Abstract
Peptidases PepI, PepL, PepW, and PepG from Lactobacillus delbrueckii subsp. lactis, which have no counterparts in Lactococcus lactis, and peptidase PepQ were examined to determine their potential to confer new peptidolytic properties to lactococci. Controllable expression of the corresponding genes (pep genes) was achieved by constructing translational fusions with the promoter of the nisA gene (PnisA). A suitable host strain, UKLc10, was constructed by chromosomal integration of the genes encoding the NisRK two-component system into the fivefold peptidase-deficient mutant IM16 of L. lactis. Recombinants of this strain were used to analyze growth, peptidase activities, peptide utilization, and intracellular protein cleavage products. After nisin induction of PnisA::pep fusions, all of the peptidases were visible as distinct bands in protein gels. Despite the fact that identical transcription and translation signals were used to express the pep genes, the relative amounts of individual peptidases varied considerably. All of the peptidases exhibited activities in extracts of recombinant UKLc10 clones, but only PepL and PepG allowed the clones to utilize specific peptide substrates as sources of essential amino acids. In milk medium, induction of pepG and induction of pepW resulted in growth acceleration. The activities of all five peptidases during growth in milk medium were revealed by high-performance liquid chromatography analyses of intracellular amino acid and peptide pools.
Lactococci are used extensively in starter cultures for cheese manufacturing. During the ripening process these organisms contribute to the development of the texture, taste, and flavor of the mature products. Among other processes, such as lipolysis and acid formation, proteolysis of milk proteins by the concerted action of indigenous milk proteins, clotting enzymes, and proteolytic activities of bacterial starter and nonstarter strains is very important. Large-scale cheese production requires a reliable and reproducible fermentation process. This requirement has resulted in fundamental genetic research on lactic acid bacteria with a special focus on the proteolytic system of Lactococcus lactis as a model system (17). Since cheese ripening is generally slow and the reactions involved are rather complex, there are economic and technological incentives for accelerating and controlling this process (9). To do this, starters may be modified by introducing appropriate genes from other food grade bacteria. New or additional peptidase activities may alter or improve the proteolytic properties of lactic acid bacteria. Therefore, we decided to express several peptidase genes from Lactobacillus delbrueckii subsp. lactis DSM7290 in Lactococcus lactis under strictly controlled conditions. To do this, we selected pepI, pepL, pepG, and pepW, which have no counterparts in L. lactis, and pepQ. Proline-specific peptidases, such as PepI, PepQ, and PepL, are believed to play a special role in casein degradation since most of the general peptidases are not able to cleave peptide bonds involving proline and the proline content of caseins is extraordinarily high (23). PepQ is a prolidase of the metalloprotease type and releases N-terminal amino acids from dipeptides containing proline in the second position (22). PepI, a proline iminopeptidase of the serine type, cleaves proline from the N termini of di- and tripeptides (14). Aminopeptidase PepL exhibits about 25.5% identity with PepI and is also able to cleave some proline-containing peptides (12). PepG, originally described as cysteine aminopeptidase (13), also exhibits endopeptidase activity with metenkephalin (Tyr-Gly-Gly-↓-Phe-Met; the arrow indicates the cleaved bond) and several N-terminally protected chromogenic p-nitroanilide (pNA) substrates. PepW, which exhibits 70% identity with PepG, was designated OrfW (13), because enzyme activity could not be demonstrated. Meanwhile, PepW could also be considered an endopeptidase, with Gly-Leu-↓-Leu-Gly as a substrate (unpublished results).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown at 37°C in Luria-Bertani medium (19), and L. lactis was routinely cultivated at 30°C in M17 medium (Difco) supplemented with 0.5% (wt/vol) glucose (GM17). Alternatively, L. lactis was grown in chemically defined medium (CDM) (15) or in milk medium containing 10% reconstituted skim milk (Difco). Cell densities in milk medium were determined by measuring the optical density at 600 nm (OD600) as described previously (15). To identify unique peptidase substrates, transformants of Salmonella typhimurium TN1547 expressing individual pep genes of L. delbrueckii subsp. lactis from appropriate plasmids (Table 1) were plated onto minimal glucose plates seeded with a few crystals of Leu- and Pro-containing peptides as described by Carter and Miller (2).
