Abstract
Pseudomonas is a large and diverse genus within the Gammaproteobacteria known for its important ecological role in the environment. These bacteria exhibit versatile features of which the ability of heterotrophic nitrification and aerobic denitrification can be applied for nitrogen removal from the wastewater. A novel denitrifying bacterium, designated JM10B5aT, was isolated from the pond water for juvenile Litopenaeus vannamei. The phylogenetic, genomic, physiological, and biochemical analyses illustrated that strain JM10B5aT represented a novel species of the genus Pseudomonas, for which the name Pseudomonas oligotrophica sp. nov. was proposed. The effects of carbon sources and C/N ratios on denitrification performance of strain JM10B5aT were investigated. In addition, the results revealed that sodium acetate was selected as the optimum carbon source for denitrification of this strain. Besides, strain JM10B5aT could exhibit complete nitrate removal at the low C/N ratio of 3. Genomic analyses revealed that JM10B5aT possessed the functional genes including napA, narG, nirS, norB, and nosZ, which might participate in the complete denitrification process. Comparative genomic analyses indicated that many genes related to aggregation, utilization of alkylphosphonate and tricarballylate, biosynthesis of cofactors, and vitamins were contained in the genome of strain JM10B5aT. These genomic features were indicative of its adaption to various niches. Moreover, strain JM10B5aT harbored the complete operons required for the biosynthesis of vibrioferrin, a siderophore, which might be conducive to the high denitrification efficiency of denitrifying bacterium at low C/N ratio. Our findings demonstrated that the strain JM10B5aT could be a promising candidate for treating wastewater with a low C/N ratio.
Keywords: Pseudomonas, 16S rRNA gene, phylogenetic analysis, genomic analysis, denitrifying bacteria, low carbon–nitrogen ratio
Introduction
Pseudomonas is widely distributed in various natural environments including water, soil, sediment, plants, animals, and clinical samples, and this genus contains a large number of species (Zou et al., 2019; Li et al., 2020; Mamtimin et al., 2021). The genus Pseudomonas was first proposed with the description of P. aeruginosa JCM 5962T by Migula (1894) and belongs to the family Pseudomonadaceae within the class Gammaproteobacteria. Recently, Lalucat et al. (2021) proposed the division of Pseudomonas into five new genera: “Halopseudomonas,” “Linyingimonas,” “Stutzerimonas,” “Alcaligenimonas,” and “Ubiquimonas” based on the phylogenetic and phylogenomic analyses, and the genus Halopseudomonas published by Rudra and Gupta (2021) has been validated. Pseudomonas strains displayed some potential roles, such as the high biodegradation capability of polycyclic aromatic hydrocarbons, sulfide oxidation, phosphate accumulation, and denitrification (Xie et al., 2016, 2021; Sun et al., 2019; Zhang R. C. et al., 2020). For example, a denitrifying phosphorus-accumulating bacterium P. stutzeri ADP-19 could remove 96.9% of nitrate and 73.3% of phosphorus under aerobic conditions (Li et al., 2021); a heterotrophic nitrifying–aerobic denitrifying bacterium P. bauzanensis DN13-1 could remove 98.8, 98.9, and 65.9% of nitrite (-N), nitrate (-N), and ammonium (-N), respectively (Zhang M. et al., 2020). Although the 16S rRNA gene is the basic tool for current bacterial classification, it cannot be used availably to differentiate closely related species of Pseudomonas (Wang et al., 2020). Thus, the taxonomy of Pseudomonas strains has evolved with the available methodologies. Up-to-date Bacterial Core Gene set (UBCG, 92 bacterial core gene sets commonly present in all bacterial genomes which are shown on the website of EZBioCloud1) together with a phylogenomic pipeline could provide the accurate phylogenomic trees for the taxonomic purpose (Na et al., 2018).
Nitrogen pollution mainly comes from the excessive use of fertilizers, poultry production, domestic sewage, industrial manufacture, and aquaculture wastewater (Rezvani et al., 2019). Excess nitrite and nitrate accumulation can cause eutrophication and pose threat to aquatic animals and human health. For instance, excess nitrate could cause the regressive, circulatory, and inflammatory damages in the post-larvae and juvenile of Macrobrachium amazonicum (Dutra et al., 2020). Among the nitrate-contaminated wastewaters, the low organic carbon-to-nitrogen (C/N) ratio wastewater is a common type such as the polluted groundwater, industrial wastewater, rural sewage, and effluent from wastewater treatment plants (Deng et al., 2016; Hong et al., 2019; Gao et al., 2020).
Biological denitrification is an effective and general method for reducing nitrate to nitrogen gas due to its high efficiency, low cost, and environmental friendliness (Pang and Wang, 2021). Heterotrophic denitrification, which utilizes the organic carbon sources as electron donors, has a higher denitrification rate compared with autotrophic denitrification (Yang et al., 2020). However, the deficiency of available carbon source is an intractable problem for heterotrophic denitrification in the treatment of wastewater with low C/N ratio. To ensure sufficient electron donors for the denitrification process, the conventional solution is to add external organic carbon (often methanol, ethanol, glucose, and acetic acid) into the wastewater with low C/N ratio (Wang et al., 2018). Nevertheless, the addition of external organic carbon may result in the risk of secondary contamination and a high cost (Ling et al., 2021). Therefore, effective and sustainable strategies to enhance heterotrophic denitrification in the treatment of wastewater with a low C/N ratio are needed.
