Abstract
Pharmaceutical products can activate immune cells, suppress their function, or change the immune responses to traditional immunologically active agonists such as those present in microbes. Therefore, the assessment of immunostimulation, immunosuppression, and immunomodulation comprises the backbone of immunotoxicity studies of new drug entities. Depending on physicochemical properties (e.g., size, charge, surface functionalities, hydrophobicity), nanoparticles can be immunostimulatory, immunosuppressive, and immunomodulatory. Various methods and experimental frameworks have been established to support preclinical translational studies of nanotechnology-based drug products. Immunophenotyping after the exposure of cells or preclinical animal models to nanoparticles can provide critical information about the changes in both the numbers of immune cells and their activation status. However, this methodology is underutilized in preclinical studies of engineered nanomaterials. Herein, we review current literature about varieties of instrumentation and methods utilized for immunophenotyping, discuss their advantages and limitations, and propose a roadmap for applying immunophenotyping to support preclinical immunological characterization of nanotechnology-based formulations.
1. Introduction
The use of nanotechnologies in the medical setting, i.e., nanomedicine, is increasingly utilized to prevent, diagnose, and treat various diseases such as cancer [1, 2]. The development of various nanomaterials (e.g., inorganics, polymers, or lipids) has helped to overcome some of the limitations of traditional drug delivery in these settings, including spatial and temporal delivery [3–5]. For example, delivery of drug treatments to the brain have long been hindered by the blood brain barrier [6, 7]. However, nanomaterials, such as polymers, can be personalized and utilized for diagnostic and therapeutic purposes in brain tumors [7]. Polymers can be conjugated to checkpoint inhibitors (anti-PD1 or anti-CTLA4) and enable the drugs to cross the blood brain barrier thus overcoming many treatment limitations in these cancers [6]. Nanoparticles can also aid in the delivery of nucleic acids in the context of both cancer vaccines and immunotherapies [8]. Moreover, nanomaterials have been utilized to directly activate different components of the innate or adaptive immune response to help stimulate antitumor immunity [8, 9]. Furthermore, drug free macromolecular therapeutics, which use biorecognition events (e.g., CD20 receptor targeting) and crosslinking to initiate therapeutic efficacy, have the potential to mitigate drug resistance (e.g., rituximab resistance) in cancer models [10]. Thus, the use of nanomaterials to treat disease is multifaceted.
However, the vast expanse of nanomaterial leads to an increasing need for techniques to characterize the efficacy and safety of such materials [2, 3]. One type of necessary nanomaterial evaluation is immunological evaluation [11, 12]. Immunological evaluation of nanomaterials is essential because some nanomaterials are designed to modify the immune system while others cause adverse effects to the immune system (immunotoxicity) [13]. Systemic administration of nanomaterial has the potential to lead to a variety of acute immune-mediated adverse effects such as hemolysis, complement activation, thrombogenicity, procoagulant activity, disseminated intravascular coagulation, inflammation, immunosuppression [12, 14]. These undesirable effects could be due to either nanotechnology platform or active pharmaceutical ingredient (API); in some cases, nanocarrier and API’s toxicities may overlap and contribute to more exaggerated adverse effects of the final product [15]. Moreover, innate immunity modulating impurities (e.g., endotoxins, beta-glucans, flagellin, CpG DNA, to name a few) may be introduced into nanoformulations during manufacturing and contribute to the overall immunotoxicity by priming the immune cells and changing their response to nanocarriers and/or APIs [16]. Therefore, the development of techniques that can assess changes to immune cell makeup and activation, such as immunophenotyping, are desirable (Figure 1).
Figure 1. Applications of flow cytometry in nanomedicine studies.
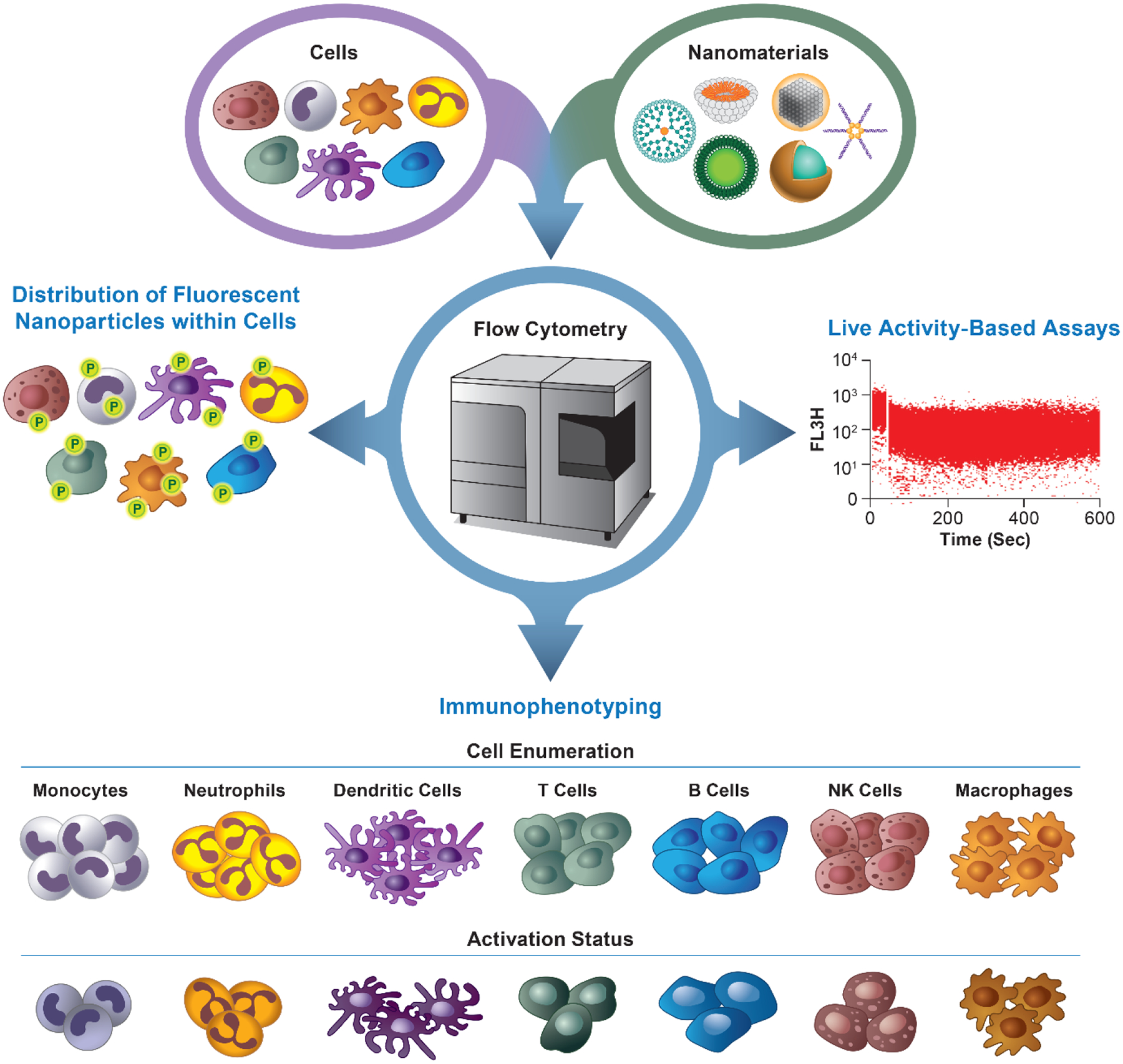
Flow cytometry can be used in a variety of applications to further nanomedicine. PBMCs that are incubated with various nanomaterials can analyzed using a flow cytometer to determine 1) the distribution of fluorescent nanomaterial within the cell population, 2) the presence of immune cell subsets based on antigen expression (immunophenotyping) along with the cellular activity, and 3) live activity of cells such as calcium (Ca2+) fluxes in cells. With the use of non-traditional cytometers, these applications can be expanded even further. NK = natural killer; P = particle
Immunophenotyping, or the identification of immune cell subsets based on antigen expression, has become an integral technique used in both the basic and clinical laboratory settings for many purposes, including the examination of cellular makeup in many diseases as well as the determination of effects of treatment, such as nanomaterials, on cell makeup and activity [17–21]. In order to perform immunophenotyping, a variety of different analytical techniques can be utilized, such as multicolor flow cytometry, hyperspectral flow cytometry, and mass cytometry—with multicolor flow cytometry currently being the most commonly used technique [19, 20]. However, given the vast array of techniques available for immunophenotyping, the consistent advancement and evolution of the cell type definitions, and the quasi-quantitative nature of flow cytometry, the universal utility of immunophenotyping is limited [18]. Nevertheless, there are constant efforts to standardize immunophenotyping panel design, sample handling, instrument setup, and data analysis [18, 22–25]. For example, the National Institute of Standards and Technology developed a Flow Cytometry Standards Consortium, supported by many corporations and groups, to develop reference standards and protocols to help establish quantitative flow cytometry measures. Moreover, while there are no universal formal guidelines, new standardized panels and techniques are regularly published by prominent research groups. In this review, we will discuss the current models and methods used for immunophenotyping as well as discuss the regulatory landscape for immunophenotyping and how to leverage this knowledge to characterize the effects of nanotechnology-based drug delivery platforms and nanoparticle-drug formulations on immune cells.
