Glu142 that flank β4. The basic residues on TGM-D3 that interact with Asp55 and Glu142 were potentially identified, as Arg198 on the N-terminal end of the HVL and Lys254 on the C-terminal tail. If proven by direct structural analysis, this would provide a remarkable demonstration of how TGM-D3 has adapted, relative to canonical CCP domains, to uniquely and specifically bind TβRII in a way that closely mimics that of the mammalian cytokine.
Though all domains of TGM are predicted to have the overall CCP fold, only TGM-D3 binds to TβRII. Sequence comparisons of TGM-D3 with the other domains of TGM show that they all contain two disulfide bonds and the HVL insertion (Fig. S12A). TGM-D3 is however unique in that the β3-β4 loop is 5 to 6 residues longer than other domains, suggesting that this loop is likely a tight β-turn in the other domains rather than a more extended turn, as in TGM-D3 (Fig. 5, A and B). This may alter the overall shape and dimensions of the C-terminal half of the protein to accommodate other binding partners. Most of the TGM-D3 residues that contribute 4-fold or greater to TβRII binding affinity, Phe235, Val236, Ile238, Tyr252, and Tyr253, are also divergent in the other domains, except for domain 1, and thus, these differences likely contribute to specific binding of TβRII by domain 3. However, Arg198 and Lys254 that may interact with TβRII Asn55 and Glu142 are divergent in domain 1 and thus may also impart D3 with specific binding to TβRII.
Though the TGM family is not fully characterized, TGM-2 and TGM-3 have been shown to also possess activity in TGF-β reporter gene assay in mouse fibroblasts and TGM-2 has been shown to possess Treg conversion activity (35). Domain 3 sequence alignments show that all TGM homologs share overall high conservation, particularly among the four β-strands, the loop connecting β2 and β3, and the extended HVL. Residues shown to contribute more than 4-fold to binding, Arg198, Phe235, Val236, Ile238, Tyr252, Tyr253, and Lys254 are also fully conserved, with the only exceptions being Phe235 which is substituted with leucine in TGM-7 and Lys254 which is substituted with serine in TGM-4 and TGM-5, proline in TGM-6, and histidine in TGM-7. Hence, it is possible that domain 3 of all TGM homologs bind TβRII, though further studies are required to determine if this is correct, and if so, how the relative affinities compare.
The structural modifications demonstrated for TGM-D3 might extend to other CCP-containing proteins in HES. HpARI, and HpBARI, for example, have three and two CCP domains, respectively (37, 38, 39) and except for domain 1 of HpARI and HpBARI, which are 63 and 64 amino acids, respectively, all are of similar length to TGM-D3 (TGM-D3, HpARI CCP2, HpARI CCP3, and HpBARI CCP2 are 86, 86, 86, and 81 residues, respectively). HpARI CCP2 and CCP3 have been shown to be responsible for binding IL-33, while CCP1 of HpARI has been shown to bind DNA (38). Hence, the protein-binding domains of HpARI appear to be of similar length to TGM-D3 and thus these might also possess modifications, relative to canonical CCP domains, that impart them with their ability to bind IL-33. It is also possible that this is so for HpBARI, though this awaits direct demonstration that domain 2 is responsible for binding the IL-33 receptor, ST2.
TGM and domain-deleted forms thereof may have therapeutic potential for treating autoimmune disorders and as TGF-β signaling antagonists, respectively. The potential of TGM for treating autoimmune disorders has already been demonstrated in an animal model of colitis (57), though further work in this area is required to determine if TGM is equally as effective in expanding suppressive Tregs in humans as in mice and to develop strategies to mitigate formation of neutralizing antibodies. TGMs that include D1-D2 or D3, and lack either D3 or D1-D2, respectively, may be used to sequester TβRI or TβRII, thus functioning as competitive receptor antagonists to block signaling. These antagonists have significant potential for attenuating both soft-tissue cancers and the tissue fibrosis that are driven by dysregulated TGF-β signaling, though as with other TGF-β antagonists, strategies to reduce adverse consequences of on-target inhibition in vital tissues such as the heart must be considered (58, 59).
There are a number of human helminth parasites, including the nematodes Necator americanus and Strongyloides stercoralis, and the flatworms Schistosoma mansoni and Taenia solium, which achieve host immunomodulation by upregulating Tregs. Though genome sequences are available, bioinformatic analyses have failed to identify any analogs of TGM, or related CCP-containing immunomodulatory proteins, such as Hp-ARI and Hp-BARI. This suggests that the expansion of the CCP-containing family in H. polygyrus is unique to this parasite and that the other parasites noted earlier upregulate Tregs by other mechanisms. Some true TGF-β family homologs, including TGH-2 from the human parasite Brugia malayi and FhTLM from the parasite Fasciola hepatica, have been implicated in the TGF-β signaling pathway, but these proteins have not yet been thoroughly characterized, either functionally or structurally (3, 4, 5).
