Abstract
Lack of CFTR (cystic fibrosis transmembrane conductance regulator) affects the transcriptome, composition, and function of large and small airway epithelia in people with advanced cystic fibrosis (CF); however, whether lack of CFTR causes cell-intrinsic abnormalities present at birth versus inflammation-dependent abnormalities is unclear. We performed a single-cell RNA-sequencing census of microdissected small airways from newborn CF pigs, which recapitulate CF host defense defects and pathology over time. Lack of CFTR minimally affected the transcriptome of large and small airways at birth, suggesting that infection and inflammation drive transcriptomic abnormalities in advanced CF. Importantly, common small airway epithelial cell types expressed a markedly different transcriptome than corresponding large airway cell types. Quantitative immunohistochemistry and electrophysiology of small airway epithelia demonstrated basal cells that reach the apical surface and a water and ion transport advantage. This single cell atlas highlights the archetypal nature of airway epithelial cells with location-dependent gene expression and function.
Keywords: airway epithelia, small airway, single-cell RNA-seq, basal cells, goblet cells, cystic fibrosis transmembrane conductance regulator
Clinical Relevance
Most chronic inflammatory airway diseases affect the small airways, yet the composition and function of cells in the small airways is not well known. This study explores the contribution of small airway epithelial cells to lung biology.
The airways of humans and other large mammals can be classified into “large” or proximal, which contain submucosal glands and cartilage, and “small” or distal (nonrespiratory and respiratory bronchioles), which are devoid of submucosal glands and cartilage and are <2 mm in diameter in humans; the early events in the pathogenesis of many respiratory diseases, including cystic fibrosis (CF), asthma, and chronic obstructive pulmonary disease (COPD), may occur in the small airways (1–7).
The cellular composition and structure of the epithelium of large airways is well known (8–11); large airways have a pseudostratified columnar epithelium consisting of basal progenitor cells (12–15), secretory (including club and goblet) cells (16–22) with various immune and nonimmune functions, and ciliated cells that clear the airways of particles and pathogens (23, 24). Airway epithelia also contain rare cell types, including pulmonary neuroendocrine cells, tuft or brush cells, and CFTR (cystic fibrosis transmembrane conductance regulator)-rich ionocytes (9, 11, 25–29).
Compared with large airways, less is known about the origins, function, and role in disease of small airway epithelia (22, 30–38). Measurement of small airway resistance in pulmonary function testing is inaccurate (1, 3, 5–7), and sampling small airways in vivo in living subjects is challenging given their small diameter (<2 mm). Other limitations to the study of small airway epithelia include interspecies differences between large mammals and small animal models; mice express different disease-relevant airway ion transporters (39), and their airway cells may follow different developmental lineages than those of humans (18, 40, 41). In addition, airway cell types vary between species: club cells are present in the trachea and bronchi of mice but only in bronchioles in humans (42), and basal cells are only present in trachea of mice but extend to both large and small airways in humans (14, 43). The role of rare cells in small airway epithelial function is also unclear; for example, ionocytes may be absent in the small airways (10).
Recent studies of human lungs with advanced CF have generated single-cell RNA-sequencing (RNA-seq) data that included large and small airway cells; these data suggest abnormal gene expression and cell lineages (10, 44); however, whether small airway epithelial cells in CF have an intrinsically abnormal transcriptome requires studying them at birth, prior to the onset of chronic inflammation and infection.
The porcine lung is a compelling model to study the composition and function of small airway epithelia. Pig and human airways have similar lung anatomy (45) and similar distribution of basal, ciliated, secretory (including club) cells along the airway tree when assessed by histopathology (42). In addition, the pig and human immune systems are closer in function and structure than that of mice (reviewed in Reference 46). Finally, CFTR-deficient pigs develop lung disease mimicking human CF (47–50).
We performed a single-cell resolution census of large and small airways in a porcine model of CF to determine the effect of CFTR on epithelial composition and gene expression. We obtained tissues at birth, before the onset of airway infection and inflammation characteristic of CF lung disease, and used airway microdissection to accurately isolate small airway tissue (51), which in newborn pigs corresponds to those of <200 μm diameter. Importantly, we found that the transcriptome of epithelial cells is not directly affected by lack of CFTR. Finally, we found that although the small and large airways share similar common cell archetypes (52, 53), their transcriptional state is highly determined by location and shows major differences that have important implications to understand cellular function and for gene therapy.
Some of the results of these studies have been previously reported in preprint form (https://doi.org/10.1101/2021.03.16.435690).
Methods
All data are available in GEO (Accession GSE150211). The aggregateBioVar analysis package is available in Bioconductor (54).
Large and small airway tissue cells from lungs of multiple newborn CFTR+/+ and CFTR−/−pigs were sequenced; single-cell RNA-seq differential expression analysis was performed accounting for subject-level variation to increase statistical rigor and decrease the false-positive rate (55).
For details regarding tissue sources, processing, single-cell RNA-seq library prep and analysis, statistical methods, imaging methodology, cell culture, and electrophysiology, please refer to the Extended Materials and Methods in the data supplement.
Results
Lack of CFTR has Minimal Effects on the Transcriptome of Large and Small Airways at Birth
We sequenced 8,928 large and 17,773 small airway cells (Figure 1) that clustered into 10 major cell types (Figures 1A and 1B), including epithelial and nonepithelial cells (Table E1 in the data supplement). Common epithelial cell types including basal, secretory, and ciliated cells were detected in both large and small airway samples and were validated by immunofluorescence confocal microscopy (Figure E1); nonepithelial cells (Figure 1C) included fibroblasts, endothelial cells, immune cells, and smooth muscle cells in both large and small airway samples. The presence of abundant nonepithelial cells in both large and small airway tissues shows that the full thickness of the epithelium was sampled. Moreover, we detected enough cells from each common cell type to perform statistically robust comparisons.
Figure 1.
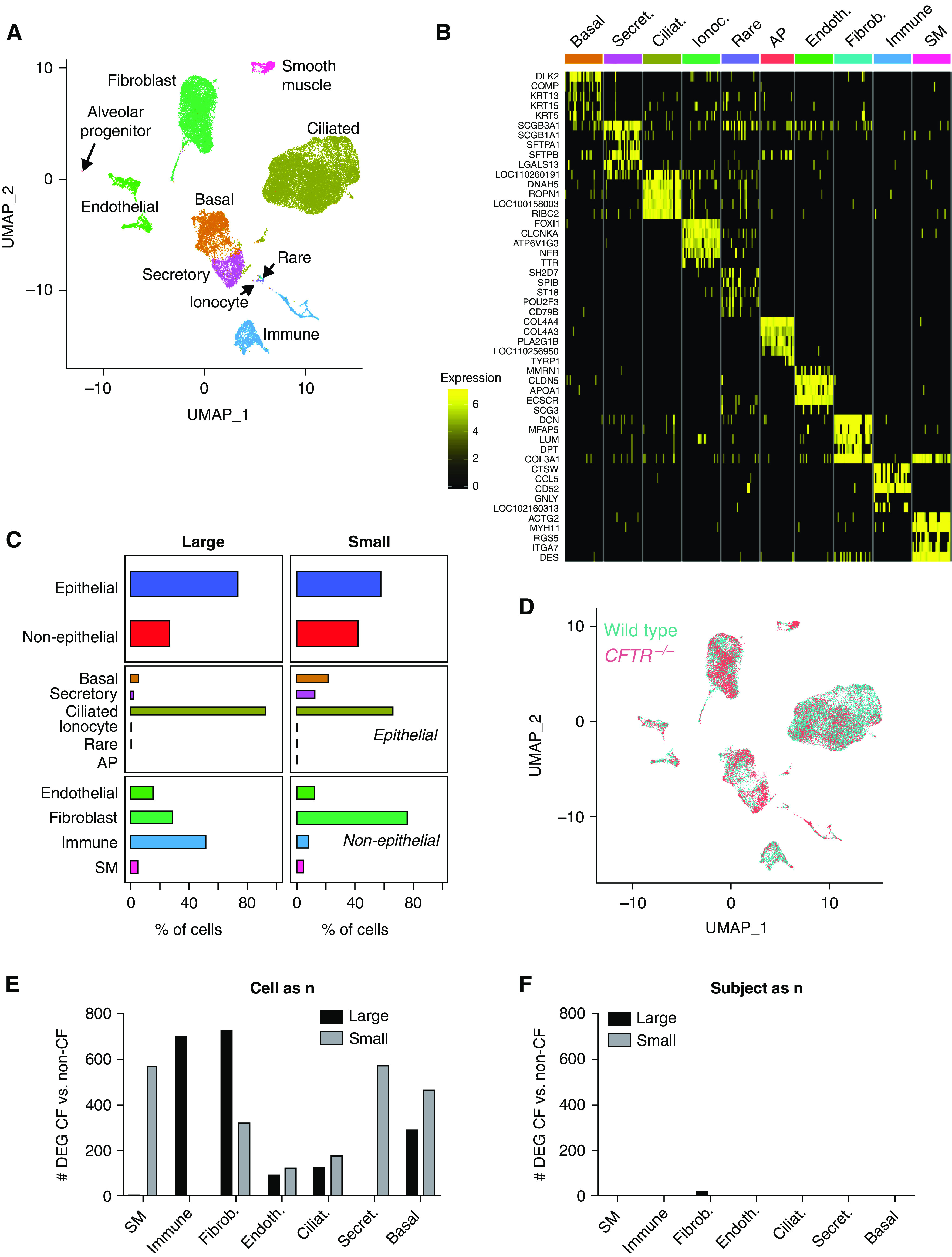
Lack of cystic fibrosis transmembrane conductance regulator (CFTR) does not affect the large and small airways single cell transcriptome in newborn piglets. (A) UMAP cluster visualization. (B) Marker genes heatmap. (C) Cellular composition in large and small airways. n = 10 large (5 CFTR+/+ and 5 CFTR−/−) and 7 small (4 CFTR+/+ and 3 CFTR−/−) airways. (D) UMAP genotype visualization. (E and F) Number of differentially expressed genes between CFTR+/+ and CFTR−/− piglets when cells (E) or subjects (F) are considered a sample/unit of analysis (“n”). AP = alveolar progenitor; CF = cystic fibrosis; Ciliat. = ciliated; Endoth. = endothelial; Fibrob. = fibroblast; Ionoc. = ionocyte; Secret. = secretory; SM = smooth muscle; UMAP = uniform manifold approximation and projection.