Accumulating approaches have been developed for the nitrate removal from the low C/N ratio wastewater, such as the system coupled with iron-based chemical reduction and autotrophic denitrification (Liu et al., 2020), the heterotrophic denitrification system amended with redox-active biochar (Wu et al., 2019), the fungal pellets immobilized bacterial bioreactor (Zheng et al., 2021), and the utilization of biodegradable and inert carriers in the sequencing batch reactors (Huang et al., 2020). These approaches are focused on generating more electron donors to enhance nitrate removal capability or providing carriers for microorganisms to create a suitable environment. In addition, some heterotrophic denitrifying bacteria that can achieve efficient denitrification at low C/N ratio have been reported, such as Acinetobacter sp., Comamonas sp., and Pseudomonas sp. (Zhang S. et al., 2020; Chen et al., 2021; Fan et al., 2021). Compared with the abovementioned approaches, these denitrifying bacteria can not only effectively and readily remove nitrate from low C/N ratio wastewater without the utilization of complex engineering but also provide the functional microorganisms possessing excellent nitrate removal ability for the wastewater treatment systems. Among the denitrifying bacteria, Pseudomonas is the dominant bacterial genus in activated sludge and biofilm reactors (Deng et al., 2018; Zhang et al., 2019). Moreover, several Pseudomonas strains possess the denitrification ability and other functions, such as phosphorus removal ability and polyhydroxybutyrate-degrading ability (Di et al., 2019; Li et al., 2021). Therefore, Pseudomonas strains would outcompete other bacteria in the practical application.
In this study, a novel denitrifying bacterium, designated JM10B5aT, was isolated from the pond water for juvenile Litopenaeus vannamei. The phylogenetic, genomic, physiological, and biochemical analyses illustrated that strain JM10B5aT represented a novel species of the genus Pseudomonas, for which the name Pseudomonas oligotrophica sp. nov. was proposed. This strain performed excellent capability for denitrification under the low C/N ratio condition. Genomic information revealed that the functional genes of napA, narG, nirS, norB, and nosZ encoding the enzymatic repertoire for completely denitrification were identified in strain JM10B5aT. Our findings demonstrated that strain JM10B5aT could be a promising candidate for treating wastewater with a low C/N ratio.
Materials and Methods
Bacterial Strains
Strain JM10B5aT was isolated from pond water for juvenile Litopenaeus vannamei collected from Jiangmen city, Guangdong Province, P. R. China (N 21° 56' 31“; E 112° 46' 16”). To isolate the aerobic denitrifying bacteria, the denitrification screening medium (DSM) was utilized. DSM was formulated as follows (per liter): sodium succinate 0.25 g, sodium citrate dihydrate 0.25 g, Na2HPO4 1.0 g, KH2PO4 1.0 g, NaNO2 0.069 g, KNO3 0.1 g, (NH4)2SO4 0.066 g, MgSO4·7H2O 0.2 g, 2.0 ml of trace element solution (TES) and 1.0 ml of mixed carbon source solution (CSS), pH 7.3, and the solid plate contained the DSM supplemented with 15.0 g/L agar. MgSO4·7H2O, TES, and CSS were all added to the autoclaved DSM after filtering using 0.22-μm filter membrane. The TES contained (per liter) EDTA-Na 10.0 g, ZnSO4·7H2O 0.5 g, MnCl2·4H2O 0.4 g, CoCl2·2H2O 0.5 g, CuSO4·5H2O 0.2 g, CaCl2 5.5 g, FeSO4·7H2O 1.1 g, and NaMoO4·2H2O 0.4 g. The CSS was described in the previous work (Zhang M. et al., 2020) and contained (per liter) D-glucose 13.8 g, D-fructose 13.8 g, D-lactose 13.8 g, sodium acetate 19.0 g, 90% lactic acid 12.8 ml, mannitol 14.0 g, ethyl alcohol 14.0 ml, glycerin 12.6 ml, sodium benzoate 9.6 g, and salicylic acid 9.2 g. Serial dilutions of 10−1 to 10−4 of pond water were made, and 0.1 ml of the 10−2, 10−3, and 10−4 dilutions was spread on DSM agar plates, respectively. Then, these plates were incubated at 30°C for 5 days. Strain JM10B5aT was isolated and stored at −80°C in the nutrient broth medium (NB) supplemented with 20% (v/v) glycerol.
P. stutzeri CGMCC 1.1803T and P. balearica CCUG 44487T were obtained from China General Microbiological Culture Collection Center (CGMCC) and Culture Collection University of Gothenburg (CCUG), respectively. These two type strains were used as the related strains for phenotypic, chemotaxonomic, and genetic analyses.
Phylogenetic Analysis
Genomic DNA of strains JM10B5aT was extracted from fresh cells, and the 16S rRNA gene sequence was amplified using the universal primers (27F/1492R) as described previously (Zhang et al., 2021). Sequencing was performed by GENEWIZ, Inc. (Suzhou, China). The BLAST algorithm2 and EzBioCloud database3 were used to search for similar sequences (Yoon et al., 2017). Pairwise identities of 16S rRNA gene sequences were calculated using the software DNAMAN version 8. Multiple alignments of the 16S rRNA sequences were performed using the software MAFFT version 7.037 under the L-INS-i iterative refinement (Katoh and Standley, 2014). The phylogenetic tree was reconstructed using the software IQ-TREE version 2.1.2 with the maximum likelihood (ML) method under the TN+F+I+G4 nucleotide substitution model (Felsenstein, 1981; Kalyaanamoorthy et al., 2017; Minh et al., 2020). Support for the inferred ML tree was inferred by the ultrafast bootstrapping with 1,000 replicates (Diep Thi et al., 2018). The visualization and annotation of the resulting phylogenetic tree were performed using the software MEGA version X (Kumar et al., 2018).
