Asthma genome-wide association studies, expression quantitative trait loci mapping, and allelic imbalance assays have identified a strong signal for risk of childhood-onset asthma at chromosome 17q21 (1, 2), with risk-associated SNPs mediating the increased expression of ORMDL3 (ORMDL sphingolipid biosynthesis regulator 3) in inflammatory cells (including CD4+ lymphocytes, mast cells, and eosinophils) and lung structural cells (airway smooth muscle cells, fibroblasts, and bronchial epithelial cells) (3–5). Transgenic mice overexpressing ORMDL3 spontaneously develop features of airway hyperresponsiveness and airway remodeling (6).
After the identification of ORMDL3 SNPs in asthma risk haplotypes, several investigations have focused on understanding the underlying mechanisms by which this gene modulates risk. The roles of ORMDL3 are pleiotropic. ORMDL family members negatively regulate de novo sphingolipid synthesis, and ORMDL3 SNPs correlate with reduced markers of de novo sphingolipid synthesis in bronchial epithelial cells and circulating cells from patients with asthma (7), which complements studies of mice treated with pharmacologic inhibitors of de novo sphingolipid synthesis or genetic deficiency of enzymes required for de novo sphingolipid synthesis (8). ORMDL3 also modulates the unfolded protein response (UPR), regulates the expression of genes relevant to inflammation and glycolysis (including the human rhinovirus coreceptor ICAM-1 [intercellular adhesion molecule 1] in response to human rhinovirus infection), regulates CD4+ T-helper cell type 2 (Th2) cytokine expression, interacts with SERCA2 (sarcoplasmic/endoplasmic reticulum calcium ATPase 2) to modulate calcium homeostasis, and induces autophagy in B cells, mast cells, and endothelial cells (9). Given the multifaceted roles of ORMDL3, a detailed analysis of its specific role in asthma-relevant structural cells in homeostatic conditions is needed before evaluating how inflammation (either allergic inflammation or viral-triggered transcriptional changes) could then superimpose additive effects to modulate asthma risk. In this issue of the Journal, Guo and colleagues (pp. 661–670) further describe how ORMDL3 may modulate asthma risk (10).
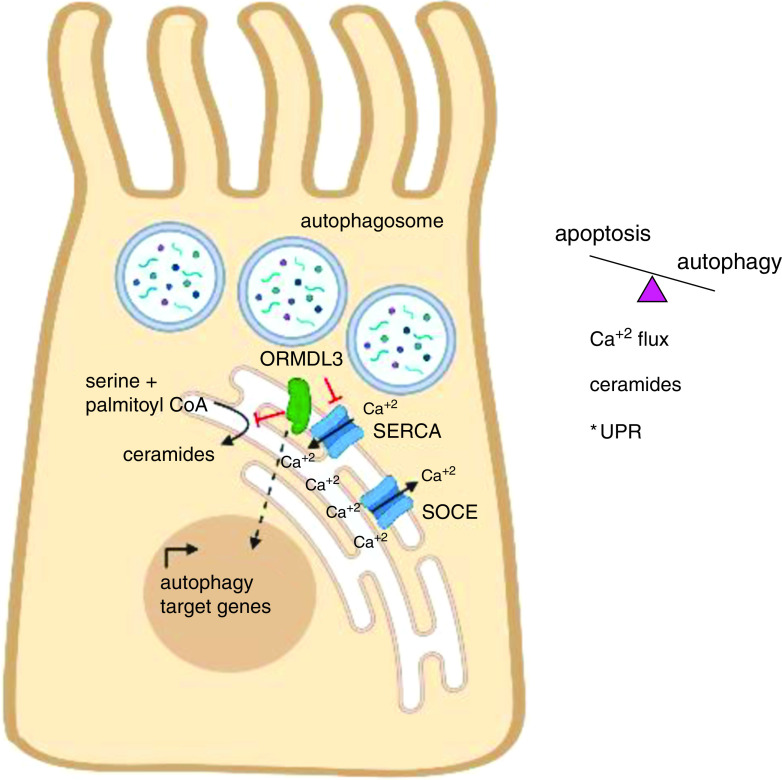
Guo and colleagues (10) have explored these results in cultured bronchial epithelial cell lines and primary bronchial epithelial cells from patients with asthma (see Figure 1). They identified a role for ORMDL3 in promoting autophagic cell death of bronchial epithelial cells in culture, with autophagosome formation enhanced by ORMDL3 overexpression and impaired by ORMDL3 gene silencing. Similarly, the expression of autophagy-related genes was enhanced in the presence of ORMDL3 overexpression and reduced by ORMDL3 silencing. Interestingly, although ORMDL3 induces autophagy via activation of ATF6 (activating transcription factor 6)-mediated UPR in B cells, mast cells, and endothelial cells, Guo and colleagues found that expression of UPR targets (mRNA and protein) was unaffected by ORMDL3 expression, indicating UPR-independent induction of autophagy, which is consistent with previous studies of bronchial epithelial cells (11). Using an unbiased mass spectrometry approach, SERCA2 was identified as an ORMDL3-interacting protein. SERCA2 has known roles in regulating calcium flux, autophagosome formation, and induction of the UPR. Guo and colleagues next determined that SERCA2 pharmacologic inhibition by thapsigargin increased autophagosome formation and expression of autophagy-related genes. Furthermore, cotransfection of ORMDL3 and SERCA2a or SERCA2b blocked autophagy mediated by isolated ORMDL3 overexpression. ORMDL3 overexpression delayed intracellular calcium mobilization, whereas ORMDL3 inhibition enhanced intracellular calcium mobilization, an effect that was partially rescued by SERCA2 inhibition. The net effect of ORMDL3 overexpression in bronchial epithelial cell lines was to increase cell death without activation of apoptosis or changes in cell proliferation. SERCA2 activation partly alleviated cell death, as did ORMDL3 gene silencing. Importantly, Guo and colleagues evaluated gene expression from bronchial epithelial cells obtained by brushings from an asthma repository. Consistent with bronchial epithelial cell lines, ORMDL3 expression correlated with the expression of autophagy-related genes ATG7 and ATG12.