The altered transcriptome of airway epithelial cells in people with advanced CF suggests abnormal cellular development in the presence of bacterial infection and chronic inflammation (44, 56); the transcriptome abnormalities partially persist in vitro. In contrast, we have previously shown minimal large airway bulk RNA-seq gene expression differences in newborn wild-type versus CFTR−/− pigs (48, 57); however, the epithelial transcriptome may be underrepresented in whole tracheal tissue, potentially leading to a high false-negative rate. We hypothesized that lack of CFTR would affect epithelial gene expression at birth only in epithelial cells that express CFTR, particularly in the small airways.
We compared large and small airways from wild-type (CFTR+/+) and CFTR−/− pigs. We stratified common epithelial cells by cell type (basal, secretory, and ciliated) and airway type (large and small) and performed differential gene expression analysis accounting for the effects of both individual cells and sample donors on gene expression. Importantly, differential expression analysis of single-cell RNA-seq data is often performed aggregating all cells from all biological sample donors according to condition (each cell is a sample, “cells as n”), which results in an inflated false-positive rate (55). We have previously shown that when single-cell RNA-seq data is analyzed accounting for biological sampling at the subject level as is standard in most biological research (each subject is a sample, “subject as n”), the false-positive rate decreases substantially with minimal effects on the false-negative rate (55). For a given cell type, gene counts were aggregated across all cells from individual biological samples to account for variation in gene expression between subjects using the Bioconductor package aggregateBioVar (54, 55) before the differential expression test.
We found that hundreds of genes were differentially expressed when not accounting for subject-level variability. A large proportion of these are expected to be false positives (55). In contrast, we found fewer than four differentially expressed genes between CFTR+/+ and CFTR−/−pigs for any cell type or region when subject-level variation was accounted for (Figures 1D–1F and Supplementary figure E2). The only consistent difference in gene expression across multiple cell types and airway regions was CFTR itself, as expected in a comparison of CFTR−/− and wild-type animals. These data suggest that, in line with previous observations, lack of CFTR activity minimally modulates expression of other genes in airway epithelial cells. Moreover, the data show that small airway epithelial cell–intrinsic gene expression is not affected by lack of CFTR at birth.
Ionocytes Were Not Detected in the Small Airways
Ionocytes are a rare airway epithelial cell type resembling renal intercalated cells (9, 11, 29) and likely participate in transepithelial ion and fluid transport. Airway ionocytes express the transcription factor FOXI1, high amounts of CFTR, and genes for vacuolar-type ATPase subunits; based on these markers, we found a small cluster of ionocyte-like cells in our single-cell RNA-seq data.
Because single-cell RNA-seq detection of ionocytes in small airway samples could be limited by uncharacterized sampling biases, we first measured ionocytes in situ with immunohistochemical staining of Barttin (BSND) (58) and FOXI1 protein expression, and for RNA expression of FOXI1. We identified cells expressing BSND and FOXI1 in large but not in small airways in newborn pigs (Figures 2A and 2B).
Figure 2.
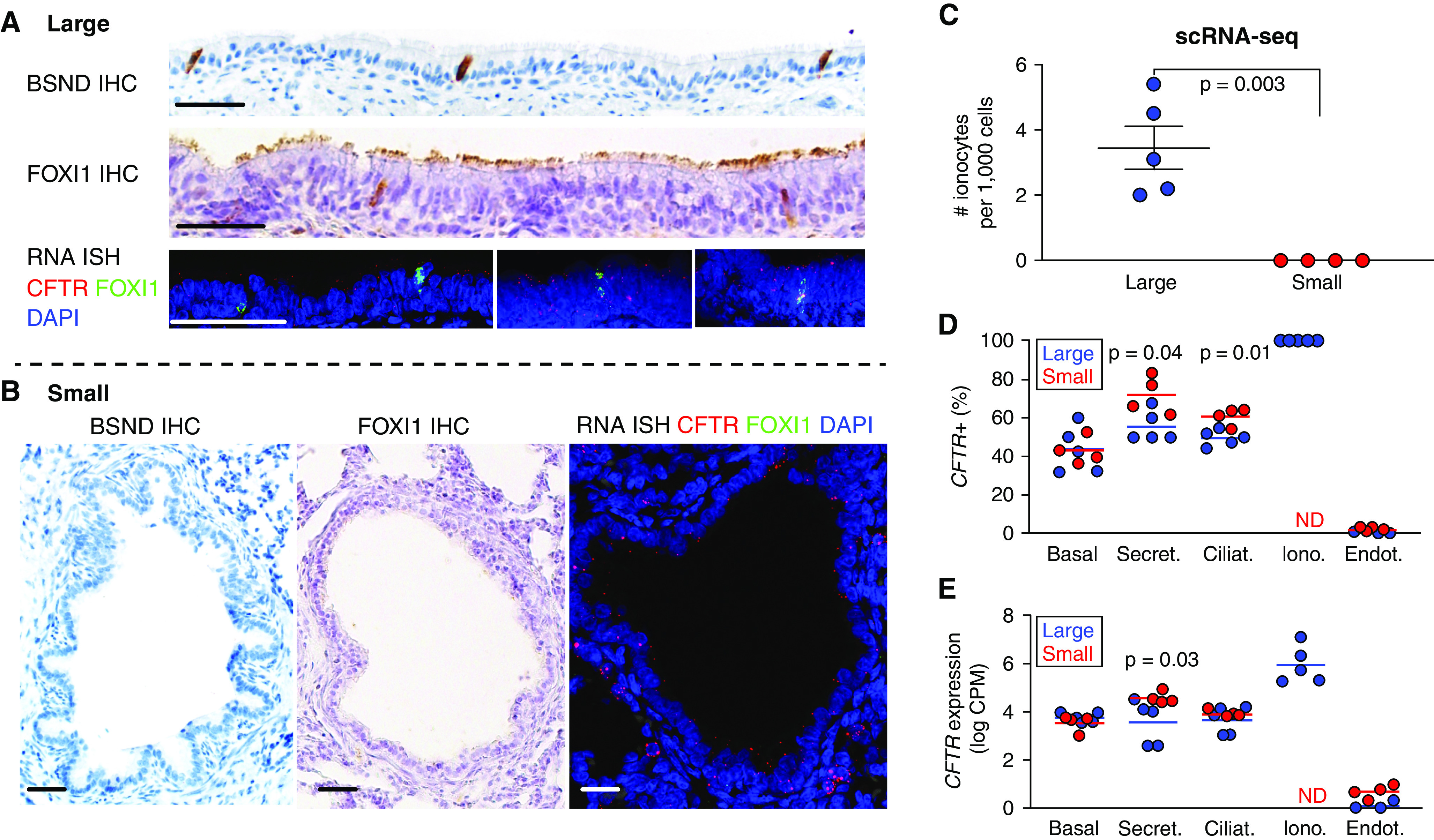
Cellular patterns of CFTR expression in the small airways of CFTR+/+ pigs. (A and B) BSND, FOXI1, and CFTR immunohistochemistry (IHC) and RNA in situ hybridization staining as indicated of large (A) and small (B) airway epithelia. Scale bars, 25 μm. (C) CFTR+/FOXI1+ (ionocyte) cell frequency per single cell RNA-sequencing (scRNA-seq) in large and small airways. (D) Percentage of cells with CFTR count greater than zero, and (E) log-transformed (counts per million) expression of CFTR. n = 5 large and 4 small airways. BSND = Barttin; CPM = counts per million; ND = not detected; RNA ISH = RNA in situ hybridization.