2. Models
As new research allows for a deeper and more detailed definition of each immune cell type, it is important to understand the current models and common cell types used in the development of immunophenotyping procedures. The most common source of immune cell populations studied ex vivo is spleen or bone marrow in rodent models and peripheral blood in humans. The major immune cell populations are divided into myeloid-derived and lymphoid-derived cells [26]. The myeloid-derived cells to be discussed here consist of the monocytes, macrophages, dendritic cells (DCs), and neutrophils which generally function as phagocytic and antigen-presenting cells [26, 27]. Furthermore, the monocytes can be subdivided into classical and non-classical monocytes, and the DCs can be subdivided into myeloid DCs (mDCs) and plasmacytoid DCs (pDCs) [25, 28, 29]. Moreover, the lymphoid population consists of T cells (cytotoxic, helper, and regulatory T cells), B cells (Naïve, memory, and transitional), natural killer (NK) cells, and NK T cells [25, 26]. In general, these broad cell types are present in both humans and mouse models. However, depending on the model identified, the definition of different cellular subtypes varies along with their presence in circulation [28, 30].
Each cellular subtype is defined by a set of extracellular and/or intracellular markers, which can be measured by cytometric methods. These cellular subsets have slight differences in markers used to define each subtype depending upon the model and literature referenced. However, for the most part, there is a consensus on the primary markers needed to define each major cell type—many of which are available in different company catalogs and/or posters [31–33]. These important markers to define major cell types are summarized in Table 1 for both human and mouse models.
Table 1. Immune cell subsets and phenotypes commonly used in research and identified by flow cytometry.
Immune cell subtypes and their phenotypes are summarized based on the currently available literature. Differences between mouse and human cells exist as determined by expression of surface markers specific to the given subset of immune cells.
| Model | Cell type | Subtype | Phenotype | References |
|---|---|---|---|---|
| Mouse | T cells | Cytotoxic T cells | CD3+, CD8+ | [31, 92] |
| Helper T cells | CD3+, CD4+ | [31, 92] | ||
| Regulatory T cells | CD3+, CD4+, CD25+, FoxP3+ | [92] | ||
| B cells | CD19+, B220+ | [31, 32, 92] | ||
| NK cells | CD3+, NKp46+ | [31, 32, 92] | ||
| Macrophages | CD11b+, F4/80+, CD68+ | [92] | ||
| DCs | mDCs | CD11b+, CD11c+, CD80+, CD86+ | [92] | |
| pDCs | CD11b−, CD11c+, B220+, Siglec- H+ | [33, 92] | ||
| Monocytes | CD11b+, CD68+, Ly6c+ | [32, 33, 92] | ||
| Neutrophils | CD11b+, Gr-1+, Ly6b+, F4/80− | [92] | ||
| Human | T cells | Cytotoxic T cells | CD3+, CD8+ | [25, 31] |
| Helper T cells | CD3+, CD4+ | [25, 31] | ||
| Regulatory T cells | CD3+, CD4+, CCR4+, CD25+, CD127low | [25] | ||
| Naïve T cells | CCR7+, CD45RA+ | [25, 93] | ||
| Effector T cells | CCR7−, CD45RA+ | [25] | ||
| Effector Memory T cells | CCR7−, CD45RA− | [25, 93] | ||
| Central Memory T cells | CCR7+, CD45RA− | [25, 93] | ||
| B cells | Naïve B cells | CD3−, CD19+, CD27− | [25, 33] | |
| Memory B cells | CD3−, CD19+, CD27+ | [25, 33] | ||
| NK cells | CD3−, CD19−, CD14−, CD20−, CD56+, CD16+/− | [25, 31, 32] | ||
| NK T cells | CD3+, CD56+ | [25] | ||
| DCs | mDCs | CD3−, CD19−, CD14−, CD20−, HLA-DR+, CD11c+ | [25, 31–33] | |
| pDCs | CD3−, CD19−, CD14−, CD20−, HLA-DR+, CD123+ | [25, 31, 33] | ||
| Monocytes | Classical | CD3−, CD19−, CD14+, CD16− | [25] | |
| Non-classical | CD3−, CD19−, CD14+, CD16+ | [25] | ||
| Neutrophils | CD15+, CD16+, CD14low | [25] |
Additionally, another aspect of defining the cellular subtype is determining the cell’s activation status. There are a variety of different activation markers that could be used to define the activation status of a cell. There are early and late activation markers, markers indicating adhesion, proliferation, degranulation, and co-stimulation and/or presentation markers [31, 33]. Table 2 lists common cellular markers found in lymphocytes and their status as phenotypic markers (P) and/or activation markers (A).
Table 2. Common surface markers present on lymphoid and myeloid cells of mouse and human origin.
Markers used in flow cytometry are summarized. P – phenotype marker; A – activation marker. The table is based on references [22, 25, 31–33, 92, 93].
| Mouse | Human | ||||||
|---|---|---|---|---|---|---|---|
| Lymphoid | Myeloid | Lymphoid | Myeloid | ||||
| Marker | Role | Marker | Role | Marker | Role | Marker | Role |
| CD3 | P | CD11c | P | CD3 | P | CD14 | P |
| CD4 | P | CD123 | P | CD4 | P | CD11c | P |
| CD8 | P | Ly6c | P | CD8 | P | CD123 | P |
| FOXP3 | P | Ly6G | P | FOXP3 | P | CD15 | P |
| CD19 | P | XCR1 | P | CD19 | P | CD16 | P |
| CD20 | P | MHCII | P | CD20 | P | HLA-DR | A |
| CD16 | P | B220 | P | IgD | P | CCR7 | A |
| CD45 | P | (CD45R) | CD16 | P | CD11b | A | |
| B220 | P | CD68 | P | CD45 | P | ||
| (CD45R) | Siglec-H | P | CD56 | P | |||
| NKp46 | P | Gr-1 | P/A | CCR4 | P | ||
| CD25 | P/A | CD11b | A | CD127 | P/A | ||
| CD45RA | A | Ly-71 | A | CD25 | P/A | ||
| CD22 | A | (F4/80) | CCR7 | P/A | |||
| CD335 | A | PD-1 | A | ||||
| Ki-67 | A | CTLA4 | A | ||||
| ICOS | A | Ki-67 | A | ||||
| ICOS | A | ||||||
| CD45RA | A | ||||||
| HLA-DR | A | ||||||
3. Methods
Many cytometric methods have been developed over the past few decades, which allow researchers to simultaneously evaluate multiple cellular features on a single cell level, as is needed for immunophenotyping. Each method established uses specific instrumentation developed by various manufacturers for specific purposes as described in the following sections. Table 3 includes examples of common types of cytometry equipment along with possible uses.
Table 3. Cytometry instrumentation and uses.
The various types of cytometry utilize different instrumentation and each instrument has the potential for different uses.
| Type of Cytometry | Instrumentation | Manufacturer | Notes |
|---|---|---|---|
| Multicolor flow cytometry | BD LSR Fortessa | BD BioSciences | Leukocyte profiles in tissues and tumors [38, 73]; Cell phenotypes and function [94] |
| BD FACS Melody™ Cell Sorter | BD BioSciences | Sorting of circulating tumor cells from peripheral blood [35] | |
| CytoFLEX Cytometer | Beckman Coulter | Detection of anti-PEG antibodies [71]; Distribution and trafficking of nanoparticles [72] | |
| NovoCyte Flow Cytometers | Agilent | Cell Apoptosis [95] | |
| MACSQuant® Analyzers | Miltenyi Biotec | Cell phenotypes and function [94] | |
| Hyperspectral flow cytometry | Aurora Spectral Cytometer | Cytek Biosciences | High dimensional immunophenotyping [96] |
| Image Cytometry | Amnis® ImageStream Imaging Flow Cytometer | Luminex | Visualize the T cell/tumor cell interface [97] |
| Celigo Image Cytometer | Nexcelom Bioscience | Viability of tumor spheroids [98] | |
| Attune™ CytPix™ Flow Cytometer | Invitrogen | Immunophenotyping, cell apoptosis [99] | |
| Mass Cytometry | Helios™ mass cytometer | Fluidigm | Immunophenotyping [64, 100] |
3.1. Multicolor flow cytometry
Currently, multicolor flow cytometry or conventional flow cytometry is the most common platform used to perform immunophenotyping and uses typical flow cytometry equipment (Table 3). In this type of flow cytometry, cells are characterized using fluorescently labeled antibodies, and the expression of proteins or detection of dyes specific to different organelles is assessed on a single cell level [19, 21, 34]. A series of lasers excite the fluorophores and other optical cell parameters and generate fluorescent and visible light signals [19, 34]. The generated light signals are then directed by different filter sets, read by specific detectors, and converted into electronic signals [19]. These electronic signals are then analyzed by a computer for a readable output [19]. The combination of visible and fluorescent light signals detected on a single cell allows the cell to be characterized and placed into a distinct cell subpopulation using different gating strategies. Furthermore, this technique allows for cell sorting, which enables flow cytometry experiments to be followed by further studies (Table 3) [19, 21, 35]. However, in this technique, the available combination of lasers, filter sets, and detectors limits the possible fluorochrome combinations due to the fluorochromes’ properties and spectral overlap [19, 20]. Therefore, although this technique can measure multiple parameters simultaneously, there is a limit to the number of fluorescent parameters which can be measured within a single panel on a particular flow cytometry instrument. However, with the development of more efficient analysis techniques, it is possible to analyze around 30 parameters at the same time [19, 36, 37].
