Abstract
miRNAs are short, noncoding RNAs that negatively and specifically regulate protein expression, the cumulative effects of which can result in broad changes to cell systems and architecture. The miRNA miR-27b is known to regulate lipid regulatory pathways in the human liver and is also induced by the hepatitis C virus (HCV). However, the functional targets of miR-27b are not well established. Herein, an activity-based protein profiling method using a serine hydrolase probe, coupled with stable isotope labeling and mass spectrometry identified direct and indirect targets of miR-27b. The hepatic lipase C (LIPC) stood out as both highly dependent on miR-27b and as a major modulator of lipid pathway misregulation. Modulation of miR-27b using both exogenous miRNA mimics and inhibitors demonstrated that transcription factors Jun, PPARα, and HNF4α, all of which also influence LIPC levels and activity, are regulated by miR-27b. LIPC was furthermore shown to affect the progress of the life cycle of HCV and to decrease levels of intracellular triglycerides, upon which HCV is known to depend. In summary, this work has demonstrated that miR-27b mediates HCV infection by downregulating LIPC, thereby reducing triglyceride degradation, which in turn increases cellular lipid levels.
Keywords: miRNA, miRNA-27b, activity-based protein profiling, hepatic lipase C, proteomics, lipid, triglyceride, lipid metabolism, lipidomics, hepatitis C virus
Abbreviations: ABPP, activity-based protein profiling; BSA, bovine serum albumin; EGFR, epidermal growth factor receptor; FP, fluorophosphonate; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; LIPC, lipase C; MS, mass spectrometry; PPAR, peroxisome proliferator activated receptor; qRT-PCR, quantitative qRT-PCR; RT, room temperature; TBS, Tris-buffered saline; TBST, Tris-buffered saline with Tween-20
Cellular metabolism exists as a set of interconnected systems and the functional output of the component enzymes of these systems relies upon multiple layers of regulatory mechanisms. In humans, miRNAs calibrate the expression of protein-coding genes by selectively recognizing sequences within mRNA transcripts and using the RNA interference silencing complex to sequester and degrade the targeted mRNA (1). As the length of base pairing is short and the complementarity imperfect, mRNAs are frequently targeted by multiple different miRNAs (1, 2), and a single miRNA will target numerous different mRNAs, typically grouped around a particular cellular function (3, 4). This ability of miRNAs to modulate cellular pathways or processes means that differential expression and dysregulation of miRNA function has the potential to either promote or inhibit diverse pathogeneses. The role of miRNA in viral disease has been demonstrated in recent years in the example of the hepatitis C virus (HCV), a small, enveloped, positive-strand RNA virus that preferentially infects the liver (5, 6, 7, 8). The role of miRNAs in HCV infection was first demonstrated via the interaction between the HCV 5′UTR and miR-122, which stabilizes the viral RNA and sequesters miR-122 away from its usual targets (9, 10). While other direct interactions have been discovered (7), a much larger number of miRNAs affect HCV by altering aspects of the cellular environment, such as its lipid metabolism (6, 7). To favor its survival and propagation, the virus perturbs the cell’s energy metabolism to remodel the host’s lipid architecture (11); this contributes to the development of long-term complications such as liver fibrosis, steatosis, and hepatocellular carcinoma (HCC) (12, 13, 14). This perturbation is realized in part by a significant alteration to the miRNA profile during viral infection (10, 15, 16).
One of these up-regulated miRNAs, miRNA-27b-3p (17), hereafter referred to as miR-27b, is one of the most abundant miRNAs in the liver (18, 19, 20), targets key factors in lipid metabolic pathways, and is considered to be a central lynchpin in the maintenance of lipid homeostasis (17, 21, 22). miR-27b induces increases in both extracellular (23) and intracellular lipids (17), including lipid deposits (24), mostly driven by increases to triglyceride levels (17, 25). miR-27b dysregulation is implicated in the progression of numerous chronic diseases, including multiple forms of cancer (26, 27, 28, 29, 30), notably playing an oncogenic role in HCC (31, 32, 33, 34). Characterizing miR-27b regulation is therefore of interest not only in the study of the role of lipid metabolism during HCV infection but also may furthermore provide insight into novel strategies to combat the effects of abnormal miRNA function, which contribute to other conditions such as atherosclerosis, cancer, and obesity-related illnesses (23, 35).
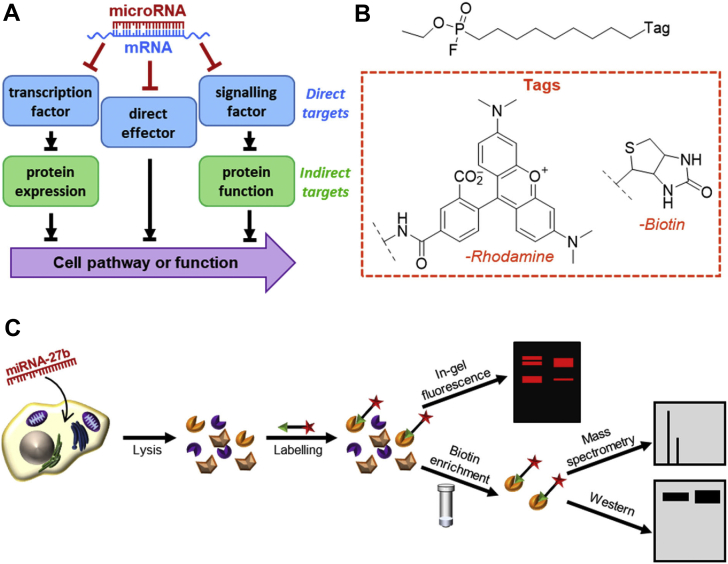
The response to a miRNA is not limited to its direct targets; rather, specific miRNA–mRNA interactions may alter key factors within larger pathways resulting in significant changes downstream of the protein originally targeted. In this work, functional effectors of miR-27b regulation of lipid metabolism are identified and the mechanisms by which their activity is controlled are explored. These functional effectors were identified by means of activity-based protein profiling (ABPP) techniques, which use small-molecule probes that covalently bind active enzymes, but not their inactive counterparts (36, 37). ABPP techniques have been previously used to gain a deeper understanding of metabolic function (38) and to characterize enzyme dysregulation during infection (39, 40, 41). As lipid metabolism is regulated in large part by the serine hydrolase class of enzymes (42), a fluorophosphonate (FP)-based probe (Fig. 1B), which broadly labels active serine hydrolases, was used to profile the effects of miRNA-27b (43). A reporter tag conjugated to the FP warhead allows enzymes bound to the probe to be identified and quantified (36, 43) (Fig. 1C).
Figure 1.
miRNAs’ functional targets and tools to identify them.A, schematic showing direct and indirect mechanisms of enzyme activity regulation by the HCV-induced miR-27. B, fluorophosphonate probe used to measure relative abundance of active lipid metabolic enzymes, with the two reporter tags used. C, activity-based protein profiling workflow.
Using this probe, differential activity profiles were established, from which the hepatic lipase C (LIPC) was confirmed as novel functional target of miR-27b. Multiple intermediary factors responsible for the miR-27b-mediated change to LIPC activity were subsequently identified. Finally, the effect of the miR-27b-induced modulation of LIPC activity was shown to be detrimental to HCV propagation. Altogether, this study has expanded our understanding of the key role played by this miRNA in liver metabolism and HCV infection and, more generally, highlighted the need for the functional profiling of miRNA activity to fully understand their roles both in normal cell function and in disease.