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Relevant properties | Reference |
|---|---|---|
| Strains | ||
| E. coli MC1061 | araD139 (ara, leu)7697 ΔlacX74 galU galK hsr strA | 3 |
| S. typhimurium TN1547 | leuBCD485 pepN90 pepA16 pepB11 supQ302 (ΔproAB pepD) pepP1 pepQ1 pepT1 pepE1 | 2 |
| L. lactis subsp. cremoris | ||
| MG1363 | Derivative of NCDO712, plasmid-free, prophage cured | 10 |
| IM16 | MG1363, pepX pepT pepO pepC pepN | 16 |
| UKLc10 | IM16, pepN::nisRK | This study |
| Plasmids | ||
| pLP712 | lac+ prt+, 33 MDa | 10 |
| pNZ8037 | Cmr, PnisA, pSH71 replicon | 5 |
| pNZ9573 | Cmr Err, pepN::nisRK, p15a replicon | 4 |
| pJUK23 | Apr, pepL, ColE1 replicon | 12 |
| pJKG8 | Kmr, pepG, pSC105 replicon | 13 |
| pJK505 | Apr, pepI, ColE1 replicon | 14 |
| pJKW2 | Kmr, pepW, pSC105 replicon | 13 |
| pMS1 | Kmr, pepQ, pSC105 replicon | 22 |
| pKS3 | Kmr, brnQ, pSC105 replicon | 21 |
| pUK200 | pNZ8037 with terminator of brnQ | This study |
| pUK200Q | pUK200, PnisA::pepQ | This study |
| pUK200I | pUK200, PnisA::pepI | This study |
| pUK200L | pUK200, PnisA::pepL | This study |
| pUK200G | pUK200, PnisA::pepG | This study |
| pUK200W | pUK200, PnisA::pepW | This study |
DNA cloning, sequencing, and PCR.
Standard techniques were used to isolate and clone DNA fragments (19) and to prepare plasmid DNA from E. coli (1) and L. lactis (7). For sequencing purposes, plasmid DNA from E. coli was purified on Nucleobond AX columns (Machery-Nagel). Restriction endonucleases and nucleic acid-modifying enzymes (Boehringer Mannheim and New England Biolabs) were used as recommended by the suppliers. E. coli and L. lactis were transformed by electroporation (8, 24) by using a Gene Pulser (Bio-Rad, Richmond, Calif.). DNA fragments were amplified by PCR performed with ULTma DNA polymerase (Perkin-Elmer). The primers used for PCR and nucleotide sequencing were purchased from MWG-Biotech. Automated sequencing with a LI-COR model 4200 sequencer (MWG-Biotech) was performed with both strands of the DNA segments. Nucleotide and amino acid sequences were analyzed by using the HUSAR (Geniusnet) and PC/Gene (Intelligenetics) programs.
Construction of the nisA translational fusion plasmid pUK200.
E. coli MC1061 was used as an intermediate host during cloning of fragments into pUK200. The terminator structure of the L. delbrueckii subsp. lactis brnQ gene, which had a free energy of 109 kJ/mol, was PCR amplified from plasmid pKS3 (21) by using primers 5′tatctagaGTCACTGTTATCTTCGCTATTTTAGCC and 5′tactcgagCTTAAAGACATTACACAAATAGTTCGAG (nucleotides in lower-case letters were added in order to introduce the underlined XbaI and XhoI sites). The 193-bp terminator fragment tailed with XbaI and XhoI sites was cloned in a directed orientation into the corresponding sites of pNZ8037, resulting in plasmid pUK200. The integrity of the terminator in plasmid pUK200 was verified by nucleotide sequencing.
Cloning of pep genes as nisA translational fusions in pUK200.