Nevertheless, creative panel design and innovative analysis techniques have allowed multicolor flow cytometry to be used beyond the traditional format. One example of the development of such a technique is called infinity flow cytometry [38]. Infinity flow cytometry uses standard flow cytometry instrumentation with an innovative computational analysis approach to implement massively parallel cytometry experiments and predict antibody expression across a multitude of cell samples [38]. In this method, samples are stained with an established panel of antibodies (backbone panel) in combination with a unique exploratory antibody and acquired using a standard flow cytometer [38]. The output data is then run through the infinity flow pipeline, which establishes relationships between the backbone and exploratory markers, predicts the missing data, and forms an augmented data matrix [38]. This infinity flow software is available at https://github.com/ebecht/infinityflow. While there are limitations to this technique, this method opens up the use of conventional flow cytometers so that more expansive panels can be designed without the limitation of instrumentation.
3.2. Hyperspectral Flow Cytometry
The development of hyperspectral cytometry overcomes many of the parameter limitations and compensation issues associated with traditional multicolor flow cytometry by using different equipment (Table 3) [19]. In contrast to traditional flow cytometry, which uses specific filter sets and detectors to read generated signals, hyperspectral flow cytometry detects the entire light spectrum for each fluorochrome [18]. However, because the entire spectrum of each fluorochrome is detected in the experimental setup, the subsequent data analysis is more sophisticated and involves reconstructing the individual fluorochrome signals based on their “fingerprints” [18–20]. Therefore, the bulk of the experiment lies in the computational analysis of signal separation but does avoid issues with spectral overlap [20, 39]. Consequently, hyperspectral cytometry has the potential of simultaneously measuring more than 40 parameters in a single cell [20].
3.3. Imaging Flow Cytometry
Imaging flow cytometry is a hybrid between traditional flow cytometry and microscopy, and therefore utilizes antibody panels tagged with fluorophores for labeling and acquisition of various cell populations at high throughput rates (traditional flow cytometry) while simultaneously acquiring fluorescent images of analyzed cells (microscopy) [19, 40–42]. It provides single-cell images using both fluorescent and brightfield capabilities which expands the data acquisition capabilities of the cytometer and results in a major advantage of the system—the ability to determine cellular distributions of the fluorescent markers allowing for the implementation of applications that require protein localization knowledge [19, 42]. However, because the system needs to collect in-focus images of each cell, these cytometers often have relatively low throughput compared to traditional flow cytometers [41]. Moreover, due to the differences in data collection, the gating strategy of imaging cytometers relies on morphologic features such as cell area, width, height, and aspect ratio rather than the forward and side scattering profiles, and these differences lead to a need for trained operators for data acquisition and analysis [40, 41]. Some examples of different image-based flow cytometers include Amnis, Celigo, and Attune CytPix (Table 3).
3.4. Mass Cytometry
Another cytometry method available for phenotyping is mass cytometry which is a combination of time-of-flight mass spectroscopy and flow cytometry. In this method, cells are labeled with heavy metal ion-tagged antibodies and subsequently passed through a mass cytometer in which the metal ions are then ionized, detected, and recorded to identify the different labeled parameters on a single cell [19, 43, 44].
Due to the inherent differences between mass cytometry and fluorescent-based cytometric systems, mass cytometry has some advantages and disadvantages when compared to other immunophenotyping techniques. One advantage of mass cytometry is the lack of spectral overlap, meaning that compensation is not necessary [19]. Additionally, the number of parameters that can be detected in a single experiment is around 50, and this number will increase as antibodies are conjugated to additional heavy metal ions [45]. Another advantage is the lack of interference when analyzed cells are exposed to and uptake materials possessing intrinsic fluorescent properties. However, one disadvantage of mass cytometry is that samples cannot be sorted for future use due to their ionization [44]. Another disadvantage is the low sensitivity of some heavy metal reporters, which makes it challenging to measure antigens expressed at low levels [20, 44]. Even though there are limitations, mass cytometry proves to be a promising technique for immunophenotyping (Table 3).
4. Regulatory landscape
4.1. Available standard practices and guidance documents for traditional fluorescence-based cytometry methods
Immunophenotyping by flow cytometry is recommended by the International Council on Harmonization (ICH) S8 for additional immunotoxicity studies; the recommendation is adopted by both the US Food and Drug Administration and European Medicine Agency [46, 47]. While there is no formal guidance regarding development and validation of flow cytometry-based methods, there are many ongoing efforts among pharmaceutical industry and regulatory agencies to standardize the methodology and streamline validation [48]. Historically, due to the broader availability of fluorescence-based, traditional cytometers, most standardization efforts focused on these instruments and relevant to them methodologies, and are detailed further below.
Flow cytometry is usually considered quasi-quantitative as accuracy is not directly measurable [18]. Therefore, there is a lack of standardization for cytometric techniques, and many errors can be introduced into a cytometry experiment through sample preparation, instrument setup, and data analysis. Furthermore, while there is formal guidance for industrial laboratory practices set forth through the Good Laboratory Practices (GLP), many laboratories that perform drug development with flow cytometry operate outside the regulated area [22]. Regardless, recommendations have been made for both the assay development and validation of flow cytometry instrumentation; some of which are detailed further below [22, 23, 49].
Early in 2011, the American Association of Pharmaceutical Scientists (AAPS) Flow Cytometry Action Program Committee released a series of publications about method validation [22, 23]. Next, the International Council for Standardization in Hematology and the International Clinical Cytometry Society established a working group to specify validation parameters for clinical cytometry-based assays [50]. This was followed by the FDA workshop and round table discussions that collectively resulted in recommendations and considerations for the validation of both instruments and methods, as well as for quality controls [51, 52]. In the US, Clinical Laboratory Improvement Amendments of 1988, and GLP Regulations are followed for conducting cytometry-based assays utilized for clinical specimen analysis and non-clinical studies, respectively [53–55]. Additional recommendations are also available from the AAPS, United States Pharmacopeia (USP), and International Society for Pharmaceutical Engineering (ISPE) [22, 56]. The installation qualification (IQ), operational qualification (OQ), and performance qualification (PQ), also known as IQ/OQ/PQ, are broadly recognized as the most important steps in the instrument validation and, among other tools, rely on vendor-specific calibration software and calibration beads for light scattering and optical alignment [48]. Method validation was also found by many to be critically important for result reproducibility within individual laboratories and inter-laboratory data comparison [23, 49]. In both research and clinical settings, there have been many attempts to guide the development and performance of cytometric protocols.
Mair and Tyznik outline a step-by-step process of designing and running a flow cytometry panel including 1) development of biological targets 2) instrument characterization and optimization 3) fluorochrome characterization 3) in silico panel design 4) antibody titration and 5) single stain controls and testing of full panel [18]. Furthermore, in Selliah et al. 2019, the authors give three distinct types of method validation, including 1) limited assay validation, 2) initial assay validation, and 3) technology transfer validation [49]. The limited assay validation gives recommendations for the minimal parameters needed for flow cytometry experiments in labs that do not follow regulations [49]. These recommendations and example experimental design steps help to create a standardized protocol for non-regulated labs [49]. The initial assay validation is a set of protocols that include those necessary for validation in regulated labs, while the technology transfer validation is for the validation of protocols that will be used across different labs. All these different validation protocols give labs guidance on what is necessary for each validation step.
While there is less available information about validating newer types of cytometry-based methods, some discussions and opinions have been released. For example, Colangelo J.L. shared Pfizer’s experience with applying mass cytometry methods for immunophenotyping and identified challenges, such as ion suppression and limited mass spectrometry calibration standards, unique to the validation of mass-cytometry methods [57]. Another recent study outlined critical quality parameters, including but not limited to sample fixation and storage, post-staining wash stability, and proposed a validation workflow as well as staining and acquisition optimization criteria for mass cytometry-based peripheral blood mononuclear cell (PBMC) profiling for clinical diagnosis of celiac disease using 33-biomarkers panel [58].