Results and discussion
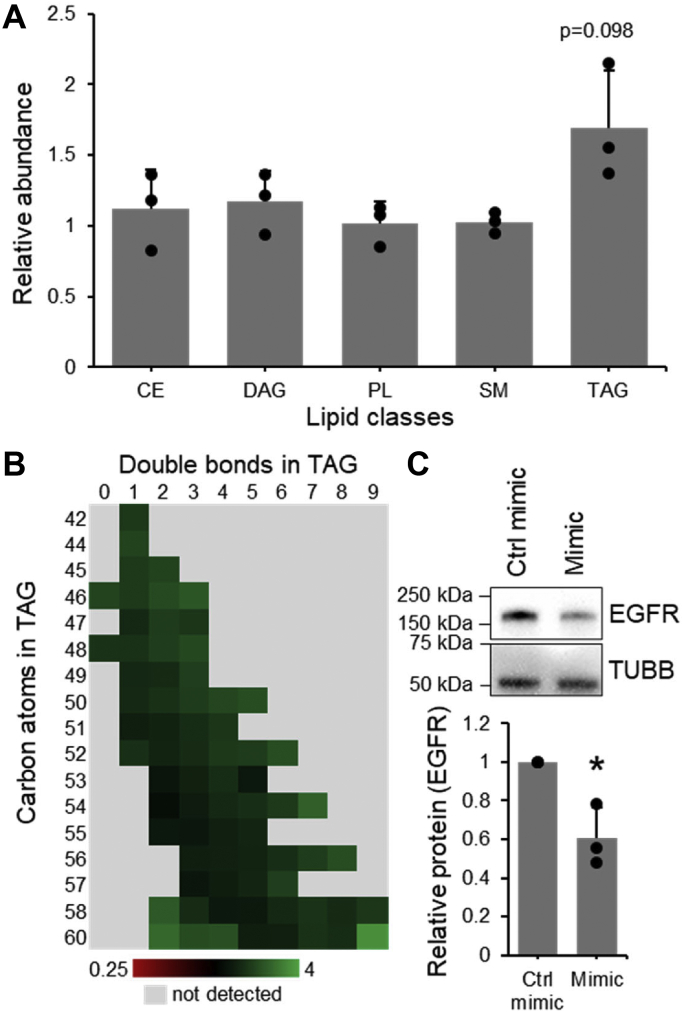
The importance of miR-27b in the regulation of hepatic lipid metabolism, both in healthy tissue and in the context of disease, has been previously established (17, 22); however, the mechanisms by which this modulation of the cell’s lipid profile is accomplished are not yet well understood. To investigate the functional consequences of miR-27b to cells’ lipid profile, lipidomic mass spectrometry was performed on cells transfected with miR-27b mimic versus a control (Tables S1–S9). A significant decrease in the expression of the established miR-27b target epidermal growth factor receptor (EGFR) (44) was used to demonstrate efficient miR-27b transfection (Fig. 2). Consistently with previous reports (17), a significant increase in the overall quantity of triglycerides present was observed (Fig. 2A), with a trend toward desaturation (Fig. 2B). No significant changes were observed in any other lipid groups (Fig. 2A), suggesting triglyceride metabolism is the primary target of regulation by miR-27b and warranting further investigation.
Figure 2.
miR-27b selectively increases the abundance of triacylglycerols. Lipid species from Huh7.5 cells transfected with miR-27b mimic or negative control were quantified. A, cumulative fold change of the lipid species in each class. B, fold change of triglycerides of different lengths and desaturation. C, Western blotting against validated miR-27b target EGFR in cells lysates reserved from lipidomics samples. Values shown are normalized to the control-transfected sample. Data represent mean values ± SD. n = 3 ∗p ≤ 0.05 using a two-tailed Student’s t test. CE, cholesterol ester; DAG, diacylglycerol; EGFR, epidermal growth factor receptor; PL, phospholipid; SM, sphingomyelin; TAG, triacylglycerol.
ABPP identifies miR-27b regulated enzymes
Prior efforts to profile miR-27b-induced changes to cell function have relied mainly on sequencing-based techniques such as ago-HITS-CLIP (10, 45, 46), which reports only on direct hydrogen-bonding interactions between miRNAs and mRNAs and does not allow the detection of any potentially more significant changes occurring downstream. Other studies have used microarrays to globally profile miR-27b′s impact, direct or indirect, on the transcriptome (47, 48). However, this type of analysis is incapable of reporting on important potential posttranslational regulation of protein function. Use of an activity-based probe measures differential enzyme activity derived not only from direct miRNA targeting or altered gene expression but also from changes to protein localization, posttranslational modifications, and protein–protein interactions, thereby providing a more complete, not to mention relevant, picture of the functional role. A significant number of enzymes responsible for the regulation of cells’ lipid profile belong to the serine hydrolase family, whose activity can be detected by the activity-based probe FP (Fig. 1, B and C).
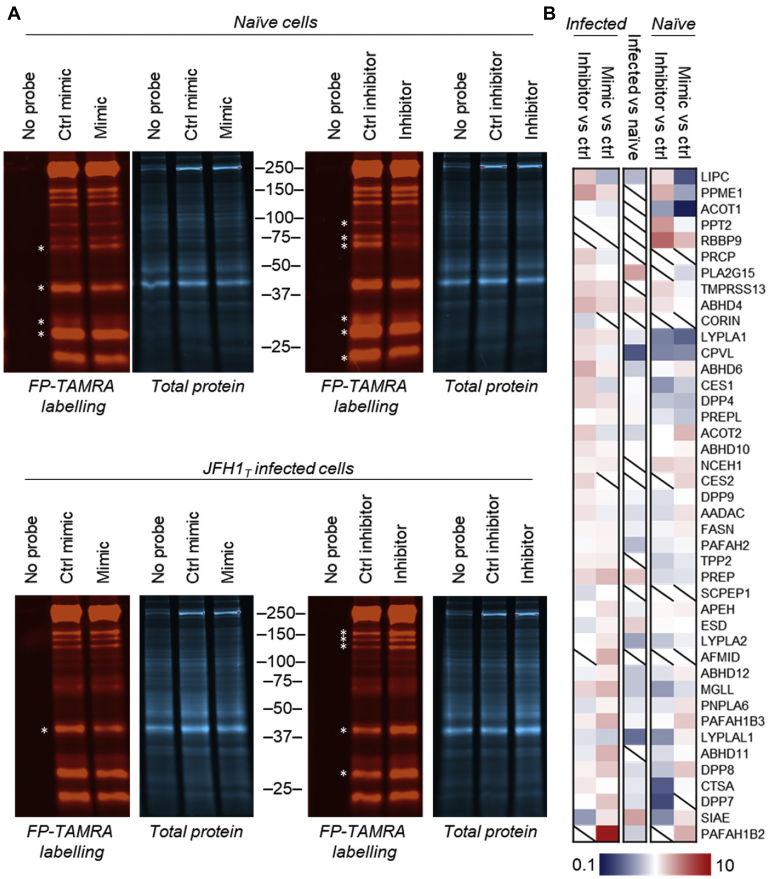
To determine whether any serine hydrolase activities are altered by miR-27b, the fluorescent FP–tetramethylrhodamine probe was used to label the proteomes of cells transfected with miR-27b mimic or inhibitor and visualized by in-gel fluorescence. Multiple differential band intensities were observed (Fig. 3A) following miR-27b overexpression or inhibition, indicating that serine hydrolases could play a role in the miR-27b-induced changes to the lipid profile. Interestingly, when this experiment was repeated in cells infected with the JFH1T strain of HCV, a distinct banding pattern was observed, suggesting that the effects of miR-27b might vary depending on the environment of the cell.
Figure 3.
Fluorophosphonate-based profiling reveals functional targets of miR-27b.A, fluorophosphonate-TAMRA (FP-TAMRA) labeled proteomes from naïve or JFH1T-infected Huh7.5 cells transfected with miR-27b mimic, inhibitor, or respective controls resolved on 10% TGX FastCast (Bio-Rad) gels. B, heatmap of mass spectrometry quantification of FP-biotin (FP-B)–labeled enzymes from naïve or JFH1T-infected Huh7.5 cells transfected with miR-27b mimic and inhibitor versus cells transfected with the respective control RNA. The geometric mean was calculated from log10-transformed fold changes of technical (n = 3) and biological (n = 3) replicates. TAMRA, tetramethylrhodamine. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Next, we determined the identity of the functional targets of miR-27b using affinity purification and HPLC-coupled mass spectrometry (MS). Differentially active serine hydrolases were identified using a biotinylated FP probe to selectively enrich labeled enzymes. Dimethyl labeling, a type of stable isotope labeling using isotopes of formaldehyde, was used to label peptides from each samples, which could then be combined and analyzed by LC-MS/MS. Relative abundances were calculated by the relative size of the peaks in M1, and peptides positively identified, following fragmentation, in M2 (49) (Tables S10–S24). Hits were ranked by prioritizing enzymes whose activity was altered, in opposing directions, by both overexpressing and inhibiting miR-27b, and who displayed this pattern in both naïve and HCV-infected cells (Fig. 3B). Lipid metabolic serine hydrolases represented a significant proportion of enriched, differentially active proteins, followed by a smaller number of signaling proteases and peptidases (Fig. 3B).
The top-ranked enzyme was LIPC, the hepatic lipase, which has not previously been associated with miR-27b. LIPC is a liver enzyme mainly localized to the cell surface of hepatocytes (50) and which predominantly hydrolyzes triglycerides, though it can also target other acyl-containing lipid species (51). The ABPP–LC-MS/MS screen showed that miR-27b significantly decreased the activity of LIPC, while the inhibitor increased activity (Fig. 3B), which suggests a role for LIPC downregulation in miR-27b-mediated lipid accumulation.
