Abstract
Population-scale studies of structural variation (SV) are growing rapidly worldwide with the development of long-read sequencing technology, yielding a considerable number of novel SVs and complete gap-closed genome assemblies. Herein, we highlight recent studies using a hybrid sequencing strategy and present the challenges toward large-scale genotyping for SVs due to the reference bias. Genotyping SVs at a population scale remains challenging, which severely impacts genotype-based population genetic studies or genome-wide association studies of complex diseases. We summarize academic efforts to improve genotype quality through linear or graph representations of reference and alternative alleles. Graph-based genotypers capable of integrating diverse genetic information are effectively applied to large and diverse cohorts, contributing to unbiased downstream analysis. Meanwhile, there is still an urgent need in this field for efficient tools to construct complex graphs and perform sequence-to-graph alignments.
Keywords: Structural variation, Long-read sequencing, Genotyping, Pan-genome
1. Introduction
Structural variation (SV) is arbitrarily defined as chromosomal genomic rearrangements greater than 50 bp, including insertions, duplications, deletions, inversions, and translocations [1], [2], [3]. Population-scale studies like Human Genome Structural Variation Consortium (HGSVC) [4], GenomeAD-SV [5], and Pan-Cancer Analysis of Whole Genomes (PCAWG) Consortium [6] have found that basic SVs are often nested together to form more complex SVs [7], [8]. Compared to the ubiquitous single nucleotide variation (SNV) and small indels, SVs are numerically fewer but larger in size, therefore having a greater impact on DNA sequences and correspondingly on gene expression and protein functionality [9], [10], [11]. As a result, SV has received extensive attention in recent studies on genome evolution [10], population diversity [12], demographic history [13], and genetic adaptation [14], bringing new insights into population genetics. In addition, SV can act as a genetic factor underlying disease risk [15] and has already been reported to be involved in the tumorigenesis of various cancers [6], [16], [17], especially nested SVs in complex genetic backgrounds [18], [19].
However, during the rapid development phase of short-read sequencing (SRS) with high base accuracy and relatively low cost, the study of SVs lagged far behind SNVs or indels [2], [3]. In contrast to these small variants, SV tends to occur in highly repetitive and polymorphic regions [12], making it more challenging to detect. Most SRS-based methods extract information of discordant read pairs (RP), split-reads (SR), and read-depth (RD) from alignment with the reference genome to infer the existence of breakpoints [20]. Nevertheless, the short readscannot span the entire repetitive sequences, leading to low-quality alignment and false-positive identification of SVs [2], [3], [21]. Even in the case of non-repetitive regions, insertions longer than short reads are easily missed because they cannot align correctly with the reference genome [12], [22]. Fortunately, the third-generation long-read sequencing (LRS) technologies, such as nanopore sequencing by Oxford Nanopore Technologies and single-molecule real-time sequencing (SMRT) by PacBio, as well as other non-sequencing-based long-range technologies [23], such as optical mapping (OM) by Bionano Genomics, have developed rapidly in recent years and revitalized SV studies. LRS is best characterized by a much longer read-length with an average size of 10 kb [2], [23], far exceeding the 100–500 bp read-length of SRS. Standard long reads generated via nanopore sequencing (R9.4.1 or R10.3 flow cell) can reach lengths of even 10–100 kb, but their accuracy (87–98%) is highly dependent on the base-calling algorithm and is inferior to that of high-fidelity (HiFi) sequencing reads, which represent the latest data type developed by PacBio with high performance (accuracy >99%, length >10 kb) [22], [23]. The advent of LRS makes it possible to span complete repetitive DNA sequences, which enables more accurate measurement of long-distance repeat elements [24], [25], resolves complex rearrangements [26], [27], and simplifies the computational complexity of de novo genome assembly [28], [29]. Taking advantage of LRS technology, researchers have successfully conducted large-scale SV studies in diverse populations worldwide [30], [31], [32], yielding a considerable number of novel SVs and complete gap-closed genome assemblies. In this review, we are concerned with technologies that produce continuous reads and do not involve optical mapping.
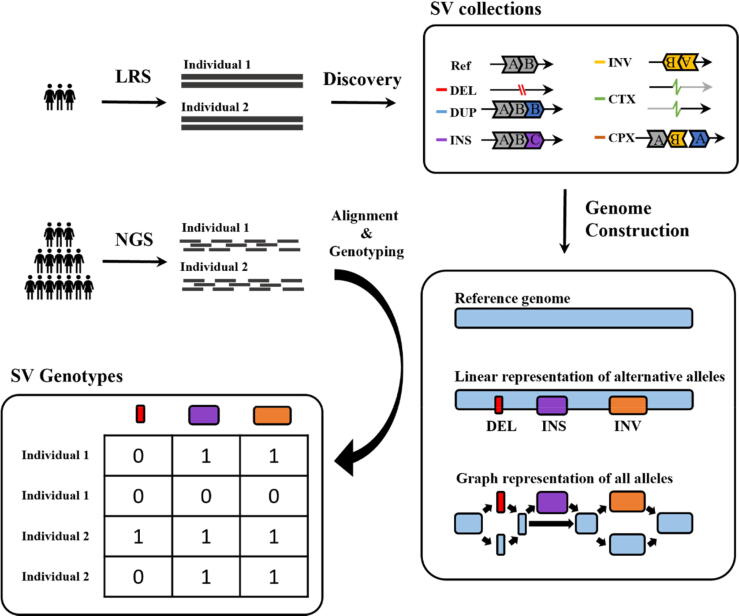
Current large-scale population studies generally use a hybrid approach combining long-read and short-read sequencing technologies to better utilize sequence-resolved SV collections [22], [33] (Fig. 1). In brief, a relatively small number of deep sequenced LRS samples are used for genome assembly and variant detection, followed by a large number of SRS samples for genotyping. On the one hand, this strategy can take advantage of LRS to accurately detect as many variant loci as possible at an economical cost. On the other hand, many large-scale whole-genome sequencing (WGS) datasets collected from valuable clinical samples and other specific populations have been established in the SRS era [34], [35], [36], [37], [38]. Detection of novel SVs identified by LRS in these SRS samples allows better estimation of the allele frequency in local populations, facilitating other genotype-based downstream analyses [12], [14], [22], [33]. As a result, genotyping of SVs in cumulative SRS samples remains a critical issue [33], [39], although large-scale SV studies using LRS samples exclusively are emerging [40], [41]. Similar to the detection strategy, traditional mapping-based genotypers extract SV signatures around the known breakpoints and determine the presence of alternative or reference alleles [39]. However, alignment against a single reference genome is biased towards the reference allele [22], [42], [43], [44], [45], so sequences containing large deviated alternative alleles are prone to mismatches or multiple alignments [46]. As reported in several population-scale SV studies, many of the SVs identified by LRS were classified as homozygous references in SRS samples [12], [14], [32], which severely impacts genotype-based population genetic studies or genome-wide association studies of complex diseases. Therefore, it is of paramount importance to explore new methods to eliminate reference bias to improve genotyping accuracy in population-scale SV studies.
Fig. 1.
An overview of the hybrid sequencing strategy. A small number of long-read sequencing (LRS) samples are used for variant detection, followed by a large number of short-read sequencing (SRS) samples for genotyping. After the discovery of SV collections, including deletions (DEL), duplications (DUP), insertions (INS), inversions (INV), inter-chromosomal translocations (CTX), and complex SVs (CPX), these variants are added to the reference to construct a linear representation of alternative alleles or a graph representation of all alleles. Two strategies are then used to perform the genotyping, aligning short reads to the primary contig along with the alternative sequences or performing a sequence-to-graph alignment.