Morphological Observations and Analyses of Physiological Characteristics
Cells and colonies of strain JM10B5aT cultivated on the nutrient broth agar medium (NA) at 30°C for 48 h were observed by a transmission electron microscope (H7650, Hitachi) and naked eyes, respectively. Oxidase activity was determined using oxidase testing strips (HKM), and catalase activity was detected by bubble production after the addition of 3.0% H2O2 (v/v) solution. Growth under an anaerobic environment was determined after 5 days of incubation on NA at 30°C in an anaerobic pouch (MGC, Mitsubishi). Hydrolysis of casein, starch, tyrosine, and Tweens 20, 40, and 80 was investigated according to the protocols described by Lanyi (1987) and Tindall et al. (2007). Cellular motility was examined using the hanging-drop method (Bernardet et al., 2002).
The temperature range for growth was measured on NA at 4, 10, 15, 20, 25, 30, 37, 40, 45, and 50°C, respectively. The pH range for growth was determined after incubation at 30°C and 180 rpm in the modified NB medium with appropriate biological buffers (50 mM): sodium citrate buffer (pH 5.0, 5.5, and 6.0), HEPES buffer (pH 6.5, 7.0, and 7.5), Tris buffer (pH 8.0, 8.5, and 9.0), and Na2CO3/NaHCO3 (pH 9.5, 10.0, 11.0, and 12.0). The pH of the medium was adjusted by adding 1.0 M HCl or 1.0 M NaOH before autoclaving. The NaCl tolerance for growth was examined in the modified NB medium (without NaCl) with different NaCl concentrations (w/v, 0, 0.5, 1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0, and 10.0 %) at 30°C and 180 rpm. After incubation for 72 h, the bacterial growth was estimated by the spectrophotometric measurement of cell density (OD600).
Additional biochemical characteristics such as the enzyme activities, acid production by fermentation, and utilization of carbon sources were carried out using API ZYM and 20NE kits (bioMérieux) and Biolog GEN III MicroPlate (Biolog) according to the manufacturers' instructions. The API strips and Biolog results were recorded every 24 h after incubation at 30°C until all reactions were steady. All API and Biolog tests were performed in duplicate under the consistent condition.
Analyses of Chemotaxonomic Characteristics
For fatty acid composition assay, the exponentially growing cells of strain JM10B5aT were harvested. Fatty acids were saponified, methylated, and extracted using the standard protocol of the Sherlock Microbial Identification System (MIDI). The prepared fatty acids were analyzed by gas chromatography (model 7890A; Agilent) using the Microbial Identification software package with the Sherlock MIDI 6.1 system and the Sherlock Aerobic Bacterial Database (TSBA 6.1).
Polar lipids were extracted using the chloroform/methanol system and separated by two-dimensional thin-layer chromatography (TLC, Silica gel 60 F254, Merck) (Minnikin et al., 1984). The plates dotted with samples were subjected to two-dimensional development with the first solvent of chloroform–methanol–water (65:25:4, v/v) and the second solvent of chloroform–methanol–acetic acid–water (80:12:15:4, v/v). Total lipids and specific functional groups were detected using the different spray staining reagents on separate TLC plates: 10% ethanolic molybdophosphoric acid, ninhydrin, molybdenum blue, and α-naphthol–sulfuric acid. The quinone was extracted from the freeze-dried cells and determined using high-performance liquid chromatography (Agilent 1200; ODS 250 × 4.6 mm × 5.0 μm; flowing phase, methanol–isopropanol, 2:1; 1.0 ml/min) according to the methods described by Collins and Jones (1981).
Genome Sequencing and Function Analysis
The genomic DNA of strain JM10B5aT was extracted using the HiPure Bacterial DNA Kit (Magen Biotech, Guangzhou) according to the manufacturer's instruction. The genome was sequenced using the Illumina NovaSeq PE150 platform in Shanghai Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). Raw reads were filtered and then de novo assembled using the software SPAdes version 3.14.1 under “–careful” mode and with a k-mer value of 127 (Bankevich et al., 2012). Available genomes of the related type strains P. stutzeri CGMCC 1.1803T and P. balearica CCUG 44487T were obtained from the GenBank database with the numbers CP002881 and CP007511, respectively. The genome of strain JM10B5aT was in draft status and was quality-checked using the software CheckM version 1.1.2 (Parks et al., 2015). Overall genome relatedness indices (OGRIs) including digital DNA–DNA hybridization values (dDDH) and average nucleotide identity values (ANI) were estimated using the genome-to-genome distance calculator version 2.1 online service with the recommended formula 24 and the software FastANI version 1.31, respectively (Meier-Kolthoff et al., 2013; Jain et al., 2018). The UBCG pipeline which could provide an accurate phylogenomic tree for the taxonomic purpose was used (Chun et al., 2018; Na et al., 2018). The genomes of strain JM10B5aT and the related type strains were annotated using the software Prokka version 1.13 (Seemann, 2014) and Rapid using Subsystem Technology (RAST) version 2.0 with the default parameters (Overbeek et al., 2014).