LIPC activity decreases along with abundance
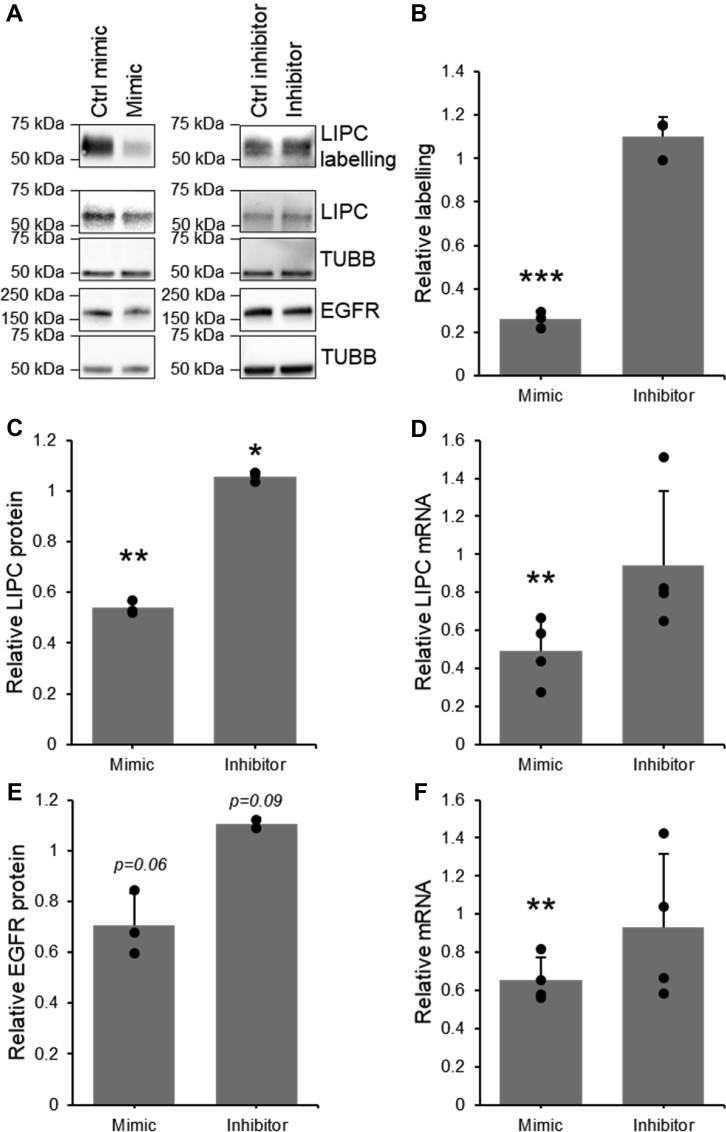
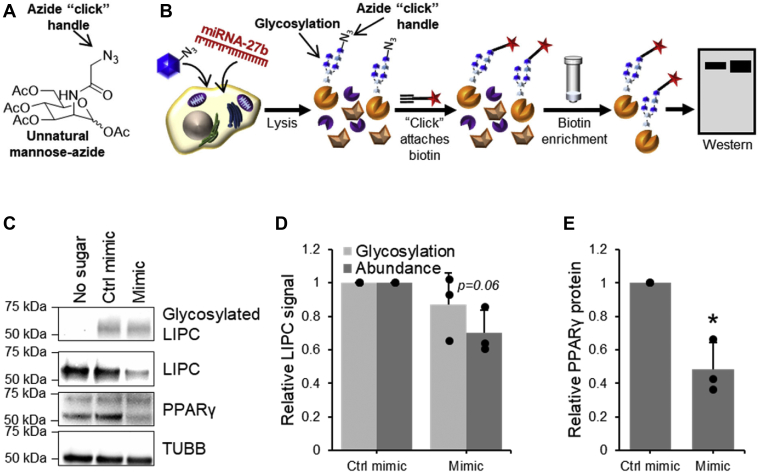
To confirm our MS-derived observations and to determine whether this regulation of LIPC activity was pretranslational or posttranslational, FP-biotin was again used to label proteomes of cells transfected with miR-27b mimic or its inhibitor, and LIPC in the enriched fraction as well as the unenriched cell lysates was quantified by Western blotting. A significant decrease in both LIPC activity and abundance in the presence of miR-27b was observed, paralleled by an insignificant increase in LIPC activity and abundance when miR-27b was inhibited (Fig. 4, A–C). Quantitative RT-PCR (qRT-PCR) was used to show a similar mimic-mediated decrease in mRNA, while the inhibitor left LIPC mRNA levels relatively unchanged (Fig. 4D). This indicates that the miR-27b-mediated decrease in LIPC activity is based largely on changes to protein expression regulated at the mRNA level, either via direct targeting of the LIPC mRNA by miR-27b or by changes to transcription. While a seed site of miR-27b was found in the 3′-UTR of LIPC (Fig. S1A), a reporter construct containing the 3′-UTR from LIPC and the luciferase gene was not targeted by miR-27 (Fig. S1B), indicating that LIPC expression was not directly targeted by miR-27b. The failure of miR-27b to target the luciferase construct may be explained by the lack of complementarity outside the seed sequence, which typically provides extra stability to miRNA–mRNA interactions (1).
Figure 4.
LIPC activity and expression are down-regulated by miR-27b. Quantification of fluorophosphonate (FP) labeling of LIPC, LIPC protein expression, and LIPC mRNA in Huh7.5 cells transfected with miR-27b mimic, inhibitor, or their respective negative controls. Results represent signal from mimic or inhibitor-transfected samples normalized to their respective controls. A, representative sample of Western blotting on FP-B labeled enriched fraction and cell lysates. B, densitometric quantification of FP-B labeling of active LIPC detected by Western blotting on the enriched fraction, n = 3. C, densitometric quantification of LIPC protein expression detected by Western blotting, n = 3. D, qRT-PCR quantification of LIPC mRNA, n = 4. E, densitometric quantification of the protein expression of positive transfection control EGFR detected by Western blotting, n = 3. F, qRT-PCR quantification of positive transfection control EGFR mRNA, n = 4. Data represent mean values ± SD. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001 using a two-tailed Student’s t test. EGFR, epidermal growth factor receptor; FP-B, FP-biotin; LIPC, lipase C; qRT-PCR, quantitative RT-PCR.
Multiple transcription factors regulate LIPC expression
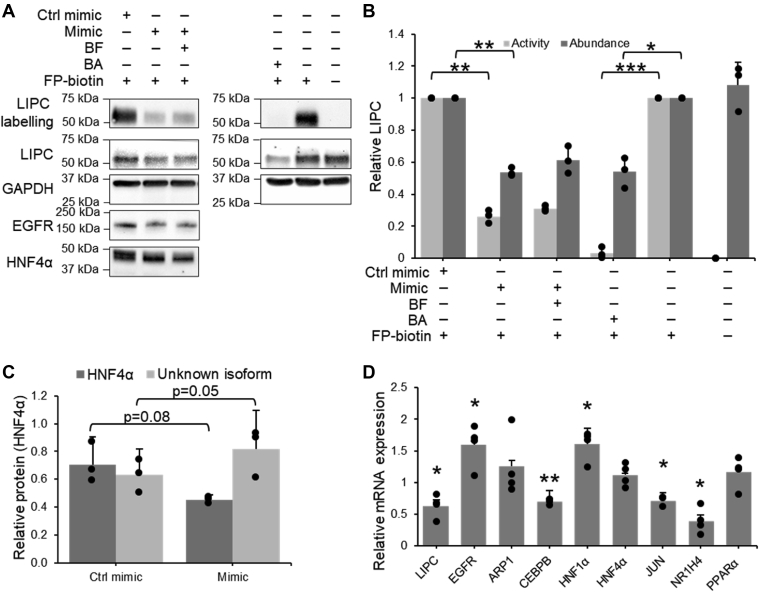
Peroxisome proliferator-activated receptors (PPARs) are a group of transcription factors, which regulate lipid metabolism and have been linked to the transcriptional regulation of LIPC (52, 53). Both PPARα and PPARγ were significantly decreased in the presence of miR-27b mimic (Fig. 5D), in agreement with previous findings (17, 22). To assess whether a reduction in PPAR activity was responsible for the decreased expression of LIPC, cells were treated with the pan-PPAR inhibitor benzamide (54) and LIPC activity was quantified by using FP-biotin to label and enrich the active serine hydrolases. Western blotting showed a large decrease in LIPC activity and expression (Fig. 5, A and B), confirming that PPARs play a role in the transcription of LIPC in the model system used. To confirm whether the miR-27b-mediated decrease in LIPC expression was due to a decrease in PPAR signaling, cells transfected with miR-27b mimic were treated with the pan-PPAR activator bezafibrate (55, 56, 57). While bezafibrate did slightly and nonsignificantly increase LIPC expression and activity, the rescue represented only a small proportion of the decrease in activity induced by miR-27b (Fig. 5, A and B), demonstrating that other transcription factors were also utilized by miR-27b to decrease LIPC abundance.
Figure 5.
miR-27b modulation of LIPC expression occurs via modulation of multiple transcription factors.A–C, quantification of changes to LIPC activity and protein expression after treatment of Huh7.5 cells with 75 μM PPAR inhibitor benzamide, or after transfection with miR-27b mimic and treatment with 75 μM PPAR activator bezafibrate. A, representative Western blot, consisting of the Western blots shown in Figure 4, expanded to include benzamide and bezafibrate treatments. B, densitometric quantification of Western blotting against LIPC in the FP-B labeled fraction and in cell lysates, n = 3. C, densitometric quantification of Western blotting against HNF4α isoforms, n = 3. D, qRT-PCR quantification of mRNA expression of LIPC-associated transcription factors in miR-27b mimic versus control-transfected cells, n = 4. Results represent signal from transfected or treated samples normalized to their respective controls. Data represent mean values ± SD. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001 using a two-tailed Student’s t test. FP-B, FP-biotin; LIPC, lipase C; PPAR, peroxisome proliferator-activated receptor; qRT-PCR, quantitative RT-PCR.