To construct translational fusions with PnisA, the pepG, pepI, pepL, pepW, and pepQ genes from L. delbrueckii subsp. lactis were PCR amplified from plasmids pJKG8, pJK505, pJUK23, pJKW2, and pMS1, respectively. The following primer pairs were used: 5′-AGTCATGAATTAACTCTGCAGGAATTGGCGG and 5′-atggatccTTAGGCTAATGAGTCCCAAGGAGCAAG for pepG, 5′-CAAATCACAGAAAAATATCTTCCATTTGGAAATTGGC and 5′-atggatccTAGTCCTGGCTGATTAACCAGTCAGACAACAGC for pepI, 5′-ataccatggATCAAACGAGGATCGTTACTTTAGACAATGG and 5′-atggatccTTACCTCCACAAATTTCCCTGCCTCAACATCACTG for pepL, 5′-attcatgaCACACGAATTAAGCCCCCAGCTGCTGGAATCC and 5′-atggatccTTAAATTAAGGAATCCCAAGGATCAAGTTCGATCGGC for pepW, and 5′-attcatgaATTTAGACAAATTACAAAACTGGCTGCAGGAAAACGGG and 5′-atggatccTTATTCCTTAACTGGCAGAACCTTCAATTCCTTGCTAGTG for pepQ (nucleotides in lower-case letters were added in order to introduce the underlined BamHI, NcoI, and RcaI sites). After the ends of the resulting PCR products were cut with the appropriate restriction enzymes, the products were inserted between the NcoI site at the ATG start codon of nisA and the BamHI site of vector pUK200. To allow for in-frame fusions with the nisA initiation codon, the primers covering the 5′ end of the pepL gene and the 5′ ends of the pepQ and pepW genes were equipped with NcoI and RcaI sites, respectively. In the case of pepG- and pepI-containing PCR products, the corresponding ends were left blunt and were joined with the linearized vector after the NcoI end of the vector was treated with the Klenow fragment. The sequences of the pep genes and their junctions with PnisA in each of the resulting pUK200 derivatives were verified by nucleotide sequencing.
Chromosomal integration of nisRK.
L. lactis IM16 was transformed with pNZ9573 (5), which is nonreplicative in members of the genus Lactococcus and contains the nisRK genes flanked by parts of the pepN sequence (5). Clones having the plasmid integrated at the chromosomal pepN locus were recovered by selecting for erythromycin resistance. Their identities were checked by performing PCR with primers which hybridized to the pepN regions flanking the nisRK integration site. To eliminate the vector by crossover between the flanking regions, single-copy integrants were grown for 100 generations in the absence of erythromycin. PCR analysis confirmed that about 50% of the erythromycin-sensitive clones obtained contained the nisRK genes integrated at the pepN locus. One of these clones, designated UKLc10, expressed the nisRK genes and allowed induction of genes under control of the nisA promotor.
Preparation and electrophoresis of cell extracts.
Cultures of UKLc10 transformants carrying appropriate derivatives of pUK200 were induced with nisin (final concentration, 0.1 or 1 ng/ml; Sigma-Aldrich) at the mid-exponential growth phase (OD600, 0.5 U) and then incubated at 30°C. Aliquots were removed at different times after induction. Further protein synthesis in the aliquots was instantaneously blocked by adding merthiolate (final concentration, 0.1 mg/ml; Sigma), and similar wet weights (about 12 OD600 units) of the killed cells were collected by centrifugation, washed with 50 mM Tris (pH 7.5), and resuspended in 180 μl of the same buffer. To disrupt the cells, the suspensions were mixed with amounts of glass beads (diameter, 0.17 mm) corresponding to the fivefold wet weight of the cells and agitated in a Vibrogen cell mill (Sauer) for 15 min at 4°C and the maximum speed. The extracts, which were obtained after debris was removed by centrifugation (15 min, 4°C, 15,000 × g), contained between 1 and 3 mg of total soluble protein/ml, as determined by the method of Spector (20). The protein patterns of the extracts were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE); the gels were stained with Coomassie brilliant blue.