The mapping-based tools for SV genotyping are biased towards the reference allele mainly because reads are aligned only to the reference genome [22]. Thus, various strategies have been tried to complement the comparison between sequencing reads and the alternative allele, two of which have been widely implemented in published genotyping tools. The first strategy still utilizes a linear reference genome, realigning short reads to a complete reference library that combines primary contigs and alternative allele sequences [12], [47]. Considering that alt-aware mapping is still a non-trivial task, these studies endeavor to filter representative sequences from the original alignment [48]. Another approach is to build a graph-based pan-genome by integrating the reference and alternative alleles and searching for the path that best matches the genetic information of the target haplotype [39]. The graphical representation can describe all nested variants in the sequence more accurately than the linear structure [49]. However, appropriate tools are required to construct the variation graph and perform sequence-to-graph alignment and genotyping [42], [43], [44]. Finally, methods based on both strategies calculate support counts for the reference or alternative allele and then estimate the genotype through probabilistic [50] or machine learning models [12]. Some of these genotypers have already been applied in population-scale SV studies and demonstrate their potential in addressing reference bias [4], [12].
Although several articles have reviewed SV calling algorithms based on LRS [2], [3], [19], [20], [22], [23], [51], [52], [53], little information is available on genotyping for SVs. In this review, we first examined population-scale SV studies using a hybrid sequencing strategy, pointing out the genotyping methods they used and the problems encountered. We are interested in the ability to genotype population-scale SVs, so we did not discuss studies analyzing a small number of target SVs [54], [55]. Then we summarized current academic efforts to resolve the reference bias, including linear and graphic representations of the alternative allele. At the end of this review, we list pan-genomic tools available for genotyping of SVs, including graph construction and sequence-to-graph alignment, in the hope of helping to develop more efficient and accurate genotypers.
2. Population-scale structural variation studies using a hybrid sequencing strategy
LRS technology has dramatically improved the sensitivity and accuracy of SV detection, facilitating large-scale SV studies in various populations worldwide [22] (Table 1). In a pioneering study of applying LRS for analyzing SVs in 2017, Huddleston et al. generated deep SMRT sequencing data from two haploid human genomes [47]. Interestingly, although nearly 90% of SVs identified by LRS were missed in the 1000 Genomes Project reports [56], 61% of these sequence-resolved SVs can be successfully genotyped by short-read sequencing data [47]. This imbalance suggests that decoupling SV genotyping from discovery allows for genotyping the majority of previously missed SVs in the human genome [47]. Therefore, this team from the University of Washington School of Medicine has carried out a subsequent intensive series of work on population-scale SV detection and genotyping using the hybrid sequencing strategy [4], [21], [54], [55], [57]. In a landmark comprehensive study in 2019, Audano et al. sequenced eleven samples from diverse populations using SMRT sequencing [12]. Combined with four additional published resources [30], [47], [58], 99,604 nonredundant SVs were identified, 15% of which were shared in more than half of the samples [12], suggesting that the current reference genome either represents minor alleles or contains assembly errors [59]. In addition to characterizing the enrichment of SVs in tandem repeat sequences and closing gaps in the reference genome, they utilized Illumina WGS data collected from 440 samples to genotype sequence-resolved insertions and deletions, finding that 55% of SVs were successfully genotyped with a missing rate <5% [12]. During this period, many researchers followed the pipeline described in the abovementioned study but still used traditional genotypers based on the analysis of alignments only against the reference genome [31], [60], such as SVTyper [61] and CNVnator [62], which are biased towards the reference allele. Although the SMRT-SV v2 genotyper developed by Audano et al. can represent both reference and alternative alleles [12], it is not scalable to larger populations due to the limitation of time-consuming alt-aware mapping [42].
Table 1.
An overview of population-scale structural variation studies using a hybrid sequencing strategy.
| Study | Discovery sample size |
Genotyping sample size |
Genotyper | Genotyping rate | Recall rate |
|---|---|---|---|---|---|
| Lu et al. (2022) [63] | 35 (20–40×)a | 35 (HGSVC) 879 (GTEx, > 25×) 445 (Geuvadis, 5×) |
danbing-tk v1.3 | – | – |
| Beyter et al. (2021) [32] | 3,622 (17×)b |
10,000 (deCODE, 34×) | GraphTyper v2.6 | – | 36% |
| Ebert et al. (2021) [4] | 35 (20–40×)a | 3,202 (1KG, 34×) | Paragraph v2.4 PanGenie v1.0 |
79% | 74% |
| Quan et al. (2021) [14] | 25 (10–20×)b | 276 (40x) | Paragraph v2.4 | 69% | 54% |
| Sirén et al. (2021) [64] | 16 (>50×)a | 2000 (MESA, 20×) 3202 (1KG, 20×) |
toil-vg | – | – |
| Yan et al. (2021) [65] | 15 (>50×)a | 2504 (1KG, 30×) | Paragraph v2.2 | 86% | 73% |
| Ouzhuluobu et al. (2020) [31] | ZF1 (70×)a | 77 (30x) | CNVnator | – | – |
| Soto etal. (2020) [60] | 2 nonhumansb | 8 (SGDP, 42×) 33 nonhumans |
SVTyper v0.7 | 96% | 45% |
| Audano et al. (2019) [12] | 15 (>50×)a | 174 (1KG, 25×) 150 (Polaris, 18×) 266 (SGDP, 15×) |
SMRT-SV v2 | 55% | 97% |
| Chaisson et al. (2019) [21] | 9 (>50×)ab | 238 (SGDP, 40×) 24 (1KG, 39×) |
SMRT-SV SVTyper |
> 92% | > 96% |
| Kronenberg et al. (2018) [57] | CHM13 (>65×)a YRI19240 (>65×)a 2 nonhumansa |
16 (SGDP) 29 nonhumans |
SVTyper v0.1 | – | – |
| Huddleston et al. (2017) [47] | CHM1 (62×)a CHM13 (66×)a | 30 (1KG, 30×) | SMRT-SV | 79% | 93% |
Genotyping rate, the proportion of SVs successfully genotyped, is usually determined by a missing rate threshold and the Hardy-Weinberg hypothesis. Recall rate, the proportion of the alternative allele presented in at least one haplotype. HGSVC, Human Genome Structural Variation Consortium. GTEx, the Genotype-Tissue Expression project. 1KG, 1000 Genomes Project. SGDP, Simons Genome Diversity Project. MESA, Multi-Ethnic Study of Atherosclerosis cohort. a Pacbio long-read sequencing. b Nanopore sequencing. – indicates that the information was not addressed in the paper.
With the advantages of LRS technology, high-quality and complete genome assemblies and an extensive collection of genetic variants have been accumulated rapidly in multiple populations, making it possible to construct a nonlinear pan-genomic model [22], [42]. The pan-genome concept was initially proposed in microbiology to describe comprehensive genetic information, including the core genome present in all strains and the dispensable genome present in specific strains [66]. Not surprisingly, researchers began introducing graph-based pan-genomic models to eliminate the reference bias and successfully scaled up the population size for genotyping from a few hundred to thousands of samples [4], [32], [63], [64], [65]. In one of the most extensive scale studies, Beyter et al. generated high-confidence SV sets discovered from 3,622 Icelanders by nanopore sequencing [32]. Combining 133,886 sequence-resolved SVs with previously discovered SNPs and indels [67], they constructed an augmented graph with a reference genome backbone using GraphTyper [68], and the resulting genotypes were utilized to explore SVs’ impact on diseases and other traits [32]. Using the same WGS dataset (median coverage 36.9×) [69], Eggertsson et al. reported that genotyping of 543,939 SVs by GraphTyper required 4.15 million CPU hours for 49,962 individuals or 483 CPU-hours per sample on average [68]. In contrast to adding genetic variations to the reference genome [67], Sirén et al. demonstrated an alternative strategy to genotype a new sample by tracing haplotype paths through the sequence graph [64]. They developed Giraffe, a haplotype-aware pangenome mapper that prioritizes alignments under supervision from known haplotypes to avoid search-space explosion caused by combinations of biologically unlikely alleles [43], [44], [49], [64], [70]. On average, it took about 194 CPU hours to genotype a sample with a median coverage of 20× by the combination of toil-vg [71] and Giraffe [64]. A similar strategy was used by Ebert et al. to integrate information from k-mer tables and genetic variation across the input panel haplotypes, which bypasses the time-consuming sequence-to-graph alignment and only took about 30 CPU hours per sample on the tested coverage of 30× [4], [72].