To identify these other factors, all known regulators of LIPC transcription were identified and investigated. After eliminating LIPC regulatory genes not expressed in the Huh7 model, seven potential regulators were left: apolipoprotein A-I regulatory protein 1 (ARP1, also abbreviated NR2F2) (53), CCAAT/enhancer-binding protein β (CEPBP) (58), hepatic nuclear factor 1α (HNF1A) (53), hepatic nuclear factor 4α (HNF4A) (53), transcription factor AP-1 (JUN) (59), bile acid receptor (NR1H4) (60), and PPARγ coactivator 1α (PPARGC1A) (53). miR-27b was overexpressed and the expression of genes associated with the regulation of LIPC were quantified by qRT-PCR. Of this list, two proteins, JUN and HNF4α, were identified as targets of interest (Fig. 5D).
HNF4α is a transcription factor that has been shown to promote the transcription of LIPC (53). miR-27b decreased HNF4α mRNA by 30% and substantially decreased expression at the molecular weight associated with the major active form of HNF4α (Fig. 5, A and C). Interestingly, blotting against HNF4α showed a second band at a lower molecular weight that increased in abundance with miR-27b treatment (Fig. 5, A and C). Though efforts to identify this protein were not successful, it is possible that this band represents an alternative splicing of the protein. This would not negate the functional effect of miR-27b on HNF4α, as these alternatively spliced forms possess less activity than the main form, and some reports indicate that they may hinder the DNA-binding activity of the main form (61). Interestingly, within the context of HCV infection, HNF4α has been shown to be proviral, and the inhibition of its activity decreased viral replication (62). The miR-27b-induced decrease in HNF4α expression discovered in this experiment therefore represents a novel potential mechanism by which miR-27b may reduce viral titers (17).
The other protein of interest, JUN, is a transcription factor implicated in the regulation of cell proliferation and survival (63), whose binding upstream of the LIPC coding sequence has been demonstrated to inhibit transcription (59). miR-27b mimic increases the expression of JUN by 60% (Fig. 5D). Altogether, this suggests that miR-27b modulation of LIPC expression proceeds from a simultaneous decrease of the positive regulators PPARα and HNF4α and an increase in the negative regulator JUN.
miR-27b does not use posttranslational glycosylation to regulate LIPC activity
As the decrease in LIPC activity observed by Western blotting was much larger than that of LIPC expression (Fig. 4, B and C), the possibility of an additional layer of posttranslational regulation of activity was investigated. As LIPC must be glycosylated to be functional (64), the possibility that miR-27b altered the glycosylation of LIPC was investigated.
The use of unnatural sugars to label protein glycosylation is well established (65, 66). Briefly, sugar monomers are synthesized to contain an unnatural moiety; in this case, an azide is introduced on the second carbon of a mannose sugar (Fig. 6A). The sugar is taken up by the cell, which incorporates it into its glycome (66). The azide, being bio-orthogonal to the remainder of the cell, can then be used as a handle to selectively attach a reporter containing an alkyne via copper-catalyzed azide-alkyne click chemistry (67).
Figure 6.
miR-27b does not decrease glycosylation of LIPC.A, structure of unnatural mannose-azide sugar used to label glycosylated LIPC. B, process of labeling and isolating glycosylated proteins. C–E, glycosylation of LIPC in Huh7.5 cells treated with 50 μM mannose-azide and transfected with either miR-27b mimic or control. Glycosylated proteins were tagged with biotin using copper-catalyzed azide-alkyne cycloaddition, enriched, and detected by Western blotting. C, representative Western blot against LIPC in the glycosylated fraction and LIPC expression. D, densitometric quantification of LIPC glycosylation and expression, n = 3. E, densitometric quantification of positive transfection control protein PPARγ. Values shown are normalized to the naïve or control-transfected sample. Data represent mean values ± SD. ∗p ≤ 0.05 using a two-tailed Student’s t test. LIPC, lipase C; PPAR, peroxisome proliferator-activated receptor.
Huh7.5 cells were transfected with miR-27b and treated with mannose-azide. After cell lysis, the azide handle on the sugar was used to attach a biotin reporter to glycosylated proteins by copper-catalyzed azide-alkyne click chemistry. Glycosylated proteins were isolated by streptavidin enrichment and the relative amount of glycosylated LIPC as compared to total LIPC was quantified by Western blotting (Fig. 6B). Interestingly, miR-27b induced no change in glycosylated LIPC, even though overall abundance decreased, indicating that the proportion of glycosylated LIPC had in fact increased (Fig. 6, C and D). Identification of other currently unknown factors that may be responsible for the activity/abundance discrepancy, such as interactions with the chaperone proteins and folding factors required for the production of active LIPC (68), is an interesting subject for future investigation.
miR-27b alteration of enzyme activity is relevant during HCV infection
Labeling by FP-biotin was repeated in cells infected with HCV and transfected with miR-27b mimic or inhibitor. Though miR-27b modulation of LIPC activity and abundance was broadly reproduced in the presence of HCV, there were a few notable changes in the magnitude of the observed changes. The decreases observed in mimic-transfected samples were inferior to that seen in experiments with HCV-naïve cells, while the inhibitor increased the expression and activity of LIPC to a much greater extent (Fig. 7, A–D). These observations are consistent with reports that HCV increases the expression of miR-27b (17); as miR-27b levels are already high, the addition of the mimic has less impact. Similarly, inhibition of miR-27b should lead to more dramatic effects when its baseline expression is higher. Altogether, these results indicate that miR-27b regulation of LIPC function occurs during HCV infection.
Figure 7.
LIPC activity and expression are downregulated by miR-27b during HCV infection. Quantification of fluorophosphonate (FP) labeling of LIPC, LIPC protein expression, and LIPC mRNA in JHF1T-infected Huh7.5 cells transfected with miR-27b mimic, inhibitor, or their respective negative controls. Results represent signal from mimic or inhibitor-transfected samples normalized to their respective controls. A, representative sample of Western blotting on FP-B labeled enriched fraction and cell lysates. B, densitometric quantification of FP-B labeling of active LIPC detected by Western blotting on the enriched fraction, n = 3. C, densitometric quantification of LIPC protein expression detected by Western blotting, n = 3. D, densitometric quantification of the protein expression of positive transfection control EGFR, n = 3. E, densitometric quantification of the protein expression of HCV CORE protein detected by Western blotting, n = 3. F, qRT-PCR quantification of LIPC mRNA, n = 4. G, qRT-PCR quantification of mRNA of positive transfection control EGFR, n = 4. H, qRT-PCR quantification of HCV CORE mRNA, n = 4. Data represent mean values ± SD. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001. EGFR, epidermal growth factor receptor; FP-B, FP-biotin; HCV, hepatitis C virus; LIPC, lipase C; qRT-PCR, quantitative RT-PCR.
LIPC activity decreases triglycerides in hepatoma cells
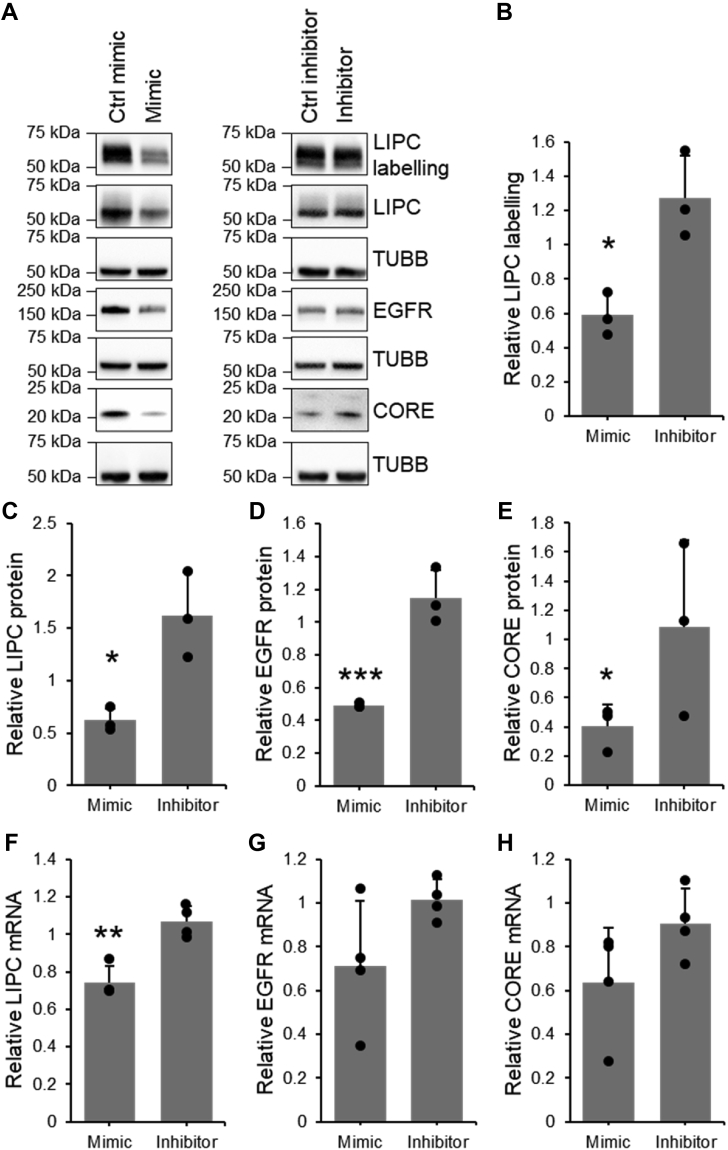
To assess the extent of the impact of the observed decrease in LIPC activity on triglyceride abundance, Huh7.5 cells were transiently transfected with a plasmid encoding the LIPC enzyme and triglyceride levels assessed using a luciferase-based assay. Soraphen A, a small molecule known to significantly decrease triglycerides in the Huh7.5 model (69), was used as a positive control. A decrease in triglyceride abundance of approximately one-third was observed during LIPC overexpression (Fig. 8A), suggesting that changes to LIPC expression and activity can function to regulate triglyceride abundance. More generally, this shows that changes to LIPC expression are relevant to the lipid architecture of hepatoma cells.