4.2. Standardization of Immunophenotyping panels
In addition to guides and standard practices for cytometry, many companies and research groups have begun developing and publishing standard immunophenotyping and panels. In an effort to standardize the immunophenotyping method, some companies have attempted to streamline the protocol. One standardization method is the development of instrumentation that allows for a centrifuge-less staining procedure, for example, the Curiox Droparray™-cell system [59]. With this system, the cells are not centrifuged but are rather washed with a laminar flow process, which allows for streamlining and standardizing the procedure and decreases cellular loss during processing [59].
Many more efforts have been put forth to develop standardized immunophenotyping panels. One effort of panel standardization has been the development of rigorously optimized multicolor immunophenotyping panels (OMIPs) published in Cytometry Part A Journal on behalf of the International Society for Advancement of Cytometry. In this Journal, there are currently seventy OMIPs of five or more colors for others to utilize [60]. The detailed information regarding the reagents used in these optimized panels allows other researchers to use the same panel or gives them a starting point for developing a new panel [60].
Additionally, the Human Immunology Project was started to try to standardize flow cytometry-based immunophenotyping in order to allow for panels to elucidate heterogeneity in human immune cells and detect changes in the immune system that are associated with disease [25]. This project aims to both create a repository for immunological data which could be mined for different biomarkers and propose standardized panels and procedures to detect T cells, B cells, macrophages, monocytes, DCs, and NK cells along with their activation status [25]. Furthermore, experts from the Human Immunology Project Consortium developed five standardized panels, each consisting of 8-colors, in order to identify the major immune cell subsets in the peripheral blood [61]. These panels identify different cell subsets and were developed in an attempt to standardize flow cytometry panels in clinical studies [61]. While these panels do not delve into the complexity of current immunophenotyping possibilities, these panels can be expanded as necessary [61]. Furthermore, it was determined that automated computational analysis methods provided by the Flow Cytometry: Critical Assessment of Population Identification Methods improved standardization of data analysis as compared to manual analysis [61, 62]. The standardization success of these panels led to the production of BD Lyoplate by BD Biosciences; lyophilized antibody cocktails can be purchased for immunophenotyping [61].
Moreover, many companies provide cytometric services and/or panels available for purchase using different immunophenotyping methods (Table 4). Many of the panels available for purchase are intended for clinical research and range from general panels to specific immune panels, as well as panels with activation markers. Beckman Coulter Life Sciences has a multitude of flow cytometry immunophenotyping panels (Duraclone panels) available that cover a broad spectrum of immune cells, including panels for rare cells (Table 4) [63]. ThermoFisher Scientific and Miltenyi Biotec also have immunophenotyping kits available for flow cytometry (Table 4). Additionally, there are CyTOF immunophenotyping panels available for purchase, including the Maxpar® Direct™ Immune Profiling Assay™ by Fluidigm (Table 4) [64]. This standardized immunophenotyping panel for CyTOF offers 30 parameters to define immune cell subtypes on the Helios™ Mass Cytometer [64]. In addition to panels available for purchase to perform immunophenotyping, there are also many companies that will design and run in-house panels which provide personalized cytometric services for flow cytometry immunophenotyping testing.
Table 4. Commercially available kits for immunophenotyping.
Panels available for purchase for different immunophenotyping techniques. Each panel includes the vendor, panel ID (or panel name), the catalog number, the markers for expression, and the fluorophores included in the panel based on the conjugation.
| Vendor | Panel ID | Catalog Number | Markers | Fluorophores |
|---|---|---|---|---|
| Beckman Coulter Life Sciences | DuraClone IM Phenotyping BASIC Tube, human | B53309 | CD45, CD3, CD4, CD8, CD19, CD14, CD16, CD56 | Krome Orange, APC-Alexa Fluor 750, APC, Alexa Fluor 700, ECD, PC7, FITC, PE |
| DuraClone IM T Cell Subsets Tube, human | B53328 | CD45RA, CD197 (CCR7), CD28, CD279 (PD-1), CD27, CD4, CD8, CD3, CD57, CD45 | FITC, PE, ECD, PC5.5, PC7, APC, Alexa Fluor 700, APC-Alexa Fluor 750, Pacific Blue, Krome Orange | |
| Duraclone IM Treg Tube, human | B53346 | CD45RA, CD25, CD39, CD4, FoxP3, CD3, Helios, CD45 | FITC, PE, PC5.5, PC7, Alexa Fluor 647, APC-Alexa Fluor 750, Pacific Blue, Krome Orange | |
| DuraClone IM TCRs Tube, human | B53340 | TCR γδ, TCR αβ, HLA-DR, TCR Vδ1, CD4, CD8, CD3, TCR Vδ2, CD45 | FITC, PE, ECD, PC7, APC, Alexa Fluor 700, APC-Alexa Fluor 750, Pacific Blue, Krome Orange | |
| DuraClone IM B cells Tube, human | B53318 | IgD, CD21, CD19, CD27, CD24, CD38, IgM, CD45 | FITC, PE, ECD, PC7, APC, APC-Alexa Fluor 750, Pacific Blue, Krome Orange | |
| DuraClone IM Dendritic Cell Tube, human | B53351 | CD16, lineage exclusion markers (CD3, CD14, CD19, CD20, CD56), CD1c, CD11c, Clec9A, CD123, HLA-DR, CD45 | FITC, PE, PC5.5, PC7, APC, APC-Alexa Fluor 700, Pacific Blue, Krome Orange | |
| DuraClone IM Granulocytes Tube, human | B88651 | CD294, CD16, CD33, CD11b, CD274, lineage exclusion markers (CD3, CD19, CD56, CD14), CD62L, CD15, CD45 | FITC, ECD, PC5.5, PC7, APC, APC-Alexa Fluor 700, APC-Alexa Fluor 750, Pacific Blue, Krome Orange | |
| DURAClone IM Innate Lymphoid Cell Tube, human | C96081 | CD294, CD1a, CD3, CD14, CD16, CD19, CD34, CD94, CD123, TCR αβ, TCR γδ, FcεR1a, CD117, CD335, CD127, CD161, CD45 | FITC, PE, PC5.5, PC7, APC, APC-Alexa Fluor 750, Krome Orange | |
| DuraClone IF T Activation Tube, human | B88649 | IFN γ, TNF α, IL-2, CD8, CD3, CD4 | FITC, PE, PC7, Alexa Fluor 700, Alexa Fluor 750, Pacific Blue | |
| DuraClone IF T Helper Cell Tube, human | C04666 | IFN γ, IL-4, CD4, CD3, IL-17A | FITC, PC7, APC, Alexa Fluor 750, Pacific Blue | |
| DURAClone IF Monocyte Activation Tube, human | C21858 | HLA-DR, TNF α, CD14, CD45 | PE, APC-Alexa Fluor 700, Pacific Blue, Krome Orange | |
| DURAClone IF Basophil Activation Tube, human | C23406 | CD203c, CD3, CD294, CD63, CD45 | PE, PC7, Alexa Fluor 647, Pacific Blue, Krome Orange | |
| DuraClone RE CLB Tube, human | B80393 | CD81, ROR1, CD79b, CD19, CD5, CD43, CD20, CD45 | FITC, PE, PC5.5, PC7, APC, APC-Alexa Fluor 750, Pacific Blue, Krome Orange | |
| DURAClone RE PC Tube, human | B80394 | CD81, CD27, CD19, CD200, CD138, CD56, CD38, CD45 | FITC, PE, PC5.5, PC7, APC, APC-Alexa Fluor 750, Pacific Blue, Krome Orange | |
| DURAClone RE ALB Tube, human | C00163 | CD58, CD34, CD10, CD19, CD38, CD20, CD45 | FITC, ECD, PC5.5, PC7, APC-Alexa Fluor 700, APC-Alexa Fluor 750, Krome Orange | |
| DURAClone SC Mesenchymal Tube, human | C34369 | CD90, CD73, CD34, CD146, CD105, CD45, CD31, CD14, CD19 | FITC, PE, ECD, PC5.5, PC7, APC-Alexa Fluor 750, Pacific Blue, Krome Orange | |
| DURAClone SC Hematopoietic Tube, human | C49589 | CD38, CD49f, CD34, CD10, CD133, CD45RA, CD90, CD45 | FITC, PE, ECD, PC7, APC, APC-Alexa Fluor 750, Pacific Blue, Krome Orange | |
| ThermoFisher Scientific | eBioscience™ Essential Human T-Cell Phenotyping Kit | A42923 | CD3, CD4, CD8, CD62L, CCR7 | APC-eFluor 780, APC, eFluor 450, FITC, PE |
| eBioscience™ Essential Human Treg Phenotyping Kit | A42925 | CD4, CD25, CD127, FoxP3 | FITC, PerCP-eFluor710, PE, eFluor450 | |
| eBioscience™ Essential Human Th1/Th17 Phenotyping Kit | A42927 | CD4, IFN γ, IL-17A | FITC, PE, APC | |
| Miltenyi Biotec | 8-Color Immunophenotyping Kit, human | 130-120-640 | CD3, CD4, CD8, CD14, CD19, CD45, CD16, CD56 | PE, VioBright 667, APC-Vio770, VioBlue, PE-Vio770, Viogreen, VioBright 515 |
| BioLegend | Biolegend’s LEGENDScreen™ Human PE Kit | 700007 | 361 human cell surface markers | PE |
| BD Biosciences | BD Lyoplate™: Human Cell Surface Marker Screening Panel A/B | 560747 | 242 human cell surface markers | Unconjugated |
| Fluidigm | Maxpar® Direct™ Immune Profiling Assay™ (CyTOF) | 201325 | CD3, CD4, CD8, CD11c, CD14, CD16, CD19, CD20, CD25, CD27, CD28, CD38, CD45, CD45RA, CD45RO, CD56, CD57, CD66b, CD123, CD127, CD161, CD294, CCR4, CCR6, CCR7, CXCR3, CXCR5, HLA-DR, IgD, TCRγδ | Not applicable |
5. Roadmap for characterization of nanotechnology-formulated products
5.1. How we can leverage current knowledge
The development of nanomaterials over the past few decades has helped to overcome the limitations of traditional drug delivery [3–5]. However, immunological techniques for characterizing the safety and efficacy of nanomaterial need further development [3, 11]. In particular, flow cytometry and immunophenotyping are underutilized techniques that could help to determine both the efficacy and safety of nanomaterials on the immune system as indicated by the International Council on Harmonization (ICH) S8 [47, 65, 66]. Immunophenotyping and flow cytometry could aid in understanding the expression of a nanomaterial target on different cell subsets or could be used to detect biomarkers of immunotoxicity [65]. While the effects of nanomaterials on immune cell populations are not always the primary endpoint of a given study, understanding the nanomaterial-driven changes in immune cell makeup and activity can help characterize nanotechnology-based formulations.