The findings presented highlight the unique nature of H. polygyrus-mediated immunomodulation through the CCP domain–containing protein TGM. They show that although TGM is structurally dissimilar to TGF-β, it nonetheless engages the same binding sites on the type I and type II receptors as mammalian TGF-β, thereby mimicking the mammalian cytokine not only functionally but also molecularly. While structural studies of CCPs have demonstrated remarkable versatility in binding partners, none of the CCP domain structures reported to date have the dramatic structural modifications found in TGM-D3. Though further studies are required, these adaptations may be restricted to not only TGM but also other immunomodulatory CCP-containing proteins in the H. polygyrus secretome. These adaptations might have arisen owing to the strong selective pressure that must exist to allow a parasite to coexist within its host.
Experimental procedures
Expression and purification of TGM domains
DNA fragments corresponding to individual domains of H. polygyrus TGM, TGM-D1, TGM-D2, TGM-D3, and TGM-D1D2 were inserted between KpnI and HindIII sites in the modified form of pET32a (EMD-Millipore) that included a KpnI site immediately following the coding sequence for the thrombin recognition sequence. The resulting constructs, which included a thioredoxin-hexahistidine tag-thrombin cleavage site-TGM domain coding cassette (Table S1), were overexpressed in BL21(DE3) cells (EMD-Millipore) cultured at 37 °C. Unlabeled samples for binding studies were produced on rich medium (LB), while 15N and 15N,13C samples for NMR studies were produced using minimal medium (M9) containing 0.1% 15NH4Cl (Cambridge Isotope Laboratories) or 0.1% 15NH4Cl and 0.3% U-13C-D-glucose (Cambridge Isotope Laboratories). Carbenicillin was included in the growth medium at 50 μg mL−1 to select for cells bearing the expression plasmid. Protein expression was induced by adding 0.8 mM IPTG when the light scattering at 600 nm reached 0.75.
Cell pellets from 3 L of culture were resuspended in 100 ml of lysis buffer (50 mM Na2HPO4, 100 mM NaCl, 5 mM imidazole, 10 μM leupeptin, 10 μM pepstatin, 1 mM benzamide, pH 8.0) and sonicated. Following centrifugation (20 min, 15000g), the pellet was washed with 50 ml of water, resuspended in 50 mM Na2HPO4, 100 mM NaCl, 5 mM imidazole, 10 μM leupeptin, 10 μM pepstatin, 1 mM benzamide, and 8 M urea, pH 8.0, and stirred overnight at 25 °C. The remaining insoluble material was removed by centrifugation, and the supernatant was loaded onto a 50-ml metal affinity column (Ni++-loaded chelating sepharose, GE Lifesciences) pre-equilibrated with 125 ml of resuspension buffer. The column was washed with 100 ml of resuspension buffer, and the bound protein was eluted by applying a linear gradient of resuspension buffer containing 0.5 M imidazole.
Protein from the eluted peak was treated with reduced glutathione (GSH) at concentration equal to 2 mM x VF/VP, where VF is the final volume of the folding buffer and VP is the volume of TGM protein to be added to the folding buffer. After a 30-min incubation at 25 °C, the protein was slowly diluted into folding buffer (0.1 M Tris, 1 mM EDTA, 0.5 mM oxidized glutathione [GSSG], pH 8.0) to a final concentration of 0.1 mg mL-1 and stirred for 12 to 16 h at 4° C. The folding mixture was concentrated using an Amicon stirred cell fitted with a 5000 MWCO ultracel filter (Millipore) and dialyzed into 25 mM Tris, pH 8.7, at 4 °C. Solid thrombin was added to a final concentration of 4 U per milligram of TGM domain and incubated overnight at 25 °C. Cleavage was stopped by the addition of 10 μM leupeptin, 10 μM pepstatin, and 100 μM PMSF, and after readjusting the pH to 8.7, the cleavage mixture was passed over a Ni++ chelating sepharose column equilibrated with water. Column flow-through and a subsequent water wash, which contained primarily the TGM domain, were collected. For the TGM-D1 and TGM-D1D2 domains, the flow-through was bound to a Source Q column (GE Lifesciences) equilibrated in 25 mM CHES, pH 9.0, and eluted with a 0 to 0.5 M NaCl gradient. For the TGM-D2 and TGM-D3 domains, the flow-through was adjusted to pH 5.0 by the addition of acetic acid, bound to a Source S column (GE Lifesciences) equilibrated in 5 mM sodium acetate, 2M Urea, pH 5.0, and eluted with a 0 to 0.5 M NaCl gradient. Masses of the TGM domains were measured by liquid chromatography electrospray ionization time-of-flight mass spectrometry (LC-ESI-TOF-MS, Bruker Micro TOF). TGM-FL was expressed in expi293 cells (Invitrogen) and purified by metal affinity chromatography as previously described (15).
Expression and purification of TGF-β receptor and growth factor constructs
The TβRII and TβRI ectodomains, and the TGF-β3 homodimer, were expressed in E. coli at 37 °C in the form of insoluble inclusion bodies, refolded, and purified as previously described (60, 61, 62). The engineered TGF-β monomer, mmTGF-β27M, which retains high affinity binding to TβRII, but has significantly improved solubility relative to TGF-β1, TGF-β2, and TGF-β3 homodimers, was produced and purified using the same procedure previously described (43). Masses were verified by LC-ESI-TOF-MS.