Figure 8.
HCV-mediated decrease in LIPC activity modulates triglycerides and promotes virion production.A, quantification of triglyceride levels in cells exogenously expressing LIPC protein, n = 3. B–D, quantification of LIPC activity, protein expression, and mRNA expression in Huh7.5 cells infected with JFH1T. B, representative sample of Western blotting on FP-B labeled enriched fraction and cell lysates. C, densitometric quantification of active LIPC in the FP-B labeled and enriched fraction and its abundance in cell lysates, n = 3. D, qRT-PCR quantification of LIPC mRNA, n = 4. E, qRT-PCR quantification of intracellular HCV RNA in Huh7.5 cells transfected with LIPC siRNA and HCV RNA in cells infected by virus secreted from siRNA-transfected cell, n = 3. F, qRT-PCR quantification of LIPC in Huh7.5 cells transfected with 10 nM LIPC siRNA, n = 3. Values shown are normalized to the naïve or control-transfected sample. Data represent mean values ± SD. ∗p ≤ 0.05, ∗∗p ≤ 0.01. FP-B, fluorophosphonate-biotin; HCV, hepatitis C virus; LIPC, lipase C; qRT-PCR, quantitative RT-PCR.
Decrease in LIPC activity is favorable to hepatitis C
Next, the link between the modulation of LIPC activity and HCV infection was examined by investigating both the effect of the virus on LIPC activity and the effect of LIPC activity on the virus. Protein lysates from Huh7.5 cells infected with the JFH1T cell culture strain of virus were labeled with FP-biotin and the enriched fraction was analyzed by LC-MS/MS (Fig. 3B) as well as by Western blotting (Fig. 8, B and C). LIPC activity was decreased by nearly 50% in infected as compared to the naïve cells, and mRNA abundance was similarly decreased (Fig. 8D). This corroborates previous findings in vivo, in which a significant decrease in LIPC mRNA was observed in liver tissue of patients suffering from HCV as compared to healthy controls (70).
To investigate the dependence of HCV infection on LIPC activity, LIPC expression was suppressed by siRNA and intracellular viral RNA was measured by qRT-PCR. The production of infectious virion was also assessed by measuring the infectivity potential of the supernatants of the infected cells (Fig. 8E). A nonsignificant increase of 40% was observed in the amount of intracellular RNA. By contrast, the levels of infectious virions produced were significantly increased by over 100%. These infectious virions exist as lipoviral particles and use the host cell’s very low-density lipoprotein machinery in their assembly and secretion (5). As LIPC plays a significant role in regulating lipoparticles (42, 71), it could be expected to likewise influence the lipoviral particle. Figure 8E replicates results from previous studies, which have shown that infectivity of secreted virions is significantly increased when LIPC abundance is decreased (72), though intracellular HCV is not affected (72, 73), which suggests that LIPC interferes with an important role in virion secretion. In this work, in vitro HCV infection induced a significant decrease in LIPC activity and abundance, and previous work showed this decrease also occurs in human infection (70).
Role of LIPC activity in pathogenesis and therapeutics
This decrease in LIPC abundance is more pronounced in HCV–HCC (70), indicating that absence of LIPC activity could furthermore contribute to the development of liver cancer. The role of LIPC in cancer has not been extensively studied; it has been suggested to have a positive effect on the anticancer immune response (74) and has been associated with increased survival in some cancers (75), though in others it has been associated with decreased survival (76). Overall, this suggests that the downregulation of LIPC activity during HCV infection via increased miR-27b performs a proviral function and that this may create an oncogenic environment, which contributes to the development of HCV-induced HCC. This is in apparent contradiction to the previously established antiviral nature of miR-27b (17); however, as miRNAs typically modulate a large number of different targets, this proviral effect is most likely overshadowed by other antiviral effects.
In this work, ABPP methods identified LIPC as a novel target of miR-27b and demonstrated how this regulatory activity is pertinent to our understanding of HCV pathogenesis and how to combat it. Findings presented herein suggest that increases to LIPC activity within the liver would decrease levels of circulating virus, thereby decreasing the spread of infection within the host and to others. This suggests the possibility of engineering an increase to LIPC activity within the liver as an antiviral strategy. Recent advances, such as delivery of mRNA in lipid nanoparticles (77), have made the organ-specific introduction of exogenous protein expression in patients possible. This increased LIPC activity could support the currently used direct-acting antivirals, which act to suppress the synthesis of new viral RNA and proteins intracellularly, by decreasing the abundance of assembled and extracellular virus.
More generally, this work has shown that functional targets of miRNA regulation cannot be predicted in their entirety based solely on in silico complementarity-based methods. Profiling changes to enzyme activity is therefore beneficial to understand the role of miRNAs in health and disease and to identify functional effectors, such as LIPC, which could form the basis of novel therapeutics.
Experimental procedures
Cell culture
Huh7.5 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% (v/v) fetal bovine serum (Wisent) and 10 mM nonessential amino acids (MilliporeSigma). The JFH1T strain of the HCV was a kind gift from the laboratory of Dr Rodney Russel (Memorial University). Huh7.5 cells were infected with JFH1T at a multiplicity of infection of 0.1 for 5 h in a one-quarter growth volume of unsupplemented DMEM, rocking plates every hour. Cells were transfected with 100 nM of hsa-miR-27b-3p mirVana mimic, mimic negative control, hsa-miR-27b-3p miRVana inhibitor, or inhibitor negative control (Thermo Fisher Scientific) immediately following infection, using Lipofectamine RNAiMax Transfection Reagent (Invitrogen), according to the manufacturer’s instructions, and lysed after 72 h. Cells treated with benzamide (Calbiochem) or bezafibrate (Cedarlane) were incubated with 75 μM compound for 24 h before lysis. Cells treated with the unnatural sugar were incubated 24 h after miRNA mimic transfection with 50 μM Ac4ManNAz for a further 48 h.
Lipidomic analysis
Huh7.5 cells transfected as aforementioned were trypsinized, spun down, and resuspended in 10 mM PBS, pH 7.4. Cells were washed by centrifugation and subsequent resuspension in PBS twice. Cells were diluted to a concentration of 3000 cells/μl and sent for analysis at Lipotype GmbH. Lipids were quantified in picomoles and normalized by the protein concentration of each sample.
Protein analysis
Proteins were harvested, prepared, and analyzed by ABPP-fluorescence, ABPP-MS, and ABPP-Western blot as previously described (78).