These studies indicate that graph-based genotyping can be effectively applied to large and diverse cohorts and promises to make an essential contribution to downstream analysis. Taking our own study as an example, we used Paragraph [39] to genotype a collection of 38,216 sequence-resolved SVs with a short-read sequencing dataset comprising 276 Chinese Tibetan and Han samples [14]. A considerable number of Tibetan-Han stratified SVs and candidate adaptive genes were inferred from unbiased genotypes, highlighting the important role of SVs in the evolutionary processes of adaptation to the Qinghai-Tibet Plateau [14]. In addition to the genotypers already applied to population-scale studies described above, researchers have made great efforts into developing algorithms to eliminate the reference bias. In the following sections, we discussed other approaches that employ linear or graph representations of the alternative allele.
3. Linear representation of the alternative allele
Traditional mapping-based genotypers, such as Delly [73], SVTyper [61], and SV2 [74], shared similar strategies with the pipeline for SV discovery [75]. These tools extract signatures of breakpoints from alignments only against the reference genome, generally including information on RP, SR, and RD, leading to a bias in favor of the reference allele [43] (Table 2). In addition to the above alignment features, some researchers extracted more sequence features near breakpoints to train a Support Vector Machine (SVM)- or Random Forest (RF)-based classifier [74], [76], expecting to improve the genotyping performance. However, these tools are not only likely to yield biased genotypes [39] but also incapable of estimating insertions [75] and are therefore not suitable for comprehensive SV studies [22].
Table 2.
An overview of structural variation genotypers based on the linear reference genome.
| Tools | Input | Feature extraction |
Genotyping model | Supported SV types |
||||
|---|---|---|---|---|---|---|---|---|
| INS | DEL | DUP | INV | TRA | ||||
| STIX [77] | SRS | RP, SR | – | × | ✓ | ✓ | ✓ | ✓ |
| muCNV [78] | SRS | RP, SR, RD | GMM | × | ✓ | ✓ | ✓ | × |
| NPSV [79] | SRS | Realignment features | SVM/RF | ✓ | ✓ | × | × | × |
| Nebula [50] | SRS | Unique and affected k-mers | GMM | ✓ | ✓ | × | ✓ | × |
| CNV-JACG [76] | SRS | RP, SR, RD, and other sequence features | RF | × | ✓ | ✓ | × | × |
| SMRT-SV [12] | SRS | Realignment features | SVM | ✓ | ✓ | × | × | × |
| SV2[74] | SRS | RP, SR, RD, HAR | SVM | × | ✓ | ✓ | × | × |
| Genome STRiP [80] | SRS | RP, SP, RD | GMM | × | ✓ | ✓ | × | × |
| SVTyper [61] | SRS | RP, SR | BM | × | ✓ | ✓ | ✓ | ✓ |
| Delly2 [73] | SRS | RP, SR, RD | BM | × | ✓ | ✓ | ✓ | ✓ |
| CNVnator [62] | SRS | RP | SGM | × | ✓ | ✓ | × | × |
| SVJedi [48] | LRS | Realignment features | BM | ✓ | ✓ | ✓ | ✓ | ✓ |
| Sniffles [81], [82] | LRS | SR, alignment events | BST | ✓ | ✓ | ✓ | ✓ | ✓ |
| svviz2 [83] | LRS | Realignment MAPQ | BM | ✓ | ✓ | ✓ | ✓ | ✓ |
RP, Read pair. SR, split-read. RD, read-depth. HAR, heterozygous allele ration. MAPQ, mapping quality. GMM, Gaussian mixture models. SVM, Support Vector Machine. RF, random forest. MLE, maximum likelihood estimation. SGM, single Gaussian models. BM, Bayesian model. BST, Binary Search Tree. SRS, short-read sequencing. LRS, long-read sequencing.
In order to minimize the reference allele bias, researchers tried to perform local realignment around known breakpoints against the alternative allele sequence [12], [48], [79], [80], [83]. Among genotypers designed for short-read sequencing data, Handsaker et al. proposed an enhanced version of Genome STRiP back in 2015, a population-based framework for genotyping SVs by aligning reads against a library containing alternative alleles [80], [84]. Genome STRiP analyzes the distribution of read-depth by fitting Gaussian mixture models (GMM) corresponding to the homozygous reference allele, the homozygous or the heterozygous alternative allele [80]. The most likely genotype is finally determined by estimating copy number likelihoods [80]. Notably, Genome STRiP is limited to genotyping of deletions and duplications. In recent studies, another two representative tools have implemented this strategy, SMRT-SV genotyper [12] and NPSV [79]. SMRT-SV is an assembly-based approach with a linear representation of both the reference and alternative alleles for each SV [21]. This genotyping method aligns all short reads against the primary contig together with assembled alternative sequences per each variant [12], using an alt-aware manner by BWA-MEM [85]. An SVM-based classifier is trained on 15 features extracted from the alignment and then used to estimate all possible genotypes [12]. In a recent study in 2021, Linderman et al. proposed NPSV, a simulation-driven approach to genotyping SVs by automatically creating sample- or variant-specific classifiers [79]. Instead of using actual data to train a genotype classifier as SMRT-SV, NPSV first generates synthetic short-read data using an SRS simulator [86] and then locally realigns these reads to the reference and alternate sequences [79]. This strategy helps generate representative training data for any putative SVs with all possible genotypes, avoiding the reference bias at the data source [79]. However, both SMART and NPSV are limited to SV genotyping of insertions and deletions, and they are not scalable to larger populations due to time-consuming alt-aware mapping.
4. Genotyping structural variation in pan-genome graphs
As discussed in the sector of population-scale studies, genotyping SVs using pan-genome graphs is still at a nascent but promising stage (Table 3). The main advantage of pan-genomic approaches is that they can more accurately represent the complex variability of the genome [22] and improve genotyping of nested SVs in complex genetic backgrounds [4], [64]. However, there is still an urgent need for efficient tools to construct complex graphs and perform sequence-to-graph alignments [42], [43], [44]. In the following sections, we summarize the characteristics of graph-based genotypers. Although Cortex [87] is an early attempt at genotyping SVs using de Bruijn graphs, it was not discussed in our review because it was mainly applied to genotyping of small variants. Pan-genomic tools for graph construction and sequence-to-graph alignment are listed in Table 4, and these tools can be helpful in combination with genotypers, as reported by Sirén et al. [64].
Table 3.