Expression and purification of biotinylated avi-tagged TβRI, TβRII, and TGM-D3
Avi-tagged TβRI, TβRII, and TGM-D3 were produced using constructs modified to include the amino acid sequence “GLNDIFEAQKIEWHE” at the C-terminus. Protein expression and purification was carried out using the same procedures described previously for the nontagged protein. Biotinylation was performed using BirA biotin ligase as previously described (63). Constructs were validated by LC-ESI-TOF-MS where addition of a single biotin increases the protein mass by 226.3 Da. Following biotinylation, the proteins were repurified using ion-exchange chromatography to remove the biotinylation reagents.
Expression and purification TGM-1 D3, TGM-1 D13, and TβRII variants
Constructs coding for H. polygyrus TGM-D3 and TβRII described previously were modified to introduce the desired substitution using site-directed mutagenesis with Phusion polymerase (ThermoFisher) as previously described (64). The resulting clones were sequenced over the entirety of their coding sequences to confirm the substitution. Constructs coding mutated forms of TGM D1-D3 (TGM D13) were generated by synthesis of coding sequences for TGM D13, identical to those described previously for TGM-1 D13, but with the desired substitution and then inserted into AscI- and ApaI-digested pSec-Tag2 as described previously (35). Desired constructs, which code for TGM D13 downstream of a signal peptide and with a C-terminal myc-tag and hexahistidine tag, were transfected into suspension cultured expi293 cells, and after 5 days, the protein was purified from the conditioned medium by capturing it on a NiNTA column (Thermo, His-Pur). The purified TGM D13 was pooled, deglycosylated with PNGase-F, concentrated, and further purified on Superdex 200 16/60 column (GE Lifesciences).
SPR measurements
SPR datasets with TGM domains binding to TβRI or TβRII were generated using a BIAcore X100 instrument (GE Lifesciences) with biotinylated avi-tagged TβRI or TβRII captured onto neutravidin-coated CM-5 sensor chips (GE Lifesciences) at a density of 50 to 150 RU. Neutravidin-coated sensor chips for capture of biotinylated avi-tag receptors were made by activating the surface of a CM-5 chip with EDC and NHS, followed by injection of neutravidin (Pierce) diluted into sodium acetate at pH 4.5 until the surface density reached 6000 to 15,000 RU. Kinetic binding assays were performed by injections of the analytes in 25 mM Hepes, pH 7.4, 150 mM NaCl, 0.005% surfactant P20 (Pierce) at 100 μl min−1. Regeneration of the surface was achieved by a 30-s injection of 1 to 4 M guanidine hydrochloride. Baseline correction was performed by subtracting the response from both the reference surface with no immobilized ligand and 5 to 10 blank buffer injections. Kinetic analyses were performed by fitting the results from a single injection series to a simple 1:1 model using the program Scrubber (Biologic Software).
SPR datasets with TGM-D3 and TβRII variants were generated in the same overall manner described previously, using either biotinylated avi-tagged TβRII or biotinylated avi-tagged TGM-D3 captured at a density of 50 to 150 RU onto neutravidin-coated CM-5 sensor chips (GE Lifesciences). Kinetic binding assays were performed by injections of the analytes in 25 mM Hepes, pH 7.4, 150 mM NaCl, 0.05% surfactant P20 (Pierce) at 100 μl min−1. Regeneration of the surface was achieved by a 30-s injection of 100 mM – 200 mM guanidine hydrochloride. Baseline correction was performed by subtracting the response from both the reference surface with no immobilized ligand and 5 to 10 blank buffer injections. Kinetic analyses were performed by fitting the results from duplicate or triplicate injection series to a simple 1:1 model using the program Scrubber (Biologic Software).
ITC experiments
ITC datasets were generated using a Microcal PEAQ-ITC instrument (Malvern Instruments). All experiments with TβRII were performed in 25 mM sodium phosphate, 50 mM NaCl, pH 6.0, at a temperature of 35 °C, while all experiments with TβRI were performed in 25 mM Hepes, 50 mM NaCl, 0.05% NaN3, pH 7.5, at a temperature of 25 °C. Proteins included in the syringe and sample cell were dialyzed against ITC buffer and concentrated as necessary prior to being loaded into either the syringe or the sample cell. Protein concentrations in the cell and syringe are indicated in Table S2. TβRII experiments were carried out with 15 2.5-μl injections with an injection duration of 5 s, a spacing of 150 s, and a reference power of 10, while TβRI experiments were carried out with 19 2.0-μl injections with an injection duration of 4 s, a spacing of 150 s, and a reference power of 10. Integration and data fitting were performed using the programs Nitpic (65), Sedphat (66, 67), and GUSSI (68).
ITC competition experiments with TβRII were performed in 25 mM sodium phosphate, 50 mM NaCl, pH 6.0, at 35 °C, while those with TβRI were performed in 25 mM Hepes, 50 mM NaCl, 0.05% NaN3, pH 7.5, at 25 °C, with exception of the TβRI TGF-β(TβRII)2 titration which was performed at 30 °C. Protein concentrations in the cell and syringe are indicated in Tables S2 and S3. The TβRII mmTGF-β27M/TGM-D3 competition experiments were performed with 13 3.0-μl injections with an injection duration of 5 s, a spacing of 150 s, and a reference power of 10, as was the TβRI TGF-β(TβRII)2 titration. The TGM-D12 TGF-β(TβRII)2(TβRI)2 titration was performed with 19 2.0-μl injections with an injection duration of 4 s, a spacing of 150 s, and a reference power of 10. The integration and data fitting were performed as stated previously.
