Abstract
Tau assembly movement from the extracellular to intracellular space may underlie transcellular propagation of neurodegenerative tauopathies. This begins with tau binding to cell surface heparan sulfate proteoglycans, which triggers macropinocytosis. Pathological tau assemblies are proposed then to exit the vesicular compartment as “seeds” for replication in the cytoplasm. Tau uptake is highly efficient, but only ∼1 to 10% of cells that endocytose aggregates exhibit seeding. Consequently, we studied fluorescently tagged full-length (FL) tau fibrils added to native U2OS cells or “biosensor” cells expressing FL tau or repeat domain. FL tau fibrils bound tubulin. Seeds triggered its aggregation in multiple locations simultaneously in the cytoplasm, generally independent of visible exogenous aggregates. Most exogenous tau trafficked to the lysosome, but fluorescence imaging revealed a small percentage that steadily accumulated in the cytosol. Intracellular expression of Gal3-mRuby, which binds intravesicular galactosides and forms puncta upon vesicle rupture, revealed no evidence of vesicle damage following tau exposure, and most seeded cells had no evidence of endolysosome rupture. However, live-cell imaging indicated that cells with pre-existing Gal3-positive puncta were seeded at a slightly higher rate than the general population, suggesting a potential predisposing role for vesicle instability. Clearance of tau seeds occurred rapidly in both vesicular and cytosolic fractions. The lysosome/autophagy inhibitor bafilomycin inhibited vesicular clearance, whereas the proteasome inhibitor MG132 inhibited cytosolic clearance. Tau seeds that enter the cell thus have at least two fates: lysosomal clearance that degrades most tau, and entry into the cytosol, where seeds amplify, and are cleared by the proteasome.
Keywords: Alzheimer’s disease, tau propagation, tauopathy, tau seed, tau aggregation
Abbreviations: AD, Alzheimer's disease; AF647, Alexa Fluor 647; FL, full-length; Gal3-Rub, galectin-3 β-galactoside–binding protein fused to mRuby3; LAMP1, lysosomal-associated membrane protein 1; LLOMe, l-leucyl-l-leucine O-methyl ester; TBST, Tris-buffered saline with Tween-20
Multiple lines of experimental evidence suggest that tau protein triggers neurodegeneration after intracellular accumulation in ordered assemblies. Myriad tauopathies are linked to distinct assembly structures and include Alzheimer’s disease (AD), frontotemporal dementia, and chronic traumatic encephalopathy, among many others (1). The transcellular propagation and faithful replication of unique assembly structures, or “strains,” appears to underlie the characteristic progression patterns of specific tauopathies (2). Tau propagation presumably involves three steps: uptake of an assembly into the cell; amplification and maintenance of the aggregated state; and exit from the cell. We originally observed that tau binding to heparan sulfate proteoglycans on the cell surface underlies uptake via macropinocytosis, and is mediated by specific heparan sulfate proteoglycans sulfation patterns (3, 4). After uptake into macropinosomes, an assembly must make contact with endogenous tau in the cytoplasm to serve as a template and amplify a specific structure. Several reports suggest that this might happen after vesicle rupture (5, 6), but the details are unclear. Finally, when tau seeds enter the cytoplasm, it is unknown how they are degraded. In this study, we used cultured cells to dynamically visualize tau uptake and seeding, to track the relationship of seeding to vesicle processing and rupture, and to determine mechanisms of seed degradation in the vesicular versus cytoplasmic compartments.
Results
Tau binds tubulin and is recruited to aggregates in the cytosol
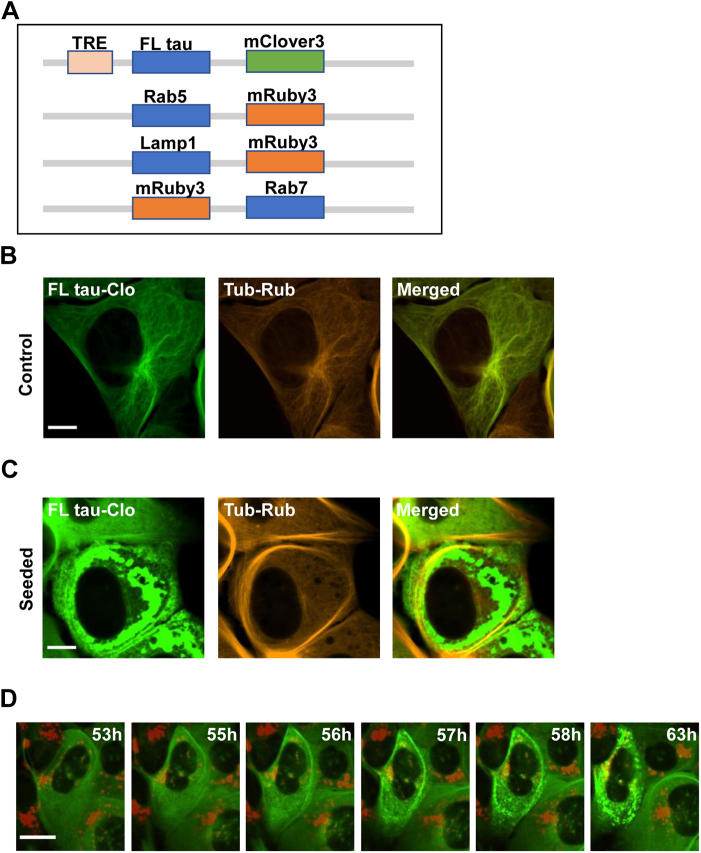
Wildtype full-length (FL) tau undergoes seeded aggregation inefficiently in cultured cells, and thus, we studied FL (2N4R) tau containing a disease-associated P301S mutation, fused to mClover3 (FL tau-Clo) (Fig. 1A). We stably expressed FL tau-Clo in U2OS cells, where it colocalized with tubulin (Fig. 1B). We then directly imaged U2OS biosensor cells that were exposed to exogenous FL wildtype (2N4R) tau fibrils covalently labeled with Alexa Fluor 647 (AF647), tracking FL tau-Clo puncta formation over time. Similar to our original observations (7), a minority of cells exhibited induced aggregation of tau, which predominated in the cytoplasm (Fig. 1C). Time-lapse imaging (IN Cell Analyzer 6000, GE) also revealed intracellular aggregation in the cytoplasm (Fig. 1D), and in imaging dynamic inclusion formation in ∼50 cells, we often observed aggregates forming simultaneously throughout the cytoplasm (Movie S1). For nascent intracellular aggregates, we observed no significant colocalization with AF647-labeled exogenous tau. This raised the question of how tau seeds traffic into the cytoplasm.
Figure 1.
Tau aggregation initiates in the cytoplasm.A, diagram of constructs used for this study. Expression of full-length (FL) tau was regulated by a tetracycline response element (TRE). B, U2OS cells expressing FL tau-Clo containing the P301S mutation and stained for tubulin, without exogenous wildtype fibril addition. FL tau-Clo colocalizes with tubulin. C, with exogenous fibril addition, tau redistributes to inclusions in the cytoplasm. D, recombinant FL tau fibrils labeled with AF647 (red) were applied to the U2OS cells expressing FL tau-Clo (green) and imaged over time to track tau aggregation (labeled). Note rapid and simultaneous development of tau inclusions throughout the cytoplasm. Scale bars = 10 μm. AF647, Alexa Fluor 647.