An overview of graph-based genotypers for structural variation.
| Tools | Graph construction | Graph Indexing strategy | Sequence-to-Graph alignment strategy |
Genotyping algorithm |
|---|---|---|---|---|
| Gramtools [49] | NDAG | vBWT | Variation-aware backward search | Coverage model |
| Minos [88] | NDAG | vBWT | Variation-aware backward search | Coverage model |
| toil-vg [71] | VG | GCSA2, GBWT, XG,snarl | SMEM seeds | Coverage model |
| PanGenie [72] | DAG | k-mer hash table | – | HMM |
| GraphTyper2 [68] | DAG | k-mer hash table | Matching k-mers as seeds | Coverage model |
| Paragraph [39] | DAG | Path families | GSSW | Coverage model |
| BayesTyper [89] | VG | Variant cluster groups | Heuristic search | Generative Model |
DAG, directed acyclic graph. NDAG, nested DAG. VG, variation graph. BWT, Burrows–Wheeler transform. vBWT, variation BWT. SMEM, super-maximal exact match. HMM, Hidden Markov Model. GSSW, graph SIMD Smith-Waterman algorithm.
Table 4.
An overview of tools for graph construction and sequence-to-graph alignment.
| Category | Tools | Graph | Output format | Description | Ref |
|---|---|---|---|---|---|
| Graph Construction | seqwish | VG | GFA | A VG building from a set of sequences and alignments between them | [96] |
| Cuttlefish | DBG | GFA FASTA |
A colored compacted DBG building from a collection of genome references | [97], [98] | |
| ODGI | VG | ODGI | A suite of tools that implements scalable algorithms | [99] | |
| Pandora | DAG | FASTA | A pan-genome graph structure and algorithms for identifying variants | [100] | |
| Simplitigs | DBG | FASTA | A compact representation of DBG | [101] | |
| Bifrost | DBG | GFA FASTA |
A parallel algorithm enabling the direct construction of the compacted DBG | [102] | |
| libbdsg | VG | GFA ODGI |
Tools allow for construction and manipulation of genome graphs with dense variation | [103] | |
| minigraph | VG | GFA | A graph-based data model to represent multiple genomes | [104] | |
| SevenBridges | DAG | – | A computationally graph genome implementation | [105] | |
| vg | VG | VG | A toolkit of computational methods for creating and manipulating VG | [90] | |
| Wheeler graphs | DBG | DOT | A framework for BWT-based data structures | [106] | |
| Graph alignment | GraphChainer | VG | GAM JSON |
A algorithm to co-linearly chain a set of seeds in an acyclic VG | [107] |
| BlastFrost | DBG | GFA FASTA |
Query Bifrost data structure for sequences of interest | [108] | |
| A* | – | ALN SAM |
A seed heuristic enabling fast and optimal sequence-to-graph alignment | [109], [110] | |
| Giraffe | VG | VG | A pangenome short-read mapper that can map to a collection of haplotypes | [64] | |
| GraphAligner | VG | GAF GAM |
A tool for aligning long reads to genome graphs | [91] | |
| SPAligner | DBG | GPA FASTA |
A tool for aligning long diverged nucleotide and amino acid sequences to assembly graphs | [111] | |
| Vargas | DAG | SAM | A heuristic-free algorithm to find the highest-scoring alignment | [112] | |
| PaSGAL | DAG | TSV | A parallel algorithm for computing sequence to graph alignments | [113] | |
| HISAT2 | DBG | SAM | A tool can align both DNA and RNA sequences using a graph Ferragina Manzini index | [114] | |
| V-ALIGN | DAG | TXT | A tool based on dynamic programming that allows gapped alignment directly on the input graph | [115] | |
VG, variation graph. DBG, de Bruijn graph. DAG, directed acyclic graph.
4.1. Pan-genome graph construction
Most graph-based genotypers construct pan-genome graphs based on the directed acyclic graph (DAG). DAG is usually ordered along the reference genome and represents variants with a bubble composed of different branches between two vertices [43]. Therefore, each path in the DAG represents a possible haplotype. Paragraph [39] and GraphTyper2 [68] are two widely used genotypers constructing DAGs from a reference genome and sequence-resolved variants. Both tools extract short reads from original alignments at breakpoints and perform local mapping to the variation-aware graph [43], which helps reduce bias toward the reference genome and improves genotype quality [39], [68]. Paragraph enables the representation of clustered SVs in the sequence graph and supports custom graph structures for genotyping more complicated events [39]. In addition, GraphTyper2 can also jointly genotype both small variants and SVs at a population scale by simultaneously encoding SNPs and indels into the pan-genome graph [68]. Nevertheless, these joint genotyping models have limitations as they cannot represent nested variants like complex SVs [68].
To genotype complex SVs in variant-dense regions containing a large number of combinations of all possible alleles, Letcher et al. applied an algorithm called recursive collapse and cluster (RCC) implemented by Gramtools and generated a nested DAG consisting of a succession of locally hierarchical subgraphs [49]. Taking advantage of the nested data structure, Gramtools helps discover previously unknown recombination patterns between genetic variants from diverged backgrounds [49], [88]. Gramtools also outputs a JSON variant call format (jVCF) to address the limitation of storing densely clustered variants in the standard VCF. Another idea about the variation graph (VG) was proposed by Garrison et al. in 2018. They combined a bidirectional sequence graph with paths that model sequences as walking through the graph [90]. Hickey et al. presented a genotyping framework toil-vg based on VG and demonstrated the best performance on actual short-read data for all SV types [71]. Instead of extracting information from original alignments, toil-vg directly aligns all short reads to the graph genome, resulting in unbiased pan-genomic analyses and representation [43], [71]. Besides, toil-vg can build graphs from the alignment of numerous de novo assemblies instead of variant collections, leading to better SV genotyping [71].
4.2. Sequence-to-graph alignment and genotyping models
Sequence-to-graph alignment is a fundamental operation for graph-based genotyping [91]. In general, classical algorithms for sequence-to-sequence alignment, such as the Smith-Waterman (SW) algorithm [92], cannot be directly applied to genome graphs. Nonetheless, Paragraph applies an extended generalization of Farrar’s striped SW algorithm [93] to local graph alignment [39], [94]. This implementation extends the recurrence relation and the corresponding scoring matrices of dynamic programming across junctions in the local graph [39], [94]. Reads aligned to a single graph location with the best mapping quality score were retained to genotype breakpoints [39]. A read is considered to support a node if its alignment overlapped the node by at least 10% of the read length, and a similar criterion is applied to the definition of supporting paths [39]. Finally, Paragraph uses an expectation–maximization algorithm to estimate genotype likelihood-based allele frequencies based on the realignment coverage of each allele [39].
Other genotypers usually use a heuristic seed-and-extend paradigm pioneered by BLAST [92]. This paradigm first finds short seed hits, usually based on practical indexing tools, and then extends these hits to obtain complete alignments [95]. A pair of matching k-mers often acts as the seed hit for graph-based genotypers [68], [72], [89]. For example, GraphTyper2 constructs a k-mer hashtable by indexing the full text of DAG and then searches for exact matches with k-mers from the read [68]. The final graph alignment is obtained by extending the longest seed through paths in the genome graph [68]. The genotype call also relies on a likelihood maximizing approach that aggregates both the original and the realignment coverage of each allele [39], [68]. Considering that graph-based whole-genome alignment is time-consuming, both Paragraph and GraphTyper2 restrict the mapping operation to local variant clusters. However, this strategy is based on the realignment of reads to local graphs and requires information from original alignments, which is still disturbed by the reference bias.