NMR sample preparation and 1D/2D experiments
Samples of TGM-D1, TGM-D2, TGM-D3, and corresponding complexes with TβRI and TβRII, for NMR were prepared at a concentration of 0.03 to 0.2 mM in 25 mM Na2HPO4, 50 mM NaCl, pH 6.0, and transferred to 5-mm susceptibility-matched microtubes (Shigemi) for data collection. NMR data were collected at 30 °C using a Bruker 600, 700, or 800 MHz spectrometer equipped with a 5-mm 1H (13C,15 N) z-gradient “TCI” cryogenically cooled probe (Bruker Biospin). One-dimensional 1H spectra were acquired with an excitation sculpting water suppression scheme (69). Two-dimensional 1H-15 N HSQC spectra were acquired with a sequence with water flipback pulses (70) and WATERGATE water suppression pulses (71). To probe conformational exchange, ZZ-exchange experiments were recorded with 15N TGM-D2 as previously described (72). NMR data were processed using nmrPipe (73) and analyzed using NMRFAM-SPARKY (74).
NMR 15N T2 calculations
15N T2 backbone amide relaxation times for TGM-D3 were measured at 310 K using the interleaved pulse sequence as described previously (75). The 15N T2 data were collected using 8 delay times ranging from 16 to 240 msec. To calculate the 15N T2 relaxation time per residue, a two-parameter decaying exponential was used to fit the relative peak intensities as a function of delay time. The sample was prepared in 25 mM MES, 50 mM NaCl, pH 6.0.
NMR backbone assignment
Backbone resonances were assigned by recording and analyzing 2-D 1H-15N HSQC and 3-D HNCACB, CBCA(CO)NH, HNCA, HN(CO)CA, HNCO, and HN(CA)CO triple resonance datasets. Proton and side chain resonances were assigned by recording and analyzing 2-D 1H-13 C CT-HSQC and 3-D CC(CO)NH, HBHACONH, HCCH-TOCSY, H(CC, CO)NH, HNHA, and HNHB datasets. NMR data were processed using nmrPipe (73) and analyzed using a combination of NMRFAM-SPARKY (74), PINE (76, 77), and PECAN (49).
NMR chemical shift perturbation calculations
Backbone resonances were assigned for TβRI and TβRII, both free and bound to TGM-D2 and TGM-D3, respectively. The absolute value of the chemical shift differences was calculated for each of the backbone nuclei (15NH, 1HN, 1Hα, 13Cα, 13Cβ, 13CO) and then normalized to the largest shift perturbation. These values were summed for each residue and then normalized according to the number of nuclei that contributed to the final shift perturbation value.
NMR structure determination of TGM-D3
The solution structure of TGM-D3 was calculated using the program NIH-XPLOR (78) with assigned 1H-1H NOEs, 1H-15N, 1Hα-13Cα and 13Cα−13CO residual dipolar couplings (RDCs), TALOS-derived phi and psi restraints (79), hydrogen bond restraints, and 3JHN-Hα J-coupling restraints as input. The 1H-1H distance restraints were derived from manually peak-picked 3D 15N-edited and 3D 13C-edited NOESY datasets using the program CCP-NMR (80), with distance restraints derived using routines provided by CCP-NMR. The RDCs were recorded using a sample with 10 mg mL-1 Pf1 phage for alignment (81) and were measured using a 2-D IPAP-HSQC (46) for 1H-15N RDCs, a 3D 13Cα-coupled HNCO for 13Cα−13CO RDCs, and a 3D Hα-coupled HN(CO)CA for 1Hα-13Cα RDCs. The 3JHN-Hα was measured from the ratio of the crosspeak to diagonal in a 3D HN-Hα experiment as described (82). Ramachandran analysis was performed using the program PROCHECK (83, 84).
TGF-β reporter bioassay
The TGF-β bioassay (cell line clone MFB-F11) developed by Tesseur et al. (52) was performed as previously described (15). Confluent cells were detached with trypsin and resuspended in DMEM with 2.5% fetal calf serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM L-glutamine at a concentration of 8 × 105 cells/ml. In 50 μl, 4 × 104 cells were added to each well of a 96-well flat-bottomed plate. Dilutions of purified proteins were then added to each well in a volume of up to 50 μl and incubated for 24 h at 37 °C. Subsequently, 20 μl of the supernatant was aspirated from each well, added to an ELISA plate (Nalge Nunc International) with 180 μl of reconstituted Sigma FastTM p-nitrophenyl phosphate substrate, and incubated at room temperature (RT) in dark for up to 18 h. Plates were read on at 405 nm on an Emax precision microplate reader (Molecular Devices). All conditions were set up in triplicate.