Figure 1.
ORMDL3 (ORMDL sphingolipid biosynthesis regulator 3) activity drives autophagic cell death in bronchial epithelial cells. ORMDL3 associates with SERCA2 (sarcoplasmic/endoplasmic reticulum calcium ATPase 2) calcium channels in the endoplasmic reticulum (ER), inhibiting SERCA2 activity. SOCE regulates calcium egress from the ER to cytosol via several possible receptor families, including IP3 receptors, voltage-gated calcium channels, and RYR family members; SERCA2 opposes this activity by pumping calcium into the ER. Overexpression of ORMDL3 or treatment with the noncompetitive SERCA inhibitor thapsigargin increases autophagosome formation and impairs intracellular calcium flux. ORMDL3 overexpression and knockdown in bronchial epithelial cell lines respectively increase or decrease the expression of autophagy-related genes. ORMDL3 intersects with several pathways regulating cell fate decisions between autophagy and apoptosis, including calcium homeostasis, de novo sphingolipid metabolism and downstream metabolites (ceramides), and the unfolded protein response (UPR). *Altered ORMDL3 expression did not modulate UPR activity in bronchial epithelial cells, in contrast to hematopoietic cells, suggesting cell-specific effects of ORMDL3 on UPR. RYR = ryanodine receptor; SOCE = store-operated calcium entry. Image created by the authors using Biorender.
The results garnered thus far in understanding the various roles of ORMDL3 are compelling in isolation and even more so when contemplating the intersections of these pathways. For example, impaired de novo sphingolipid synthesis (due to elevated ORMDL3 expression by high-risk SNPs) could directly regulate cell fate determination between apoptosis and autophagy (12). De novo sphingolipid synthesis also activates the UPR to induce autophagy in some cell types. Disruptions in calcium homeostasis can cause protein misfolding to activate the UPR (13). Furthermore, calcium homeostasis can act as a rheostat to determine cell fate between apoptosis and autophagy (14). When taking into account coordinately regulated genes in the high-risk 17q21 haplotypes, such as GSDMB (gasdermin B), even more permutations regarding cell fate decisions are possible. SNPs in GSDMB, a member of the pore-forming gasmodermin gene family, form a risk haplotype associated with ORMDL3 (1, 2). GSDMB is upregulated by a respiratory viral infection in concert with ORMDL3 and mediates cell death via pyroptosis (15). The high-risk GSDMB SNPs cause the generation of a splice variant that lacks the capacity to induce pyroptosis (16), potentially shifting the balance of cell fate in light of increased autophagy conferred by elevated ORMDL3 expression. Interestingly, the overexpression of a pyroptosis-resistant splice variant of human GSDMB in transgenic mice phenocopies ORMDL3 transgenic mice (15).
The results from Guo and colleagues suggest that one effect of elevated ORMDL3 expression afforded by high-risk SNPs is to increase the extent of bronchial epithelial cell autophagy in homeostatic conditions, culminating in increased bronchial epithelial cell death. The results from Guo and colleagues could suggest that pharmacologic intervention of ORMDL3 may modulate autophagy in patients who are asthma-prone and carry the high-risk haplotype. It is of future interest to address the effect of diverse inflammatory stimuli (including allergic inflammation, environmental tobacco exposure, and viral infections), which increase ORMDL3 expression and have been implicated in the risk of childhood asthma mediated by 17q21 SNPs in genetic studies (reviewed in Reference 3). Exploring the intersection of impaired pyroptosis by GSDMB splice variants and elevated rates of autophagy conferred by increased ORMDL3 expression could be a future area of interest in understanding mechanisms driving childhood-onset asthma and airway remodeling. Airway remodeling is a disease process that current asthma therapeutics have not yet been able to prevent, so identifying mechanisms mediating this process could provide future targets for therapeutic development.