Internalized tau aggregates traffic to the endolysosomal system
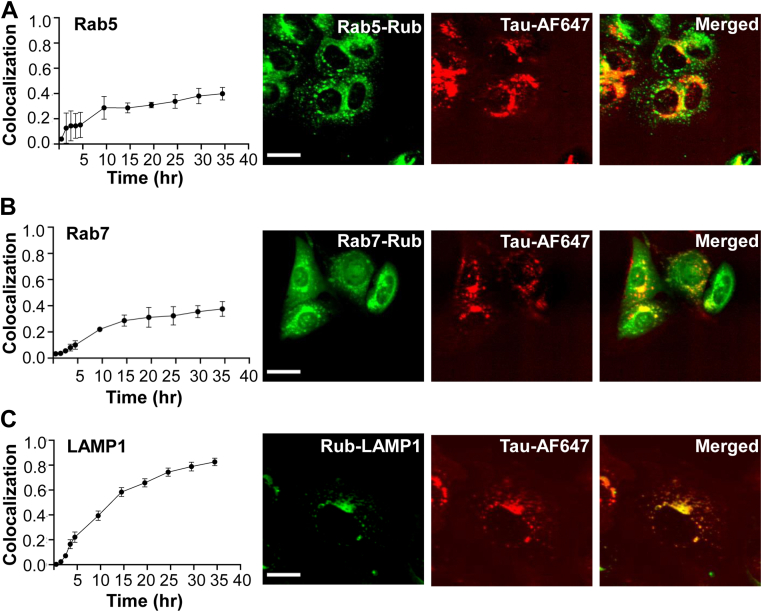
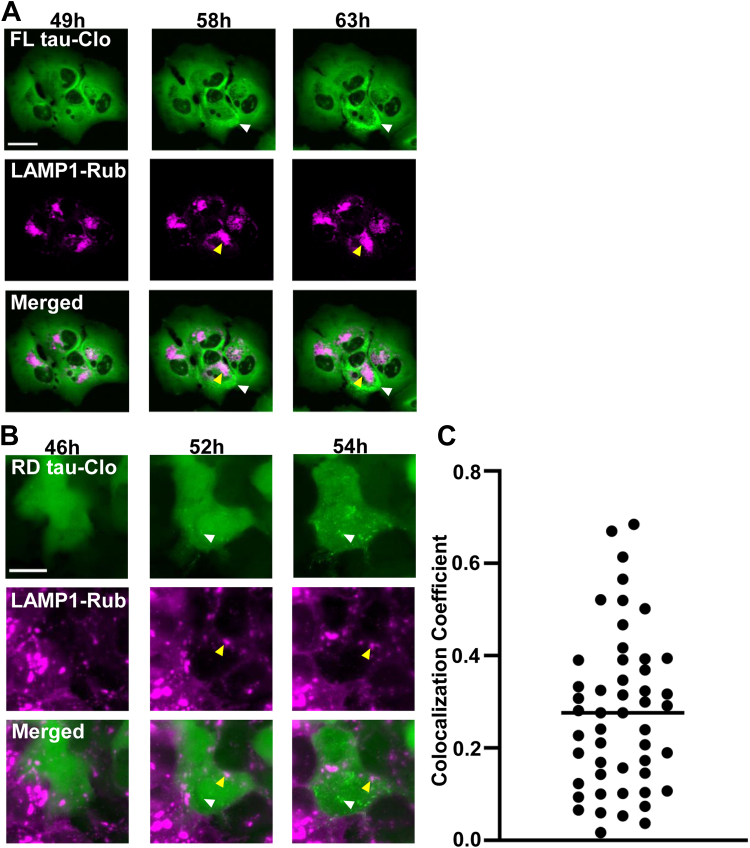
To study the fate of internalized tau, we exposed U2OS to FL tau fibrils labeled with AF647, imaging them repeatedly over 2 days with high-content microscopy. We first tested for colocalization with vesicles by stably expressing mRuby3 fusions to Rab5 (to mark early endosomes, Figs. 1A and 2A), Rab7 (to mark late endosomes, Figs. 1A and 2B), and lysosomal-associated membrane protein 1 (LAMP1) (to mark lysosomes, Figs. 1A and 2C). Tau progressively colocalized with these markers, especially LAMP1. We concluded that most internalized tau entered the endolysosomal pathway. When we tracked seeding in U2OS biosensor cells expressing FL tau-Clo using LAMP1, we observed no significant colocalization of emergent tau puncta with the lysosome, which was largely in a separate compartment versus the induced FL-tau-Clo aggregates (Fig. 3A). FL tau seeding is inefficient enough to make capture of large numbers of seeding events relatively difficult. We therefore performed the same experiment using repeat domain tau-Clo containing a P301S mutation (RD tau-Clo) to image a high number of seeded cells (Fig. 3B). Similar to our observation with FL tau, we observed no significant colocalization between RD tau-Clo aggregates and LAMP1 (Fig. 3C). Thus, although most internalized tau wound up in the lysosome, this seemed unlikely to be the primary location of seeding.
Figure 2.
Exogenous tau fibrils colocalize with the lysosome. U2OS cells were treated with FL tau fibrils labeled with AF647. Markers of early (Rab5), late (Rab7), and lysosome (LAMP1) were expressed as mRuby (Rub) fusions to track colocalization with internalized fibrils (Fig. 1A). Cells were tracked over time using high-content microscopy. Colocalization ratio refers to the fractional colocalization of the two fluorescent signals. Representative endpoint images are shown. A, colocalization of tau with Rab5-Rub increases over time but never exceeds ∼0.4 over 36 h. B, colocalization of tau with Rub-Rab7 reaches ∼0.4 over 36 h. C, colocalization with Lamp1-Rub reaches ∼0.8 over 36 h. n > 200 cells were imaged iteratively for each condition. Error bars represent SD. Scale bars = 30 μm. AF647, Alexa Fluor 647; LAMP1, lysosomal-associated membrane protein 1.
Figure 3.
Tau inclusion formation is independent of the lysosome.A, U2OS cells expressing FL tau-Clo and LAMP1-Rub were treated with exogenous fibrils. Cells were observed over time for coincidence of LAMP1-Rub signal and FL tau-Clo inclusion formation. A representative image shows the seeded tau (white arrowhead) and LAMP1 signal (magenta). No colocalization was observed between tau aggregates (white arrow) and lysosome (yellow arrow). Time after tau fibril addition is indicated above each column. The images are representative of five seeded cells studied. Scale bars = 20 μm. B, U2OS cells expressing RD tau-Clo and LAMP1-Rub were treated with tau fibril and followed over time for coincidence of LAMP1 signal and RD tau-Clo aggregation. No colocalization was observed between tau aggregates (white arrowhead) and the LAMP1 marking lysosome (yellow arrowhead). The images are representative of 50 seeded cells. Scale bars = 20 μm. C, quantification of colocalization of LAMP1-Rub with RD tau-Clo aggregates. The graph shows colocalization coefficient (Mander’s overlap coefficient) from 50 seeded cells, and the bar indicates the mean. LAMP1, lysosomal-associated membrane protein 1; RD, repeat domain.