Peptidase assays.
The activities of PepI, PepL, and PepG were determined with the chromogenic compounds Pro-pNA, Leu-pNA, and Ala-Ala-pNA (Bachem) as described previously (11–13). Cell extracts were used in the assays at the following dilutions: 1:1,000 for PepI, 1:50 for PepL, and 1:10 for PepG. Enzyme specific activities were expressed in nanomoles of nitrophenol released from the chromogenic substrates per milligram of protein per minute.
PepQ and PepW were assayed by measuring the release of l-leucine from the PepQ substrate Leu-Pro or the PepW substrate Gly-Leu-Leu-Gly in an l-amino acid oxidase-dependent reaction as described by Stucky et al. (22). The optimal conditions for the PepW assay were 37°C and pH 6.6 (unpublished data). The concentration of the final reaction product, formazan, was determined at 492 nm with a Spectramax reader (Molecular Devices). Specific activities were expressed in nanomoles of l-leucine released from the peptides per milligram of protein per minute.
HPLC analysis of intracellular amino acids and peptides.
Transformants of UKLc10(pLP712) carrying appropriate pUK200 derivatives were grown in milk medium. Cells harvested at various times were disrupted as described previously (16). After centrifugation for 1 h at 4°C and 50,000 × g, the resulting extracts were adjusted to pH 2.7 with NaOH. Amino acids and peptides were derivatized with o-phthaldialdehyde (OPA) (Sigma-Aldrich) by using the recommendations of the manufacturer. Derivatized products were separated by reverse-phase high-performance liquid chromatography (HPLC) by using a LiChrospher RP-18 column (125 by 4 mm; Merck). Amino acids and peptides were eluted with a linear 80 to 20% 50 mM NaPO4 (pH 5.5) gradient in methanol at 30°C and a flow rate of 1 ml/min. OPA derivatives were detected at 330 nm for 65 min.
RESULTS AND DISCUSSION
Design of nisin-inducible PnisA::pep fusion strains.
In order to perform expression studies with L. lactis under defined and comparable conditions, the pepI, pepQ, pepL, pepG, and pepW genes of L. delbrueckii subsp. lactis were translationally fused to the initiation codon of the nisin structural gene (nisA) in plasmid pUK200. This vector was derived from pNZ8037 (6) by inserting the strong terminator of the L. delbrueckii subsp. lactis brnQ gene (21), which ensured efficient transcription termination of cloned pep genes.
To facilitate sensitive detection of effects caused by additional peptidases in L. lactis, we used the fivefold pep-deficient mutant IM16 (16) as the host. As a precondition for nisin inducibility of the PnisA::pep fusions, the nisR and nisK functions, which are required for nisin signal transduction, were introduced into this strain. To avoid undesired effects related to plasmid selection and stability, we integrated nisRK into the chromosome of IM16 by using the pNZ9573 plasmid, which is nonreplicative in L. lactis. This plasmid contains a fragment of L. lactis chromosomal DNA covering the 5′ and 3′ ends and flanking sequences of the pepN gene, interrupted by insertion of nisRK. After transformation of IM16 with pNZ9573 and selection for erythromycin-resistant clones containing the entire plasmid integrated into the chromosome, we searched for recombinants which had lost the vector part due to a subsequent crossover event. The resulting derivative of strain IM16, containing nisRK at the chromosomal pepN locus, was designated UKLc10.
Controlled expression of pep genes in L. lactis UKLc10.