We then determined the abundance of cells coexpressing CFTR and FOXI1 (forkhead box I1) in wild-type pig single-cell RNA-seq data (Figure 2C) from large and small airways. We found 2 to 6 ionocytes per 1,000 cells in large airways and none in the small airways; using a Bayesian model, we computed a 95% posterior probability of fewer than 1.49 ionocytes per 1,000 cells in the small airways. Given the absence of CFTR-rich ionocytes in small airways, we expected CFTR expression in other epithelial cells compensating for the lack of ionocyte CFTR. We determined the fraction of cells with detectable CFTR (Figure 2D) and the average degree of CFTR expression (Figure 2E and Table E1) for each cell type; because CFTR is expressed at low amounts in most cell types, yet background transcripts may contaminate single-cell RNA-seq data, we show endothelial cells, which rarely contained CFTR transcripts, as comparison. CFTR was detected most often in ionocytes (100%, by definition), secretory cells (50–70%) followed by ciliated, and lastly basal cells. We measured increased detection of CFTR in secretory and ciliated cells in small airways compared with large airways (secretory: 72% ± 4.9% vs. 55% ± 3.5% respectively; ciliated: 61% ± 2.3% vs. 49% ± 1.8% respectively). We also detected increased CFTR expression in small airway secretory cells compared with large airways (4.6 ± 0.12 vs. 3.6 ± 0.4 log2CPM, respectively). The measured detectability is similar to that observed using single-cell RNA in situ hybridization and single-cell quantitative PCR in human cells, suggesting our sequencing depth allowed precise estimates of CFTR expression (10). These data show that CFTR expression in both the large and small airways occurs in a majority of secretory and ciliated cells, in addition to higher degrees of CFTR expression in ionocytes in the large airways.
Cell Type–Specific Gene Expression Varies Geographically in Airway Epithelia
Cells with similar morphology and canonical gene expression biomarkers are generally assigned the same cell identity, but their gene expression profile and physiology may vary owing to regulation by the cellular microenvironment or because of developmental origin and history (59–61). We therefore compared the transcriptional profile of cell types identified in the large airways to their corresponding cell types in small airways, accounting for subjects as the unit of analysis, as in our CFTR−/− versus wild-type comparison. We expected few differentially expressed genes between large and small airways on the basis of prior data (51) including AQP4 (aquaporin 4), SFTPD (surfactant protein D), and ITGA9 (integrin α 9) expected to be expressed in small airways. We were surprised by the large number and magnitude of differences between corresponding cell types in small and large airways even when using our statistical framework. Figure 3A shows the results of differential expression analysis (Table E2). We discovered 406 differentially expressed genes (Adjusted P value < 0.05 and log2 fold change < −1 or >1) between small and large airways in basal cells, 746 in secretory cells, and 2,546 genes in ciliated cells. Although we found large expression differences in common cell types in the large versus small airways, this was not the case for cell types localized deeper from the epithelial surface, such as endothelial and smooth muscle cells (Figure E3). Only 113 shared genes were differentially expressed among all three common epithelial cell types (Table E2, epithelial intersection). Genes expressed at higher degrees in all large airway epithelia–common cell types included well-characterized genes involved in goblet cell metaplasia including AGR2 (anterior gradient 2), which is a protein disulfide isomerase involved in the epithelial allergic response and mucin production (62), and CLCA1 (chloride channel accessory 1), which participates in MAPK signaling driving mucus expression and in TMEM16A-mediated chloride transport (63).
Figure 3.
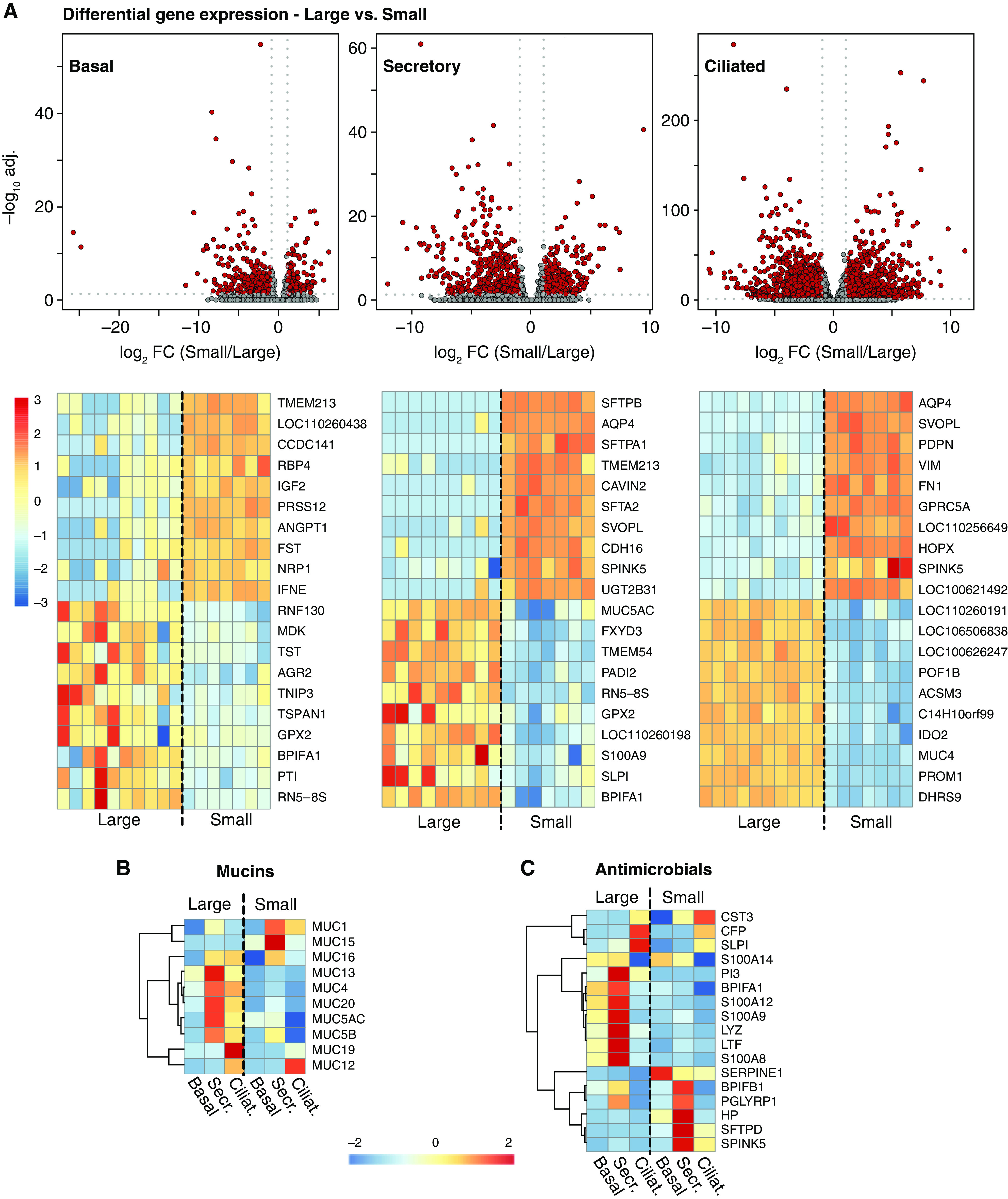
Cell type–specific gene expression varies between large and small airways. (A) Volcano plots and top 20 cell-specific differentially expressed genes in basal, secretory, and ciliated large and small airway epithelial cells. (B) Mucin and (C) antimicrobial genes heatmaps: normalized and centered average gene expression in large and small airway for cell types indicated, genes detected at 2 or higher CPM. n = 10 large and 7 small airway. CFTR+/+ and CFTR −/− pig samples were grouped together. adj. = adjusted; FC = fold change.
Given their importance for airway physiology in health and disease, we examined expression of mucin genes in large and small airways (Figures 3B and E4 and Table E1) (38). Expression of the secreted mucins MUC5AC and MUC5B was site specific. In particular, we observed the previously described transition of secretory cells from goblet-like phenotype (e.g., MUC5AC-rich) proximally in large airways to club-like phenotype (e.g., MUC5AC-low) distally in small airways (22, 64). MUC5B expression was also higher in the large airways. Small airway secretory cells expressed higher amounts of tethered mucins MUC1 and MUC15 and lower amounts of MUC13 and MUC20 compared with large airways. Together with differential expression of goblet cell genes CLCA1 and AGR2, these data suggest an upstream site-specific expression program promoting secreted mucins in the large airways.