Assessment of Nitrogen Removal Characteristics
The results of the API 20NE test preliminarily indicated that strain JM10B5aT was capable of reducing nitrate and nitrite. Furthermore, the genome annotation revealed that strain JM10B5aT possessed the functional genes including napA, narG, nirS, norB, and nosZ that participated in the denitrification process. These results suggested that strain JM10B5aT had the potential capability of complete denitrification. To investigate the optimum carbon source of denitrification for strain JM10B5aT, the carbon source was replaced in the DSM-1 (DSM with KNO3 0.36 g/L as the sole nitrogen source) or DSM-2 (DSM with NaNO2 0.25 g/L as the sole nitrogen source) by sodium acetate, sodium succinate, sodium citrate, glucose, sucrose, and starch, respectively, with the C/N ratio of 10 (the molar mass of carbon to the molar mass of nitrogen). Furthermore, the initial C/N ratios of 2, 3, 4, and 5 were controlled by changing the addition amount of the optimum carbon source in the DSM-1. Before the experiments, strain JM10B5aT was incubated in NB at 30°C and 180 rpm for 24 h. The suspension was centrifuged and then washed 3 times with sterile physiological saline to remove the residual medium. The biomass was resuspended in sterile water with the initial OD600 adjusting to 1.0. The bacterial suspension was inoculated into a conical flask containing sterile medium with the inoculum amount of 4.0% and incubated at 30°C statically. Samples taken from flasks after 48 h of incubation were used for determining the OD600 and chemical analyses. The medium without bacterial inoculation was used as control treatment. All the experiments were performed in biological quadruplicate.
Analytical Methods and Statistical Analyses
The concentrations of -N and -N were measured using N-(1-naphthyl) ethylenediamine dihydrochloride and UV spectrophotometry, respectively, according to the standard analytical procedures (Association et al., 2005). All the data were presented as mean and standard error (SE). One-way ANOVA (multiple range test) was performed with Tukey's HSD test (P < 0.05) using SPSS Statistics 19.
Results and Discussion
16S rRNA Gene Sequence Analyses and Chemotaxonomic Characterization
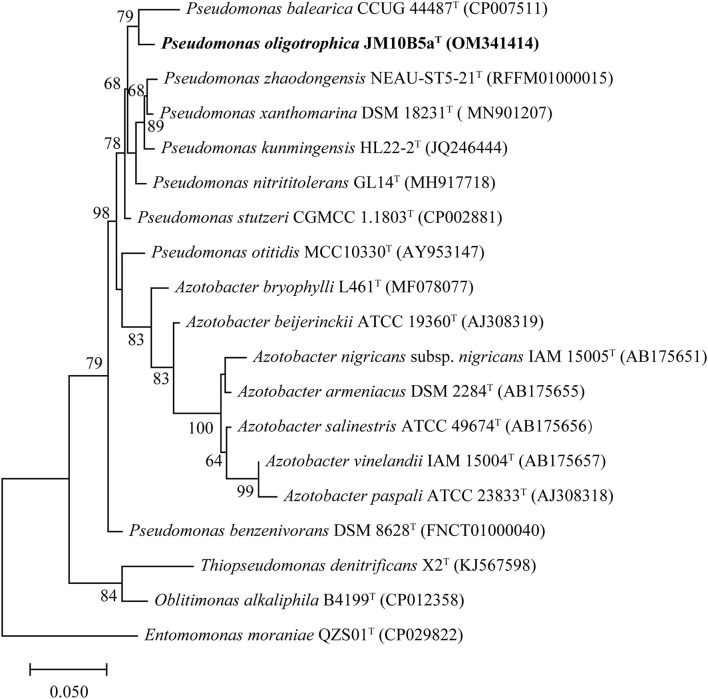
The almost complete 16S rRNA gene sequence of strain JM10B5aT obtained by amplification (1,375 bp) was included in the complete 16S rRNA gene sequence assembled from genomic sequences (1,537 bp). The sequence comparison showed that strain JM10B5aT fell into the genus Pseudomonas and shared the highest similarity with P. stutzeri CGMCC 1.1803T (98.0%). All the other type strains showed similarities lower than 98.0% with strain JM10B5aT. Based on phylogenetic analysis using the ML algorithm, strain JM10B5aT was stably located in the genus Pseudomonas and formed a clade with P. balearica CCUG 44487T (the similarity to JM10B5aT was 97.0%) at the 79.0% bootstrap confidence level (Figure 1). The similarity and phylogenetic analysis based on the 16S rRNA gene sequences indicated that strain JM10B5aT should represent a novel member of the genus Pseudomonas. Therefore, the type strains P. stutzeri CGMCC 1.1803T and P. balearica CCUG 44487T were purchased from the culture collection centers and used as references for further comparisons of phenotypic and chemotaxonomic characteristics.
Figure 1.
ML tree based on 16S rRNA gene sequences of strain JM10B5aT and the related type strains of genus Pseudomonas. Type strain Entomomonas moraniae QZS01T was used as an outgroup. There were a total of 1,303 positions used to construct the phylogenetic tree. Bootstrap values higher than 60% were shown at branch points. Bar, 0.05 represents the number of substitutions per site.