Expression of the five peptidases studied was induced in UKLc10 transformed with the PnisA::pep fusion plasmids and was monitored by performing SDS-PAGE with cell extracts. The intensities of the peptidase bands increased with time after induction and depended on the nisin concentration used. All of the peptidases were clearly detectable in extracts prepared 15 min after we added 0.1 ng (data not shown) or 1 ng (Fig. 1A) of nisin per ml. The absolute amounts of the individual peptidases varied widely. PepG, PepI, and PepW were heavily overexpressed compared with PepQ and PepL, which produced only tiny bands in the gel. Since the PnisA::pep fusion plasmids were identical except for the pep genes, the differences probably were due to posttranscriptional effects, such as mRNA stability, translation efficiency, or protein stability. Cell extracts (Fig. 1A) were also used to measure specific peptidase activities with appropriate substrates. PepI, PepL, and PepG activities were determined by monitoring the release of p-nitroaniline from chromogenic substrates, whereas in the case of PepQ and PepW, cleavage of nonchromogenic peptides was estimated by detecting liberated amino acids after reactions with l-amino acid oxidase. Consistent with the protein profiles shown in Fig. 1A, the activities of all five peptidases were detectable 15 min after nisin was added and increased with time after induction (Fig. 1B). Although the slopes of the resulting kinetics lines depended on the nisin concentration used, the correlations between the induction rate and the nisin concentration were different for the individual constructs. Only in the case of PepG did a 10-fold-higher nisin concentration actually result in a 10-fold increase in the induction rate. For the remaining peptidases only two- to fourfold increases were observed. Remarkably, the specific activities, as well as the induction rates, obtained with proline iminopeptidase PepI were 2 to 3 orders of magnitude higher than the values obtained with the other peptidases, which might indicate that PepI has a special physiological role in lactic acid bacteria.
FIG. 1.
Induction of PnisA::pep fusions. Transformants of strain UKLc10 harboring plasmids pUK200W (PepW), pUK200G (PepG), pUK200L (PepL), pUK200Q (PepQ), and pUK200I (PepI) were grown in GM17. At an OD600 of 0.5 U, expression of the PnisA::pep fusions was induced by adding nisin, and cell extracts were prepared from culture aliquots at zero time and 15, 30, 60, 90, and 120 min after induction. (A) Extracts from cultures induced with 1 ng of nisin/ml were analyzed by SDS-PAGE on 12% polyacrylamide gels. The arrowheads indicate the expected gel positions of individual peptidases. The molecular masses of protein markers (M) (in kilodaltons) are indicated on the left. (B) Specific peptidase activities determined in extracts obtained from cultures induced with 0.1 ng of nisin/ml (○) and 1 ng of nisin/ml (●).
Growth experiments.
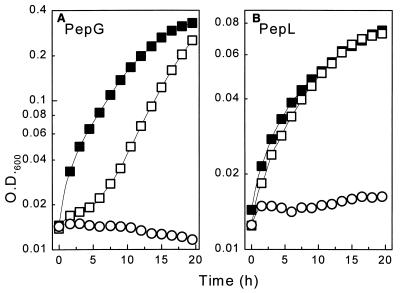
Transformants of strain UKLc10 expressing the various PnisA::pep fusions were grown in CDM, GM17, and milk medium. As a precondition for the growth experiments performed with CDM, unique peptide substrates which were specifically cleaved by the individual peptidases had to be identified. This was done by performing plating assays (2) with transformants of the multiple-peptidase-deficient strain S. typhimurium TN1547, which expressed the pepI, pepQ, pepL, pepG, and pepW genes from plasmids pJK505, pMS1, pJUK23, pJKG8, and pJKW2, respectively. We identified peptides as unique substrates in the plating assays, and for growth experiments performed with L. lactis UKLc10 we used peptides which could not serve as sources of essential amino acids (Glu, Gln, Leu, Val, Ile, Met, His) (18) required by UKLc10 but had the potential to release such amino acids when the heterologous pepI, pepQ, pepL, pepG, and pepW genes from L. delbrueckii subsp. lactis were expressed. No substrates fulfilling these criteria were identified for PepQ. PepI, in spite of its high activity, did not mediate growth of the transformants of UkLc10 on any of the Pro-His-Leu, Pro-His-Gly, and Pro-Val-Gly substrates tested, nor did PepW mediate growth on Gly-Leu-Leu-Gly. This suggested that cleavage of these substrates was rather inefficient in UKLc10 transformants and that strong overproduction of the peptidases probably contributed to the depletion of essential amino acids. Expression of pepL and pepG, in contrast, allowed utilization of specific peptides. Induction of pepG resulted in significant growth of UKLc10(pUK200G) on Leu-Leu-Leu (Fig. 2A), and PepL allowed UKLc10(pUK200L) to grow with Leu-Gly-Pro at almost the same rate as with the free amino acid leucine (Fig. 2B). Remarkably, both of these peptides are potentially present in the sequence of bovine β-casein (11). This was the first direct evidence that physiological activity of Lactobacillus peptidases occurs in the genus Lactococcus.