5.2. Nanomaterial effects on immune cell phenotypes
Nanomaterials can be designed to have different effects on the immune system. Indeed, some nanomaterials are specifically intended to modify the immune cell phenotypes and cell activation status, while others do not have that purpose and modify immune cells through an adverse effect [13]. One example of nanomaterial which is designed to modify immune cell function are nanoparticles that deliver small inhibiting RNA against a voltage-gated potassium (Kv1.3) channel specifically to memory (CD45RO+) T cells [67]. The treatment of immune cells with these nanoparticles changed both immune cell makeup and activation as detected by flow cytometry [67]. The treatment of T cells with the nanoparticles decreased the overall percentage of memory cells in the population as well as decreased the expression of CD40 ligand, tumor necrosis factor-α, and interferon-γ in the treated memory T cells as measured by flow cytometry [67]. Another example of nanomaterial designed to modify the immune is a series of nanoparticles designed to decrease different inflammatory cytokines of innate immune cells [68]. It was determined, through using flow cytometry, ELISA, and transcriptional activity cell array, that the physicochemical properties of the nanoparticles differentially affected the expression of molecular markers, cytokines, and transcription factor activity of bone marrow-derived macrophages [68]. Furthermore, a study by Simón-Vázquez et al. showed that human lymphocytes treated with various metal oxide nanoparticles activated mitogen-activated protein kinase and nuclear factor kappa-light-chain-enhancer of activated B cells signaling pathways and changed gene expression using real-time PCR [69]. Given the use of such nanomaterials to modulate the immune system, more techniques, such as immunophenotyping, to measure such modulation should be developed.
5.3. Flow cytometry use in biomedical nanotechnology
In addition to the analysis of nanoparticles’ effects on immune cells, flow cytometry was successfully used to detect markers of nanoparticle immunoreactivity. For example, Liptrott et al. applied flow cytometry for the analysis of nanoparticle-plasma protein interactions, aka protein corona, as well as physical characteristics of the nanoparticles [70]. Forward scatter and side scatter of the nanoparticle samples were used to determine polydispersity of the nanoparticles, changes in characteristics of the nanoparticles in varying matrices, and the formation of the protein coronas [70]. Moreover, Roffler’s team proposed using flow cytometry for the detection of anti-PEG IgG and IgM antibodies in human plasma response to the exposure to PEG and PEGylated nanomaterials [71]. In this experimental setup, beads were incubated with human plasma, stained with fluorescent anti-IgG or IgM secondary antibodies, and then analyzed via flow cytometry which determined the presence of anti-PEG antibodies based on the mean florescence intensity [71]. Furthermore, Garcia Romeu et al. used flow cytometry to detect nanoparticle distribution within cellular organelles [72]. Flow cytometry was also used to determine the effect of nanoparticle size on the kinetics and trafficking of nanoparticles to organelles such as lysosomes [72].
Not only is cytometry used in nanoparticle characterization, but it is also used to assess the effect of nanomaterials in the preclinical setting. He et al. used nanoscale coordination polymer nanoparticles to combine photodynamic therapy and chemotherapy to increase antitumor immunity both in vitro and in vivo in colorectal cancer models [73]. In vitro, flow cytometry was used to assess which nanoparticle/treatment components cause immunogenic cell death and apoptosis and necrosis [73]. Additionally, leukocyte profiles of tumors harvested from the treated murine tumor models were analyzed by flow cytometry [73]. In another study, flow cytometry was instrumental at detecting and analyzing CD4+ and CD8+ T-lymphocytes infiltrating tumors and correlating the cell number to the anti-tumor effect seen after the immunization with PDS0101 (ImmunoMAPK-RDOTAP/HPV-16 E6 and E7 Peptides) liposome-based vaccine developed by PDS Biotechnology [74]. Furthermore, Barth et al. applied flow cytometry to assess the in vivo efficacy of calcium phosphosilicate nanoparticles containing indocyanine green as a photosensitizer for photodynamic therapy in a leukemia model as well as to detect possible off target effects [75].
Over the past decade, there have been many efforts to identify and validate biomarkers for the routine clinical monitoring of hematological and solid tumors progression and evaluating the efficacy of applied anti-cancer therapies, especially in the immune-oncology field [76–81]. In addition, there is a growing interest in methods allowing to personalize new therapies. Such efforts require powerful research tools, and flow cytometry became one of the most successful analytical approaches for the single-cell analysis to both detect and monitor immune cell phenotypes in patients with hematological malignancies, solid tumors undergoing immunotherapies, metastatic and residual stages of cancer progression. The increasing need in simultaneous analysis of multiple cellular markers pushed the technology beyond conventional flow cytometry with mass cytometry and full spectrum flow cytometry being among the most popular[76–82].
Monitoring the efficacy and understanding the mechanism of action of nanoparticle-based vaccines are additional clinical characterization areas commonly relying on flow cytometry. For example, clinical assessment of efficacy and safety of lipid-nanoparticles formulated mRNA SARS-Co-V2 vaccines relied on the flow cytometry for monitoring Th1 vs. Th2 responses of CD4+ lymphocytes following immunization [83, 84]. Likewise, detection of tumor-infiltrating CD4+ and CD8+ lymphocytes and their characterization in terms of exhaustion and activation status are among the study endpoints in the ongoing Phase II clinical trial of liposome-based HPV vaccine utilizing Versamune (PDS0101) platform [85].
Another interesting, yet less traditional application of flow cytometry is the finding of quantum-dots-labeled bacteriophages in complex media for the rapid and sensitive detection of bacterial contamination [86]. This approach is of interest to bioterrorism surveillance, agriculture safety and medical diagnosis fields.
5.4. What challenges exist for nanomaterials and possible solutions
Many challenges exist in the characterization and implementation of nanomaterials for biomedical applications. Formulations of nanomaterials are often complex and can lead to unexpected biological interactions [87, 88]. In order to characterize any immunological interactions and/or effects of the nanomaterials on the immune system, immunophenotyping could be utilized. However, there are challenges when it comes to using immunophenotyping to determine the effect of the nanomaterials. One challenge of implementing immunophenotyping to characterize the effects of nanomaterials is that the most common immunophenotyping technique, multicolor flow cytometry, is limited in its ability to test cells exposed to nanomaterials possessing inherent fluorescent properties due to possible optical interferences of the said nanomaterials. Some nanomaterials are designed with optical and/or fluorescent components, while others have components that inherently interfere with certain visible or fluorescent wavelengths [89–91]. Metallic nanoparticles, metal oxides, and metal-containing formulations (e.g., superparamagnetic iron oxide nanoparticles and arsenic trioxide nanoparticles), as well as nanomaterials containing traces of metal catalysts utilized during synthesis or accidently introduced during manufacturing (e.g., iron catalyst in carbon nanotubes formulations, and tungsten nanoparticles in recombinant protein therapeutics), may interfere with mass cytometry-based immunophenotyping. Therefore, depending on the nanomaterial studied, the use of different immunophenotyping techniques and selection of appropriate antibody tags, such as fluorophores for multicolor flow cytometry and heavy metals for CyTOF, may need to be considered. Otherwise, established flow cytometry immunophenotyping panels would need to be tailored to compensate for the optical components of the nanomaterial.