The predominant cellular fatty acids (>10%) of strain JM10B5aT were C16:0 (22.0%), summed feature 3 (C16:1 ω6c and/or C16:1 ω7c, 21.3%), and summed feature 8 (C18:1 ω6c and/or C18:1 ω7c, 29.1%), which were also detected in other Pseudomonas species (Mamtimin et al., 2021). Comparative fatty acid profiles between strain JM10B5aT and the two related type strains are shown in Table 1. The fatty acid profile of JM10B5aT was similar to that of other related strains, although there were minor quantitative differences observed. The predominant respiratory quinone of strain JM10B5aT was ubiquinone-9 (Q-9) which was consistent with that of other members of the genus Pseudomonas (Zou et al., 2019; Mamtimin et al., 2021). The major polar lipids were phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and diphosphatidylglycerol (DPG), which were consistent with the previously published data for Pseudomonas species (Zou et al., 2019; Li et al., 2020). In addition, minor amounts of two unidentified aminophospholipids (APLs) and three unidentified phospholipids (PLs) were also present (Supplementary Figure 1).
Table 1.
Cellular fatty acid profiles of strain JM10B5aT and the closely related type strains of genus Pseudomonas.
| Fatty acid | JM10B5aT | P. stutzeri CGMCC 1.1803T | P. balearica CCUG 44487T |
|---|---|---|---|
| Straight-chain saturated | |||
| C12:0 | 8.3 | 8.1 | 8.3 |
| C14:0 | 1.4 | 1.0 | 0.9 |
| C16:0 | 22.0 | 20.8 | 23.1 |
| Hydroxy | |||
| C10:0 3-OH | 2.8 | 2.8 | 3.2 |
| C12:0 3-OH | 4.0 | 3.8 | 4.1 |
| Branched saturated | |||
| C17:0 cyclo | 5.2 | 2.0 | 6.0 |
| iso C17:0 | TR | TR | 0.8 |
| Unsaturated | |||
| C19:0 cyclo ω8c | 4.2 | 2.3 | 5.5 |
| Summed Feature 3* | 21.3 | 22.8 | 18.9 |
| Summed Feature 8* | 29.1 | 34.1 | 27.4 |
All data were obtained from this study. Constituents <0.5% in three strains were not shown. TR, trace (<0.5%).
Summed features contain two or more fatty acids that cannot be separated by the MIDI system; summed feature 3 comprises C16:1 ω6c and/or C16:1 ω7c; summed feature 8 comprises C18:1 ω6c and/or C18:1 ω7c.
Morphological Observations and Physiological Characterization
The morphological features of strain JM10B5aT were studied on NA medium and formed irregular, filmy sheet, non-transparent, and ecru white colonies after 48 h of incubation at 30°C (Supplementary Figure 2A). Cells of strain JM10B5aT were observed to be gram-stain-negative, aerobic, rod-shaped (0.6–0.7 × 1.4–1.5 μm), facultative anaerobic, and motile with a single polar flagellum (Supplementary Figure 2B). Growth was determined at 10–45°C (optimum, 25–30°C), at pH 5.5–11.0 (optimum, 6.0), and in 0–4.0% (w/v) NaCl (optimum, 0.5–2.0%). The activities of catalase and oxidase were positive, and the further details differentiating strain JM10B5aT from the two related type strains are shown in Table 2. For instance, strain JM10B5aT could not grow in the NB with a NaCl concentration of 5.0 while the two related type strains could grow; the optimum pH of strain JM10B5aT was 6.0, which was inconsistent with that of P. stutzeri CGMCC 1.1803T and P. balearica CCUG 44487T of 6.5–7.5 and 8.5–9.5, respectively; the growth of strain JM10B5aT was weaker than that of the two related type strains at 45°C. Besides, stain JM10B5aT could be distinguished from the reference type strains by positive for adipic acid, trisodium citrate, esterase (C4), and lipase (C14) in the API 20NE and ZYM tests; and positive for D-galacturonic acid, D-glucuronic acid, gentiobiose, L-arginine, L-aspartic acid, L-galactonic acid lactone, mucic acid, quinic acid, α-hydroxybutyric acid, and γ-aminobutyric acid in the Biolog GNE III MicroPlate system. Importantly, strain JM10B5aT exhibited the capability for complete denitrification that liberated copious amounts of nitrogen gas from nitrate. The denitrification characteristic of this strain was consistent with that of P. stutzeri and P. balearica strains (Bennasar et al., 1996; Li et al., 2021).
Table 2.
Differential characteristics of strain JM10B5aT and the closely related type strains of genus Pseudomonas.