FIG. 2.
Effects of PepG and PepL on peptide utilization by UKLc10. Transformants of UKLc10 harboring pUK200G (A) or pUK200L (B) were grown in CDM supplemented with all of the essential amino acids except leucine. Leucine was supplied either as a free amino acid in the presence of nisin (■) or in the form of specific peptides in the presence (□) and in the absence (○) of nisin. The specific substrates used were Leu-Leu-Leu for UKLc10(pUK200G) and Leu-Gly-Pro for UKLc10(pUK200L). To induce expression of pepG and pepL, nisin was used at a concentration of 1 ng/ml.
Addition of nisin considerably reduced the growth rates of control cultures containing free leucine. At least part of this effect was independent of the presence of the pep genes, because growth inhibition was also observed after induction of controls which contained only unmodified vector pUK200 (data not shown). Growth inhibition by induction of PnisA alone was not observed during growth of UKLc10(pUK200) in rich medium (GM17), suggesting that under limiting conditions excessive transcription from PnisA may exhaust the cells. In the case of pepL, growth was inhibited further by the production of the peptidase itself, indicating that high levels of PepL may be harmful to cells.
Also in GM17, induction of each of the PnisA::pep fusions with 1 ng of nisin per ml led to clear reductions in the growth rates and final cell densities of the UKLc10 cultures. This was most obvious in the cases of pepQ and pepW, whereas overexpression of pepG had only a minor effect. With lower nisin concentrations (concentrations up to 0.1 ng/ml), growth inhibition was not detected in GM17.
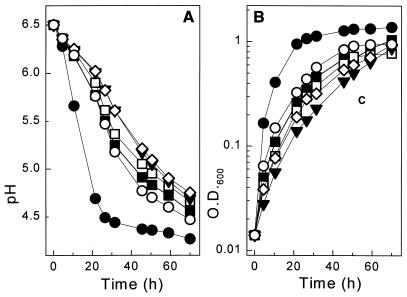
In milk medium, no inhibition was observed during growth of the corresponding transformants, which also contained the pLP712 plasmid in order to enable lactose utilization and protease production. As estimated from cell density and acidification measurements, induction of the pepI, pepL, and pepQ fusions had no significant effect, whereas expression of pepG resulted in a three- to fivefold increase in the growth rate in milk medium (Fig. 3). Weaker but reproducible growth stimulation was also observed with the pepW fusion. This is of particular interest since both PepG and PepW exhibit endopeptidase activity.
FIG. 3.
Expression of PnisA::pep fusions during growth in milk medium. Transformants of strain UKLc10(pLP712) harboring pUK200 (■), pUK200G (●), pUK200I (□), pUK200L (▾), pUK200Q (◊), and pUK200W (○) were grown in milk medium in the presence of nisin (1 ng/ml). Acidification (A) and cell densities (B) of the cultures were measured.
Analysis of protein cleavage products.