Because large and small airways may be exposed to different amounts and types of bacteria, fungi, and viruses, we expected expression of secreted antimicrobials to be different in these two sites. Overall, secretory cells expressed most antimicrobials (Figure 3C). Our data show that compared with small airways, large airway secretory cells express higher amounts of LYZ (lysozyme), LTF (lactoferrin), and S100A8, S100A9, and S100A12 (calprotectin genes). In contrast, small airway secretory cells express high amounts of SFTPD, SERPINE1 (plasminogen activator-inhibitor 1), PGLYRP1 (peptidoglycan recognition protein 1), HP (haptoglobin) and SPINK5 (serine peptidase inhibitor kazal type 5). We also analyzed expression of genes associated with responses to bacteria and viruses and found differential expression between large and small airway epithelial cells (Figure E5). Taken together, these data suggest location-specific regulation of secreted innate antimicrobial molecules in large and small airways. Surfactants are key regulators of epithelial surface tension and are required to maintain alveolar and airway patency (reviewed in Reference 65). BPIFA1 (BPI fold containing family A member 1, also known as SPLUNC) has antimicrobial, surfactant, and smooth muscle signaling functions (66–69) and is expressed at high amounts in secretory and ciliated cells of the large airway. Expression of BPIFA1 in small airway secretory cells is decreased almost 50-fold compared with large airways (Figure 3 and Table E2). Instead, and perhaps as an alternative to BPIFA1, small airway secretory cells expressed high amounts of the surfactants SFTPB (surfactant protein B) (500-fold higher than large airways) and SFTPA1 and SFTA2 (surfactant protein A1 and surfactant-associated 2) (170- and 60-fold higher, respectively); these three genes were among the top six differentially expressed genes with the criteria used.
Small Airway Basal Cells Express Barrier-Forming Claudins and Apical Membrane Ion Transporters and Reach the Apical Epithelial Surface
We were surprised by the high degrees of expression of transepithelial ion apical transporter genes in small airway basal cells, as basal cells are not believed to participate in apical-basolateral ion transport. This was particularly striking for genes such as the amiloride-sensitive sodium channel (ENaC) SCNN1B and SCNN1G, which are considered apical surface sodium channels in airway epithelia (70, 71), and CLDN1 (the barrier-forming claudin 1) (Tables E1 and E2) (72, 73); these 3 genes were among the top 20 genes highly expressed in small versus large airway basal cells. We therefore hypothesized that basal cells in the small airways reach both the basement membrane and the apical surface.
We performed a detailed examination of the abundance and localization of basal cells in small and large airway epithelia. p63 (tumor protein 63) is a well-characterized basal cell marker (14, 61, 74–76). Figure 4A shows that in the cuboidal monolayer of distal respiratory bronchioles, there were p63+ cells that contact the lumen. Interestingly, some p63+ cells also appeared to have cilia (Figures 4A and E6), which contrasts with the prior evidence that basal cells first differentiate into secretory cells before differentiation into ciliated cells in the large airways. Using quantitative immunohistochemistry (Figure 4B), we found that although small airway epithelia had less p63+ cells per surface unit, most small airway p63+ cells contacted both the basement membrane and the airway lumen. In contrast, almost none of large airway basal cells contact the lumen.
Figure 4.
Small airway basal cells are surface cells. (A) IHC at various airway levels (large airways: trachea-bronchus; small airways: bronchioles). Scale bars, 25 μm. Arrows: surface p63+ cells, red inset (100× magnification) surface p63+ cell with and without visible cilia. n = 5 large and 4 small airways. (B) Immunohistochemical quantification of basal cells and their position in large and small airways. Lg. = large; Sm. = small.
Small Airway Epithelia Have Distinct Transepithelial Ion Conductance
Our data show that the expression of the anion transporter CFTR is markedly different in small airway epithelia (devoid of ionocytes) compared with large airway epithelia and suggest distinct mechanisms of transepithelial ion transport and water movement in small airways. We therefore investigated the expression patterns of other ion transporters, ion transporter regulators, water channels, and barrier-forming claudin genes in small airway epithelia.
The amiloride-responsive transepithelial voltage of airway epithelia decreases in a proximal-distal manner (32, 33, 77, 78). We expected that expression of genes coding for the amiloride-sensitive sodium channel could be similar (based on our previous in vitro data [51]) or differ (based on References 10 and 21) in large and small airway epithelia. Small airway epithelial cells expressed higher amounts of the ENaC genes SCNN1B and SCNN1G (Figures 5A and E7 and Table E2); this correlated with lower concentrations of BPIFA1 (SPLUNC), which inhibits ENaC expression and function (67, 79) and suggests that ENaC might be more active in small airways. Next, we investigated expression of transcellular water channels and tight junction genes (80) (Figure 5B and Table E1). Only AQP3, -4, and -5, respectively, were consistently detected in at least one cell type. AQP3 is the predominant aquaporin expressed in large airway cells and was expressed at lower amounts in small airway secretory and ciliated cells which instead expressed large amounts of AQP4. AQP4 was a top differentially expressed gene at nearly 200-fold higher amounts in small airway cells (Table E2). In addition, small airway secretory cells expressed nearly twice as many total aquaporin transcripts than any large airway epithelial cell type (Table E1). These data suggest that small airway epithelia are more permissive to osmotic water movement than large airway epithelia. Finally, we compared the expression of claudins, which regulate epithelial tight junction water permeability and ion selectivity (72, 81, 82). In the large airways, claudins are primarily expressed by surface ciliated and secretory cells. We found that small airway epithelial cells expressed the barrier-forming claudins 1, 4, and 10 (isoform 10b) at higher amounts than large airway cells (Figure 5C). CLDN18 (Claudin 18) was detected only in type II alveolar progenitor cells, consistent with the literature (83). The differential expression of ion transporters and claudins, and the different cellular architecture of large and small airway epithelia suggest that regulation of ion transport and barrier function is site specific.
Figure 5.
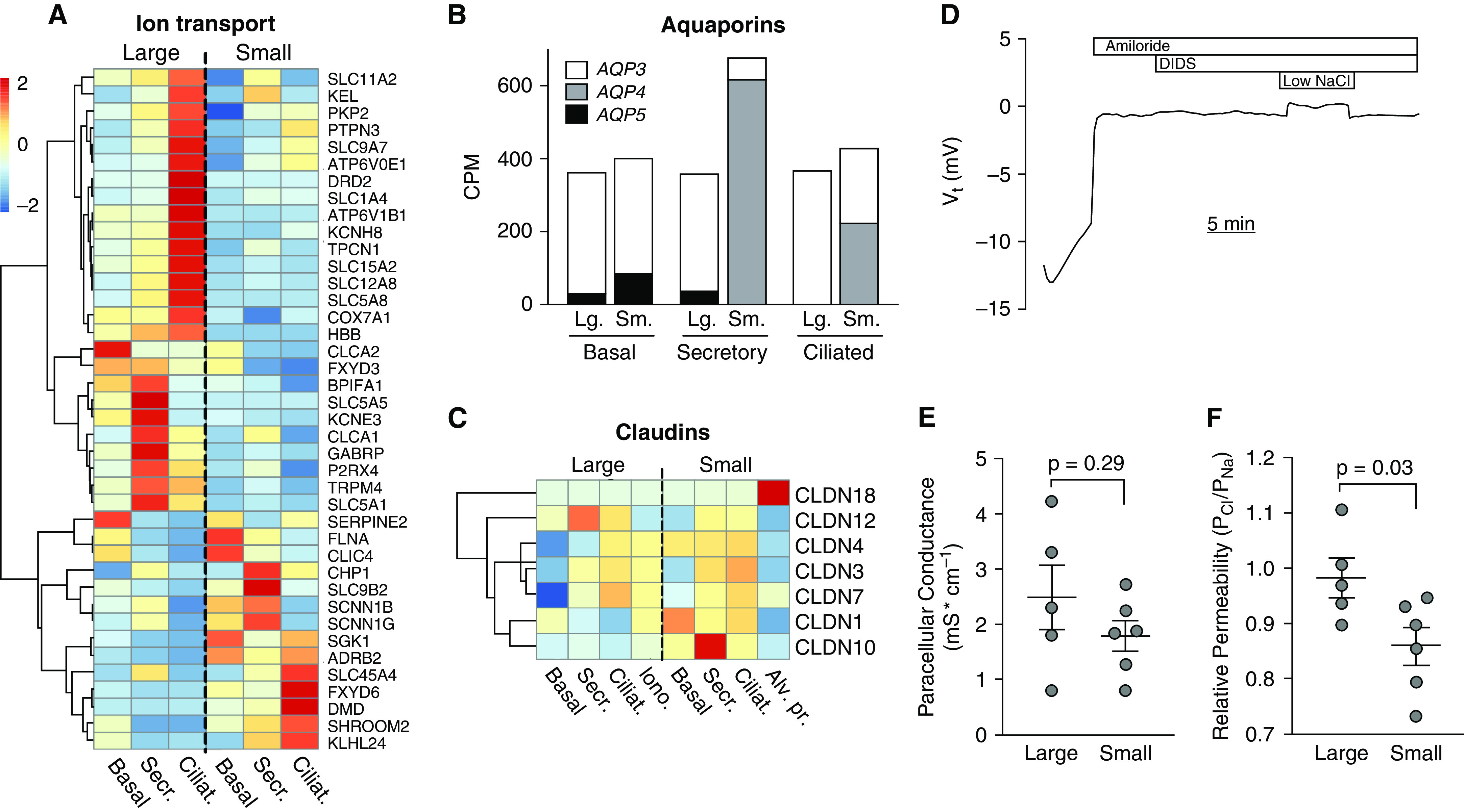
Ion and water transporter gene expression and paracellular ion selectivity differs in small and large cultured airway epithelia. (A) Ion transport and regulator genes heatmap: normalized and centered average gene expression in large and small airway for cell types indicated. (B) Aquaporin 3, 4, and 5 average CPM gene expression in common large and small airway epithelia cell types. (C) Claudin genes heatmap: normalized and centered average gene expression in large and small airways for cell types indicated, genes detected at 2 or more CPM. n = 10 large and 7 small airways. (D) Ussing chamber measurements in CFTR−/− pig airway epithelia treated with amiloride and DIDS (4,4′-diisothiocyanotostilbene-2,2′-disulfonic acid) to block transcellular ion transport. (E) Shows paracellular conductance, and (F) shows relative paracellular permeability to sodium and chloride. n = 5 large and 6 small airway samples.