In fact, a complete alignment is usually not necessary for genotyping of target SVs. Some researchers suggested that a traversal list of variants supported by each read is sufficient for genotyping [50], [89]. BayesTyper, which is also a k-mer based method, adopts a kind of pseudo-alignment model [89]. This method compares the unbiased distribution of k-mers from sequencing reads to the k-mer profile along paths representing the most likely haplotypes [89]. The posterior distribution over all possible genotypes is estimated according to the counts of k-mers in the reads based on a generative model [89]. However, approaches based solely on the k-mer counts cannot reliably genotype variants in repetitive regions because unique k-mers may not exist for the variants [4], [72]. In a recent study, Ebler et al. proposed PanGenie, which integrates information from k-mer tables and genetic variation across the input panel haplotypes [72]. They utilized information from known haplotype sequences to infer genotypes based on neighboring variants, therefore avoiding the inability to genotype in the absence of unique k-mers [72]. Since PanGenie and BayesTyper bypass the time-consuming alignment step, they are much faster than the remaining mapping-based methods.
5. Summary and outlook
The rapid development of LRS in recent years has revitalized SV studies. Taking advantage of LRS technology, researchers have successfully conducted large-scale SV studies in diverse populations worldwide [30], [31], [32], yielding a considerable number of novel SVs and complete gap-closed genome assemblies. However, genotyping SVs in a large-scale short-read sequencing cohort remains challenging. Traditional mapping-based genotypers are biased towards the reference allele [22]. Therefore, researchers have made great efforts to eliminate the reference bias by representing both the reference and the alternative allele using a linear or graph genome. Notably, most recent population-scale studies of SVs have used pan-genomic models to eliminate reference bias and successfully scaled up the population size for genotyping from a few hundred to thousands of samples [4], [32], [63], [64], [65], facilitating other genotype-based downstream analyses. Recently, the Telomer-to-Telomere (T2T) Consortium and the Human Pangenome Reference Consortium have successively announced their exciting progress in constructing complete and error-free T2T assemblies of all chromosomes as well as full-spectrum genomic variant collections [116], [117], [118], which will further promote the application of pan-genomic approaches in population genetic studies.
Genotyping SVs using pan-genome graphs is still at a nascent stage. There is still an urgent need in this field for efficient tools to construct complex graphs and perform sequence-to-graph alignments. For example, complex SVs often occur in repetitive regions and are nested with other small variants. Despite the potential for reliable genotyping of complex SVs by bidirectional variation graphs and nested DAGs, complex SVs have not been comprehensively analyzed in population-scale studies. Little is known about their contribution to genetic evolution or their interaction with other variants. The same problem is faced by mosaic and low-frequency SVs, which have been reported to be risk factors for neurological diseases [82], [119]. Besides, it remains unclear whether pan-genomic approaches will become mainstream in clinical diagnostics. Some researchers argue that graph-based genotyping relies on single-base resolution breakpoints, making it more suitable for studying common variants rather than somatic or pathogenic variants [120]. In addition, graph-based genotyping approaches are not entirely mature, with competing implementations and data formats [22]. There is an urgent need for a benchmark to evaluate the genotyping performance of graph-based genotypers with uniform criteria.
CRediT authorship contribution statement
Cheng Quan: Writing - original draft, Visualization. Hao Lu: Writing - original draft, Writing - review & editing. Yiming Lu: Writing - original draft, Writing - review & editing, Supervision. Gangqiao Zhou: Writing - review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded by the General Program (31771397 and 81672369) of the Natural Science Foundation of China (www.nsfc.gov.cn), the National Key R&D Program of China (No. 2017YFA0504301), and the Chinese Key Project for Infectious Diseases (No. 2018ZX10732202 and 2017ZX10203205).
Contributor Information
Yiming Lu, Email: ylu.phd@gmail.com.
Gangqiao Zhou, Email: zhougq114@126.com.
References
- 1.Alkan C., Coe B.P., Eichler E.E. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sedlazeck F.J., Lee H., Darby C.A., Schatz M.C. Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat Rev Genet. 2018;19:329–346. doi: 10.1038/s41576-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 3.Mahmoud M., Gobet N., Cruz-Dávalos D., Mounier N., Dessimoz C., Sedlazeck F.J. Structural variant calling: the long and the short of it. Genome Biol. 2019;20:246. doi: 10.1186/s13059-019-1828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ebert P., Audano P.A., Zhu Q., Rodriguez-Martin B., Porubsky D., Bonder M., et al. Haplotype-resolved diverse human genomes and integrated analysis of structural variation. Science. 2021;372 doi: 10.1126/science.abf7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins R.L., Brand H., Karczewski K.J., Zhao X., Alföldi J., Francioli L.C., et al. A structural variation reference for medical and population genetics. Nature. 2020;581:444–451. doi: 10.1038/s41586-020-2287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y., Roberts N.D., Wala J.A., Shapira O., Schumacher S.E., Kumar K., et al. Patterns of somatic structural variation in human cancer genomes. Nature. 2020;578:112–121. doi: 10.1038/s41586-019-1913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin J., Yang X., Kosters W., Xu T., Jia Y., Wang S., et al. Mako: A graph-based pattern growth approach to detect complex structural variants. Genom Proteom Bioinform. 2021 doi: 10.1016/j.gpb.2021.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchis-Juan A., Stephens J., French C.E., Gleadall N., Mégy K., Penkett C., et al. Complex structural variants in Mendelian disorders: identification and breakpoint resolution using short- and long-read genome sequencing. Genome Med. 2018;10:95. doi: 10.1186/s13073-018-0606-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jakubosky D., D’Antonio M., Bonder M.J., Smail C., Donovan M., Greenwald W.W., et al. Properties of structural variants and short tandem repeats associated with gene expression and complex traits. Nat Commun. 2020;11:2927. doi: 10.1038/s41467-020-16482-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fudenberg G., Pollard K.S. Chromatin features constrain structural variation across evolutionary timescales. Proc Natl Acad Sci. 2019;116:201808631. doi: 10.1073/pnas.1808631116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang C., Scott A.J., Davis J.R., Tsang E.K., Li X., Kim Y., et al. The impact of structural variation on human gene expression. Nat Genet. 2017;49:692–699. doi: 10.1038/ng.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Audano P.A., Sulovari A., Graves-Lindsay T.A., Cantsilieris S., Sorensen M., Welch A.E., et al. Characterizing the major structural variant alleles of the human genome. Cell. 2019;176:663–675.e19. doi: 10.1016/j.cell.2018.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almarri M.A., Bergström A., Prado-Martinez J., Yang F., Fu B., Dunham A.S., et al. Population structure, stratification, and introgression of human structural variation. Cell. 2020 doi: 10.1016/j.cell.2020.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quan C., Li Y., Liu X., Wang Y., Ping J., Lu Y., et al. Characterization of structural variation in Tibetans reveals new evidence of high-altitude adaptation and introgression. Genome Biol. 2021;22:159. doi: 10.1186/s13059-021-02382-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L., Abel H.J., Das I., Larson D.E., Ganel L., Kanchi K.L., et al. Association of structural variation with cardiometabolic traits in Finns. Am J Hum Genetics. 2021;108:583–596. doi: 10.1016/j.ajhg.