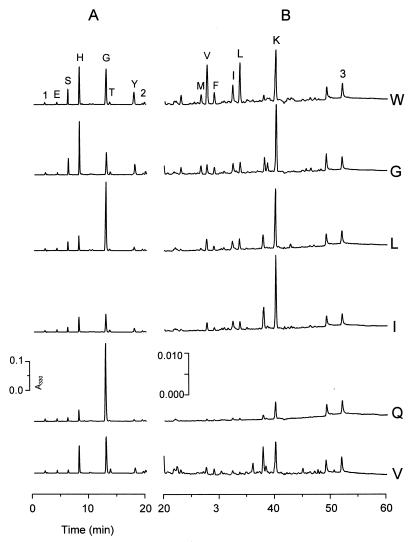
During growth in milk medium, expression of individual PnisA::pep fusions was induced in transformants of UKLc10(pLP712), and intracellular amino acid and peptide pools were analyzed by HPLC. The profiles which were obtained after expression of each of the five fusions significantly differed from the profile of control strain UKLc10(pLP712, pUK200) (Fig. 4). The differences were most pronounced for endopeptidases PepW and PepG, which was expected due to the potential of these enzymes to deliver new substrates for the resident aminopeptidases of the host strain. Amino acids whose intracellular concentrations were clearly higher after expression of individual peptidases from L. delbrueckii subsp. lactis were Gly for PepQ; Ile, Leu, and Lys for PepI and PepL; Leu, Val, Ile, Met, Phe, Ser, Tyr, Lys, and His for PepW; and Ile, Met, Ser, Tyr, Lys, and His for PepG. This finding provided an attractive method for modulating specific intracellular amino acid pools in L. lactis.
FIG. 4.
Effects of PnisA::pep expression on the patterns of intracellular amino acids and peptides. Transformants of strain UKLc10(pLP712) harboring pUK200W (W), pUK200G (G), pUK200L (L), pUK200I (I), pUK200Q (Q), or unmodified vector pUK200 (V) were grown in milk medium in the presence of nisin (1 ng/ml). Cells harvested during exponential growth of the cultures (pH 5.5) were used for HPLC analysis of OPA-derivatized amino acids and peptides. Two sections of the HPLC profiles, corresponding to 0 to 20 min (A) and 20 to 60 min (B), are shown with different y-axis scales. Amino acid peaks are indicated by one-letter designations. The signal of γ-aminobutyrate (peak 2), which was used as an internal standard, and two peaks of the OPA reagent (peaks 1 and 3) are indicated. A330, absorbance at 330 nm.
ACKNOWLEDGMENTS
We are grateful to Ingrid van Alen-Boerrigter and Brigitte Rosenberg for valuable assistance and support.
This work was conducted as a part of the STARLAB project (contract ERBBIO4CT960016) of the European Community.
REFERENCES
- 1.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter T H, Miller C G. Aspartate-specific peptidases in Salmonella typhimurium: mutants deficient in peptidase E. J Bacteriol. 1984;159:453–459. doi: 10.1128/jb.159.2.453-459.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 4.de Ruyter P G, Kuipers O P, Beerthuyzen M M, van Alen-Boerrigter I, de Vos W M. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J Bacteriol. 1996;178:3434–3439. doi: 10.1128/jb.178.12.3434-3439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Ruyter P G, Kuipers O P, de Vos W M. Controlled gene expression systems for Lactococcus lactis with the food grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Ruyter P G, Kuipers O P, Meijer W C, de Vos W M. Food-grade controlled lysis of Lactococcus lactis for accelerated cheese ripening. Nat Biotechnol. 1997;15:976–979. doi: 10.1038/nbt1097-976. [DOI] [PubMed] [Google Scholar]
- 7.de Vos W M, Vos P, de Haard H, Boerrigter I. Cloning and expression of the Lactococcus lactis subsp. cremoris SK11 gene encoding an extracellular serine proteinase. Gene. 1989;85:169–176. doi: 10.1016/0378-1119(89)90477-0. [DOI] [PubMed] [Google Scholar]
- 8.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox P F, Wallace J M, Morgan S, Lynch E J, Niland E J, Tobin J. Acceleration of cheese ripening. Antonie Leeuwenhoek. 1996;70:271–297. doi: 10.1007/BF00395937. [DOI] [PubMed] [Google Scholar]