Data availability
The assigned chemical shifts for the TGM-D2 bound form of TβRI, the TGM-D3 bound form of TβRII, TβRII-bound TGM-D3, and unbound TGM-D3 have been deposited to BioMagResBank under accession codes 51083, 51084, 51085, and 51086, respectively. The structures, and accompanying restraints, of TGM-D3 have been deposited to the RCSB PDB under accession code 7SXB.
Supporting information
This article contains supporting information.
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors would like to thank Mike Delk for assistance with the NMR instrumentation and Henry McSorley and Matthew Whitley for providing valuable comments on the manuscript.
Author contributions
A. M., C-. H. B., D. J. S., R. M. M., and A. P. H. conceptualization; A. M., C-. H. B., and C. S. H. validation; A. M., C-. H. B., and A. P. H. formal analysis; A. M., C-. H. B., K. C., and T. C. investigation; A. M., R. M. M., and A. P. H. writing-original draft; A. M., D. J. S., R. M. M., and A. P. H. writing-review and editing; A. M. and C-. H. B. visualization; C-. H. B., C. S. H., and T. C. resources; R. M. M. and A. P. H. supervision; A. P. H. project administration; A. P. H. funding acquisition.
Funding and additional information
This research was supported by the NIH (GM58670 and AI53915 awarded to A. H.), the Wellcome Trust through an Investigator Award to RMM (Ref 219530), and the Wellcome Trust core-funded Wellcome Centre for Integrative Parasitology (Ref: 104111). Training support for A. M. was provided by the NIH (AI157069). Molecular graphics and analyses were performed with UCSF Chimera, which is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco supported by NIGMS P41-GM103311. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Wolfgang Peti
Footnotes
Present address for Danielle J. Smyth: Division of Cell Signalling and Immunology, University of Dundee, UK.
Supporting information
References
- 1.Hotez P.J., Brindley P.J., Bethony J.M., King C.H., Pearce E.J., Jacobson J. Helminth infections: the great neglected tropical diseases. J. Clin. Invest. 2008;118:1311–1321. doi: 10.1172/JCI34261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pullan R.L., Smith J.L., Jasrasaria R., Brooker S.J. Global numbers of infection and disease burden of soil transmitted helminth infections in 2010. Parasit. Vectors. 2014;7:37. doi: 10.1186/1756-3305-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maizels R.M., Smits H.H., McSorley H.J. Modulation of host immunity by helminths: the expanding repertoire of parasite effector molecules. Immunity. 2018;49:801–818. doi: 10.1016/j.immuni.2018.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryan S.M., Eichenberger R.M., Ruscher R., Giacomin P.R., Loukas A. Harnessing helminth-driven immunoregulation in the search for novel therapeutic modalities. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiedemann M., Voehringer D. Immunomodulation and immune escape strategies of gastrointestinal helminths and schistosomes. Front. Immunol. 2020;11:572865. doi: 10.3389/fimmu.2020.572865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maizels R.M., Smith K.A. Regulatory T cells in infection. Adv. Immunol. 2011;112:73–136. doi: 10.1016/B978-0-12-387827-4.00003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White M.P.J., McManus C.M., Maizels R.M. Regulatory T-cells in helminth infection: induction, function and therapeutic potential. Immunology. 2020;160:248–260. doi: 10.1111/imm.13190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Logan J., Navarro S., Loukas A., Giacomin P. Helminth-induced regulatory T cells and suppression of allergic responses. Curr. Opin. Immunol. 2018;54:1–6. doi: 10.1016/j.coi.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 9.Vignali D.A., Collison L.W., Workman C.J. How regulatory T cells work. Nat. Rev. Immunol. 2008;8:523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith K.A., Filbey K.J., Reynolds L.A., Hewitson J.P., Harcus Y., Boon L., et al. Low-level regulatory T-cell activity is essential for functional type-2 effector immunity to expel gastrointestinal helminths. Mucosal Immunol. 2016;9:428–443. doi: 10.1038/mi.2015.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W., Jin W., Hardegen N., Lei K.J., Li L., Marinos N., et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanjabi S., Oh S.A., Li M.O. Regulation of the immune response by TGF-beta: from conception to autoimmunity and infection. Cold Spring Harb. Perspect. Biol. 2017;9:a022236. doi: 10.1101/cshperspect.a022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng Y., Laouar Y., Li M.O., Green E.A., Flavell R.A. TGF-beta regulates in vivo expansion of Foxp3-expressing CD4+CD25+ regulatory T cells responsible for protection against diabetes. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4572–4577. doi: 10.1073/pnas.0400810101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grainger J.R., Smith K.A., Hewitson J.P., McSorley H.J., Harcus Y., Filbey K.J., et al. Helminth secretions induce de novo T cell Foxp3 expression and regulatory function through the TGF-beta pathway. J. Exp. Med. 2010;207:2331–2341. doi: 10.1084/jem.20101074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnston C.J.C., Smyth D.J., Kodali R.B., White M.P.J., Harcus Y., Filbey K.J., et al. A structurally distinct TGF-beta mimic from an intestinal helminth parasite potently induces regulatory T cells. Nat. Commun. 