Furthermore, the use of flow cytometry for direct nanoparticle characterization (without attachment to cells) is challenging due to the detection limit of typical flow cytometry instrumentation and the small size of the nanoparticles. In order to be able to use flow cytometry to characterize nanoscale particles many optimization steps are required as detailed in the study by Garcia Romeu et al. [72]. However, new cytometers such as the Flow NanoAnalyzer by NanoFCM have been designed for the purpose of detection and characterization of both synthetic nanoparticles and natural nanomaterials such as exosomes. The high resolution and detection of objects with a wide size range from 7 to 1000 nm by this new generation cytometer was made possible by combining light scattering and fluorescent detection (https://www.nanofcm.com/). The development of these types of cytometers could extend the methodology for nanoparticle physicochemical characterization.
6. Conclusion
Immunophenotyping is an underutilized technique with much potential to characterize the effect of nanomaterials on immune cell makeup and activity. There are many techniques that could be utilized to perform immunophenotyping, including, but not limited to, multicolor flow cytometry, hyperspectral flow cytometry, and mass cytometry [19]. However, the ever-evolving definitions of immune cell subsets make immunophenotyping challenging as the immunophenotyping panels may need to be updated as new definitions come about [18]. Additionally, most immunophenotyping techniques’ qualitative nature leads to the difficulty in standardizing experimental protocols. Nanoparticle characterization involves additional set of challenges related to nanoparticle intrinsic fluorescent properties or composition that create interference with fluorescence-based or mass spectrometry-based flow cytometry approaches, respectively. Nevertheless, there are many universal efforts to develop references and standards that can be used across different facilities including those characterizing nanomaterials [24, 25, 61]. Therefore, as the technology and techniques improve, immunophenotyping has an even greater potential to be a standard protocol to evaluate the efficacy and safety of nanomaterials.
Acknowledgments.
The study was funded in whole by federal funds from the National Cancer Institute, National Institutes of Health, under contract 75N91019D00024. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. We thank Allen Kane for the help with figure preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest. The authors declare no conflict of interest.
References
- [1].Soares S, Sousa J, Pais A, Vitorino C, Nanomedicine: principles, properties, and regulatory issues, Frontiers in chemistry, 6 (2018) 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Patra JK, Das G, Fraceto LF, Campos EVR, del Pilar Rodriguez-Torres M, Acosta-Torres LS, Diaz-Torres LA, Grillo R, Swamy MK, Sharma S, Nano based drug delivery systems: recent developments and future prospects, Journal of nanobiotechnology, 16 (2018) 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].McNeil SE, Characterization of nanoparticles intended for drug delivery, Springer; 2018. [Google Scholar]
- [4].Irvine DJ, Dane EL, Enhancing cancer immunotherapy with nanomedicine, Nat Rev Immunol, 20 (2020) 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Desai N, Challenges in development of nanoparticle-based therapeutics, AAPS J, 14 (2012) 282–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Galstyan A, Markman JL, Shatalova ES, Chiechi A, Korman AJ, Patil R, Klymyshyn D, Tourtellotte WG, Israel LL, Braubach O, Ljubimov VA, Mashouf LA, Ramesh A, Grodzinski ZB, Penichet ML, Black KL, Holler E, Sun T, Ding H, Ljubimov AV, Ljubimova JY, Blood-brain barrier permeable nano immunoconjugates induce local immune responses for glioma therapy, Nat Commun, 10 (2019) 3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ljubimov VA, Ramesh A, Davani S, Danielpour M, Breunig JJ, Black KL, Neurosurgery at the crossroads of immunology and nanotechnology. New reality in the COVID-19 pandemic, Adv Drug Deliv Rev, 181 (2021) 114033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Riley RS, June CH, Langer R, Mitchell MJ, Delivery technologies for cancer immunotherapy, Nat Rev Drug Discov, 18 (2019) 175–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li S, Luo M, Wang Z, Feng Q, Wilhelm J, Wang X, Li W, Wang J, Cholka A, Fu YX, Sumer BD, Yu H, Gao J, Prolonged activation of innate immune pathways by a polyvalent STING agonist, Nat Biomed Eng, 5 (2021) 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yang J, Li L, Kopeček J, Biorecognition: A key to drug-free macromolecular therapeutics, Biomaterials, 190–191 (2019) 11–23. [DOI] [PubMed] [Google Scholar]
- [11].Grossman JH, Crist RM, Clogston JD, Early Development Challenges for Drug Products Containing Nanomaterials, AAPS J, 19 (2017) 92–102. [DOI] [PubMed] [Google Scholar]
- [12].Dobrovolskaia MA, Germolec DR, Weaver JL, Evaluation of nanoparticle immunotoxicity, Nature nanotechnology, 4 (2009) 411–414. [DOI] [PubMed] [Google Scholar]
- [13].Dobrovolskaia MA, McNeil SE, Immunological properties of engineered nanomaterials, Nat Nanotechnol, 2 (2007) 469–478. [DOI] [PubMed] [Google Scholar]
- [14].Halamoda-Kenzaoui B, Vandebriel R, Howarth A, Siccardi M, David C, Liptrott N, Santin M, Borgos S, Bremer-Hoffman S, Caputo F, Methodological needs in the quality and safety characterisation of nanotechnology-based health products: Priorities for method development and standardisation, Journal of Controlled Release, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dobrovolskaia MA, McNeil SE, Strategy for selecting nanotechnology carriers to overcome immunological and hematological toxicities challenging clinical translation of nucleic acid-based therapeutics, Expert Opin Drug Deliv, 12 (2015) 1163–1175. [DOI] [PubMed] [Google Scholar]
- [16].Holley CK, Dobrovolskaia MA, Innate Immunity Modulating Impurities and the Immunotoxicity of Nanobiotechnology-Based Drug Products, Molecules, 26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Oughton JA, Kerkvliet NI, Immune cell phenotyping using flow cytometry, Current protocols in toxicology, 23 (2005) 18.18. 11–18.18. 24. [DOI] [PubMed] [Google Scholar]
- [18].Mair F, Tyznik AJ, High-Dimensional Immunophenotyping with Fluorescence-Based Cytometry: A Practical Guidebook, Methods Mol Biol, 2032 (2019) 1–29. [DOI] [PubMed] [Google Scholar]
- [19].McKinnon KM, Flow Cytometry: An Overview, Curr Protoc Immunol, 120 (2018) 5 1 1–5 1 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gondhalekar C, Rajwa B, Patsekin V, Ragheb K, Sturgis J, Robinson JP, Alternatives to current flow cytometry data analysis for clinical and research studies, Methods, 134–135 (2018) 113–129. [DOI] [PubMed] [Google Scholar]
- [21].Litwin V, Marder P, Flow cytometry in drug discovery and development, John Wiley & Sons; 2011. [Google Scholar]
- [22].Green CL, Brown L, Stewart JJ, Xu Y, Litwin V, Mc Closkey TW, Recommendations for the validation of flow cytometric testing during drug development: I instrumentation, J Immunol Methods, 363 (2011) 104–119. [DOI] [PubMed] [Google Scholar]
- [23].O’Hara DM, Xu Y, Liang Z, Reddy MP, Wu DY, Litwin V, Recommendations for the validation of flow cytometric testing during drug development: II assays, J Immunol Methods, 363 (2011) 120–134. [DOI] [PubMed] [Google Scholar]
- [24].Le Lann L, Jouve PE, Alarcon-Riquelme M, Jamin C, Pers JO, P.F.C.S. Group, P.C. Consortium, Standardization procedure for flow cytometry data harmonization in prospective multicenter studies, Sci Rep, 10 (2020) 11567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Maecker HT, McCoy JP, Nussenblatt R, Standardizing immunophenotyping for the Human Immunology Project, Nat Rev Immunol, 12 (2012) 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Owen JA, Punt J, Stranford SA, Kuby immunology, Freeman WH New York, NY, USA: 2013. [Google Scholar]
- [27].Kawamoto H, Minato N, Myeloid cells, The international journal of biochemistry & cell biology, 36 (2004) 1374–1379. [DOI] [PubMed] [Google Scholar]
- [28].Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, Leenen PJ, Liu Y-J, MacPherson G, Randolph GJ, Nomenclature of monocytes and dendritic cells in blood, Blood, The Journal of the American Society of Hematology, 116 (2010) e74–e80. [DOI] [PubMed] [Google Scholar]
- [29].Wu L, Liu Y-J, Development of dendritic-cell lineages, Immunity, 26 (2007) 741–750. [DOI] [PubMed] [Google Scholar]
- [30].Mestas J, Hughes CC, Of mice and not men: differences between mouse and human immunology, The Journal of Immunology, 172 (2004) 2731–2738. [DOI] [PubMed] [Google Scholar]
- [31].BD Biosciences, Human and Mouse CD Marker Handbook, https://www.bdbiosciences.com, 2011.