MS
MS analysis was performed by Dr Gleb G. Mirinov and Dr Zoran Minic, John L. Holmes Mass Spectrometry Facility, Department of Chemistry and Biomolecular Sciences, University of Ottawa. Digested peptides were analyzed by HPLC-MS/MS using a Dionex Ultimate 3000 nano-HPLC system with an Acclaim PepMap RSLC 75 mm ID × 150 mm length separation column (Thermo Fisher Scientific), coupled with Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific). Five milliliters of sample were injected and separated by the following gradient (solution A: 0.1% formic acid in H2O; solution B: 80% acetonitrile, 0.1% formic acid in H2O) with the flow of 200 nl/min, such that 0.0 to 80.0 min increased between 0 to 40% B, 80.0 to 80.1 min rose to 40 to 80% B, 80.1 to 90.0 min rose to 80% B, 90.0 to 90.1 min decreased between 80 to 2% B, and 90.1 to 115.0 min maintained 2% B. Peptides were ionized using nano-ESI with spray voltage in positive mode at 2000 V. Ion transfer tube temperature was 275 °C and the S-lens radio frequency level was 60. Survey scans were performed on peptide precursors between 300 and 1500 m/z at 60K resolution (at 200 m/z). The ion count target was set to 2 × 105 and the maximum injection time 50 ms. Precursor peptides for tandem MS analyses were isolated by quadrupole at 0.7 Th. Collision-induced dissociation fragmentation was performed with collision energy of 35% and 5% step and normal scan MS analysis in the ion trap. Precursors with charge state 2 to 6 were sampled for MS2, and the MS2 ion count target was set to 104 with a max injection time of 35 ms. Duration of dynamic exclusion was set to 60 s with 10 ppm tolerance around precursor ions and their isotopes. The instrument was run in 4 s cycles in top speed mode. Proteome Discoverer (version 1.4.1.14, Thermo Fisher Scientific) was used to process the raw data, and MS2 spectra were searched against a UniProt/SwissProt database (July 28, 2018) for Homo sapiens (Human) (http://www.uniprot.org) with SEQUEST HT engine. Peptides were generated from a tryptic digestion containing up to two missed cleavages. Fixed modifications accounted for were of carbamidomethylation of cysteines and variable modifications were oxidation of methionines and protein N-terminal acetylation. Precursor mass tolerance was 10 ppm and product ions were searched at 0.6 Da tolerances. The XCorr of peptides were ≥1.5 and percolator q-value ≤ 0.01. A target decoy validation with false discovery rate of 1% was used to validate peptide spectral matches. Signal from treated samples were normalized over control transfected samples and the geometric mean of this ratio calculated as the fold-change.
Western blotting
Blots were blocked for 30 min in 2.5% (m/v) bovine serum albumin (BSA) in Tris-buffered saline with Tween-20 (TBST) (10 mM Tris, 0.15 M NaCl, pH 8, 0.05% Tween-20). Blots were incubated with primary antibodies PAFAH1B3 1:200 (Santa Cruz; sc-393612), PAFAH1B1 1:500 (Abcam; ab2607), LIPC 1:500 (Santa Cruz; sc-21740), ACOT1/2 1:500 (Santa Cruz; sc-373917), GAPDH 1:10,000 (Ambion; AM4300), β-tubulin 1:4000 (Abcam; ab6046), and PPARG 1:500 (Abcam; ab178860) in 2.5% BSA in TBST overnight at 4 °C. Blots were incubated for 1 h at room temperature (RT) with the appropriate secondary antibody, peroxidase AffiniPure goat antimouse immunoglobulin G (H + L) (Jackson ImmunoResearch Laboratories; 115-035-062) or peroxidase AffiniPure Donkey anti-rabbit immunoglobulin G (H + L) (Jackson ImmunoResearch Laboratories; 711-035-152), at 1:20,000 in 2.5% BSA in TBST. Densitometric analysis was performed using ImageLab 6.0 software (Bio-Rad) and significance was calculated using two-tailed paired t tests.
siRNA knockdowns
Expression knockdown was performed using Silencer Select Pre-Designed siRNAs against ACOT1/2 (s54802; ThermoFisher) negative control number 1 (ThermoFisher), PAFAH1B3 (s224162; ThermoFisher), and LIPC (s8202) at 10 nM using RNAiMax according to the manufacturer’s instructions. Fort-eight hours after transfection, cells were infected with JFH1T at a multiplicity of infection of 0.1. RNA was harvested 48 h after infection, and the conditioned media was used to infect naïve cells. RNA from conditioned media–infected cells was harvested after 48 h.
RNA harvest, purification, and quantification
Cells were lysed and RNA purified using the RNeasy Plus Mini Kit (Qiagen), according to the manufacturer’s instructions. Any sample containing live JFH1T virus was vortexed for 30 s after harvesting and incubated for 5 min at RT to destroy the virus before proceeding to genomic DNA elimination. Purified RNA was quantified by NanoDrop 1000 (Thermo Fisher Scientific). Complementary DNA was synthesized using the iScript Reverse Transcription kit (Bio-Rad) according to the manufacturer’s instructions. Quantitative PCR was performed using the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad), according to the manufacturer’s instructions. Results were normalized using 18S RNA quantification and significance was calculated using two-tailed paired t tests.
3′-UTR assays
The LIPC 3′UTR dual luciferase construct and negative control was purchased from Genecopoeia. The miR-27b-3p seed site was mutated using KOD extreme polymerase (MilliporeSigma) using the following protocol: 120 s at 98 °C, (30 s at 95 °C, 60 s at 55 °C, 440 s at 72 °C 18 times), and 300 s at 72 °C. DNA was digested with 0.6 U/μl ANZA DpnI (Invitrogen) for 45 min, followed by heat inactivation for 15 min at 80 °C. Mutagenesis was confirmed by sequencing at McGill University and Génome Québec Innovation Centre. Cells were transfected with 1 μg construct per milliliter of media, using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer’s instruction. After 24 h, cells were transfected with 100 nM miR-27b-3p mirVana mimic using Lipofectamine RNAiMax Transfection Reagent (Invitrogen) according to the manufacturer’s instructions. Cells were lysed after 48 h in Passive Lysis Buffer (Promega). Luciferase assays were performed as previously described (79) using a SpectraMax L (Molecular Devices). Luciferase signal was normalized over protein concentration quantified by DC assay (Bio-Rad).
Triglyceride assays
A pCMV6-LIPC plasmid was purchased (Origene; RC215870) and 0.4 μg/ml transfected into Huh7.5 using Lipofectamine 2000, according to manufacturer’s instructions. Cells were lysed after 72 h and triglycerides quantified using the Triglyceride-Glo assay kit (Promega), according to the manufacturer’s instructions. Triglyceride concentration was normalized to protein concentration of cell lysates which were lysed in the absence of lipase, measured by bicinchoninic acid assay (Pierce) according to the manufacturer’s instructions. Results were normalized to the positive control and significance was calculated using two-tailed paired t tests.
Data availability
All data are contained within the text or the Supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This study was supported by funding from a Natural Sciences and Engineering Research Council (NSERC) grant 210719 and a Canadian Institutes of Health Research (CIHR) grants 159532, 159600. Mass Spectrometry was performed at the John L. Holmes Mass Spectrometry Facility at the University of Ottawa. Lipidomics analysis was performed by Lipotype GmbH, Tatzberg, Dresden, Germany.
Author contributions
G. F. D. and J. P. P. conceptualization; G. F. D. and J. P. P. methodology; G. F. D. and J. P. P. formal analysis; G. F. D., R. F., M. B., and T. S. data curation; G. F. D. and J. P. P. writing–original draft.
Funding and additional information
G. F. D. was supported by a scholarship from the Fonds de recherche du Québec, Nature et Technologies. R. F. was supported by an NSERC CGS graduate scholarship.