We have previously shown that small airway epithelia cultured at the air–liquid interface have higher CFTR conductance than and similar ENaC current to large airway epithelia. Based on our findings in this study, we hypothesized that the paracellular conductance of small airway epithelia at the air–liquid interface in vitro would be different than that of large airway epithelia. We minimized transcellular ion transport using epithelia from CFTR−/− pigs and treated them with the apical ENaC sodium channel blocker amiloride and DIDS (4,4′-diisothiocyanotostilbene-2,2′-disulfonic acid), which blocks most non-CFTR Cl− channels in the apical membrane (Figure 5D). We then performed Na+ and Cl− dilution potential assays and recorded the potential difference that arose by diluting apical NaCl as previously described (84). We found that contrary to our hypothesis, large and small airway epithelia had similar paracellular conductance (Figure 5E). However, we found that small airway epithelia had a lower PCl/PNa compared with large airways (Figure 5F); these data suggest that the claudin channels in small airway epithelia are more permeable to cations than to anions, whereas large airway epithelia have similar paracellular permeability to cations and anions.
Discussion
The transcriptome of a cell may be determined by cell-intrinsic properties such as its epigenome, or by the extracellular environment (59–61). Understanding the relative contribution of cell versus environment is key to interpreting how a transcriptome may be affected by a disease. Our previous work investigating the transcriptome in a porcine model of CF suggested that the transcriptional abnormalities observed in airway epithelia of adults with CF were secondary to chronic infection and inflammation as they were not present at birth in the large airways (48, 56, 57); however, whether the small airways were affected at birth remained unclear. In this study, we performed a single-cell RNA-seq census of large and small porcine airway epithelia at birth, using microdissection-based sampling techniques. Our study provides key insights supported by several methodological strengths.
CFTR Does Not Regulate Expression of Other Airway Epithelial Genes at Birth
CFTR coexpression with other genes in humans and animal models varies by cell type and inflammation or disease state (53). But does CFTR itself regulate expression of other genes via protein–protein interactions or via ion transport-mediated transcriptional regulation? Our data strikingly show that knocking out CFTR does not modify gene expression except for CFTR itself. This finding was enabled by two methodological advantages: 1) by sampling CFTR−/− pigs at birth, we were able to measure the cell-intrinsic transcriptome of CFTR−/− airway epithelia before the onset of chronic infection and inflammation; and 2) by performing single-cell RNA-seq differential expression analysis accounting for interindividual variability (54, 55), we optimize the false-positive rate. We conclude that whereas lack of CFTR ultimately results in chronic infection and epithelial inflammation that modulate many genes, CFTR itself does not directly regulate other genes. However, we cannot exclude whether mutant CFTR protein (e.g., the F508 del mutation) induces aberrant gene expression at birth through mechanisms related to protein processing (e.g., aggresome formation) (85).
CFTR Expression Varies in the Large and Small Airways
We found that large and small airway epithelia share similar common (basal, secretory/goblet, and ciliated) cell types; in contrast, ionocytes were absent in small airway epithelia at birth. This conclusion was enabled by including in situ immunohistochemical imaging for both RNA and protein ionocyte markers in tissue. Because ion transport is critically important in the airways both at birth and throughout life, how does it differ in the absence of ionocytes? The human large airway epithelium contains approximately 40,000 lumen-reaching cells/mm2 (86), of which 0.5% are ionocytes. Assuming uniform distribution of ionocytes on the airway surface, we expect an estimated average distance of approximately 70 μm between ionocytes; this cell specialization may not be optimal in the fractal small airways, as it may result in airways without ionocytes. We speculate that 1) if ionocytes are primarily secretory, small airways with ionocytes would be prone to obstruction by secreted fluid; or 2) alternatively, if ionocytes primarily aid fluid absorption, they may be absent in small airways given that lungs can continue to carry out adequate respiration if a few small airways are obstructed, but not if a large airway is.
Cell Type–associated Gene Expression and Function Is Topography Dependent
We found that cell types sharing the same gene expression markers and morphology may have very different gene expression profiles when in two different locations; the magnitude of hundreds of differentially expressed genes between corresponding large and small airway cell archetypes (52, 53) was striking. These findings were enabled by the use of microdissection of small airway tissues and large airway epithelial scrapings, which minimized sampling of cell types overrepresented in gross tissue biopsies (e.g., alveolar cells) or that confound comparison of surface epithelial cells (e.g., submucosal gland cells), in addition to confirming results with immunohistochemistry or electrophysiology assays.
The branching pattern and anatomical configuration of the mammalian airway tree results in a very different environment in the lumen of the large and small airways. Two aspects of our findings led us to speculate that location-dependent gene expression in airway epithelial common cell archetypes depends on the local luminal microenvironment: 1) the types of genes upregulated in large airways (secreted mucins and antimicrobial peptides) are key for clearance of inhaled or aspirated particles and microorganism to which the large airways are constantly exposed, whereas genes upregulated in small airways (ion transporters, water channels, barrier-forming claudins, and surfactants) are key for small airway patency; and 2) cell types more likely to interact with the epithelial apical surface (epithelial cells, immune cells, and fibroblasts) differed more in large versus small airways than smooth muscle and endothelial cells. Other microenvironmental factors may also drive large versus small airway epithelial archetypal differences. For example, large airways are exposed to wider respiratory cycle-driven fluctuations in CO2 and O2 concentration, humidity, and temperature than the small airways. We detected higher expression of mucin and antimicrobial genes involved in responses to inhaled allergens and pathogens in large airway epithelia; in contrast, we detected higher expression of various ion transporters, water channels, barrier-forming claudin genes, and surfactants in small airway epithelia.
We found important differences in the configuration of cell types in small versus large airway epithelia. Specifically, the ion transporter and barrier-forming claudin gene expression pattern of small airway basal cells led us to discover that at least some small airway basal cells reach the apical surface; we speculate that they participate in transepithelial ion transport. This finding has important practical implications. Gene therapy for lung diseases ideally targets pulmonary stem cells. Airway epithelial basal cells are important progenitor/stem cells and were considered difficult to target via aerosolization given their localization beneath the airway surface. Our data suggest that some small airway basal cells may be directly targeted by gene therapy vectors via aerosolization.
Finally, taken together, our data lead us to speculate that ion and water transport in large and small airway epithelia are mediated by similar cell archetypes, but are finely tuned by differences in gene expression: 1) small airway epithelia express more aquaporins, so changes in regulated ion transport are followed by transcellular fluid secretion or absorption; 2) higher expression of CFTR, whose activation is regulated, and lower paracellular anion conductance gives small airways an advantage for fluid secretion; and 3) higher expression and function of ENaC allows higher Na+ absorption, whereas higher relative paracellular cation conductance facilitates paracellular reflux and secretion of Na+; this would provide support for higher CFTR-mediated secretion when CFTR activity is high and higher ENaC-mediated absorption when CFTR activity is low. Overall, the data suggest that small airway epithelia have an advantage over large airway epithelia for both rapid absorption and secretion of ions and water. This is consistent with the notion that small airways need to be “wet enough to be pliable” yet “dry enough to remain patent” as described by Shamsuddin and Quinton (33).
Limitations
Our study is limited by potential biases in proportional sampling of various cell types owing to differential isolation, viability, or lysis sensitivity of specific cell types; however, our conclusions do not depend on relative proportions of cell types as determined by single-cell RNA sequencing, and we only directly compare cell type proportions by using immunohistochemistry in situ (e.g., ionocytes and basal cells). Moreover, we focused our analysis only on surface epithelial cells from trachea (which may differ transcriptionally from bronchial cells [87, 88]) and small airways; we did not include submucosal gland cells, which contribute to water and electrolyte transport in large airways in vivo, and did not analyze immune cells (which play a key role in inflammation and development) included in our dataset in depth. Finally, we do not perform a time course to detect the first single-cell transcriptomic changes in CF lung disease.