| Characteristic | JM10B5aT | P. stutzeri CGMCC 1.1803T | P. balearica CCUG 44487T |
|---|---|---|---|
| Growth | |||
| NaCl concentration (optimum, w/v) | 0–4.0 (0.5–2.0) | 0–5.0 (0.5) | 0–5.0 (1.0) |
| pH range (optimum) | 5.5–11.0 (6.0) | 6.0–11.0 (8.5–9.5) | 5.5–11.0 (6.5–7.5) |
| Temperature (optimum, °C) | 10–45 (25–30) | 10–45 (30–37) | 10–45 (30–37) |
| 45°C | w | + | + |
| API ZYM and 20NE tests | |||
| Adipic acid | + | + | – |
| Trisodium citrate | + | + | – |
| Esterase (C4) | + | – | – |
| Lipase (C14) | + | + | – |
| Biolog (GEN III) tests | |||
| Dextrin | – | + | + |
| D-Galacturonic acid | + | – | – |
| D-Glucuronic acid | + | w | – |
| D-Maltose | – | + | + |
| D-Mannitol | – | + | – |
| D-Saccharic acid | + | – | – |
| Formic acid | – | + | – |
| Gentiobiose | + | – | – |
| L-Arginine | + | – | – |
| L-Aspartic acid | + | – | + |
| L-Galactonic acid lactone | + | – | – |
| L-Pyroglutamic acid | – | + | – |
| Mucic acid | + | – | – |
| Quinic acid | + | – | – |
| α-Hydroxy-butyric acid | + | – | + |
| γ-Amino butyric acid | + | – | – |
| G + C content (%) | 67.2 | 63.6§ | 64.7§ |
| Genome size (Mb) | 4.0 | 4.5§ | 4.4§ |
All data were obtained from this study unless indicated otherwise.
Data from draft genomes in the NCBI genome database.
+, positive; –, negative; w, weakly positive.
Genome Data and Comparative Genomic Analysis
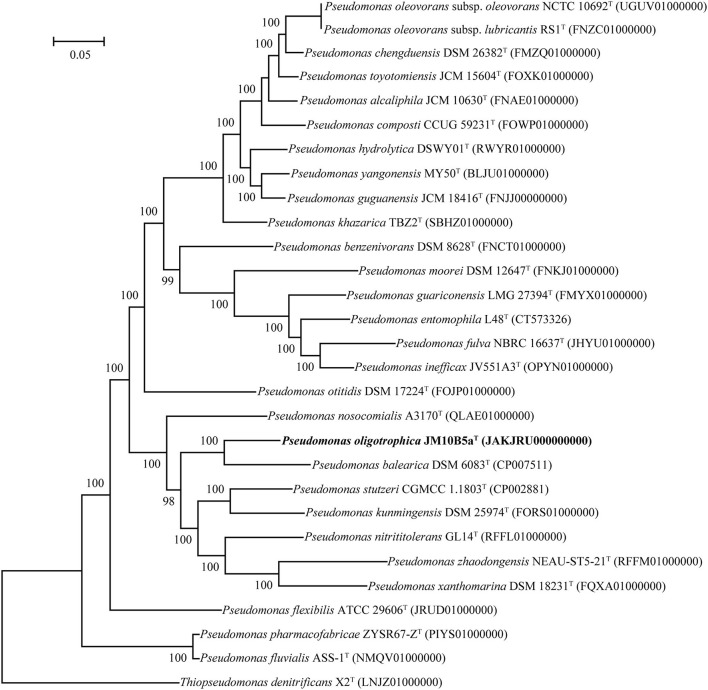
The draft genome of strain JM10B5aT was assembled into 20 contigs (>500 bp) with a genome size of 3,978,222 bp, and the estimated completeness and contamination were 100 and 0.1%, respectively, indicating that this genome was high quality according to the standards (Parks et al., 2015). Genomes of strains P. stutzeri CGMCC 1.1803T and P. balearica CCUG 44487T were obtained from the GenBank database with the numbers CP002881 and CP007511, respectively. Based on the Prokka annotation, a total of 3,744 genes, 3,679 protein-coding sequences (CDS), 60 tRNA genes, and 3 rRNA genes were found in the genome of strain JM10B5aT. The genomic DNA G + C content of JM10B5aT was 67.2%. Compared with the genomes of related type strains, JM10B5aT had ANI values of 78.9–88.1% and dDDH values of 20.8–30.8% (Supplementary Table 1), which were all below 95.0 and 70% cutoff commonly used to define a bacterial species, respectively (Stackebrandt and Goebel, 1994; Richter and Rossello-Mora, 2009). To further determine the taxonomic position of strain JM10B5aT, the phylogenomic tree based on the 92 bacterial core gene sets was reconstructed (Na et al., 2018). As shown in Figure 2, strain JM10B5aT and P. balearica CCUG 44487T formed a clade with 100% bootstrap value, which was consistent with the phylogenetic tree based on the 16S rRNA gene sequences. Therefore, the comprehensive analyses of OGRIs and phylogenomic tree further indicated that strain JM10B5aT should represent a novel species within the genus of Pseudomonas.
Figure 2.
Phylogenomic tree based on 92 bacterial core gene sets of strain JM10B5aT and the related type strains of genus Pseudomonas. Type strain Thiopseudomonas denitrificans X2T was used as an outgroup. Bootstrap values higher than 70% were shown at branch points. Bar, 0.05 represents the number of substations per site.