Tau seeding is independent of the lysosome
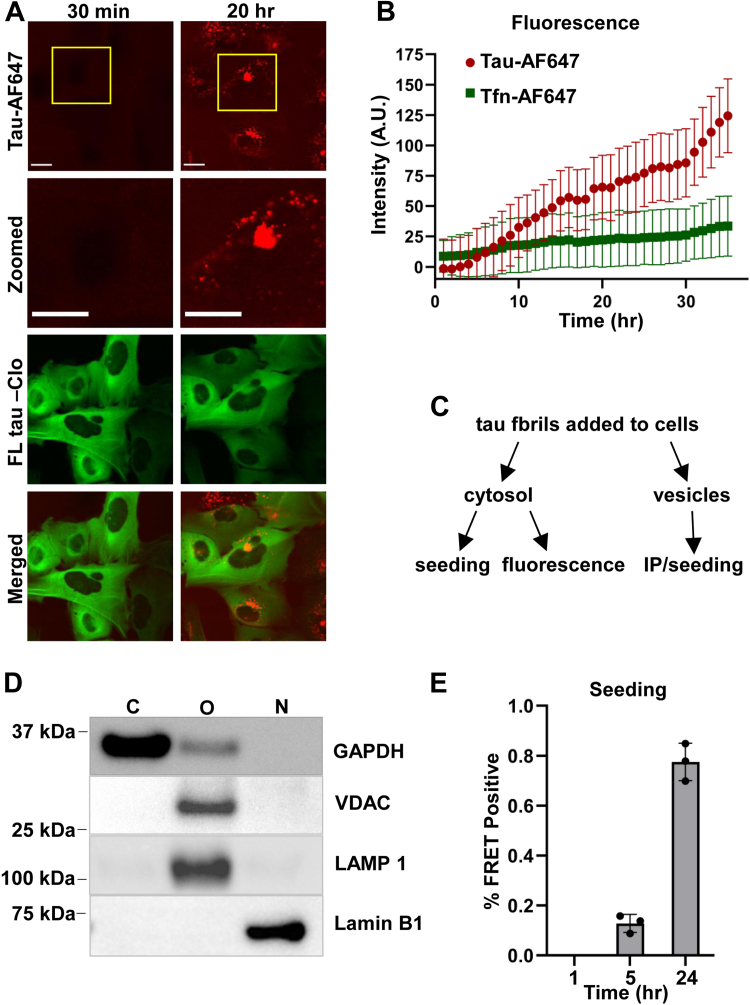
Since we did not observe significant colocalization of newly formed tau puncta with vesicle markers, we hypothesized that tau seeds might be released into the cytoplasm from a vesicular pool. After exposing U2OS cells to FL tau fibrils labeled with AF647, we detected diffuse fluorescence in the cytoplasm at 20 h (Fig. 4A). We next used live-cell imaging to monitor hundreds of cells exposed to FL tau fibrils tagged with AF647. We observed a small but steady increase of cytosolic AF647 signal, whereas transferrin-AF647 did not increase in signal following a similar exposure protocol (Fig. 4B). To test our observations by a different approach, we used cell fractionation based on differential centrifugation to measure tau seed levels in cytosol versus organelle (vesicle) fractions (Fig. 4C). We confirmed the accuracy of the fractionations using Western blot against GAPDH (cytosol), voltage-dependent anion channel (organelle), LAMP1 (organelle), and lamin B1 (nucleus) (Fig. 4D). We attempted to quantify tau protein levels via Western blot, ELISA, and mass spectrometry, but they were too low for reliable measurements. We next monitored seeding activity in the cytoplasm by transducing lysate into a well-characterized biosensor assay based on a next-generation cell line, v2L (8, 9). We observed a steady increase in cytosol seeding activity over time (Fig. 4E). Tau seeds thus steadily moved from vesicles to the cytosol.
Figure 4.
Exogenous tau enters the cytosol. U2OS cells expressing FL tau-Clo (green) were exposed to tau fibrils labeled with AF647 (red), and fluorescence was tracked over time using live-cell imaging. A, representative images illustrate the increase in the cytosolic AF647 fluorescence between 30 min and 20 h. Zoomed-in regions are marked in yellow box and pictured in the second row. Quantification was based on imaging n >500 cells. Scale bars = 20 μm. B, tau-AF647 signal in the cytosol was compared with transferrin (Tfn)-AF647 added to cells. Tau signal increased over time, whereas Tfn signal did not. Fluorescence is indicated in arbitrary units (A.U.). Error bars represent SD. C, diagram indicating steps used to quantify exogenous tau seeding in different fractions. Immunoprecipitation was used to measure tau seeding in vesicle fractions, as crude lysate was toxic. D, fractions were analyzed by Western blot to assess fractionation fidelity. Antibodies against the indicated proteins were used to probe different cell fractions. C: cytosolic; O: organelle; and N: nuclear. E, cytosol tau seeding was measured by transducing v2L biosensors with the cytosol fraction from untransfected U2OS cells (after tau fibril addition). Representative graph shows the increase in cytosol seeding over time. The background seeding value from a negative control (cytosol from untreated cells) was subtracted from each data point. The seeding assay was performed on three independent biological replicates, and each individual measurement was carried out in triplicate. Error bars indicate SD. AF647, Alexa Fluor 647.
Tau fibrils do not damage vesicles
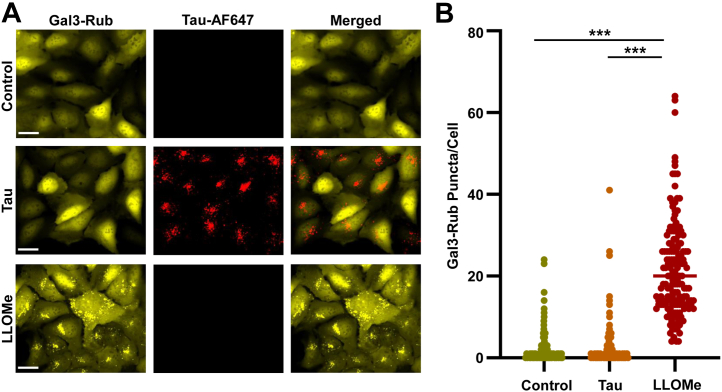
Prior studies have proposed that tau-mediated damage to vesicles might allow leakage into the cytoplasm to initiate seeding (6, 10). To test this hypothesis, we expressed the galectin-3 β-galactoside–binding protein fused to mRuby3 (Gal3-Rub), to observe the relationship of vesicle rupture and tau seeding. β-galactosides localize to the outer leaflet of the cell membrane and are present in the lumen of endocytic vesicles. Gal3-Rub expressed intracellularly is normally diffusely distributed. However, if an endosome is damaged, Gal3-Rub binds β-galactosides, creating puncta (11, 12). We stably expressed Gal3-Rub in U2OS cells and tracked puncta formation using high-content microscopy. Gal3-Rub diffusely distributed in most cells (Fig. 5A). Following exposure of cells to FL tau fibrils tagged with AF647, we observed tau in every cell. However, we observed no change in Gal3-Rub puncta formation (Fig. 5A). This was confirmed by quantitation of hundreds of cells (Fig. 5B). We contrasted this with l-leucyl-l-leucine O-methyl ester (LLOMe; Cayman Chemical) treatment to disrupt vesicles, which strongly induced Gal3-Rub puncta formation (Fig. 5B). In summary, we observed no detectable change in overall vesicle permeability upon tau exposure, suggesting that tau fibrils do not significantly damage endocytic vesicles.