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortés-Ciriano I., Lee J., Xi R., Jain D., Jung Y.L., Yang L., et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nat Genet. 2020;52:331–341. doi: 10.1038/s41588-019-0576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aganezov S., Goodwin S., Sherman R.M., Sedlazeck F.J., Arun G., Bhatia S., et al. Comprehensive analysis of structural variants in breast cancer genomes using single-molecule sequencing. Genome Res. 2020;30:1258–1273. doi: 10.1101/gr.260497.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hadi K., Yao X., Behr J.M., Deshpande A., Xanthopoulakis C., Tian H., et al. Distinct classes of complex structural variation uncovered across thousands of cancer genome graphs. Cell. 2020;183:197–210.e32. doi: 10.1016/j.cell.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshitaka S., Suzuko Z., Yutaka S., Masahide S., Ayako S. Application of long-read sequencing to the detection of structural variants in human cancer genomes. Comput Struct Biotechnol J. 2021;19:4207–4216. doi: 10.1016/j.csbj.2021.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho S.S., Urban A.E., Mills R.E. Structural variation in the sequencing era. Nat Rev Genet. 2019:1–19. doi: 10.1038/s41576-019-0180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaisson M.J., Sanders A.D., Zhao X., Malhotra A., Porubsky D., Rausch T., et al. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat Commun. 2019;10:1784. doi: 10.1038/s41467-018-08148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coster W., Weissensteiner M.H., Sedlazeck F.J. Towards population-scale long-read sequencing. Nat Rev Genet. 2021;22:1–16. doi: 10.1038/s41576-021-00367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Logsdon G.A., Vollger M.R., Eichler E.E. Long-read human genome sequencing and its applications. Nat Rev Genetics. 2020:1–18. doi: 10.1038/s41576-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magi A., Semeraro R., Mingrino A., Giusti B., D’Aurizio R. Nanopore sequencing data analysis: state of the art, applications and challenges. Brief Bioinform. 2017;19:1256–1272. doi: 10.1093/bib/bbx062. [DOI] [PubMed] [Google Scholar]
- 25.Lu T-Y, Consortium T, Munson KM, Lewis AP, Zhu Q, Tallon LJ, et al. Profiling variable-number tandem repeat variation across populations using repeat-pangenome graphs. Nat Commun 2021;12:4250. https://doi.org/10.1038/s41467-021-24378-0. [DOI] [PMC free article] [PubMed]
- 26.Stancu M., van Roosmalen M.J., Renkens I., Nieboer M.M., Middelkamp S., de Ligt J., et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat Commun. 2017;8:1326. doi: 10.1038/s41467-017-01343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens Z., Wang C., Iyer R.K., Kocher J.-P. Detection and visualization of complex structural variants from long reads. BMC Bioinf. 2018;19:508. doi: 10.1186/s12859-018-2539-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jain M., Koren S., Miga K.H., Quick J., Rand A.C., Sasani T.A., et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol. 2018;36:338–345. doi: 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miga K.H., Koren S., Rhie A., Vollger M.R., Gershman A., Bzikadze A., et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature. 2020;585:79–84. doi: 10.1038/s41586-020-2547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi L., Guo Y., Dong C., Huddleston J., Yang H., Han X., et al. Long-read sequencing and de novo assembly of a Chinese genome. Nat Commun. 2016;7:12065. doi: 10.1038/ncomms12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ouzhuluobu H.Y., Lou H., Cui C., Deng L., Gao Y., et al. De novo assembly of a Tibetan genome and identification of novel structural variants associated with high altitude adaptation. Natl Sci Rev. 2020;7:391–402. doi: 10.1093/nsr/nwz160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beyter D., Ingimundardottir H., Oddsson A., Eggertsson H.P., Bjornsson E., Jonsson H., et al. Long-read sequencing of 3,622 Icelanders provides insight into the role of structural variants in human diseases and other traits. Nat Genet. 2021;53:1–8. doi: 10.1038/s41588-021-00865-4. [DOI] [PubMed] [Google Scholar]
- 33.Coster W., Broeckhoven C. Newest methods for detecting structural variations. Trends Biotechnol. 2019;37:973–982. doi: 10.1016/j.tibtech.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Lan T., Lin H., Zhu W., Laurent T., Yang M., Liu X., et al. Deep whole-genome sequencing of 90 Han Chinese genomes. GigaScience. 2017;6:1–7. doi: 10.1093/gigascience/gix067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergström A., McCarthy S.A., Hui R., Almarri M.A., Ayub Q., Danecek P., et al. Insights into human genetic variation and population history from 929 diverse genomes. Science. 2020;367 doi: 10.1126/science.aay5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mallick S., Li H., Lipson M., Mathieson I., Gymrek M., Racimo F., et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016;538:201–206. doi: 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Byrska-Bishop M, Evani US, Zhao X, Basile AO, Abel HJ, Regier AA, et al. High coverage whole genome sequencing of the expanded 1000 Genomes Project cohort including 602 trios. Biorxiv 2021:2021.02.06.430068. https://doi.org/10.1101/2021.02.06.430068. [DOI] [PMC free article] [PubMed]
- 38.Campbell P.J., Getz G., Korbel J.O., Stuart J.M., Jennings J.L., Stein L.D., et al. Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen S., Krusche P., Dolzhenko E., Sherman R.M., Petrovski R., Schlesinger F., et al. Paragraph: a graph-based structural variant genotyper for short-read sequence data. Genome Biol. 2019;20:291. doi: 10.1186/s13059-019-1909-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi J, Jia Z, Zhao X, Sun J, Liang F, Park M, et al. Structural variant selection for high-altitude adaptation using single-molecule long-read sequencing. Biorxiv 2021:2021.03.27.436702. https://doi.org/10.1101/2021.03.27.436702.
- 41.Wu Z., Jiang Z., Li T., Xie C., Zhao L., Yang J., et al. Structural variants in the Chinese population and their impact on phenotypes, diseases and population adaptation. Nat Commun. 2021;12:6501. doi: 10.1038/s41467-021-26856-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherman R.M., Salzberg S.L. Pan-genomics in the human genome era. Nat Rev Genet. 2020;21:243–254. doi: 10.1038/s41576-020-0210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eizenga J.M., Novak A.M., Sibbesen J.A., Heumos S., Ghaffaari A., Hickey G., et al. Pangenome graphs. Annu Rev Genom Hum G. 2020;21:139–162. doi: 10.1146/annurev-genom-120219-080406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Outten J., Warren A. Methods and developments in graphical pangenomics. J Indian I Sci. 2021;101:485–498. doi: 10.1007/s41745-021-00255-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miga K.H., Wang T. The need for a human pangenome reference sequence. Annu Rev Genom Hum G. 2021;22:1–22. doi: 10.1146/annurev-genom-120120-081921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen N.-C., Solomon B., Mun T., Iyer S., Langmead B. Reference flow: reducing reference bias using multiple population genomes. Genome Biol. 2021;22:8. doi: 10.1186/s13059-020-02229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huddleston J., Chaisson M., Steinberg K., Warren W., Hoekzema K., Gordon D., et al. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res. 2017;27:677–685. doi: 10.1101/gr.214007.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lecompte L., Peterlongo P., Lavenier D., Lemaitre C. SVJedi: Genotyping structural variations with long reads. Bioinform Oxf Engl. 2020 doi: 10.1093/bioinformatics/btaa527. [DOI] [PubMed] [Google Scholar]
- 49.Letcher B., Hunt M., Iqbal Z. Gramtools enables multiscale variation analysis with genome graphs. Genome Biol. 2021;22:259. doi: 10.1186/s13059-021-02474-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khorsand P, Hormozdiari F. Nebula: ultra-efficient mapping-free structural variant genotyper. Nucleic Acids Res 2021:gkab025. https://doi.org/10.1093/nar/gkab025. [DOI] [PMC free article] [PubMed]
- 51.Schmidt M, Kutzner A. State-of-the-art structural variant calling: What went conceptually wrong and how to fix it? Biorxiv 2021:2021.01.12.426317. https://doi.org/10.1101/2021.01.12.426317.