2017;8:1741. doi: 10.1038/s41467-017-01886-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White M.P., Smyth D.J., Cook L., Ziegler S.F., Levings M.K., Maizels R.M. The parasite cytokine mimic Hp-TGM potently replicates the regulatory effects of TGF-beta on murine CD4(+) T cells. Immunol. Cell Biol. 2021;99:848–864. doi: 10.1111/imcb.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cook L., Reid K.T., Hakkinen E., de Bie B., Tanaka S., Smyth D.J., et al. Induction of stable human FOXP3(+) Tregs by a parasite-derived TGF-beta mimic. Immunol. Cell Biol. 2021;99:833–847. doi: 10.1111/imcb.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groppe J., Hinck C.S., Samavarchi-Tehrani P., Zubieta C., Schuermann J.P., Taylor A.B., et al. Cooperative assembly of TGF-beta superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol. Cell. 2008;29:157–168. doi: 10.1016/j.molcel.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 19.Wrana J.L., Attisano L., Wieser R., Ventura F., Massague J. Mechanism of activation of the TGF-beta receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 20.Wrana J.L., Attisano L., Carcamo J., Zentella A., Doody J., Laiho M., et al. TGF beta signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 21.Shull M.M., Ormsby I., Kier A.B., Pawlowski S., Diebold R.J., Yin M., et al. Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaartinen V., Voncken J.W., Shuler C., Warburton D., Bu D., Heisterkamp N., et al. Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nat. Genet. 1995;11:415–421. doi: 10.1038/ng1295-415. [DOI] [PubMed] [Google Scholar]
- 23.Sanford L.P., Ormsby I., Gittenberger-de Groot A.C., Sariola H., Friedman R., Boivin G.P., et al. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development. 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly A., Houston S.A., Sherwood E., Casulli J., Travis M.A. Regulation of innate and adaptive immunity by TGFbeta. Adv. Immunol. 2017;134:137–233. doi: 10.1016/bs.ai.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Ihara S., Hirata Y., Koike K. TGF-beta in inflammatory bowel disease: a key regulator of immune cells, epithelium, and the intestinal microbiota. J. Gastroenterol. 2017;52:777–787. doi: 10.1007/s00535-017-1350-1. [DOI] [PubMed] [Google Scholar]
- 26.Kim K.K., Sheppard D., Chapman H.A. TGF-beta1 signaling and tissue fibrosis. Cold Spring Harb. Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a022293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu H.H., Chen D.Q., Wang Y.N., Feng Y.L., Cao G., Vaziri N.D., et al. New insights into TGF-beta/Smad signaling in tissue fibrosis. Chem. Biol. Interact. 2018;292:76–83. doi: 10.1016/j.cbi.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 28.Massague J. TGFbeta in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seoane J., Gomis R.R. TGF-beta family signaling in tumor suppression and cancer progression. Cold Spring Harb. Perspect. Biol. 2017;9 doi: 10.1101/cshperspect.a022277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mariathasan S., Turley S.J., Nickles D., Castiglioni A., Yuen K., Wang Y., et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tauriello D.V.F., Palomo-Ponce S., Stork D., Berenguer-Llergo A., Badia-Ramentol J., Iglesias M., et al. TGFbeta drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554:538–543. doi: 10.1038/nature25492. [DOI] [PubMed] [Google Scholar]
- 32.Akhurst R.J. Targeting TGF-beta signaling for therapeutic gain. Cold Spring Harb. Perspect. Biol. 2017;9 doi: 10.1101/cshperspect.a022301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derynck R., Turley S.J., Akhurst R.J. TGFbeta biology in cancer progression and immunotherapy. Nat. Rev. Clin. Oncol. 2021;18:9–34. doi: 10.1038/s41571-020-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirkitadze M.D., Barlow P.N. Structure and flexibility of the multiple domain proteins that regulate complement activation. Immunol. Rev. 2001;180:146–161. doi: 10.1034/j.1600-065x.2001.1800113.x. [DOI] [PubMed] [Google Scholar]
- 35.Smyth D.J., Harcus Y., White M.P.J., Gregory W.F., Nahler J., Stephens I., et al. TGF-beta mimic proteins form an extended gene family in the murine parasite Heligmosomoides polygyrus. Int. J. Parasitol. 2018;48:379–385. doi: 10.1016/j.ijpara.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hewitson J.P., Ivens A.C., Harcus Y., Filbey K.J., McSorley H.J., Murray J., et al. Secretion of protective antigens by tissue-stage nematode larvae revealed by proteomic analysis and vaccination-induced sterile immunity. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osbourn M., Soares D.C., Vacca F., Cohen E.S., Scott I.C., Gregory W.F., et al. HpARI protein secreted by a helminth parasite suppresses interleukin-33. Immunity. 2017;47:739–751.e735. doi: 10.1016/j.immuni.