Figure 5.
Tau fibrils do not damage vesicles. U2OS cells were stably transfected with Gal3-Rub to mark a loss of vesicle integrity. A, images of untreated cells (control); cells treated with FL tau-AF647 fibrils; cells treated with LLOMe. No obvious effect of fibril treatment on Gal3-Rub puncta formation was observed, whereas LLOMe produced many positive puncta. Scale bars = 30 μm. B, hundreds of cells were imaged after fixation at 24 h using high-content microscopy, and image analysis was used to quantify the frequency of Gal3-Rub puncta. There was no difference between control cells and those treated with tau fibrils, whereas LLOMe strongly induced Gal3-Rub puncta formation. n = 160 to 200 cells for each condition. p < 0.001 for effect of LLOMe (t test). Gal3-Rub, galectin-3 β-galactoside–binding protein fused to mRuby3; LLOMe, l-leucyl-l-leucine O-methyl ester.
Vesicle lysis slightly increases tau seeding
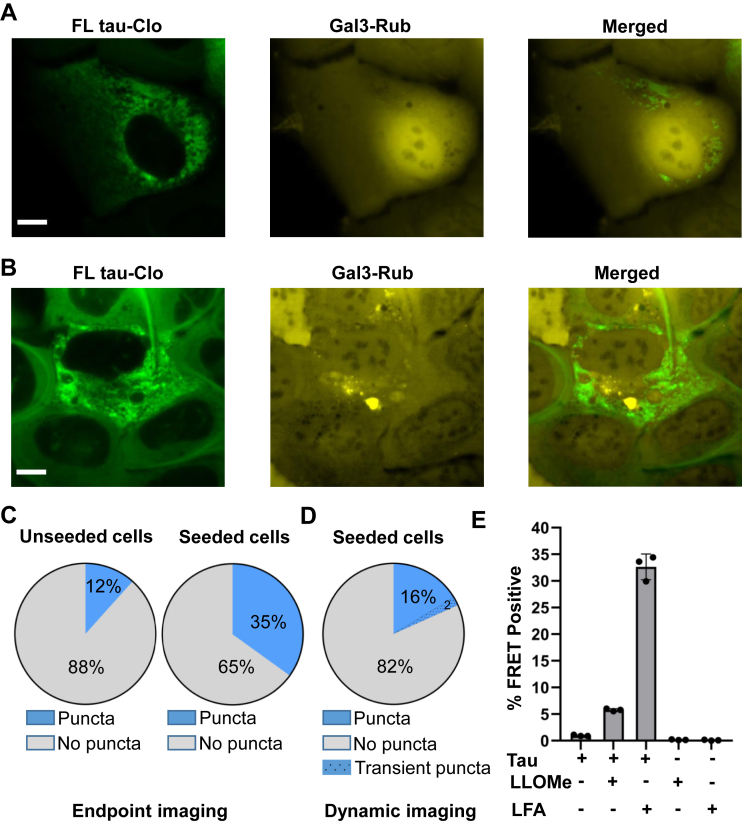
We used live-cell imaging to track seeding into cells using expression of FL tau-Clo as a biosensor. We observed seeding into cells with and without coincident Gal3-Rub–positive puncta (Fig. 6, A and B). Seeding efficiency onto FL tau is relatively low; so to more easily quantify Gal3-Rub puncta in relation to seeding events, we used the RD tau-clo biosensor. This enabled recording of hundreds of seeding events. We began by observing in an endpoint assay that ∼12% of untreated cells exhibited Gal3-Rub puncta (Fig. 6C). By contrast, ∼35% of cells with seeding exhibited coincident Gal3-Rub puncta, whereas ∼65% did not (Fig. 6C). The higher association of Gal3-Rub puncta with seeded cells led us to test the relationship more rigorously. We used dynamic imaging of cells in culture to identify those that developed tau inclusions after exogenous seeding. We then tracked the cells backward ∼12 h prior to the appearance of tau inclusions to determine the percentage that showed no Gal3-Rub puncta at any time (82%), transient Gal3-Rub puncta (2%), or pre-existing puncta (16%) (Fig. 6D). The large majority of seeded cells did not show any prior evidence of vesicle rupture. The number with pre-existing evidence of vesicle instability (18% total) was slightly higher than the total we observed in untreated cells (12%). To directly compare the seeding efficiency after direct rupture of vesicles, we tested the effect of vesicle rupture by LLOMe versus seed transduction via Lipofectamine 2000 (Thermo Fisher Scientific). We observed a modest increase in seeding after LLOMe treatment (from ∼1% to ∼5%); however, Lipofectamine treatment, which presumably delivers aggregates directly to the cytoplasm, increased seeding to ∼32% (Fig. 6E). We concluded that while vesicle integrity may influence the frequency of seeding events, this is not the primary determinant.
Figure 6.
Gal3-positive cells are partially associated with increased seeding. U2OS cells expressing Gal3-Rub were exposed to full-length (FL) tau fibrils and imaged either in endpoint assays (with cells expressing FL tau-Clo) or via live-cell imaging (with cells expressing RD tau-Clo). A and B, representative image showing seeded cells expressing FL tau-Clo (green) without (A) or with (B) coincident Gal3-Rub puncta (yellow). There was no obvious colocalization of tau and Gal3-Rub puncta. Scale bars = 10 μm. C, endpoint assay of cells expressing FL tau-Clo with or without exposure to tau fibrils. First chart shows ∼12% of Gal3-Rub puncta in unseeded cells (n = 206 cells). Second chart shows that in cells with evident FL tau-Clo puncta, the incidence of Gal3-Rub puncta increased to ∼35% (n = 166 cells). D, dynamic imaging of cells expressing tau RD-Clo and Gal3-Rub exposed to fibrils, in which cells exhibiting inclusions were tracked backward in time to determine the presence or the absence of Gal3-Rub puncta. Approximately 82% of seeded cells showed no Gal3-Rub puncta, whereas ∼18% of seeded cells showed pre-existing (16%) or transient (2%) Gal3-Rub puncta (n = 202 cells). E, comparison of seeding between unmodified application of fibrils, LLOMe, or Lipofectamine 2000 (LFA)-mediated fibril exposure. LLOMe modestly increased seeding, whereas LFA strongly increased seeding. Error bars represent SD. Each data point represents a sample analyzed in technical triplicate. Gal3-Rub, galectin-3 β-galactoside–binding protein fused to mRuby3; LLOMe, l-leucyl-l-leucine O-methyl ester; RD, repeat domain.