Our data show how the function and gene expression profile of an airway epithelial cell archetype varies depending on its cellular microenvironment. Our study highlights that important proximal–distal patterns of cellular composition and gene expression observed in gut and kidney analysis (89, 90) also apply to the airways with important implications for lung disease pathophysiology and for therapy of lung disease.
Acknowledgments
Acknowledgment
The authors thank Linda Powers, Mallory Stroik, Nicholas Gansemer, Christian Brommel, Keyan Zarei, Jason Ratcliff, Michael Chimenti, and the University of Iowa Institute for Human Genetics Genomics Facility for excellent technical support.
Footnotes
Supported in part by National Heart, Lung, and Blood Institute K01HL140261 (A.A.P.), P01HL051670 (D.K.M., D.A.S., M.J.W., and J.Z.), P01HL091842 (D.A.S., M.J.W., J.Z., and A.A.P.), T32HL007638 (G.S.R.-I., M.J.W., and J.Z.), and T32GM007337 (G.S.R.-I.); Cystic Fibrosis Foundation University of Iowa RDP (D.A.S., M.J.W., J.Z., and A.A.P.), PEZZUL20A1-KB (A.A.P.), and LI19XX0 (X.L.); Gilead Sciences Research Scholars Program in Cystic Fibrosis (I.M.T.); National Institute of Diabetes and Digestive and Kidney Diseases P30DK054759 (J.Z.); and the University of Iowa Physician Scientist Training Program (A.A.P.).
Author Contributions: Conceptualization: A.L.T., X.L., J.Z., and A.A.P. Methodology: A.L.T., X.L., and W.Y. Formal analysis: A.L.T., X.L., W.Y., D.K.M., I.M.T., and A.A.P. Investigation: A.L.T., X.L., R.V., W.Y., H.G., S.E.M., G.S.R.-I., D.K.M., I.M.T., and A.A.P. Resources: A.L.T., X.L., G.S.R.-I., D.K.M., D.A.S., and A.A.P. Data curation: A.L.T., X.L., W.Y., and A.A.P. Writing, original draft: A.L.T., J.Z., and A.A.P. Writing, review and editing: All authors. Visualization: A.L.T. and A.A.P. Supervision: M.J.W., J.Z., and A.A.P. Funding acquisition: D.A.S., M.J.W., J.Z., and A.A.P.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2021-0499OC on March 2, 2022
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1. Hogg JC, Macklem PT, Thurlbeck WM. Site and nature of airway obstruction in chronic obstructive lung disease. N Engl J Med . 1968;278:1355–1360. doi: 10.1056/NEJM196806202782501. [DOI] [PubMed] [Google Scholar]
- 2. Hyde DM, Hamid Q, Irvin CG. Anatomy, pathology, and physiology of the tracheobronchial tree: emphasis on the distal airways. J Allergy Clin Immunol . 2009;124:S72–S77. doi: 10.1016/j.jaci.2009.08.048. [DOI] [PubMed] [Google Scholar]
- 3. Macklem PT, Mead J. Resistance of central and peripheral airways measured by a retrograde catheter. J Appl Physiol . 1967;22:395–401. doi: 10.1152/jappl.1967.22.3.395. [DOI] [PubMed] [Google Scholar]
- 4. Macklem PT, Mead J. The physiological basis of common pulmonary function tests. Arch Environ Health . 1967;14:5–9. doi: 10.1080/00039896.1967.10664685. [DOI] [PubMed] [Google Scholar]
- 5. Hogg JC, Paré PD, Hackett TL. The contribution of small airway obstruction to the pathogenesis of chronic obstructive pulmonary disease. Physiol Rev . 2017;97:529–552. doi: 10.1152/physrev.00025.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carr TF, Altisheh R, Zitt M. Small airways disease and severe asthma. World Allergy Organ J . 2017;10:20. doi: 10.1186/s40413-017-0153-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown R, Woolcock AJ, Vincent NJ, Macklem PT. Physiological effects of experimental airway obstruction with beads. J Appl Physiol . 1969;27:328–335. doi: 10.1152/jappl.1969.27.3.328. [DOI] [PubMed] [Google Scholar]
- 8. Jackson ND, Everman JL, Chioccioli M, Feriani L, Goldfarbmuren KC, Sajuthi SP, et al. Single-cell and population transcriptomics reveal pan-epithelial remodeling in type 2-high asthma. Cell Rep . 2020;32:107872. doi: 10.1016/j.celrep.2020.107872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature . 2018;560:319–324. doi: 10.1038/s41586-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Okuda K, Dang H, Kobayashi Y, Carraro G, Nakano S, Chen G, et al. Secretory cells dominate airway CFTR expression and function in human airway superficial epithelia. Am J Respir Crit Care Med . 2021;203:1275–1289. doi: 10.1164/rccm.202008-3198OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Plasschaert LW, Žilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature . 2018;560:377–381. doi: 10.1038/s41586-018-0394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. Basal cells are a multipotent progenitor capable of renewing the bronchial epithelium. Am J Pathol . 2004;164:577–588. doi: 10.1016/S0002-9440(10)63147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. In vivo differentiation potential of tracheal basal cells: evidence for multipotent and unipotent subpopulations. Am J Physiol Lung Cell Mol Physiol . 2004;286:L643–L649. doi: 10.1152/ajplung.00155.2003. [DOI] [PubMed] [Google Scholar]
- 14. Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci USA . 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boers JE, Ambergen AW, Thunnissen FB. Number and proliferation of basal and parabasal cells in normal human airway epithelium. Am J Respir Crit Care Med . 1998;157:2000–2006. doi: 10.1164/ajrccm.157.6.9707011. [DOI] [PubMed] [Google Scholar]
- 16. Chen G, Korfhagen TR, Xu Y, Kitzmiller J, Wert SE, Maeda Y, et al. SPDEF is required for mouse pulmonary goblet cell differentiation and regulates a network of genes associated with mucus production. J Clin Invest . 2009;119:2914–2924. doi: 10.1172/JCI39731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Danahay H, Pessotti AD, Coote J, Montgomery BE, Xia D, Wilson A, et al. Notch2 is required for inflammatory cytokine-driven goblet cell metaplasia in the lung. Cell Rep . 2015;10:239–252. doi: 10.1016/j.celrep.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 18. Hayashi T, Ishii A, Nakai S, Hasegawa K. Ultrastructure of goblet-cell metaplasia from Clara cell in the allergic asthmatic airway inflammation in a mouse model of asthma in vivo. Virchows Arch . 2004;444:66–73. doi: 10.1007/s00428-003-0926-8. [DOI] [PubMed] [Google Scholar]
- 19. Pezzulo AA, Tudas RA, Stewart CG, Buonfiglio LGV, Lindsay BD, Taft PJ, et al. HSP90 inhibitor geldanamycin reverts IL-13- and IL-17-induced airway goblet cell metaplasia. J Clin Invest . 2019;129:744–758. doi: 10.1172/JCI123524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chinet TC, Gabriel SE, Penland CM, Sato M, Stutts MJ, Boucher RC, et al. CFTR-like chloride channels in non-ciliated bronchiolar epithelial (Clara) cells. Biochem Biophys Res Commun . 1997;230:470–475. doi: 10.1006/bbrc.1996.5939. [DOI] [PubMed] [Google Scholar]
- 21. Van Scott MR, Hester S, Boucher RC. Ion transport by rabbit nonciliated bronchiolar epithelial cells (Clara cells) in culture. Proc Natl Acad Sci USA . 1987;84:5496–5500. doi: 10.1073/pnas.84.15.5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zuo WL, Shenoy SA, Li S, O’Beirne SL, Strulovici-Barel Y, Leopold PL, et al. Ontogeny and biology of human small airway epithelial club cells. Am J Respir Crit Care Med . 2018;198:1375–1388. doi: 10.1164/rccm.201710-2107OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. You Y, Huang T, Richer EJ, Schmidt JE, Zabner J, Borok Z, et al. Role of f-box factor foxj1 in differentiation of ciliated airway epithelial cells. Am J Physiol Lung Cell Mol Physiol . 2004;286:L650–L657. doi: 10.1152/ajplung.00170.2003. [DOI] [PubMed] [Google Scholar]