Edited by Ronald Wek
Supporting information
References
- 1.O'Brien J.J. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun W., Julie Li Y.-S., Huang H.-D., Shyy J.Y.-J., Chien S. microRNA: a master regulator of cellular processes for bioengineering systems. Annu. Rev. Biomed. Eng. 2010;12:1–27. doi: 10.1146/annurev-bioeng-070909-105314. [DOI] [PubMed] [Google Scholar]
- 3.Cai Y., Yu X., Hu S., Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7:147–154. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayes J., Peruzzi P.P., Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Neufeldt C.J., Cortese M., Acosta E.G., Bartenschlager R. Rewiring cellular networks by members of the Flaviviridae family. Nat. Rev. Microbiol. 2018;16:125–142. doi: 10.1038/nrmicro.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singaravelu R., Russell R.S., Tyrrell D.L., Pezacki J.P. Hepatitis C virus and microRNAs: miRed in a host of possibilities. Curr. Opin. Virol. 2014;7:1–10. doi: 10.1016/j.coviro.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Shrivastava S., Steele R., Ray R., Ray R.B. MicroRNAs: role in hepatitis C virus pathogenesis. Genes Dis. 2015;2:35–45. doi: 10.1016/j.gendis.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong C.-M., Tsang F.H., Ng I.O.L. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat. Rev. Gastroenterol. Hepatol. 2018;15:137–151. doi: 10.1038/nrgastro.2017.169. [DOI] [PubMed] [Google Scholar]
- 9.Jopling C.L., Norman K.L., Sarnow P. Positive and negative modulation of viral and cellular mRNAs by liver-specific microRNA miR-122. Cold Spring Harb. Symp. Quant. Biol. 2006;71:369–376. doi: 10.1101/sqb.2006.71.022. [DOI] [PubMed] [Google Scholar]
- 10.Luna J.M., Scheel T.K.H., Danino T., Shaw K.S., Mele A., Fak J.J., et al. Hepatitis C virus RNA functionally sequesters miR-122. Cell. 2015;160:1099–1110. doi: 10.1016/j.cell.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herker E., Ott M. Unique ties between hepatitis C virus replication and intracellular lipids. Trends Endocrinol. Metab. 2011;22:241–248. doi: 10.1016/j.tem.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pekow J.R., Bhan A.K., Zheng H., Chung R.T. Hepatic steatosis is associated with increased frequency of hepatocellular carcinoma in patients with hepatitis C-related cirrhosis. Cancer. 2007;109:2490–2496. doi: 10.1002/cncr.22701. [DOI] [PubMed] [Google Scholar]
- 13.Bartosch B.B. Hepatitis C virus-induced hepatocarcinogenesis. J. Hepatol. 2009;51:810–820. doi: 10.1016/j.jhep.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Bassendine M.F., Sheridan D.A., Bridge S.H., Felmlee D.J., Neely R.D.G. Lipids and HCV. Semin. Immunopathol. 2013;35:87–100. doi: 10.1007/s00281-012-0356-2. [DOI] [PubMed] [Google Scholar]
- 15.Li Q., Lowey B., Sodroski C., Krishnamurthy S., Alao H., Cha H., et al. Cellular microRNA networks regulate host dependency of hepatitis C virus infection. Nat. Commun. 2017;8:1789. doi: 10.1038/s41467-017-01954-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singaravelu R., O'Hara S., Jones D.M., Chen R., Taylor N.G., Srinivasan P., et al. MicroRNAs regulate the immunometabolic response to viral infection in the liver. Nat. Chem. Biol. 2015;11:988–993. doi: 10.1038/nchembio.1940. [DOI] [PubMed] [Google Scholar]
- 17.Singaravelu R., Chen R., Lyn R.K., Jones D.M., O'Hara S., Rouleau Y., et al. Hepatitis C virus induced up-regulation of microRNA-27: a novel mechanism for hepatic steatosis. Hepatology. 2014;59:98–108. doi: 10.1002/hep.26634. [DOI] [PubMed] [Google Scholar]
- 18.Lizio M., Harshbarger J., Shimoji H., Severin J., Kasukawa T., Sahin S., et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015;16:22. doi: 10.1186/s13059-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lizio M., Abugessaisa I., Noguchi S., Kondo A., Hasegawa A., Hon C.C., et al. Update of the FANTOM web resource: expansion to provide additional transcriptome atlases. Nucleic Acids Res. 2018;47:D752–D758. doi: 10.1093/nar/gky1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Rie D., Abugessaisa I., Alam T., Arner E., Arner P., Ashoor H., et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017;35:872–878. doi: 10.1038/nbt.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kida K., Nakajima M., Mohri T., Oda Y., Takagi S., Fukami T., et al. PPARα is regulated by miR-21 and miR-27b in human liver. Pharm. Res. 2011;28:2467–2476. doi: 10.1007/s11095-011-0473-y. [DOI] [PubMed] [Google Scholar]
- 22.Vickers K.C., Shoucri B.M., Levin M.G., Wu H., Pearson D.S., Osei-Hwedieh D., et al. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology. 2013;57:533–542. doi: 10.1002/hep.25846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie W., Li L., Zhang M., Cheng H.-P., Gong D., Lv Y.-C., et al. MicroRNA-27 prevents atherosclerosis by suppressing lipoprotein lipase-induced lipid accumulation and inflammatory response in apolipoprotein E knockout mice. PLoS One. 2016;11 doi: 10.1371/journal.pone.0157085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castaño C., Kalko S., Novials A., Párrizas M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc. Natl. Acad. Sci. U. S. A. 2018;115:12158–12163. doi: 10.1073/pnas.1808855115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murata Y., Yamashiro T., Kessoku T., Jahan I., Usuda H., Tanaka T., et al. Up-regulated MicroRNA-27b promotes adipocyte differentiation via induction of acyl-CoA thioesterase 2 expression. Biomed. Res. Int. 2019;2019 doi: 10.1155/2019/2916243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu B., Chen W., Cao G., Dong Z., Xu J., Luo T., et al. MicroRNA-27b inhibits cell proliferation in oral squamous cell carcinoma by targeting FZD7 and Wnt signaling pathway. Arch. Oral Biol. 2017;83:92–96. doi: 10.1016/j.archoralbio.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 27.Chen X., Cui Y., Xie X., Xing Y., Yuan Z., Wei Y. Functional role of miR-27b in the development of gastric cancer. Mol. Med. Rep. 2018;17:5081–5087. doi: 10.3892/mmr.2018.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen D., Si W., Shen J., Du C., Lou W., Bao C., et al. MiR-27b-3p inhibits proliferation and potentially reverses multi-chemoresistance by targeting CBLB/GRB2 in breast cancer cells. Cell Death Dis. 2018;9:188. doi: 10.1038/s41419-017-0211-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo Y., Yu S.-Y., Chen J.-J., Qin J., Qiu Y.-E., Zhong M., et al. MiR-27b directly targets Rab3D to inhibit the malignant phenotype in colorectal cancer. Oncotarget. 2018;9:3830–3841. doi: 10.18632/oncotarget.23237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang H., Ai-Jun J., Ji-Zong Z., Jian-Bo H., Liang Z., Yong-Xiang Y., et al. Clinicopathological significance of miR-27b targeting Golgi protein 73 in patients with hepatocellular carcinoma. Anticancer Drugs. 2019;30:186–194. doi: 10.1097/CAD.0000000000000711. [DOI] [PubMed] [Google Scholar]
- 31.Zhu H.-T., Liu R.-B., Liang Y.-Y., Hasan A.M.E., Wang H.-Y., Shao Q., et al. Serum microRNA profiles as diagnostic biomarkers for HBV-positive hepatocellular carcinoma. Liver Int. 2017;37:888–896. doi: 10.1111/liv.13356. [DOI] [PubMed] [Google Scholar]
- 32.Sun X.-F., Sun J.-P., Hou H.-T., Li K., Liu X., Ge Q.-X. MicroRNA-27b exerts an oncogenic function by targeting Fbxw7 in human hepatocellular carcinoma. Tumor Biol. 2016;37:15325–15332. doi: 10.1007/s13277-016-5444-9. [DOI] [PubMed] [Google Scholar]
- 33.He S., Zhang J., Lin J., Zhang C., Sun S. Expression and function of microRNA-27b in hepatocellular carcinoma. Mol. Med. Rep. 2016;13:2801–2808. doi: 10.3892/mmr.2016.4851. [DOI] [PubMed] [Google Scholar]
- 34.Zhuo L., Liu J., Wang B., Gao M., Huang A. Differential miRNA expression profiles in hepatocellular carcinoma cells and drug-resistant sublines. Oncol. Rep. 2013;29:555–562. doi: 10.3892/or.2012.2155. [DOI] [PubMed] [Google Scholar]
- 35.Chen W.-J., Yin K., Zhao G.-J., Fu Y.-C., Tang C.-K. The magic and mystery of microRNA-27 in atherosclerosis. Atherosclerosis. 2012;222:314–323. doi: 10.1016/j.atherosclerosis.2012.01.020. [DOI] [PubMed] [Google Scholar]
- 36.Barglow K.T., Cravatt B.F. Activity-based protein profiling for the functional annotation of enzymes. Nat. Methods. 2007;4:822–827. doi: 10.1038/nmeth1092. [DOI] [PubMed] [Google Scholar]
- 37.Cravatt B.F., Wright A.T., Kozarich J.W. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 38.Hunerdosse D., Nomura D.K. Activity-based proteomic and metabolomic approaches for understanding metabolism. Curr. Opin. Biotechnol. 2014;28:116–126. doi: 10.1016/j.copbio.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strmiskova M., Desrochers G.F., Shaw T.A., Powdrill M.H., Lafreniere M.A., Pezacki J.P. Chemical methods for probing virus–host proteomic interactions. ACS Infect. Dis. 2016;2:773–786. doi: 10.1021/acsinfecdis.6b00084. [DOI] [PubMed] [Google Scholar]
- 40.Desrochers G.F., Pezacki J.P. ABPP and host–virus interactions. Curr. Top. Microbiol. Immunol. 2019;420:131–154. doi: 10.1007/82_2018_139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharifzadeh S., Shirley J.D., Carlson E.E. In: Activity-Based Protein Profiling Methods to Study Bacteria: The Power of Small-Molecule Electrophiles BT - Activity-Based Protein Profiling. Cravatt B.F., Hsu K.-L., Weerapana E., editors. Springer International Publishing; Cham: 2019. pp. 23–48. [DOI] [PubMed] [Google Scholar]