Footnotes
Supported by the National Institute of Environmental Health Sciences (P42 ES027723), the National Heart, Lung, and Blood Institute (1R01 HL128502-01A1), and ALA/AAAAI Allergic Respiratory Disease Research Award to M.L.C.
Originally Published in Press as DOI: 10.1165/rcmb.2022-0023ED on April 4, 2022
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature . 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 2. Sharma S, Zhou X, Thibault DM, Himes BE, Liu A, Szefler SJ, et al. A genome-wide survey of CD4(+) lymphocyte regulatory genetic variants identifies novel asthma genes. J Allergy Clin Immunol . 2014;134:1153–1162. doi: 10.1016/j.jaci.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stein MM, Thompson EE, Schoettler N, Helling BA, Magnaye KM, Stanhope C, et al. A decade of research on the 17q12-21 asthma locus: piecing together the puzzle. J Allergy Clin Immunol . 2018;142:749–764.e3. doi: 10.1016/j.jaci.2017.12.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hao K, Bossé Y, Nickle DC, Paré PD, Postma DS, Laviolette M, et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet . 2012;8:e1003029. doi: 10.1371/journal.pgen.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miller M, Tam AB, Cho JY, Doherty TA, Pham A, Khorram N, et al. ORMDL3 is an inducible lung epithelial gene regulating metalloproteases, chemokines, OAS, and ATF6. Proc Natl Acad Sci USA . 2012;109:16648–16653. doi: 10.1073/pnas.1204151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miller M, Rosenthal P, Beppu A, Mueller JL, Hoffman HM, Tam AB, et al. ORMDL3 transgenic mice have increased airway remodeling and airway responsiveness characteristic of asthma. J Immunol . 2014;192:3475–3487. doi: 10.4049/jimmunol.1303047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ono JG, Kim BI, Zhao Y, Christos PJ, Tesfaigzi Y, Worgall TS, et al. Decreased sphingolipid synthesis in children with 17q21 asthma-risk genotypes. J Clin Invest . 2020;130:921–926. doi: 10.1172/JCI130860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Worgall TS, Veerappan A, Sung B, Kim BI, Weiner E, Bholah R, et al. Impaired sphingolipid synthesis in the respiratory tract induces airway hyperreactivity. Sci Transl Med . 2013;5:186ra67. doi: 10.1126/scitranslmed.3005765. [DOI] [PubMed] [Google Scholar]
- 9. James B, Milstien S, Spiegel S. ORMDL3 and allergic asthma: from physiology to pathology. J Allergy Clin Immunol . 2019;144:634–640. doi: 10.1016/j.jaci.2019.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guo F, Hao Y, Zhang L, Croteau-Chonka DC, Thibault D, Kothari P, et al. Asthma susceptibility gene ORMDL3 promotes autophagy in human bronchial epithelium. Am J Respir Cell Mol Biol . 2022;66:661–670. doi: 10.1165/rcmb.2021-0305OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hsu KJ, Turvey SE. Functional analysis of the impact of ORMDL3 expression on inflammation and activation of the unfolded protein response in human airway epithelial cells. Allergy Asthma Clin Immunol . 2013;9:4. doi: 10.1186/1710-1492-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Young MM, Kester M, Wang HG. Sphingolipids: regulators of crosstalk between apoptosis and autophagy. J Lipid Res . 2013;54:5–19. doi: 10.1194/jlr.R031278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carreras-Sureda A, Pihán P, Hetz C. The unfolded protein response: at the intersection between endoplasmic reticulum function and mitochondrial bioenergetics. Front Oncol . 2017;7:55. doi: 10.3389/fonc.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sukumaran P, Nascimento Da Conceicao V, Sun Y, Ahamad N, Saraiva LR, Selvaraj S, et al. Calcium signaling regulates autophagy and apoptosis. Cells . 2021;10:2125. doi: 10.3390/cells10082125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raby BA, Weiss ST. Diversity and the splice of life: mapping the 17q12-21.1 locus for variants associated with early-onset asthma in African American individuals. Am J Respir Crit Care Med . 2021;203:401–403. doi: 10.1164/rccm.202010-3802ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gui H, Levin AM, Hu D, Sleiman P, Xiao S, Mak ACY, et al. Mapping the 17q12-21.1 locus for variants associated with early-onset asthma in African Americans. Am J Respir Crit Care Med . 2021;203:424–436. doi: 10.1164/rccm.202006-2623OC. [DOI] [PMC free article] [PubMed] [Google Scholar]