- [32].Abcam, Immune cell markers poster, www.abcam.com, 2021.
- [33].ThermoFisher Scientific, Immune cell guide: Human and mouse antigens, www.thermofisher.com, 2017.
- [34].Kasili PM, Vo-Dinh T, Hyperspectral imaging system using acousto-optic tunable filter for flow cytometry applications, Cytometry A, 69 (2006) 835–841. [DOI] [PubMed] [Google Scholar]
- [35].Yu L, Sa S, Wang L, Dulmage K, Bhagwat N, Yee SS, Sen M, Pletcher CH Jr, Moore JS, Saksena S, An integrated enrichment system to facilitate isolation and molecular characterization of single cancer cells from whole blood, Cytometry Part A, 93 (2018) 1226–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Payne K, Li W, Salomon R, Ma CS, OMIP-063: 28-Color Flow Cytometry Panel for Broad Human Immunophenotyping, Cytometry A, 97 (2020) 777–781. [DOI] [PubMed] [Google Scholar]
- [37].Liechti T, Roederer M, OMIP-058: 30-Parameter Flow Cytometry Panel to Characterize iNKT, NK, Unconventional and Conventional T Cells, Cytometry A, 95 (2019) 946–951. [DOI] [PubMed] [Google Scholar]
- [38].Becht E, Tolstrup D, Dutertre C-A, Morawski PA, Campbell DJ, Ginhoux F, Newell EW, Gottardo R, Headley MB, High-throughput single-cell quantification of hundreds of proteins using conventional flow cytometry and machine learning, Science Advances, 7 (2021) eabg0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fienberg HG, Nolan GP, High-Dimensional Single Cell Analysis, Springer; 2014. [Google Scholar]
- [40].Basiji DA, Ortyn WE, Liang L, Venkatachalam V, Morrissey P, Cellular image analysis and imaging by flow cytometry, Clinics in laboratory medicine, 27 (2007) 653–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Stavrakis S, Holzner G, Choo J, DeMello A, High-throughput microfluidic imaging flow cytometry, Current opinion in biotechnology, 55 (2019) 36–43. [DOI] [PubMed] [Google Scholar]
- [42].Zuba-Surma EK, Ratajczak MZ, Analytical capabilities of the ImageStream cytometer, Methods in cell biology, 102 (2011) 207–230. [DOI] [PubMed] [Google Scholar]
- [43].Olsen LR, Leipold MD, Pedersen CB, Maecker HT, The anatomy of single cell mass cytometry data, Cytometry A, 95 (2019) 156–172. [DOI] [PubMed] [Google Scholar]
- [44].Spitzer MH, Nolan GP, Mass Cytometry: Single Cells, Many Features, Cell, 165 (2016) 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cosma A, Nolan G, Gaudilliere B, Mass cytometry: The time to settle down, Cytometry A, 91 (2017) 12–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].European Medicines Agency, ICH Topic S 8 Immunotoxicity Studies for Human Pharmaceuticals, https://www.ema.europa.eu/, May 2006.
- [47].U.S. Food and Drug Administration, Guidance for Industry S8 Immunotoxicity Studies for Human Pharmaceuticals, https://www.fda.gov/, April 2006.
- [48].Du L, Grover A, Ramanan S, Litwin V, The evolution of guidelines for the validation of flow cytometric methods, International Journal of Laboratory Hematology, 37 (2015) 3–10. [DOI] [PubMed] [Google Scholar]
- [49].Selliah N, Eck S, Green C, Oldaker T, Stewart J, Vitaliti A, Litwin V, Flow Cytometry Method Validation Protocols, Curr Protoc Cytom, 87 (2019) e53. [DOI] [PubMed] [Google Scholar]
- [50].Wood B, Jevremovic D, Bene MC, Yan M, Jacobs P, Litwin V, I.I.W. Group, Validation of cell-based fluorescence assays: practice guidelines from the ICSH and ICCS - part V - assay performance criteria, Cytometry B Clin Cytom, 84 (2013) 315–323. [DOI] [PubMed] [Google Scholar]
- [51].Landgren O, Gormley N, Turley D, Owen RG, Rawstron A, Paiva B, Barnett D, Arroz M, Wallace P, Durie B, Yuan C, Dogan A, Stetler-Stevenson M, Marti GE, Flow cytometry detection of minimal residual disease in multiple myeloma: Lessons learned at FDA-NCI roundtable symposium, Am J Hematol, 89 (2014) 1159–1160. [DOI] [PubMed] [Google Scholar]
- [52].Food and Drug Administration, Notice: Clinical Flow Cytometry in Hematologic Malignancies; Public Workshop; Request for Comments, National Archives, https://www.federalregister.gov/, 2013.
- [53].Code of Federal Regulations Title 42 Chapter IV Subchapter G Part 493, National Archives, https://www.ecfr.gov/, 2022.
- [54].Centers for Medicare and Medicaid Services, Clinical Laboratory Improvement Amendments (CLIA), https://www.cms.gov/, 2021.
- [55].Code of Federal Regulations Title 21 Chapter I Subchapter A Part 58, National Archives, https://www.ecfr.gov/, 2022.
- [56].Bansal SK, Layloff T, Bush ED, Hamilton M, Hankinson EA, Landy JS, Lowes S, Nasr MM, Jean PAS, Shah VP, Qualification of analytical instruments for use in the pharmaceutical industry: a scientific approach, Aaps Pharmscitech, 5 (2004) 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Colangelo JL, The Validation of Quantitative Mass Spectrometry Assays for Clinical Chemistry Assessments in Animal Models, Toxicol Pathol, 45 (2017) 977–982. [DOI] [PubMed] [Google Scholar]
- [58].Estevam J, Krutzik P, Vander Tuig J, McAuliffe WJ, Smithson G, Development and validation of a high-parameter mass cytometry workflow to decipher immunomodulatory changes in celiac disease, Cytometry B Clin Cytom, 100 (2021) 92–102. [DOI] [PubMed] [Google Scholar]
- [59].Curiox Biosystems, Centrifuge-Less Immunostaining of Suspension Cells for Flow Cytometry Analysis using the Droparray™ System, 2017.
- [60].Mahnke Y, Chattopadhyay P, Roederer M, Publication of optimized multicolor immunofluorescence panels, Cytometry A, 77 (2010) 814–818. [DOI] [PubMed] [Google Scholar]
- [61].Finak G, Langweiler M, Jaimes M, Malek M, Taghiyar J, Korin Y, Raddassi K, Devine L, Obermoser G, Pekalski ML, Pontikos N, Diaz A, Heck S, Villanova F, Terrazzini N, Kern F, Qian Y, Stanton R, Wang K, Brandes A, Ramey J, Aghaeepour N, Mosmann T, Scheuermann RH, Reed E, Palucka K, Pascual V, Blomberg BB, Nestle F, Nussenblatt RB, Brinkman RR, Gottardo R, Maecker H, McCoy JP, Standardizing Flow Cytometry Immunophenotyping Analysis from the Human ImmunoPhenotyping Consortium, Sci Rep, 6 (2016) 20686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Aghaeepour N, Finak G, C.A.P.C. Flow, D. Consortium, Hoos H, Mosmann TR, Brinkman R, Gottardo R, Scheuermann RH, Critical assessment of automated flow cytometry data analysis techniques, Nat Methods, 10 (2013) 228–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Beckman Coulter Life Sciences, DURAClone Panels, https://www.beckman.com/, 2021.
- [64].Fluidigm, Helios, a CyTOF System, https://www.fluidigm.com/, 2022.