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chauche C., Vacca F., Chia S.L., Richards J., Gregory W.F., Ogunkanbi A., et al. A truncated form of HpARI stabilizes IL-33, amplifying responses to the cytokine. Front. Immunol. 2020;11:1363. doi: 10.3389/fimmu.2020.01363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vacca F., Chauche C., Jamwal A., Hinchy E.C., Heieis G., Webster H., et al. A helminth-derived suppressor of ST2 blocks allergic responses. Elife. 2020;9 doi: 10.7554/eLife.54017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inman G.J., Nicolas F.J., Callahan J.F., Harling J.D., Gaster L.M., Reith A.D., et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 41.Willems E., Cabral-Teixeira J., Schade D., Cai W., Reeves P., Bushway P.J., et al. Small molecule-mediated TGF-beta type II receptor degradation promotes cardiomyogenesis in embryonic stem cells. Cell Stem Cell. 2012;11:242–252. doi: 10.1016/j.stem.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pellaud J., Schote U., Arvinte T., Seelig J. Conformation and self-association of human recombinant transforming growth factor-beta3 in aqueous solutions. J. Biol. Chem. 1999;274:7699–7704. doi: 10.1074/jbc.274.12.7699. [DOI] [PubMed] [Google Scholar]
- 43.Kim S.K., Barron L., Hinck C.S., Petrunak E.M., Cano K.E., Thangirala A., et al. An engineered transforming growth factor beta (TGF-beta) monomer that functions as a dominant negative to block TGF-beta signaling. J. Biol. Chem. 2017;292:7173–7188. doi: 10.1074/jbc.M116.768754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerhard Wider D.N., Kurt W. Studies of slow conformational equilibria in macromolecules by exchange of heteronuclear longitudinal 2-spin-order in a 2D difference correlation experiment. J. Biomol. NMR. 1991;1:5. [Google Scholar]
- 45.Eberhardt E.S., Loh S.N., Hinck A.P., Raines R.T. Solvent effects on the energetics of prolyl peptide bond isomerization. J. Am. Chem. Soc. 1992;114:5437–5439. doi: 10.1021/ja00039a072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ottiger M., Delaglio F., Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 47.Hinck A.P., Walker K.P., 3rd, Martin N.R., Deep S., Hinck C.S., Freedberg D.I. Sequential resonance assignments of the extracellular ligand binding domain of the human TGF-beta type II receptor. J. Biomol. NMR. 2000;18:369–370. doi: 10.1023/a:1026775321886. [DOI] [PubMed] [Google Scholar]
- 48.Ilangovan U., Deep S., Hinck C.S., Hinck A.P. Sequential resonance assignments of the extracellular domain of the human TGFbeta type II receptor in complex with monomeric TGFbeta3. J. Biomol. NMR. 2004;29:103–104. doi: 10.1023/B:JNMR.0000019468.50957.42. [DOI] [PubMed] [Google Scholar]
- 49.Eghbalnia H.R., Wang L., Bahrami A., Assadi A., Markley J.L. Protein energetic conformational analysis from NMR chemical shifts (PECAN) and its use in determining secondary structural elements. J. Biomol. NMR. 2005;32:71–81. doi: 10.1007/s10858-005-5705-1. [DOI] [PubMed] [Google Scholar]
- 50.Holm L. Using dali for protein structure comparison. Methods Mol. Biol. 2020;2112:29–42. doi: 10.1007/978-1-0716-0270-6_3. [DOI] [PubMed] [Google Scholar]
- 51.Holm L. DALI and the persistence of protein shape. Protein Sci. 2020;29:128–140. doi: 10.1002/pro.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tesseur I., Zou K., Berber E., Zhang H., Wyss-Coray T. Highly sensitive and specific bioassay for measuring bioactive TGF-beta. BMC Cell Biol. 2006;7:15. doi: 10.1186/1471-2121-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hewitson J.P., Harcus Y., Murray J., van Agtmaal M., Filbey K.J., Grainger J.R., et al. Proteomic analysis of secretory products from the model gastrointestinal nematode Heligmosomoides polygyrus reveals dominance of venom allergen-like (VAL) proteins. J. Proteomics. 2011;74:1573–1594. doi: 10.1016/j.jprot.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuniga J.E., Groppe J.C., Cui Y., Hinck C.S., Contreras-Shannon V., Pakhomova O.N., et al. Assembly of TbetaRI:TbetaRII:TGFbeta ternary complex in vitro with receptor extracellular domains is cooperative and isoform-dependent. J. Mol. Biol. 2005;354:1052–1068. doi: 10.1016/j.jmb.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 55.Radaev S., Zou Z., Huang T., Lafer E.M., Hinck A.P., Sun P.D. Ternary complex of transforming growth factor-beta1 reveals isoform-specific ligand recognition and receptor recruitment in the superfamily. J. Biol. Chem. 2010;285:14806–14814. doi: 10.1074/jbc.M109.079921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang T., David L., Mendoza V., Yang Y., Villarreal M., De K., et al. TGF-beta signalling is mediated by two autonomously functioning TbetaRI:TbetaRII pairs. EMBO J. 2011;30:1263–1276. doi: 10.1038/emboj.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smyth D.J., Ren B., White M.P.J., McManus C., Webster H., Shek V., et al. Oral delivery of a functional algal-expressed TGF-beta mimic halts colitis in a murine DSS model. J. Biotechnol. 2021;340:1–12. doi: 10.1016/j.jbiotec.2021.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitra M.S., Lancaster K., Adedeji A.O., Palanisamy G.S., Dave R.A., Zhong F., et al. A potent pan-TGFbeta neutralizing monoclonal antibody elicits cardiovascular toxicity in mice and cynomolgus monkeys. Toxicol. Sci. 2020;175:24–34. doi: 10.1093/toxsci/kfaa024. [DOI] [PubMed] [Google Scholar]