- 52.Bizjan B., Katsila T., Tesovnik T., Šket R., Debeljak M., Matsoukas M., et al. Challenges in identifying large germline structural variants for clinical use by long read sequencing. Comput Struct Biotechnol J. 2019;18:83–92. doi: 10.1016/j.csbj.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Z., Roberts R., Mercer T.R., Xu J., Sedlazeck F.J., Tong W. Towards accurate and reliable resolution of structural variants for clinical diagnosis. Genome Biol. 2022;23:68. doi: 10.1186/s13059-022-02636-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Logsdon G.A., Vollger M.R., Hsieh P., Mao Y., Liskovykh M.A., Koren S., et al. The structure, function and evolution of a complete human chromosome 8. Nature. 2021:1–7. doi: 10.1038/s41586-021-03420-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsieh P., Vollger M.R., Dang V., Porubsky D., Baker C., Cantsilieris S., et al. Adaptive archaic introgression of copy number variants and the discovery of previously unknown human genes. Science. 2019;366:eaax2083. doi: 10.1126/science.aax2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kronenberg Z.N., Fiddes I.T., Gordon D., Murali S., Cantsilieris S., Meyerson O.S., et al. High-resolution comparative analysis of great ape genomes. Science. 2018;360:eaar6343. doi: 10.1126/science.aar6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seo J.-S., Rhie A., Kim J., Lee S., Sohn M.-H., Kim C.-U., et al. De novo assembly and phasing of a Korean human genome. Nature. 2016;538:243–247. doi: 10.1038/nature20098. [DOI] [PubMed] [Google Scholar]
- 59.Yang X., Lee W.-P., Ye K., Lee C. One reference genome is not enough. Genome Biol. 2019;20:104. doi: 10.1186/s13059-019-1717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soto D.C., Shew C., Mastoras M., Schmidt J.M., Sahasrabudhe R., Kaya G., et al. Identification of structural variation in chimpanzees using optical mapping and nanopore sequencing. Genes-Basel. 2020;11:276. doi: 10.3390/genes11030276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chiang C., Layer R.M., Faust G.G., Lindberg M.R., Rose D.B., Garrison E.P., et al. SpeedSeq: Ultra-fast personal genome analysis and interpretation. Nat Methods. 2015;12:966–968. doi: 10.1038/nmeth.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abyzov A., Urban A.E., Snyder M., Gerstein M. CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu T.-Y., Chaisson M. The motif composition of variable-number tandem repeats impacts gene expression. BioRxiv. 2022 doi: 10.1101/2022.03.17.484784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sirén J., Monlong J., Chang X., Novak A.M., Eizenga J.M., Markello C., et al. Pangenomics enables genotyping of known structural variants in 5202 diverse genomes. Science. 2021;374:abg8871. doi: 10.1126/science.abg8871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yan S.M., Sherman R.M., Taylor D.J., Nair D.R., Bortvin A.N., Schatz M.C., et al. Local adaptation and archaic introgression shape global diversity at human structural variant loci. Elife. 2021;10:e67615. doi: 10.7554/elife.67615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tettelin H., Masignani V., Cieslewicz M.J., Donati C., Medini D., Ward N.L., et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eggertsson H.P., Jonsson H., Kristmundsdottir S., Hjartarson E., Kehr B., Masson G., et al. Graphtyper enables population-scale genotyping using pangenome graphs. Nat Genet. 2017;49:1654–1660. doi: 10.1038/ng.3964. [DOI] [PubMed] [Google Scholar]
- 68.Eggertsson H.P., Kristmundsdottir S., Beyter D., Jonsson H., Skuladottir A., Hardarson M.T., et al. GraphTyper2 enables population-scale genotyping of structural variation using pangenome graphs. Nat Commun. 2019;10:5402. doi: 10.1038/s41467-019-13341-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jónsson H., Sulem P., Kehr B., Kristmundsdottir S., Zink F., Hjartarson E., et al. Whole genome characterization of sequence diversity of 15,220 Icelanders. Sci Data. 2017;4:170115. doi: 10.1038/sdata.2017.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sirén J., Garrison E., Novak A.M., Paten B., Durbin R. Haplotype-aware graph indexes. Bioinformatics. 2020;36:400–407. doi: 10.1093/bioinformatics/btz575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hickey G., Heller D., Monlong J., Sibbesen J.A., Sirén J., Eizenga J., et al. Genotyping structural variants in pangenome graphs using the vg toolkit. Genome Biol. 2020;21:35. doi: 10.1186/s13059-020-1941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ebler J, Clarke WE, Rausch T, Audano PA, Houwaart T, Korbel J, et al. Pangenome-based genome inference. Biorxiv 2020:2020.11.11.378133. https://doi.org/10.1101/2020.11.11.378133.
- 73.Rausch T., Zichner T., Schlattl A., Stütz A.M., Benes V., Korbel J.O. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Antaki D., Brandler W.M., Sebat J. SV2: accurate structural variation genotyping and de novo mutation detection from whole genomes. Bioinformatics. 2017;34:1774–1777. doi: 10.1093/bioinformatics/btx813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chander V., Gibbs R.A., Sedlazeck F.J. Evaluation of computational genotyping of structural variation for clinical diagnoses. GigaScience. 2019;8 doi: 10.1093/gigascience/giz110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhuang X., Ye R., So M.-T., Lam W.-Y., Karim A., Yu M., et al. A random forest-based framework for genotyping and accuracy assessment of copy number variations. Nar Genom Bioinform. 2020;2:lqaa071. doi: 10.1093/nargab/lqaa071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chowdhury M., Pedersen B.S., Sedlazeck F.J., Quinlan A.R., Layer R.M. Searching thousands of genomes to classify somatic and novel structural variants using STIX. Nat Methods. 2022;19:445–448. doi: 10.1038/s41592-022-01423-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jun G., Sedlazeck F., Zhu Q., English A., Metcalf G., Kang H., et al. muCNV: genotyping structural variants for population-level sequencing. Bioinformatics. 2021 doi: 10.1093/bioinformatics/btab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Linderman M.D., Paudyal C., Shakeel M., Kelley W., Bashir A., Gelb B.D. NPSV: A simulation-driven approach to genotyping structural variants in whole-genome sequencing data. GigaScience. 2021;10:giab046. doi: 10.1093/gigascience/giab046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Handsaker R.E., Doren V., Berman J.R., Genovese G., Kashin S., Boettger L.M., et al. Large multiallelic copy number variations in humans. Nat Genet. 2015;47:296–303. doi: 10.1038/ng.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sedlazeck F.J., Rescheneder P., Smolka M., Fang H., Nattestad M., von Haeseler A., et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 2018;15:461–468. doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smolka M., Paulin L.F., Grochowski C.M., Mahmoud M., Behera S., Gandhi M., et al. Comprehensive structural variant detection: from mosaic to population-level. BioRxiv. 2022 doi: 10.1101/2022.04.04.487055. [DOI] [Google Scholar]
- 83.Spies N., Zook J.M., Salit M., Sidow A. svviz: a read viewer for validating structural variants. Bioinform Oxf Engl. 2015;31:3994–3996. doi: 10.1093/bioinformatics/btv478. [DOI] [PubMed] [Google Scholar]
- 84.Handsaker R.E., Korn J.M., Nemesh J., McCarroll S.A. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat Genet. 2011;43:269–276. doi: 10.1038/ng.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM 2013.