Distinct mechanisms of tau seed clearance in vesicles versus cytosol
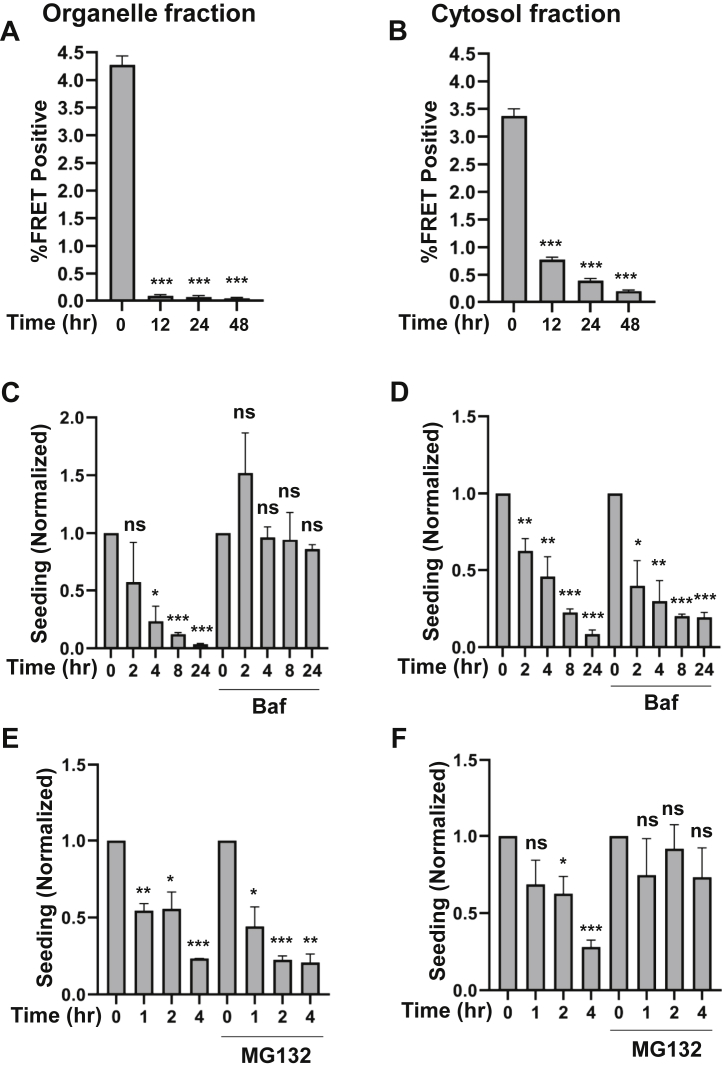
Our observations were consistent with escape of tau seeds from the vesicular compartment to the cytosol, where seeding occurs. The persistence of tau in each compartment would thus determine the relative efficiency of seeding. Consequently, we evaluated the kinetics of tau seed clearance using purification of seeds from each fraction, coupled with detection using standard v2L biosensor cells (9). We exposed U2OS cells to recombinant FL tau fibrils, followed by fractionation of cells into organelle (vesicle) versus cytosol fractions. We used immunoprecipitation to test for seeding within the organelle fraction (to avoid toxicity on biosensor cells) and directly assayed the cytosol fraction. In the organelle fraction, tau had virtually disappeared by 12 h (Fig. 7A). In the cytosol fraction, clearance was slightly slower, with tau seeding detectable even at 48 h (Fig. 7B). We attempted to use ELISA and mass spectrometry to monitor tau clearance directly, but levels were too low for accurate measurement without using inordinately large amounts of recombinant tau in cultured cells.
Figure 7.
Differential degradation of tau in cytosol versus vesicles. Native U2OS cells were exposed to tau fibrils overnight, then treated with trypsin, washed, and incubated for indicated times. Cells were then subjected to biochemical fractionation to isolate tau in the cytosol versus organelle (vesicle) fractions. For organelle fractions, seeds were extracted by immunoprecipitation to avoid cytotoxicity of vesicle contents. The respective fractions were transduced with Lipofectamine 2000 into V2L biosensor cells. A and B, time course of tau seeding activity present in cytosol (A) versus organelle (B) fractions revealed degradation in both. A t test was performed to compare samples with the corresponding untreated control (0 h). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. C and D, bafilomycin (Baf; 200 nM) was added to block lysosomal acidification. This slowed loss of tau seeding activity in the organelle fraction but had no effect on seeding within the cytosol. A t test was performed to compare samples with the corresponding untreated control (0 h). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. E and F, MG132 (10 μM) was added to block proteasome activity. E, MG132 had no effect on seeding in the organelle fraction, (F) but slowed cytoplasm tau degradation. Assays were performed in technical triplicate. Data are representative of three similar experiments for MG132 and bafilomycin treatment for cytosolic fractions and two replicates for organelle fractions. Error bars represent SEM. A t test was performed to compare samples with the corresponding untreated control (0 h). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
We next tested the effect of inhibitors of the lysosome/autophagy (bafilomycin Sigma–Aldrich) and proteasome (MG132, Sigma-Aldrich) on seed clearance over 4 h, a maximum time point picked to minimize secondary effects. Bafilomycin halted degradation in the organelle fraction (Fig. 7C) but had no effect on the cytosol fraction (Fig. 7D). By contrast, MG132 had no effect on organelle clearance (Fig. 7E) but prevented clearance of tau seeds from the cytosol (Fig. 7F). We observed a rapid seed clearance in the organelle fraction. In the cytosol, there appeared to be a rapid clearance with t1/2 of ∼4 h and a slower phase of 12 to 24 h. Taken together, the data suggested two paths of seed clearance for tau: in the vesicle, seeds are degraded via the lysosome, whereas in the cytoplasm, seeds are cleared rapidly via the proteasome.
Discussion
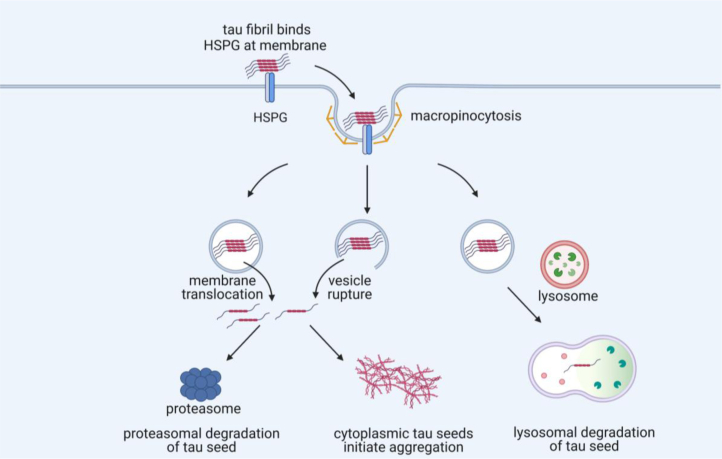
It is unknown how a tau assembly propagates a unique structure from the outside to the inside of a cell. This study has investigated the trafficking kinetics of tau seeds into the cytosol and mechanisms of degradation. For ease of labeling and tracking, we used recombinant heparin-induced FL tau fibrils, which undoubtedly lack the same seed conformation that occurs in AD, or post-translational modifications. Second, rather than primary neurons, which might more accurately reflect disease processes, we used an immortalized cell line, U2OS, because it is highly adherent, has a large cell body, and is useful for live-cell imaging over days. We observed that most tau taken up by the cell traffics to the endolysosomal system, where it is degraded fairly rapidly. A small percentage of assemblies entered the cytosol to seed intracellular aggregation, which appeared to occur often simultaneously at multiple sites. Seeding increased after experimental rupture of endosomes; however, this did not appear to be the predominant mechanism, and tau assemblies did not measurably disrupt vesicles. Most assemblies appeared to be degraded via bafilomycin-sensitive mechanisms in the lysosome, whereas cytosolic seeds were degraded primarily by the proteasome. In summary, our data indicate two major routes into the cell for a tau seed: one into the lysosomal degradation pathway, and the other to the cytosol, where recruitment of native tau and template-based replication occur (Fig. 8).