- 24. Horani A, Ustione A, Huang T, Firth AL, Pan J, Gunsten SP, et al. Establishment of the early cilia preassembly protein complex during motile ciliogenesis. Proc Natl Acad Sci USA . 2018;115:E1221–E1228. doi: 10.1073/pnas.1715915115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bankova LG, Dwyer DF, Yoshimoto E, Ualiyeva S, McGinty JW, Raff H, et al. The cysteinyl leukotriene 3 receptor regulates expansion of IL-25-producing airway brush cells leading to type 2 inflammation. Sci Immunol . 2018;3:eaat9453. doi: 10.1126/sciimmunol.aat9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sui P, Wiesner DL, Xu J, Zhang Y, Lee J, Van Dyken S, et al. Pulmonary neuroendocrine cells amplify allergic asthma responses. Science . 2018;360:eaan8546. doi: 10.1126/science.aan8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Perniss A, Schmidt P, Soultanova A, Papadakis T, Dahlke K, Voigt A, et al. Development of epithelial cholinergic chemosensory cells of the urethra and trachea of mice. Cell Tissue Res . 2021;385:21–35. doi: 10.1007/s00441-021-03424-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krasteva G, Canning BJ, Papadakis T, Kummer W. Cholinergic brush cells in the trachea mediate respiratory responses to quorum sensing molecules. Life Sci . 2012;91:992–996. doi: 10.1016/j.lfs.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 29. Scudieri P, Musante I, Venturini A, Guidone D, Genovese M, Cresta F, et al. Ionocytes and CFTR chloride channel expression in normal and cystic fibrosis nasal and bronchial epithelial cells. Cells . 2020;9:E2090. doi: 10.3390/cells9092090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tilley AE, O’Connor TP, Hackett NR, Strulovici-Barel Y, Salit J, Amoroso N, et al. Biologic phenotyping of the human small airway epithelial response to cigarette smoking. PLoS One . 2011;6:e22798. doi: 10.1371/journal.pone.0022798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vucic EA, Chari R, Thu KL, Wilson IM, Cotton AM, Kennett JY, et al. DNA methylation is globally disrupted and associated with expression changes in chronic obstructive pulmonary disease small airways. Am J Respir Cell Mol Biol . 2014;50:912–922. doi: 10.1165/rcmb.2013-0304OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shamsuddin AK, Quinton PM. Native small airways secrete bicarbonate. Am J Respir Cell Mol Biol . 2014;50:796–804. doi: 10.1165/rcmb.2013-0418OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shamsuddin AK, Quinton PM. Surface fluid absorption and secretion in small airways. J Physiol . 2012;590:3561–3574. doi: 10.1113/jphysiol.2012.230714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zuo WL, Rostami MR, Shenoy SA, LeBlanc MG, Salit J, Strulovici-Barel Y, et al. Cell-specific expression of lung disease risk-related genes in the human small airway epithelium. Respir Res . 2020;21:200. doi: 10.1186/s12931-020-01442-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. O’Beirne SL, Shenoy SA, Salit J, Strulovici-Barel Y, Kaner RJ, Visvanathan S, et al. Ambient pollution-related reprogramming of the human small airway epithelial transcriptome. Am J Respir Crit Care Med . 2018;198:1413–1422. doi: 10.1164/rccm.201712-2526OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang J, Zuo WL, Fukui T, Chao I, Gomi K, Lee B, et al. Smoking-dependent distal-to-proximal repatterning of the adult human small airway epithelium. Am J Respir Crit Care Med . 2017;196:340–352. doi: 10.1164/rccm.201608-1672OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hackett NR, Butler MW, Shaykhiev R, Salit J, Omberg L, Rodriguez-Flores JL, et al. RNA-seq quantification of the human small airway epithelium transcriptome. BMC Genomics . 2012;13:82. doi: 10.1186/1471-2164-13-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Okuda K, Chen G, Subramani DB, Wolf M, Gilmore RC, Kato T, et al. Localization of secretory mucins muc5ac and muc5b in normal/healthy human airways. Am J Respir Crit Care Med . 2019;199:715–727. doi: 10.1164/rccm.201804-0734OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shah VS, Meyerholz DK, Tang XX, Reznikov L, Abou Alaiwa M, Ernst SE, et al. Airway acidification initiates host defense abnormalities in cystic fibrosis mice. Science . 2016;351:503–507. doi: 10.1126/science.aad5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pardo-Saganta A, Law BM, Gonzalez-Celeiro M, Vinarsky V, Rajagopal J. Ciliated cells of pseudostratified airway epithelium do not become mucous cells after ovalbumin challenge. Am J Respir Cell Mol Biol . 2013;48:364–373. doi: 10.1165/rcmb.2012-0146OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reader JR, Tepper JS, Schelegle ES, Aldrich MC, Putney LF, Pfeiffer JW, et al. Pathogenesis of mucous cell metaplasia in a murine asthma model. Am J Pathol . 2003;162:2069–2078. doi: 10.1016/S0002-9440(10)64338-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Plopper CG, Hill LH, Mariassy AT. Ultrastructure of the nonciliated bronchiolar epithelial (Clara) cell of mammalian lung. III. A study of man with comparison of 15 mammalian species. Exp Lung Res . 1980;1:171–180. doi: 10.3109/01902148009069646. [DOI] [PubMed] [Google Scholar]
- 43. Rock JR, Gao X, Xue Y, Randell SH, Kong YY, Hogan BL. Notch-dependent differentiation of adult airway basal stem cells. Cell Stem Cell . 2011;8:639–648. doi: 10.1016/j.stem.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carraro G, Langerman J, Sabri S, Lorenzana Z, Purkayastha A, Zhang G, et al. Transcriptional analysis of cystic fibrosis airways at single-cell resolution reveals altered epithelial cell states and composition. Nat Med . 2021;27:806–814. doi: 10.1038/s41591-021-01332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Judge EP, Hughes JM, Egan JJ, Maguire M, Molloy EL, O’Dea S. Anatomy and bronchoscopy of the porcine lung. A model for translational respiratory medicine. Am J Respir Cell Mol Biol . 2014;51:334–343. doi: 10.1165/rcmb.2013-0453TR. [DOI] [PubMed] [Google Scholar]
- 46. Pabst R. The pig as a model for immunology research. Cell Tissue Res . 2020;380:287–304. doi: 10.1007/s00441-020-03206-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Meyerholz DK, Stoltz DA, Namati E, Ramachandran S, Pezzulo AA, Smith AR, et al. Loss of cystic fibrosis transmembrane conductance regulator function produces abnormalities in tracheal development in neonatal pigs and young children. Am J Respir Crit Care Med . 2010;182:1251–1261. doi: 10.1164/rccm.201004-0643OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pezzulo AA, Tang XX, Hoegger MJ, Abou Alaiwa MH, Ramachandran S, Moninger TO, et al. Reduced airway surface pH impairs bacterial killing in the porcine cystic fibrosis lung. Nature . 2012;487:109–113. doi: 10.1038/nature11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rogers CS, Stoltz DA, Meyerholz DK, Ostedgaard LS, Rokhlina T, Taft PJ, et al. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science . 2008;321:1837–1841. doi: 10.1126/science.1163600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stoltz DA, Meyerholz DK, Pezzulo AA, Ramachandran S, Rogan MP, Davis GJ, et al. Cystic fibrosis pigs develop lung disease and exhibit defective bacterial eradication at birth. Sci Transl Med . 2010;2:29ra31. doi: 10.1126/scitranslmed.3000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li X, Tang XX, Vargas Buonfiglio LG, Comellas AP, Thornell IM, Ramachandran S, et al. Electrolyte transport properties in distal small airways from cystic fibrosis pigs with implications for host defense. Am J Physiol Lung Cell Mol Physiol . 2016;310:L670–L679. doi: 10.1152/ajplung.00422.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Montoro DT, Haber AL, Rood JE, Regev A, Rajagopal J. A synthesis concerning conservation and divergence of cell types across epithelia. Cold Spring Harb Perspect Biol . 2020;12:a035733. doi: 10.1101/cshperspect.a035733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schupp JC, Khanal S, Gomez JL, Sauler M, Adams TS, Chupp GL, et al. Single-cell transcriptional archetypes of airway inflammation in cystic fibrosis. Am J Respir Crit Care Med . 2020;202:1419–1429. doi: 10.1164/rccm.202004-0991OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ratcliff J TA, Thurman AL, Chimenti M, Pezzulo AA.aggregateBioVar: differential gene expression analysis for multi-subject scrna-seqBioconductor; 2020https://github.com/jasonratcliff/aggregateBioVar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thurman AL, Ratcliff JA, Chimenti MS, Pezzulo AA. Differential gene expression analysis for multi-subject single cell RNA sequencing studies with aggregateBioVar. Bioinformatics . 2021;37:3243–3251. doi: 10.1093/bioinformatics/btab337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zabner J, Scheetz TE, Almabrazi HG, Casavant TL, Huang J, Keshavjee S, et al. CFTR DeltaF508 mutation has minimal effect on the gene expression profile of differentiated human airway epithelia. Am J Physiol Lung Cell Mol Physiol . 2005;289:L545–L553. doi: 10.1152/ajplung.00065.2005. [DOI] [PubMed] [Google Scholar]