- 10.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jimenez-Flores R, Kang Y C, Richardson T. Cloning and sequence analysis of bovine beta-casein cDNA. Biochem Biophys Res Commun. 1987;142:617–621. doi: 10.1016/0006-291x(87)90318-4. [DOI] [PubMed] [Google Scholar]
- 12.Klein J R, Dick A, Schick J, Matern H T, Henrich B, Plapp R. Molecular cloning and DNA sequence analysis of pepL, a leucyl aminopeptidase gene from Lactobacillus delbrueckii subsp. lactis DSM7290. Eur J Biochem. 1995;228:570–578. [PubMed] [Google Scholar]
- 13.Klein J R, Schick J, Henrich B, Plapp R. Lactobacillus delbrueckii subsp. lactis DSM7290 pepG gene encodes a novel cysteine aminopeptidase. Microbiology. 1997;143:527–537. doi: 10.1099/00221287-143-2-527. [DOI] [PubMed] [Google Scholar]
- 14.Klein J R, Schmidt U, Plapp R. Cloning, heterologous expression, and sequencing of a novel proline iminopeptidase gene, pepI, from Lactobacillus delbrueckii subsp. lactis DSM 7290. Microbiology. 1994;140:1133–1139. doi: 10.1099/13500872-140-5-1133. [DOI] [PubMed] [Google Scholar]
- 15.Mierau I, Haandrikman A J, Velterop O, Tan P S, Leenhouts K L, Konings W N, Venema G, Kok J. Tripeptidase gene (pepT) of Lactococcus lactis: molecular cloning and nucleotide sequencing of pepT and construction of a chromosomal deletion mutant. J Bacteriol. 1994;176:2854–2861. doi: 10.1128/jb.176.10.2854-2861.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mierau I, Kunji E R, Leenhouts K J, Hellendoorn M A, Haandrikman A J, Poolman B, Konings W N, Venema G, Kok J. Multiple-peptidase mutants of Lactococcus lactis are severely impaired in their ability to grow in milk. J Bacteriol. 1996;178:2794–2803. doi: 10.1128/jb.178.10.2794-2803.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mierau I, Kunji E R, Venema G, Kok J. Casein and peptide degradation in lactic acid bacteria. Biotechnol Genet Eng Rev. 1997;14:279–301. doi: 10.1080/02648725.1997.10647945. [DOI] [PubMed] [Google Scholar]
- 18.Poolman B, Kunji E R, Hagting A, Juillard V, Konings W N. The proteolytic pathway of Lactococcus lactis. Soc Appl Bacteriol Symp Ser. 1995;24:65S–75S. [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Spector T. Refinement of the Coomassie blue method of protein quantitation. A simple and linear spectrophotometric assay for less than or equal to 0.5 to 50 micrograms of protein. Anal Biochem. 1978;86:142–146. doi: 10.1016/0003-2697(78)90327-5. [DOI] [PubMed] [Google Scholar]
- 21.Stucky K, Hagting A, Klein J R, Matern H, Henrich B, Konings W N, Plapp R. Cloning and characterization of brnQ, a gene encoding a low-affinity, branched-chain amino acid carrier in Lactobacillus delbrueckii subsp. lactis DSM7290. Mol Gen Genet. 1995;249:682–690. doi: 10.1007/BF00418038. [DOI] [PubMed] [Google Scholar]
- 22.Stucky K, Klein J R, Schüller A, Matern H, Henrich B, Plapp R. Cloning and DNA sequence analysis of pepQ, a prolidase gene from Lactobacillus delbrueckii subsp. lactis DSM7290 and partial characterization of its product. Mol Gen Genet. 1995;247:494–500. doi: 10.1007/BF00293152. [DOI] [PubMed] [Google Scholar]
- 23.Visser S. Proteolytic enzymes and their relation to cheese ripening and flavour: an overview. J Dairy Sci. 1993;76:329–350. [Google Scholar]
- 24.Wells J M, Wilson P W, Le Page R W. Improved cloning vectors and transformation procedure for Lactococcus lactis. J Appl Bacteriol. 1993;74:629–636. doi: 10.1111/j.1365-2672.1993.tb05195.x. [DOI] [PubMed] [Google Scholar]