Figure 8.
Model of tau trafficking. External tau fibrils bind to heparan sulfate proteoglycans (HSPG), which lead to their internalization via macropinocytosis. Most tau within vesicles is directed toward lysosomal degradation. Some tau in this pathway escapes degradation and instead enters the cytosol. This could potentially occur by vesicle rupture, but most tau seeds appear to enter the cytosol in the absence of this process through an unknown mechanism, potentially membrane translocation. Once the fibril enters the cytosol, it serves as a template for replication with endogenous tau to amplify the aggregate. Proteasome-mediated degradation in the cytosol accounts for clearance of most tau, although long-term clearance mechanisms could involve other mechanisms.
Cytoplasmic tau seeding
In cultured cells, despite widespread tau uptake by macropinocytosis, seeding is relatively inefficient. To overcome this problem in experimental systems, we (8) and others (13) have used lipid-based transduction reagents to improve cytoplasmic delivery of tau assemblies. In biosensor cells expressing FL tau-Clo, we observed seeding in ~1% of exposed cells, despite ∼100% of the cells taking up labeled tau assemblies into the endolysosomal system. We expressed FL tau-Clo constructs that bind the cytoplasmic tubulin network. Within hours after tau seed exposure, however, we observed simultaneous evolution of tau puncta throughout the cytoplasm. The simultaneous appearance of puncta within the cell suggests that there may be a regulatory pathway controlling this process. In data not presented, we tested for a role for the cell cycle but did not observe any. We have concluded that seeds not visible by microscopy escaped the endolysosomal compartment to recruit endogenous FL tau-Clo away from the tubulin network. Taken together, our results imply that, following macropinocytosis, only a small subset of tau assemblies wind up in the cytosol, where they serve as templates for amplification.
Endolysosome rupture only partially explains tau seeding
Prior studies have suggested that tau enters the cytosol by rupturing vesicles (6, 10). However, using formation of Gal3-Rub puncta as a marker of vesicle integrity, we found no evidence that tau aggregates directly induce rupture. Neither did we observe FL tau-Clo puncta formation in close proximity to Gal3-stained vesicles. Finally, based on live-cell imaging of hundreds of cells, in the majority of cases (∼65–80%), we observed no evidence of vesicle rupture in association with seeding events. However, when we specifically tested the relationship of Gal3-Rub puncta to subsequent cell seeding, we observed a slight increase in the percentage of cells that exhibited pre-existing evidence of Gal3-Rub binding (∼16%), relative to cells that did not have evidence of seeding (∼12% of the total overall). Notably, 82% of seeded cells showed no evidence of preceding vesicle rupture. Pre-existing vesicle instability might slightly predispose toward tau seeding, but is clearly not a major determinant. Genes associated with the endolysosomal system have been linked to AD (14). When we treated cells with LLOMe to rupture lysosomes following loading with tau, we only saw a ∼5× increase in seeding, from 1% to 5%. In contrast, treatment with Lipofectamine 2000 increased seeding ∼35×. This implies that the route by which tau enters the cytosol may impact its seeding efficiency and argues against simple vesicle rupture as a primary mode. The contrast of our findings to those of others (6, 10) may reflect our use of different systems and purified recombinant tau fibrils. Importantly, a recent study using Human embryonic kidney 293 cells and primary neurons also concluded that tau does not damage vesicles (15). We are continuing to investigate the molecular mechanism by which assemblies might cross the vesicle membrane in the absence of rupture, potentially via direct translocation.
Cytoplasmic tau seeds are degraded by the proteasome
Our data suggest trafficking of tau seeds into two compartments: the endolysosome and the cytoplasm. The relative amount of tau protein in both compartments was too low to measure via Western blot, ELISA, or even mass spectrometry, but we readily quantified tau seeding activity using biosensor cells. We determined that tau seeds within the vesicle compartment are rapidly cleared. This was blocked by bafilomycin, consistent with lysosomal degradation. By contrast, seeds within the cytosol appeared to have a rapid and slow phase of degradation. The rapid phase, with a half time of ∼4 h, was blocked by proteasome inhibition with MG132. The persistence of seeds in the cytosol for up to 48 h after cell exposure suggests that a small subset may be protected from degradation, with a longer half-life. The role of autophagy in aggregate degradation has been proposed by many studies (16, 17). Our data suggest a more nuanced interpretation, with the proteasome rapidly degradeing exogenous seeds that enter the cytosol. But we cannot exclude a role for autophagy for slower clearance. Since the proteasome must digest single unfolded proteins, we propose that disassembly factors could be important. One candidate is valosin-containing protein (p97/VCP), which has been directly implicated in dominantly inherited tauopathy (18).
Tau traffics to distinct pools
Our data indicate that two populations of intracellular tau result from macropinocytosis. Most tau enters the endolysomal system for rapid degradation, resulting in relatively inefficient seeding overall. We observed a steady increase of tau in the cytosol over time both by imaging and fractionation studies. Cytosol seeding thus appears to be derived from tau leakage out of the endolysosomal system, whether by vesicle rupture or other mechanisms such as membrane translocation (15, 19, 20). Our conclusions that there exist two paths for tau into the cell, with one leading to intracellular seed amplification, may help clarify the molecular mechanisms of tauopathy and may explain the identification of endolysomal genes as AD risk factors (14).
Experimental procedures
Tau fibrillization, labeling, and imaging
Recombinant FL (2N4R) wildtype tau was purified and fibrillized as described previously (3). To label tau, 8 μM purified tau fibrils were incubated with 0.025 mg AF647 succinimidyl ester dye (Invitrogen) for 1 h at room temperature and quenched with 100 mM glycine for 1 h at room temperature. The solution was dialyzed overnight into PBS using dialysis cassettes (Thermo Fisher Scientific) to remove unbound dye. Labeled fibrils were stored at 4 °C for short term (days to weeks) or at −80 °C for the longer term. For live-cell imaging assays, labeled fibrils were added to the U2OS cells and imaged using an IN Cell Analyzer 6000 (GE).
Cell lines
Lentiviral delivery vectors were used to create U2OS cells that stably overexpressed proteins of interest. To generate the cell lines, cells were plated at 30,000 cells/well in a 12-well plate. After 24 h, ∼200 μl conditioned media containing lentivirus were added to the wells. Cells were grown for 3 days and then replated in a 10 cm dish, grown to confluency, and stored in liquid nitrogen until use. Protein expression was confirmed by epifluorescence microscopy. The stable U2OS cell lines generated for this study are listed: Tet off tau FL(P301S)-mCerulean3/mClover3: FL 2N4R human tau with P301S mutation tagged to mCerulean3 and mClover3 expressed under Tet-regulated promoter Tet off tau FL(P301S)-Cerulean3/mClover3; Galectin3-mRuby3: FL 2N4R human tau with P301S mutation fused to mCerulean3 or mClover3, both expressed under Tet-regulated promoter; Galectin-3 is fused to mRuby3; Galectin3-mRuby3: Galectin-3 is fused to mRuby3.