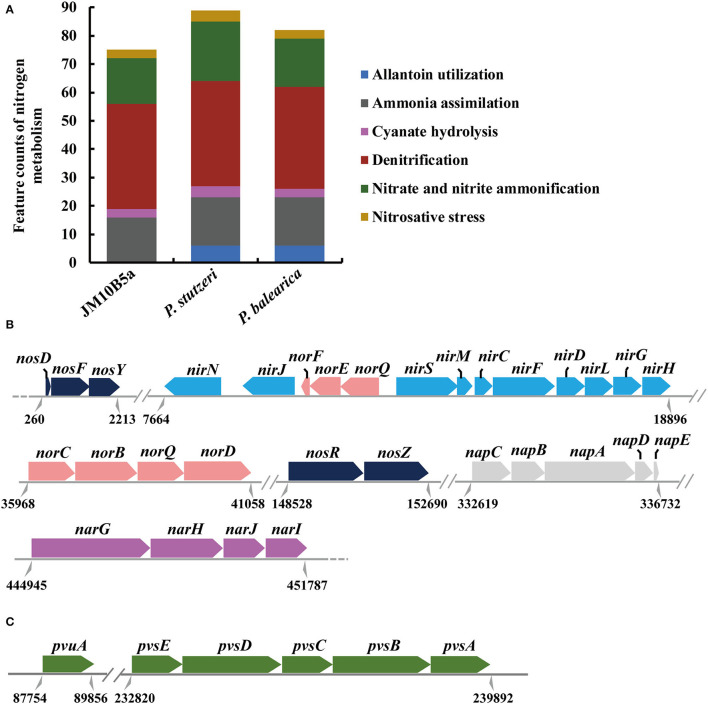
According to the subsystem category distribution of RAST annotation, there were 190, 128, 402, and 336 genes associated with “RNA metabolism,” “DNA metabolism,” “amino acid and derivatives,” and “carbohydrates,” respectively, which might be important to the bacterial growth. In addition, 75 genes were identified in the nitrogen metabolism, including the functional genes related to denitrification and assimilation of nitrite and nitrate (Figures 3A,B). Nitrate reductase catalyzing the reduction in nitrate to nitrite plays an important role in the nitrogen cycle (Kuypers et al., 2018). Previous studies have shown that bacterial dissimilatory nitrate reduction could be catalyzed by two different enzymes: a membrane-bound nitrate reductase (NAR, the catalytic subunit NarG encoded by narG) and periplasmic nitrate reductase (NAP, the catalytic subunit NapA encoded by napA) (Moreno-Vivian et al., 1999). Genes narG and napA were both contained in strain JM10B5aT, which was consistent with Pseudomonas sp. JQ-H3 and Paracoccus denitrificans (Moreno-Vivian et al., 1999; Wang et al., 2019). The dissimilatory reduction in nitrite to nitric oxide can be catalyzed by two unrelated enzymes: a cytochrome cd1 nitrite reductase (cd1-NIR, encoded by nirS) or a Cu-containing nitrite reductase (Cu-NIR, encoded by nirK), which were widespread among bacteria and archaea (Kuypers et al., 2018). Gene nirS rather than nirK was identified in strain JM10B5aT, which was consistent with Pseudomonas sp. JQ-H3, P. bauzanensis DN13-1, and P. stutzeri T13 (Wang et al., 2019; Zhang M. et al., 2020; Feng et al., 2021). In addition, genes norB encoding for nitric oxide reductase (NorB, the key enzyme for reducing nitric oxide to nitrous oxide) and nosZ encoding for nitrous oxide reductase (NosZ, the key enzyme for reducing nitrous oxide to nitrogen gas) were also identified. Therefore, it was speculated that strain JM10B5aT could perform the complete denitrification pathway: -N → -N → NO → N2O → N2. Hence, strain JM10B5aT was a novel denitrifying bacterium.
Figure 3.
Genomic analyses based on the RAST annotation. (A) Comparative analyses of the functional genes assigned to nitrogen metabolisms of strain JM10B5aT and the related type strains of genus Pseudomonas. (B) Relative arrangements of the denitrification genes on the draft genome of strain JM10B5aT. (C) The biosynthetic gene cluster for vibrioferrin on the draft genome of strain JM10B5aT. Parallel double lines indicate a break in locus organization among scaffolds, and dotted black lines indicate where unrelated continuity loci are not shown. Numbers below the line symbolize the locations.
In addition, comparative genomic analyses showed that strain JM10B5aT encoded 126 and 147 functional genes more than P. balearica CCUG 44487T and P. stutzeri CGMCC 1.1803T, respectively, and 67 genes of them were the shared differential genes (Supplementary Table 2). These genes were related to type I secretion system for aggregation, utilization of alkylphosphonate and tricarballylate, histidine ABC transporter, LysR family transcriptional regulator, and many other proteins. Most of these proteins were mainly affiliated with dehydratase, transferase, and biosynthesis of cofactors and vitamins. The genes encoding arginase and galactarate dehydratase made strain JM10B5aT performing the positive for L-arginine and mucic acid (Table 2). Moreover, strain JM10B5aT was found to harbor the complete operon pvsABCDE and vibrioferrin receptor gene pvuA (Figure 3C) that are required for the production of a siderophore vibrioferrin. This siderophore was shown to be responsible for vibrioferrin-mediated iron uptake in the terrestrial bacteria Azotobacter vinelandii, Pseudomonas sp., and the marine bacterium Vibrio parahaemolyticus (Yamamoto et al., 1994; Baars et al., 2016; Stanborough et al., 2018; Sood et al., 2019). It was reported that siderophore had a very high affinity to iron with the formation of Fe-siderophore which could facilitate the transport of iron through the membrane directly (Stintzi et al., 2000). Moreover, the denitrification efficiency might be enhanced with the improvement of iron transport from extracellular to intracellular (Jiang et al., 2020). Therefore, the biosynthesis of vibrioferrin might partly explain that the strain JM10B5aT performed the high denitrification efficiency under the low C/N ratio condition.