- 86.Huang W., Li L., Myers J.R., Marth G.T. ART: a next-generation sequencing read simulator. Bioinformatics. 2012;28:593–594. doi: 10.1093/bioinformatics/btr708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Iqbal Z., Caccamo M., Turner I., Flicek P., McVean G. De novo assembly and genotyping of variants using colored de Bruijn graphs. Nat Genet. 2012;44:226–232. doi: 10.1038/ng.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hunt M, Letcher B, Malone K, Nguyen G, Hall, Colquhoun R, et al. Minos: variant adjudication and joint genotyping of cohorts of bacterial genomes. Biorxiv 2021:2021.09.15.460475. https://doi.org/10.1101/2021.09.15.460475. [DOI] [PMC free article] [PubMed]
- 89.Consortium T, Sibbesen J., Maretty L., Krogh A. Accurate genotyping across variant classes and lengths using variant graphs. Nat Genet. 2018;50:1054–1059. doi: 10.1038/s41588-018-0145-5. [DOI] [PubMed] [Google Scholar]
- 90.Garrison E., Sirén J., Novak A.M., Hickey G., Eizenga J.M., Dawson E.T., et al. Variation graph toolkit improves read mapping by representing genetic variation in the reference. Nat Biotechnol. 2018;36:875–879. doi: 10.1038/nbt.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rautiainen M., Marschall T. GraphAligner: rapid and versatile sequence-to-graph alignment. Genome Biol. 2020;21:253. doi: 10.1186/s13059-020-02157-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smith T.F., Waterman M.S. Comparison of biosequences. Adv Appl Math. 1981;2:482–489. doi: 10.1016/0196-8858(81)90046-4. [DOI] [Google Scholar]
- 93.Farrar M. Striped Smith-Waterman speeds database searches six times over other SIMD implementations. Bioinformatics. 2007;23:156–161. doi: 10.1093/bioinformatics/btl582. [DOI] [PubMed] [Google Scholar]
- 94.Zhao M., Lee W.-P., Garrison E.P., Marth G.T. SSW library: an SIMD Smith-Waterman C/C++ library for use in genomic applications. PLoS ONE. 2013;8:e82138. doi: 10.1371/journal.pone.0082138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ghaffaari A., Marschall T. Fully-sensitive seed finding in sequence graphs using a hybrid index. Bioinformatics. 2019;35:i81–i89. doi: 10.1093/bioinformatics/btz341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Garrison E, Guarracino A. Unbiased pangenome graphs. Biorxiv 2022:2022.02.14.480413. https://doi.org/10.1101/2022.02.14.480413.
- 97.Khan J., Patro R. Cuttlefish: fast, parallel and low-memory compaction of de Bruijn graphs from large-scale genome collections. Bioinformatics. 2021;37:i177–i186. doi: 10.1093/bioinformatics/btab309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Khan J, Kokot M, Deorowicz S, Patro R. Scalable, ultra-fast, and low-memory construction of compacted de Bruijn graphs with Cuttlefish 2. Biorxiv 2021:2021.12.14.472718. https://doi.org/10.1101/2021.12.14.472718. [DOI] [PMC free article] [PubMed]
- 99.Guarracino A, Heumos S, Nahnsen S, Prins P, Garrison E. ODGI: understanding pangenome graphs. Biorxiv 2021:2021.11.10.467921. https://doi.org/10.1101/2021.11.10.467921.
- 100.Colquhoun R.M., Hall M.B., Lima L., Roberts L.W., Malone K.M., Hunt M., et al. Pandora: nucleotide-resolution bacterial pan-genomics with reference graphs. Genome Biol. 2021;22:267. doi: 10.1186/s13059-021-02473-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Břinda K., Baym M., Kucherov G. Simplitigs as an efficient and scalable representation of de Bruijn graphs. Genome Biol. 2021;22:96. doi: 10.1186/s13059-021-02297-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Holley G., Melsted P. Bifrost: highly parallel construction and indexing of colored and compacted de Bruijn graphs. Genome Biol. 2020;21:249. doi: 10.1186/s13059-020-02135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Eizenga J.M., Novak A.M., Kobayashi E., Villani F., Cisar C., Heumos S., et al. Efficient dynamic variation graphs. Bioinformatics. 2020;36:5139–5144. doi: 10.1093/bioinformatics/btaa640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li H., Feng X., Chu C. The design and construction of reference pangenome graphs with minigraph. Genome Biol. 2020;21:265. doi: 10.1186/s13059-020-02168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rakocevic G., Semenyuk V., Lee W.-P., Spencer J., Browning J., Johnson I.J., et al. Fast and accurate genomic analyses using genome graphs. Nat Genet. 2019;51:354–362. doi: 10.1038/s41588-018-0316-4. [DOI] [PubMed] [Google Scholar]
- 106.Gagie T., Manzini G., Sirén J. Wheeler graphs: A framework for BWT-based data structures. Theor Comput Sci. 2017;698:67–78. doi: 10.1016/j.tcs.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ma J, Cáceres M, Salmela L, Mäkinen V, Tomescu AI. GraphChainer: Co-linear Chaining for Accurate Alignment of Long Reads to Variation Graphs. Biorxiv 2022:2022.01.07.475257. https://doi.org/10.1101/2022.01.07.475257.
- 108.Luhmann N., Holley G., Achtman M. BlastFrost: fast querying of 100,000s of bacterial genomes in Bifrost graphs. Genome Biol. 2021;22:30. doi: 10.1186/s13059-020-02237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ivanov P, Bichsel B, Vechev M. Fast and Optimal Sequence-to-Graph Alignment Guided by Seeds. Biorxiv 2021:2021.11.05.467453. https://doi.org/10.1101/2021.11.05.467453.
- 110.Ivanov P., Bichsel B., Mustafa H., Kahles A., Rätsch G., Vechev M. AStarix: fast and optimal sequence-to-graph alignment. Lect Notes Comput Sc. 2020:104–119. doi: 10.1007/978-3-030-45257-5_7. [DOI] [Google Scholar]
- 111.Dvorkina T., Antipov D., Korobeynikov A., Nurk S. SPAligner: alignment of long diverged molecular sequences to assembly graphs. BMC Bioinf. 2020;21:306. doi: 10.1186/s12859-020-03590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Darby C.A., Gaddipati R., Schatz M.C., Langmead B. Vargas: heuristic-free alignment for assessing linear and graph read aligners. Bioinformatics. 2020;36:3712–3718. doi: 10.1093/bioinformatics/btaa265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jain C, Dilthey A, Misra S, Zhang H, Aluru S. Accelerating Sequence Alignment to Graphs. Biorxiv 2019:651638. https://doi.org/10.1101/651638.
- 114.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kavya V., Tayal K., Srinivasan R., Sivadasan N. Sequence alignment on directed graphs. J Comput Biol. 2019;26:53–67. doi: 10.1089/cmb.2017.0264. [DOI] [PubMed] [Google Scholar]
- 116.Nurk S., Koren S., Rhie A., Rautiainen M., Bzikadze A.V., Mikheenko A., et al. The complete sequence of a human genome. Science. 2022;376:44–53. doi: 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Aganezov S., Yan S.M., Soto D.C., Kirsche M., Zarate S., Avdeyev P., et al. A complete reference genome improves analysis of human genetic variation. Science. 2022;376 doi: 10.1126/science.abl3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang T., Antonacci-Fulton L., Howe K., Lawson H.A., Lucas J.K., Phillippy A.M., et al. The Human Pangenome Project: a global resource to map genomic diversity. Nature. 2022;604:437–446. doi: 10.1038/s41586-022-04601-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sekar S, Tomasini L, Proukakis C, Bae T, Manlove L, Jang Y, et al. Complex mosaic structural variations in human fetal brains. Genome Res 2020:gr.262667.120. https://doi.org/10.1101/gr.262667.120. [DOI] [PMC free article] [PubMed]
- 120.Layer RM, Sedlazeck FJ, Pedersen BS, Quinlan AR. Mining Thousands of Genomes to Classify Somatic and Pathogenic Structural Variants. Biorxiv 2021:2021.04.21.440844. https://doi.org/10.1101/2021.04.21.440844.