Measuring nonvesicular cytoplasmic tau signal
To segment cells within an image, we calculated the image gradient of the FITC channel using the Sobel method and normalized this output by the gradient magnitude. We then subtracted the resulting normalized image gradient from the original image to enhance cell edges. To remove the remaining image noise, we applied a Gaussian filter with sigma value of 5. Last, we calculated the Rosin threshold from the filtered image and used this threshold value to segment cell boundaries. To segment vesicle boundaries from the Cy5 channel, we applied a lower and upper intensity threshold. For the lower, we calculated the Rosin threshold and ignored all pixels below this value. For the upper (to avoid artifacts from dying cells), we calculated the top 1% intensity and ignored pixels above this value. To compare tau signal inside and outside the vesicles, we calculated tau signal from the Cy5 channel from inside cell and inside vesicle boundaries or signal inside the cell but outside vesicle boundaries. We then calculated the median intensity for all pixels within these regions. After calculating the median intensity outside the vesicles from the first time point, we background subtracted this value from all subsequent time points.
Seeding assay
V2L Biosensor cells (9) expressing tau RD containing a disease-associated mutation (P301S) fused to mCerulean3 (Cer) or mClover3 (Clo) (tau RD-Clo/Cer) were plated at a density of 10,000 cells/well in a 96-well plate. Recombinant tau fibrils were sonicated for 30 s at a setting of 65 (Qsonica Sonicator) and applied to cells at 50 μl per well. Cells were then incubated 48 h. Tau (80 nM) was added directly to the cells after sonication (for naked seeding). Alternatively, Lipofectamine 2000 was used to transduce tau (5 nM). After 48 h, cells were harvested with 0.05% trypsin, fixed in 2% paraformaldehyde for 10 min, and then resuspended in flow cytometry buffer (Hank’s balanced salt solution plus 1% fetal bovine serum and 1 mM EDTA). We quantified FRET as described previously using an LSRFortessa flow cytometer (21). For each dataset, three technical replicates were used. For each experiment, a minimum of ∼5000 single cells per replicate were analyzed. Data analysis was performed using FlowJo (BD Biosciences), version 10, software and GraphPad Prism, version 8 (GraphPad Software, Inc).
Cell culture and treatments
Human U2OS (American Type Culture Collection) and human embryonic kidney 293T cells were cultured in McCoy’s 5A and Dulbecco's modified Eagle's medium (Gibco) supplemented with GlutaMAX and 10% fetal bovine serum. Cells were incubated in humidified air with 5% CO2 at 37 °C and were subcultured every 3 to 4 days. For vesicle permeabilization, 1 mM LLOMe was added to the media for 6 h. Proteasome inhibitor MG132 (Sigma) dissolved in dimethylsulfoxide was used at 10 μM final concentration, and the autophagy inhibitor, bafilomycin, dissolved in dimethylsulfoxide was used at 200 nM final concentration.
Western blot
Lysates were mixed with 4× SDS buffer and run on a NuPAGE 10% Bis–Tris Gel at 100 V. The gel was then transferred onto Immobilon P membrane for 1 h at 20 V using a semidry transfer apparatus (Bio-Rad). The membrane was blocked with 5% Blotto (Bio-Rad) in Tris-buffered saline with Tween-20 (TBST) for 1.5 h before primary (GAPDH: 1:5000 dilution [R&D Systems], lamin B1: 1:1000 dilution [Cell Signaling Technology], LAMP1: 1:1000 dilution [Thermo Fisher Scientific], voltage-dependent anion channel: 1:500 dilution [Thermo Fisher Scientific]) antibody was added at specific dilutions and incubated on a shaker overnight at 4 °C. The membrane was then washed three times with TBST at 10 min intervals. It was reprobed with (goat anti-rabbit/mice) secondary antibody for 1.5 h at room temperature. The membrane was washed four times with TBST and exposed to Enhanced Chemiluminescence Prime Western blot detection kit (GE Lifesciences) for 2 min. Blots were imaged with a Syngene digital imager.
Cellular fractionation
Fractionation was guided by published protocols (22, 23). Cells were trypsinized and resuspended in 5 ml of culture medium. They were centrifuged at 500g for 10 min at 4 °C, the supernatant was discarded, and the pellet was washed with 500 μl of ice-cold PBS. About ∼250 μl (depending on pellet size) of ice-cold lysis buffer (150 mM NaCl, 50 mM Hepes [pH 7.4], 25 μg/ml digitonin, and 1 M hexylene glycol) was added to the pellet and then incubated on end-over-end rotator for 10 to 15 min at 4 °C. The sample was centrifuged at 2000g for 10 min at 4 °C. The supernatant was collected, and the pellet was processed for the next step. The supernatant was clarified by centrifugation at 18,000g for 20 min. Supernatant was collected as the cytosol fraction.
The pellet was washed with wash buffer (150 mM NaCl, 50 mM Hepes [pH 7.4]) and then 250 μl of ice-cold buffer (150 mM NaCl, 50 mM Hepes [pH 7.4], 1% IGEPAL [v:v], 1 M hexylene glycol) and resuspended by vortexing. It was incubated on ice for 30 min and then centrifuged at 7000g for 10 min at 4 °C. The supernatant was collected. This fraction contains the proteins from all membrane-bound organelles (endosomes, mitochondria, endoplasmic reticulum, Golgi, etc) except nuclei. The pellet was washed with wash buffer and then 250 μl of ice-cold buffer (150 mM NaCl, 50 mM Hepes [pH 7.4], 0.5% sodium deoxycholate [w:v], 0.1% sodium dodecyl sulfate [w:v], 1 M hexylene glycol, and 7 μl of benzonase [25,000 units/ml]) was added. It was incubated on an end-over-end rotator for 30 min at 4 °C to allow complete solubilization of nuclei and digestion of genomic DNA. It was then centrifuged at 7800g for 10 min at 4 °C. The supernatant was collected. This fraction contains the nuclear proteins. 1× Roche EDTA-free cOmplete protease inhibitor was added to all buffers.
Seed degradation assay
The U2OS cells were incubated with tau fibrils for 16 h. The next day, cells were washed with PBS for 2 min, and a 30 s 0.05% trypsin wash was used to digest attached tau, followed by quenching with a 3 min wash with McCoy’s 5A media. Cells were then incubated for the defined amount of time in media without tau, trypsinized, and fractionated to collect the cytosolic or organelle fractions, which were used for seeding assays.
Immunoprecipitation
About 75 μl of Dynabeads Protein A (Thermo Fisher Scientific) were washed according to the manufacturer’s protocol and incubated with 15 μg of tau polyclonal antibody raised against tau RD TauA (9), for 1 h at room temperature. The beads were washed with PBS with Tween-20. The beads were added to 75 μg of lysate (organelle fraction) and incubated with rotation overnight at 4 °C. The next day, beads were washed with PBS with Tween-20 and protein was eluted in low pH elution buffer (Pierce). The reaction was neutralized with 1:10 1 M Tris (pH 8.5) in a final volume of 120 μl.