- 59.Tauriello D.V.F., Sancho E., Batlle E. Overcoming TGFbeta-mediated immune evasion in cancer. Nat. Rev. Cancer. 2022;22:25–44. doi: 10.1038/s41568-021-00413-6. [DOI] [PubMed] [Google Scholar]
- 60.Zuniga J.E., Ilangovan U., Mahlawat P., Hinck C.S., Huang T., Groppe J.C., et al. The TbetaR-I pre-helix extension is structurally ordered in the unbound form and its flanking prolines are essential for binding. J. Mol. Biol. 2011;412:601–618. doi: 10.1016/j.jmb.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walker R.G., Czepnik M., Goebel E.J., McCoy J.C., Vujic A., Cho M., et al. Structural basis for potency differences between GDF8 and GDF11. BMC Biol. 2017;15:19. doi: 10.1186/s12915-017-0350-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang T., Hinck A.P. Production, isolation, and structural analysis of ligands and receptors of the TGF-beta superfamily. Methods Mol. Biol. 2016;1344:63–92. doi: 10.1007/978-1-4939-2966-5_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cull M.G., Schatz P.J. Biotinylation of proteins in vivo and in vitro using small peptide tags. Methods Enzymol. 2000;326:430–440. doi: 10.1016/s0076-6879(00)26068-0. [DOI] [PubMed] [Google Scholar]
- 64.Liu H., Naismith J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keller S., Vargas C., Zhao H., Piszczek G., Brautigam C.A., Schuck P. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal. Chem. 2012;84:5066–5073. doi: 10.1021/ac3007522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brautigam C.A., Zhao H., Vargas C., Keller S., Schuck P. Integration and global analysis of isothermal titration calorimetry data for studying macromolecular interactions. Nat. Protoc. 2016;11:882–894. doi: 10.1038/nprot.2016.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao H., Piszczek G., Schuck P. SEDPHAT--a platform for global ITC analysis and global multi-method analysis of molecular interactions. Methods. 2015;76:137–148. doi: 10.1016/j.ymeth.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brautigam C.A. Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 2015;562:109–133. doi: 10.1016/bs.mie.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 69.Hwang T.L., Shaka A.J. Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. Ser. A. 1995;112:4. [Google Scholar]
- 70.Grzesiek S., Bax A. The importance of not saturating water in protein NMR. Application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc. 1993;115:12593–12594. [Google Scholar]
- 71.Pitto M., Saudek V., Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 72.Farrow N.A., Zhang O.W., Formankay J.D., Kay L.E. A heteronuclear correlation experiment for simultaneous determination of N-15 longitudinal decay and chemical-exchange rates of systems in slow equilibrium. J. Biomol. Nmr. 1994;4:727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 73.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 74.Lee W., Tonelli M., Markley J.L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kay L.E., Torchia D.A., Bax A. Backbone dynamics of proteins as studied by N-15 inverse detected heteronuclear Nmr-spectroscopy - application to staphylococcal nuclease. Biochemistry-Us. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 76.Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 77.Bahrami A., Assadi A.H., Markley J.L., Eghbalnia H.R. Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schwieters C.D., Kuszewski J.J., Tjandra N., Clore G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 79.Shen Y., Delaglio F., Cornilescu G., Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Skinner S.P., Goult B.T., Fogh R.H., Boucher W., Stevens T.J., Laue E.D., et al. Structure calculation, refinement and validation using CcpNmr analysis. Acta Crystallogr. D Biol. Crystallogr. 2015;71:154–161. doi: 10.1107/S1399004714026662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hansen M.R., Hanson P., Pardi A. Pf1 filamentous phage as an alignment tool for generating local and global structural information in nucleic acids. J. Biomol. Struct. Dyn. 2000;17:365–369. doi: 10.1080/07391102.2000.10506642. [DOI] [PubMed] [Google Scholar]
- 82.Vuister G.W., Bax A. Quantitative J correlation: a new approach for measuring homonuclear three-bond J(HNH.alpha.) coupling constants in 15N-enriched proteins. J. Am. Chem. Soc. 1993;115:5. [Google Scholar]
- 83.Laskowski R.A., Rullmannn J.A., MacArthur M.W., Kaptein R., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 84.Morris A.L., MacArthur M.W., Hutchinson E.G., Thornton J.M. Stereochemical quality of protein structure coordinates. Proteins. 1992;12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The assigned chemical shifts for the TGM-D2 bound form of TβRI, the TGM-D3 bound form of TβRII, TβRII-bound TGM-D3, and unbound TGM-D3 have been deposited to BioMagResBank under accession codes 51083, 51084, 51085, and 51086, respectively. The structures, and accompanying restraints, of TGM-D3 have been deposited to the RCSB PDB under accession code 7SXB.