- 57. Bartlett JA, Ramachandran S, Wohlford-Lenane CL, Barker CK, Pezzulo AA, Zabner J, et al. Newborn cystic fibrosis pigs have a blunted early response to an inflammatory stimulus. Am J Respir Crit Care Med . 2016;194:845–854. doi: 10.1164/rccm.201510-2112OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rehman T, Thornell IM, Pezzulo AA, Thurman AL, Romano Ibarra GS, Karp PH, et al. TNFα and IL-17 alkalinize airway surface liquid through CFTR and pendrin. Am J Physiol Cell Physiol . 2020;319:C331–C344. doi: 10.1152/ajpcell.00112.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang G, Lou HH, Salit J, Leopold PL, Driscoll S, Schymeinsky J, et al. Characterization of an immortalized human small airway basal stem/progenitor cell line with airway region-specific differentiation capacity. Respir Res . 2019;20:196. doi: 10.1186/s12931-019-1140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Morrisey EE, Cardoso WV, Lane RH, Rabinovitch M, Abman SH, Ai X, et al. Molecular determinants of lung development. Ann Am Thorac Soc . 2013;10:S12–S16. doi: 10.1513/AnnalsATS.201207-036OT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rao W, Wang S, Duleba M, Niroula S, Goller K, Xie J, et al. Regenerative metaplastic clones in copd lung drive inflammation and fibrosis. Cell . 2020;181:848–864.e18. doi: 10.1016/j.cell.2020.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schroeder BW, Verhaeghe C, Park SW, Nguyenvu LT, Huang X, Zhen G, et al. AGR2 is induced in asthma and promotes allergen-induced mucin overproduction. Am J Respir Cell Mol Biol . 2012;47:178–185. doi: 10.1165/rcmb.2011-0421OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sala-Rabanal M, Yurtsever Z, Nichols CG, Brett TJ. Secreted CLCA1 modulates TMEM16A to activate Ca(2+)-dependent chloride currents in human cells. eLife . 2015;4:e05875. doi: 10.7554/eLife.05875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Widdicombe JH.Airway epitheliumGranger DN, Granger JP, editors. Colloquium series on integrated systems physiology: from molecule to function to disease. San Rafael, CA: Morgan & Claypool; 2013 [Google Scholar]
- 65. Whitsett JA, Wert SE, Weaver TE. Alveolar surfactant homeostasis and the pathogenesis of pulmonary disease. Annu Rev Med . 2010;61:105–119. doi: 10.1146/annurev.med.60.041807.123500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Garland AL, Walton WG, Coakley RD, Tan CD, Gilmore RC, Hobbs CA, et al. Molecular basis for pH-dependent mucosal dehydration in cystic fibrosis airways. Proc Natl Acad Sci USA . 2013;110:15973–15978. doi: 10.1073/pnas.1311999110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Garcia-Caballero A, Rasmussen JE, Gaillard E, Watson MJ, Olsen JC, Donaldson SH, et al. SPLUNC1 regulates airway surface liquid volume by protecting ENaC from proteolytic cleavage. Proc Natl Acad Sci USA . 2009;106:11412–11417. doi: 10.1073/pnas.0903609106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ahmad S, Tyrrell J, Walton WG, Tripathy A, Redinbo MR, Tarran R. Short palate, lung, and nasal epithelial clone 1 has antimicrobial and antibiofilm activities against the burkholderia cepacia complex. Antimicrob Agents Chemother . 2016;60:6003–6012. doi: 10.1128/AAC.00975-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wu T, Huang J, Moore PJ, Little MS, Walton WG, Fellner RC, et al. Identification of BPIFA1/SPLUNC1 as an epithelium-derived smooth muscle relaxing factor. Nat Commun . 2017;8:14118. doi: 10.1038/ncomms14118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. McDonald FJ, Price MP, Snyder PM, Welsh MJ. Cloning and expression of the beta- and gamma-subunits of the human epithelial sodium channel. Am J Physiol . 1995;268:C1157–C1163. doi: 10.1152/ajpcell.1995.268.5.C1157. [DOI] [PubMed] [Google Scholar]
- 71. Smith JJ, Karp PH, Welsh MJ. Defective fluid transport by cystic fibrosis airway epithelia. J Clin Invest . 1994;93:1307–1311. doi: 10.1172/JCI117087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Flynn AN, Itani OA, Moninger TO, Welsh MJ. Acute regulation of tight junction ion selectivity in human airway epithelia. Proc Natl Acad Sci USA . 2009;106:3591–3596. doi: 10.1073/pnas.0813393106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gan H, Wang G, Hao Q, Wang QJ, Tang H. Protein kinase D promotes airway epithelial barrier dysfunction and permeability through down-regulation of claudin-1. J Biol Chem . 2013;288:37343–37354. doi: 10.1074/jbc.M113.511527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Daniely Y, Liao G, Dixon D, Linnoila RI, Lori A, Randell SH, et al. Critical role of p63 in the development of a normal esophageal and tracheobronchial epithelium. Am J Physiol Cell Physiol . 2004;287:C171–C181. doi: 10.1152/ajpcell.00226.2003. [DOI] [PubMed] [Google Scholar]
- 75. Hawkins FJ, Suzuki S, Beermann ML, Barillà C, Wang R, Villacorta-Martin C, et al. Derivation of airway basal stem cells from human pluripotent stem cells. Cell Stem Cell . 2021;28:79–95.e8. doi: 10.1016/j.stem.2020.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zuo W, Zhang T, Wu DZ, Guan SP, Liew AA, Yamamoto Y, et al. p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature . 2015;517:616–620. doi: 10.1038/nature13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ballard ST, Schepens SM, Falcone JC, Meininger GA, Taylor AE. Regional bioelectric properties of porcine airway epithelium. J Appl Physiol (1985) . 1992;73:2021–2027. doi: 10.1152/jappl.1992.73.5.2021. [DOI] [PubMed] [Google Scholar]
- 78. Chen JH, Stoltz DA, Karp PH, Ernst SE, Pezzulo AA, Moninger TO, et al. Loss of anion transport without increased sodium absorption characterizes newborn porcine cystic fibrosis airway epithelia. Cell . 2010;143:911–923. doi: 10.1016/j.cell.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Rollins BM, Garcia-Caballero A, Stutts MJ, Tarran R. SPLUNC1 expression reduces surface levels of the epithelial sodium channel (ENaC) in Xenopus laevis oocytes. Channels (Austin) . 2010;4:255–259. doi: 10.4161/chan.4.4.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kreda SM, Gynn MC, Fenstermacher DA, Boucher RC, Gabriel SE. Expression and localization of epithelial aquaporins in the adult human lung. Am J Respir Cell Mol Biol . 2001;24:224–234. doi: 10.1165/ajrcmb.24.3.4367. [DOI] [PubMed] [Google Scholar]
- 81. Krug SM, Günzel D, Conrad MP, Lee IF, Amasheh S, Fromm M, et al. Charge-selective claudin channels. Ann N Y Acad Sci . 2012;1257:20–28. doi: 10.1111/j.1749-6632.2012.06555.x. [DOI] [PubMed] [Google Scholar]
- 82. Van Itallie CM, Anderson JM. The role of claudins in determining paracellular charge selectivity. Proc Am Thorac Soc . 2004;1:38–41. doi: 10.1513/pats.2306013. [DOI] [PubMed] [Google Scholar]
- 83. Sweerus K, Lachowicz-Scroggins M, Gordon E, LaFemina M, Huang X, Parikh M, et al. Claudin-18 deficiency is associated with airway epithelial barrier dysfunction and asthma. J Allergy Clin Immunol . 2017;139:72–81.e1. doi: 10.1016/j.jaci.2016.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Thornell IM, Rehman T, Pezzulo AA, Welsh MJ. Paracellular bicarbonate flux across human cystic fibrosis airway epithelia tempers changes in airway surface liquid pH. J Physiol . 2020;598:4307–4320. doi: 10.1113/JP280120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins. J Cell Biol . 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Korhonen LK, Holopainen E, Paavolainen M. Some histochemical characteristics of tracheobronchial tree and pulmonary neoplasms. Acta Histochem . 1969;32:57–73. [PubMed] [Google Scholar]
- 87. Mertens TCJ, van der Does AM, Kistemaker LE, Ninaber DK, Taube C, Hiemstra PS. Cigarette smoke differentially affects IL-13-induced gene expression in human airway epithelial cells. Physiol Rep . 2017;5:e13347. doi: 10.14814/phy2.13347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Pezzulo AA, Starner TD, Scheetz TE, Traver GL, Tilley AE, Harvey BG, et al. The air-liquid interface and use of primary cell cultures are important to recapitulate the transcriptional profile of in vivo airway epithelia. Am J Physiol Lung Cell Mol Physiol . 2011;300:L25–L31. doi: 10.1152/ajplung.00256.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lindgren D, Eriksson P, Krawczyk K, Nilsson H, Hansson J, Veerla S, et al. Cell-type-specific gene programs of the normal human nephron define kidney cancer subtypes. Cell Rep . 2017;20:1476–1489. doi: 10.1016/j.celrep.2017.07.043. [DOI] [PubMed] [Google Scholar]
- 90. Wang Y, Song W, Wang J, Wang T, Xiong X, Qi Z, et al. Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J Exp Med . 2020;217:e20191130. doi: 10.1084/jem.20191130. [DOI] [PMC free article] [PubMed] [Google Scholar]