- 42.Long J.Z., Cravatt B.F. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem. Rev. 2011;111:6022–6063. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y., Patricelli M.P., Cravatt B.F. Activity-based protein profiling: the serine hydrolases. Proc. Natl. Acad. Sci. U. S. A. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiyomaru T., Seki N., Inoguchi S., Ishihara T., Mataki H., Matsushita R., et al. Dual regulation of receptor tyrosine kinase genes EGFR and c-Met by the tumor-suppressive microRNA-23b/27b cluster in bladder cancer. Int. J. Oncol. 2015;46:487–496. doi: 10.3892/ijo.2014.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Helwak A., Kudla G., Dudnakova T., Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore M.J., Scheel T.K.H., Luna J.M., Park C.Y., Fak J.J., Nishiuchi E., et al. miRNA–target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat. Commun. 2015;6:8864. doi: 10.1038/ncomms9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rogler C.E., LeVoci L., Ader T., Massimi A., Tchaikovskaya T., Norel R., et al. MicroRNA-23b cluster microRNAs regulate transforming growth factor-beta/bone morphogenetic protein signaling and liver stem cell differentiation by targeting Smads. Hepatology. 2009;50:575–584. doi: 10.1002/hep.22982. [DOI] [PubMed] [Google Scholar]
- 48.Poon V.Y., Gu M., Ji F., VanDongen A.M., Fivaz M. miR-27b shapes the presynaptic transcriptome and influences neurotransmission by silencing the polycomb group protein Bmi1. BMC Genomics. 2016;17:777. doi: 10.1186/s12864-016-3139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boersema P.J., Raijmakers R., Lemeer S., Mohammed S., Heck A.J.R. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 50.Kuusi T., Nikklä E.A., Virtanen I., Kinnunen P.K.J. Localization of the heparin-releasable lipase in situ in the rat liver. Biochem. J. 1979;181:245–246. doi: 10.1042/bj1810245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Connelly P.W. The role of hepatic lipase in lipoprotein metabolism. Clin. Chim. Acta. 1999;286:243–255. doi: 10.1016/s0009-8981(99)00105-9. [DOI] [PubMed] [Google Scholar]
- 52.Rakhshandehroo M., Knoch B., Müller M., Kersten S. Peroxisome proliferator-activated receptor alpha target genes. PPAR Res. 2010;2010:612089. doi: 10.1155/2010/612089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rufibach L.E., Duncan S.A., Battle M., Deeb S.S. Transcriptional regulation of the human hepatic lipase (LIPC) gene promoter. J. Lipid Res. 2006;47:1463–1477. doi: 10.1194/jlr.M600082-JLR200. [DOI] [PubMed] [Google Scholar]
- 54.Lee G., Elwood F., McNally J., Weiszmann J., Lindstrom M., Amaral K., et al. T0070907, a selective ligand for peroxisome proliferator-activated receptor γ, functions as an antagonist of biochemical and cellular activities. J. Biol. Chem. 2002;277:19649–19657. doi: 10.1074/jbc.M200743200. [DOI] [PubMed] [Google Scholar]
- 55.Berger J., Moller D.E. The mechanisms of action of PPARs. Annu. Rev. Med. 2002;53:409–435. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- 56.Tenenbaum A., Motro M., Fisman E.Z. Dual and pan-peroxisome proliferator-activated receptors (PPAR) co-agonism: the bezafibrate lessons. Cardiovasc. Diabetol. 2005;4:14. doi: 10.1186/1475-2840-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tenenbaum A., Fisman E.Z. Balanced pan-PPAR activator bezafibrate in combination with statin: comprehensive lipids control and diabetes prevention? Cardiovasc. Diabetol. 2012;11:140. doi: 10.1186/1475-2840-11-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Deursen D., Botma G.-J., Jansen H., Verhoeven A.J.M. Down-regulation of hepatic lipase expression by elevation of cAMP in human hepatoma but not adrenocortical cells. Mol. Cell. Endocrinol. 2008;294:37–44. doi: 10.1016/j.mce.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 59.Oka K., Ishimura-Oka K., Chu M., Chan L. Transcription of the human hepatic lipase gene is modulated by multiple negative elements in HepG2 cells. Gene. 1996;180:69–80. doi: 10.1016/s0378-1119(96)00408-8. [DOI] [PubMed] [Google Scholar]
- 60.Sirvent A., Verhoeven A.J.M., Jansen H., Kosykh V., Darteil R.J., Hum D.W., et al. Farnesoid X receptor represses hepatic lipase gene expression. J. Lipid Res. 2004;45:2110–2115. doi: 10.1194/jlr.M400221-JLR200. [DOI] [PubMed] [Google Scholar]
- 61.Drewes T., Senkel S., Holewa B., Ryffel G.U. Human hepatocyte nuclear factor 4 isoforms are encoded by distinct and differentially expressed genes. Mol. Cell. Biol. 1996;16:925–931. doi: 10.1128/mcb.16.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Levy G., Habib N., Guzzardi M.A., Kitsberg D., Bomze D., Ezra E., et al. Nuclear receptors control pro-viral and antiviral metabolic responses to hepatitis C virus infection. Nat. Chem. Biol. 2016;12:1037–1045. doi: 10.1038/nchembio.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mirzaei H., Khodadad N., Karami C., Pirmoradi R., Khanizadeh S. The AP-1 pathway; a key regulator of cellular transformation modulated by oncogenic viruses. Rev. Med. Virol. 2020;30 doi: 10.1002/rmv.2088. [DOI] [PubMed] [Google Scholar]
- 64.Wolle J., Jansen H., Smith L.C., Chan L. Functional role of N-linked glycosylation in human hepatic lipase: asparagine-56 is important for both enzyme activity and secretion. J. Lipid Res. 1993;34:2169–2176. [PubMed] [Google Scholar]
- 65.Lopez Aguilar A., Briard J.G., Yang L., Ovryn B., Macauley M.S., Wu P. Tools for studying glycans: recent advances in chemoenzymatic glycan labeling. ACS Chem. Biol. 2017;12:611–621. doi: 10.1021/acschembio.6b01089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saxon E., Bertozzi C.R. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 67.Worrell B.T., Malik J.A., Fokin V.V. Direct evidence of a dinuclear copper intermediate in Cu(I)-catalyzed azide-alkyne cycloadditions. Science. 2013;340:457–460. doi: 10.1126/science.1229506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Doolittle M.H., Ben-Zeev O., Bassilian S., Whitelegge J.P., Péterfy M., Wong H. Hepatic lipase maturation: a partial proteome of interacting factors. J. Lipid Res. 2009;50:1173–1184. doi: 10.1194/jlr.M800603-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singaravelu R., Desrochers G.F., Srinivasan P., O'Hara S., Lyn R.K., Müller R., et al. Soraphen A: a probe for investigating the role of de novo lipogenesis during viral infection. ACS Infect. Dis. 2015;1:130–134. doi: 10.1021/acsinfecdis.5b00019. [DOI] [PubMed] [Google Scholar]
- 70.Wu J.-M., Skill N.J., Maluccio M.A. Evidence of aberrant lipid metabolism in hepatitis C and hepatocellular carcinoma. HPB (Oxford) 2010;12:625–636. doi: 10.1111/j.1477-2574.2010.00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brunham L.R., Hayden M.R. Human genetics of HDL: insight into particle metabolism and function. Prog. Lipid Res. 2015;58:14–25. doi: 10.1016/j.plipres.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 72.Shimizu Y., Hishiki T., Sugiyama K., Ogawa K., Funami K., Kato A., et al. Lipoprotein lipase and hepatic triglyceride lipase reduce the infectivity of hepatitis C virus (HCV) through their catalytic activities on HCV-associated lipoproteins. Virology. 2010;407:152–159. doi: 10.1016/j.virol.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 73.Shinohara Y., Imajo K., Yoneda M., Tomeno W., Ogawa Y., Fujita K., et al. Hepatic triglyceride lipase plays an essential role in changing the lipid metabolism in genotype 1b hepatitis C virus replicon cells and hepatitis C patients. Hepatol. Res. 2013;43:1190–1198. doi: 10.1111/hepr.12072. [DOI] [PubMed] [Google Scholar]
- 74.Stoll G.G. Metabolic enzymes expressed by cancer cells impact the immune infiltrate. Oncoimmunology. 2019;8 doi: 10.1080/2162402X.2019.1571389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Curtis C., Shah S.P., Chin S.-F., Turashvili G., Rueda O.M., Dunning M.J., et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang J.-Y., Zhang W.-L., Xing Y.-N., Hou W.-B., Yin S.-C., Wang Z.-N., et al. Increased expression of LIPC is associated with aggressive phenotype of Borrmann type 4 gastric cancer. J. Gastrointest. Surg. 2020;25:900–910. doi: 10.1007/s11605-020-04550-5. [DOI] [PubMed] [Google Scholar]
- 77.Witzigmann D., Kulkarni J.A., Leung J., Chen S., Cullis P.R., van der Meel R. Lipid nanoparticle technology for therapeutic gene regulation in the liver. Adv. Drug Deliv. Rev. 2020;159:344–363. doi: 10.1016/j.addr.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Filip R., Desrochers G.F., Lefebvre D.M., Reed A., Singaravelu R., Cravatt B.F., et al. Profiling of microRNA targets using activity-based protein profiling: linking enzyme activity to MicroRNA-185 function. Cell Chem. Biol. 2021;28:202–212.e6. doi: 10.1016/j.chembiol.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dyer B.W., Ferrer F.A., Klinedinst D.K., Rodriguez R. A noncommercial dual luciferase enzyme assay system for reporter gene analysis. Anal. Biochem. 2000;282:158–161. doi: 10.1006/abio.2000.4605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the text or the Supporting information.