- [65].Wang X, Lebrec H, Immunophenotyping: application to safety assessment, Toxicologic pathology, 45 (2017) 1004–1011. [DOI] [PubMed] [Google Scholar]
- [66].Lebrec H, O’Lone R, Freebern W, Komocsar W, Moore P, Survey: immune function and immunotoxicity assessment in dogs, J Immunotoxicol, 9 (2012) 1–14. [DOI] [PubMed] [Google Scholar]
- [67].Chimote AA, Hajdu P, Kottyan LC, Harley JB, Yun Y, Conforti L, Nanovesicle-targeted Kv1.3 knockdown in memory T cells suppresses CD40L expression and memory phenotype, J Autoimmun, 69 (2016) 86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Casey LM, Kakade S, Decker JT, Rose JA, Deans K, Shea LD, Pearson RM, Cargo-less nanoparticles program innate immune cell responses to toll-like receptor activation, Biomaterials, 218 (2019) 119333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Simón-Vázquez R, Lozano-Fernández T, Dávila-Grana A, González-Fernández A, Metal oxide nanoparticles interact with immune cells and activate different cellular responses, International journal of nanomedicine, 11 (2016) 4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Liptrott NJ, Giardiello M, Hunter JW, Tatham L, Tidbury LR, Siccardi M, Rannard S, Owen A, Flow cytometric analysis of the physical and protein-binding characteristics of solid drug nanoparticle suspensions, Nanomedicine (Lond), 10 (2015) 1407–1421. [DOI] [PubMed] [Google Scholar]
- [71].Fang J-L, Beland FA, Tang Y, Roffler SR, Flow cytometry analysis of anti-polyethylene glycol antibodies in human plasma, Toxicology Reports, 8 (2021) 148–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Garcia Romeu H, Deville S, Salvati A, Time- and Space-Resolved Flow-Cytometry of Cell Organelles to Quantify Nanoparticle Uptake and Intracellular Trafficking by Cells, Small, 17 (2021) e2100887. [DOI] [PubMed] [Google Scholar]
- [73].He C, Duan X, Guo N, Chan C, Poon C, Weichselbaum RR, Lin W, Core-shell nanoscale coordination polymers combine chemotherapy and photodynamic therapy to potentiate checkpoint blockade cancer immunotherapy, Nat Commun, 7 (2016) 12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Rumfield CS, Pellom ST, Morillon YM II, Schlom J, Jochems C, Immunomodulation to enhance the efficacy of an HPV therapeutic vaccine, Journal for immunotherapy of cancer, 8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Barth BM, Altinoğlu EI, Shanmugavelandy SS, Kaiser JM, Crespo-Gonzalez D, DiVittore NA, McGovern C, Goff TM, Keasey NR, Adair JH, Targeted indocyanine-green-loaded calcium phosphosilicate nanoparticles for in vivo photodynamic therapy of leukemia, ACS nano, 5 (2011) 5325–5337. [DOI] [PubMed] [Google Scholar]
- [76].Amor C, Feucht J, Leibold J, Ho Y-J, Zhu C, Alonso-Curbelo D, Mansilla-Soto J, Boyer JA, Li X, Giavridis T, Senolytic CAR T cells reverse senescence-associated pathologies, Nature, 583 (2020) 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Wei SC, Anang N-AA, Sharma R, Andrews MC, Reuben A, Levine JH, Cogdill AP, Mancuso JJ, Wargo JA, Pe’er D, Combination anti–CTLA-4 plus anti–PD-1 checkpoint blockade utilizes cellular mechanisms partially distinct from monotherapies, Proceedings of the National Academy of Sciences, 116 (2019) 22699–22709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Bauman JE, Ohr J, Gooding WE, Ferris RL, Duvvuri U, Kim S, Johnson JT, Soloff AC, Wallweber G, Winslow J, Phase I Study of ficlatuzumab and cetuximab in cetuximab-resistant, recurrent/metastatic head and neck cancer, Cancers, 12 (2020) 1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Bendall SC, Nolan GP, From single cells to deep phenotypes in cancer, Nat Biotechnol, 30 (2012) 639–647. [DOI] [PubMed] [Google Scholar]
- [80].Cunningham RA, Holland M, McWilliams E, Hodi FS, Severgnini M, Detection of clinically relevant immune checkpoint markers by multicolor flow cytometry, Journal of Biological Methods, 6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Carozza JA, Böhnert V, Nguyen KC, Skariah G, Shaw KE, Brown JA, Rafat M, von Eyben R, Graves EE, Glenn JS, Smith M, Li L, Extracellular cGAMP is a cancer cell-produced immunotransmitter involved in radiation-induced anti-cancer immunity, Nat Cancer, 1 (2020) 184–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Bonilla DL, Reinin G, Chua E, Full Spectrum Flow Cytometry as a Powerful Technology for Cancer Immunotherapy Research, Frontiers in Molecular Biosciences, 7 (2020) 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, An mRNA vaccine against SARS-CoV-2—preliminary report, New England Journal of Medicine, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Anderson EJ, Rouphael NG, Widge AT, Jackson LA, Roberts PC, Makhene M, Chappell JD, Denison MR, Stevens LJ, Pruijssers AJ, Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults, New England Journal of Medicine, 383 (2020) 2427–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].U.S. National Library of Medicine, A Vaccine (PDS0101) and Chemoradiation for the Treatment of Stage IB3-IVA Cervical Cancer, the IMMUNOCERV Trial; ClinicalTrials.gov Identifier: NCT04580771, ClinicalTrials.gov, 2021.
- [86].Yim PB, Clarke ML, McKinstry M, De Paoli Lacerda SH, Pease LF 3rd, Dobrovolskaia MA, Kang H, Read TD, Sozhamannan S, Hwang J, Quantitative characterization of quantum dot-labeled lambda phage for Escherichia coli detection, Biotechnol Bioeng, 104 (2009) 1059–1067. [DOI] [PubMed] [Google Scholar]
- [87].Sun Q, Bai X, Sofias AM, van der Meel R, Ruiz-Hernandez E, Storm G, Hennink WE, De Geest B, Kiessling F, Yu HJ, Lammers T, Shi Y, Cancer nanomedicine meets immunotherapy: opportunities and challenges, Acta Pharmacol Sin, 41 (2020) 954–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Shi J, Kantoff PW, Wooster R, Farokhzad OC, Cancer nanomedicine: progress, challenges and opportunities, Nat Rev Cancer, 17 (2017) 20–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Yang S, Li Y, Fluorescent hybrid silica nanoparticles and their biomedical applications, Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology, 12 (2020) e1603. [DOI] [PubMed] [Google Scholar]
- [90].Zhang Y, Zhang K, Wang J, Tian Z, Li AD, Photoswitchable fluorescent nanoparticles and their emerging applications, Nanoscale, 7 (2015) 19342–19357. [DOI] [PubMed] [Google Scholar]
- [91].Yang G, Liu Y, Zhao C-X, Quantitative comparison of different fluorescent dye-loaded nanoparticles, Colloids and Surfaces B: Biointerfaces, (2021) 111923. [DOI] [PubMed] [Google Scholar]
- [92].Hensel JA, Khattar V, Ashton R, Ponnazhagan S, Characterization of immune cell subtypes in three commonly used mouse strains reveals gender and strain-specific variations, Lab Invest, 99 (2019) 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Beeton C, Chandy KG, Potassium channels, memory T cells, and multiple sclerosis, Neuroscientist, 11 (2005) 550–562. [DOI] [PubMed] [Google Scholar]
- [94].Veluchamy JP, Delso-Vallejo M, Kok N, Bohme F, Seggewiss-Bernhardt R, Van Der Vliet HJ, De Gruijl TD, Huppert V, Spanholtz J, Standardized and flexible eight colour flow cytometry panels harmonized between different laboratories to study human NK cell phenotype and function, Scientific reports, 7 (2017) 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Cerignoli F, Abassi YA, Lamarche BJ, Guenther G, Santa Ana D, Guimet D, Zhang W, Zhang J, Xi B, In vitro immunotherapy potency assays using real-time cell analysis, PLoS One, 13 (2018) e0193498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Ferrer-Font L, Pellefigues C, Mayer JU, Small SJ, Jaimes MC, Price KM, Panel Design and Optimization for High-Dimensional Immunophenotyping Assays Using Spectral Flow Cytometry, Curr Protoc Cytom, 92 (2020) e70. [DOI] [PubMed] [Google Scholar]
- [97].Fousek K, Watanabe J, Joseph SK, George A, An X, Byrd TT, Morris JS, Luong A, Martínez-Paniagua MA, Sanber K, Navai SA, Gad AZ, Salsman VS, Mathew PR, Kim HN, Wagner DL, Brunetti L, Jang A, Baker ML, Varadarajan N, Hegde M, Kim YM, Heisterkamp N, Abdel-Azim H, Ahmed N, CAR T-cells that target acute B-lineage leukemia irrespective of CD19 expression, Leukemia, 35 (2021) 75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Kessel S, Cribbes S, Bonasu S, Rice W, Qiu J, Chan LL, Real-time viability and apoptosis kinetic detection method of 3D multicellular tumor spheroids using the Celigo Image Cytometer, Cytometry A, 91 (2017) 883–892. [DOI] [PubMed] [Google Scholar]
- [99].ThermoFisher Scientific, Attune Flow Cytometer Sample Data, n.d.
- [100].Heck S, Bishop CJ, Ellis RJ, Immunophenotyping of Human Peripheral Blood Mononuclear Cells by Mass Cytometry, Methods Mol Biol, 1979 (2019) 285–303. [DOI] [PubMed] [Google Scholar]