Data availability
All data generated and analyzed during this study are included in this article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by the Crowley Foundation, the Cure Alzheimer's Foundation, the Rainwater Charitable Foundation, the Hamon Foundation, and the National Institutes of Health (grant no.: WU-16-376-MOD-5). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We also thank the Moody Foundation Flow Cytometry Core Facility and the Sequencing Core Facility at UT Southwestern.
Author contributions
M. I. D. and S. K. methodology; S. K., A. R. V., and D. A. D. investigation; S. K., A. R. V., D. A. D., V. A. P., O. M. K., and C. L. W. resources; M. I. D. and S. K. writing–original draft.
Edited by Paul Fraser
Supporting information
Recombinant FL tau fibrils labeled with Alexa Fluor 647 were applied to the U2OS cells expressing FL Tau-Clo (green) and imaged using a GE IN Cell Analyzer 6000 to track tau aggregation formation over time. The images taken at different time points post fibril addition are used to make this movie (same cell is shown in Fig. 1D). Rapid and simultaneous development of tau inclusions throughout the cytoplasm is evident.
References
- 1.Lee V.M.-Y., Goedert M., Trojanowski J.Q. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- 2.Vaquer-Alicea J., Diamond M.I. Propagation of protein aggregation in neurodegenerative diseases. Annu. Rev. Biochem. 2019;88:785–810. doi: 10.1146/annurev-biochem-061516-045049. [DOI] [PubMed] [Google Scholar]
- 3.Holmes B.B., DeVos S.L., Kfoury N., Li M., Jacks R., Yanamandra K., et al. Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E3138–E3147. doi: 10.1073/pnas.1301440110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stopschinski B.E., Holmes B.B., Miller G.M., Manon V.A., Vaquer-Alicea J., Prueitt W.L., et al. Specific glycosaminoglycan chain length and sulfation patterns are required for cell uptake of tau versus α-synuclein and β-amyloid aggregates. J. Biol. Chem. 2018;293:10826–10840. doi: 10.1074/jbc.RA117.000378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J.J., Nathaniel D.L., Raghavan P., Nelson M., Tian R., Tse E., et al. Compromised function of the ESCRT pathway promotes endolysosomal escape of tau seeds and propagation of tau aggregation. J. Biol. Chem. 2019;294:18952–18966. doi: 10.1074/jbc.RA119.009432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calafate S., Flavin W., Verstreken P., Moechars D. Loss of Bin1 promotes the propagation of tau pathology. Cell Rep. 2016;17:931–940. doi: 10.1016/j.celrep.2016.09.063. [DOI] [PubMed] [Google Scholar]
- 7.Frost B., Jacks R.L., Diamond M.I. Propagation of tau misfolding from the outside to the inside of a cell. J. Biol. Chem. 2009;284:12845–12852. doi: 10.1074/jbc.M808759200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holmes B.B., Furman J.L., Mahan T.E., Yamasaki T.R., Mirbaha H., Eades W.C., et al. Proteopathic tau seeding predicts tauopathy in vivo. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E4376–E4385. doi: 10.1073/pnas.1411649111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hitt B.D., Vaquer-Alicea J., Manon V.A., Beaver J.D., Kashmer O.M., Garcia J.N., et al. Ultrasensitive tau biosensor cells detect no seeding in Alzheimer’s disease CSF. Acta Neuropathol. Commun. 2021;9:99. doi: 10.1186/s40478-021-01185-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falcon B., Noad J., McMahon H., Randow F., Goedert M. Galectin-8–mediated selective autophagy protects against seeded tau aggregation. J. Biol. Chem. 2018;293:2438–2451. doi: 10.1074/jbc.M117.809293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paz I., Sachse M., Dupont N., Mounier J., Cederfur C., Enninga J., et al. Galectin-3, a marker for vacuole lysis by invasive pathogens. Cell. Microbiol. 2010;12:530–544. doi: 10.1111/j.1462-5822.2009.01415.x. [DOI] [PubMed] [Google Scholar]
- 12.Siew J.J., Chern Y. Microglial lectins in health and neurological diseases. Front. Mol. Neurosci. 2018;11:158. doi: 10.3389/fnmol.2018.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo J.L., Lee V.M.-Y. Seeding of normal tau by pathological tau conformers drives pathogenesis of Alzheimer-like tangles. J. Biol. Chem. 2011;286:15317–15331. doi: 10.1074/jbc.M110.209296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao S., Casey A.E., Sargeant T.J., Mäkinen V.-P. Genetic variation within endolysosomal system is associated with late-onset Alzheimer’s disease. Brain. 2018;141:2711–2720. doi: 10.1093/brain/awy197. [DOI] [PubMed] [Google Scholar]
- 15.Tuck B.J., Miller L.V.C., Wilson E.L., Katsinelos T., Cheng S., Vaysburd M., et al. Tau assemblies enter the cytosol in a cholesterol sensitive process essential to seeded aggregation. bioRxiv. 2021 doi: 10.1101/2021.06.21.449238. [preprint] [DOI] [Google Scholar]
- 16.Nixon R.A. The role of autophagy in neurodegenerative disease. Nat. Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 17.Festa B.P., Barbosa A.D., Rob M., Rubinsztein D.C. The pleiotropic roles of autophagy in Alzheimer’s disease: from pathophysiology to therapy. Curr. Opin. Pharmacol. 2021;60:149–157. doi: 10.1016/j.coph.2021.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darwich N.F., Phan J.M., Kim B., Suh E., Papatriantafyllou J.D., Changolkar L., et al. Autosomal dominant VCP hypomorph mutation impairs disaggregation of PHF-tau. Science. 2020;370 doi: 10.1126/science.aay8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katsinelos T., Zeitler M., Dimou E., Karakatsani A., Müller H.-M., Nachman E., et al. Unconventional secretion mediates the trans-cellular spreading of tau. Cell Rep. 2018;23:2039–2055. doi: 10.1016/j.celrep.2018.04.056. [DOI] [PubMed] [Google Scholar]
- 20.Polanco J.C., Hand G.R., Briner A., Li C., Götz J. Exosomes induce endolysosomal permeabilization as a gateway by which exosomal tau seeds escape into the cytosol. Acta Neuropathol. 2021;141:235–256. doi: 10.1007/s00401-020-02254-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolay S., Diamond M.I. Alzheimer’s disease risk modifier genes do not affect tau aggregate uptake, seeding or maintenance in cell models. FEBS Open Bio. 2020;10:1912–1920. doi: 10.1002/2211-5463.12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baghirova S., Hughes B.G., Hendzel M.J., Schulz R. Sequential fractionation and isolation of subcellular proteins from tissue or cultured cells. MethodsX. 2015;2:440–445. doi: 10.1016/j.mex.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCaig W.D., Deragon M.A., Haluska R.J., Jr., Hodges A.L., Patel P.S., LaRocca T.J. Cell fractionation of U937 cells in the absence of high-speed centrifugation. J. Vis. Exp. 2019 doi: 10.3791/59022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Recombinant FL tau fibrils labeled with Alexa Fluor 647 were applied to the U2OS cells expressing FL Tau-Clo (green) and imaged using a GE IN Cell Analyzer 6000 to track tau aggregation formation over time. The images taken at different time points post fibril addition are used to make this movie (same cell is shown in Fig. 1D). Rapid and simultaneous development of tau inclusions throughout the cytoplasm is evident.
Data Availability Statement
All data generated and analyzed during this study are included in this article.