Effect of Carbon Source on Denitrification by Strain JM10B5aT
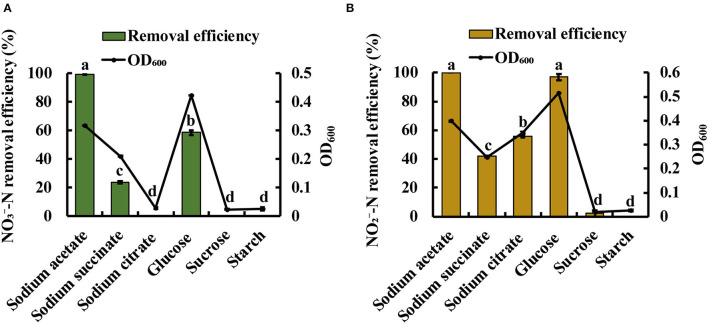
Heterotrophic denitrifying bacteria require organic carbon for cell growth and as the electron donor in the denitrification process (Rajta et al., 2020). Thus, carbon source was considered to be an important factor influencing denitrification. As expected, significant differences were observed using different carbon sources. As shown in Figure 4A, the strain JM10B5aT could grow well with the OD600 of 0.42 when glucose was used as the sole carbon source. However, it exhibited a higher -N removal efficiency of 99.5% when sodium acetate served as carbon source than that of 58.6% when glucose served as carbon source. As shown in Figure 4B, strain JM10B5aT could grow quite well when glucose served as the carbon source (OD600 of 0.52), but it performed the relatively high -N removal of 100% when sodium acetate served as the carbon source. These results indicated that sodium acetate was the optimum carbon source for the performance of denitrification for strain JM10B5aT, which was consistent with Bacillus pumilus, Arthrobacter sp., and Streptomyces lusitanus (Elkarrach et al., 2021). Therefore, sodium acetate was employed in the following experiments.
Figure 4.
Growth (OD600) and -N and -N removal performance of strain JM10B5aT under various common carbon sources. (A) -N as the sole nitrogen source. (B) -N as the sole nitrogen source. Values are mean ± SE (standard error) for four replicates.
Effect of Carbon/Nitrogen Ratio on Nitrate Removal by Strain JM10B5aT
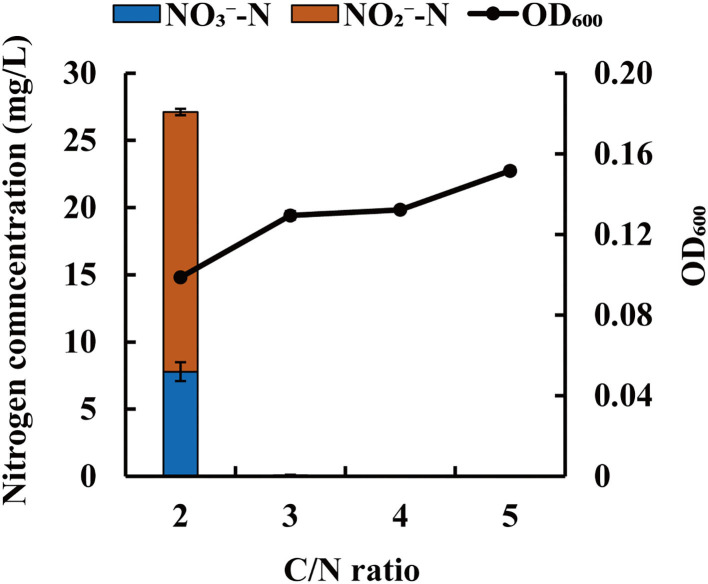
C/N ratio is a measure of the electron donor to acceptor ratio in the biological denitrification process. This factor can influence the denitrification efficiency and accumulation of -N. To investigate the influence of C/N ratio on denitrification of strain JM10B5aT, the initial concentration of -N was fixed at ~50.0 mg/L and the different initial C/N ratios (2, 3, 4, and 5) were controlled. As shown in Figure 5, strain JM10B5aT grew poorly with the OD600 range of 0.10–0.15 at these low C/N ratios. The -N was reduced completely at the C/N ratios of 3–5 which was distinctly higher than that of 84.4% at the C/N ratio of 2. Furthermore, there was no -N accumulation observed at the C/N ratios of 3–5, but the -N concentration of 19.3 mg/L was detected at the C/N ratio of 2. The low -N removal efficiency and -N accumulation at the C/N ratio of 2 were mainly due to the limited carbon source that could not provide sufficient energy for bacterial growth and electron donors for denitrification (Fan et al., 2021). These results were consistent with previous reports that denitrification efficiency could decrease under the extremely low carbon concentration (Zhao et al., 2018). Numerous studies have suggested that the optimum C/N ratios for most heterotrophic denitrifying bacteria were in the range of 8–15 (Guo et al., 2016; Liu et al., 2018; Zhao et al., 2018). For example, the C/N ratios of 10 and 15 were the most suitable for strains P. stutzeri XL-2 and P. taiwanensis J to achieve efficient nitrate removal, respectively (He et al., 2018; Zhao et al., 2018). In addition, some denitrifying bacteria could achieve complete denitrification at the relatively low C/N ratios. For example, Rout et al. (2017) used Bacillus cereus to remove nitrogen from domestic wastewater with 7.5 as the optimum C/N ratio; Zhang S. et al. (2020) reported that strain Comamonas sp. YSF15 could achieve complete denitrification at C/N of 3. Our results showed that strain JM10B5aT could achieve the complete nitrate removal without accumulation of nitrite at the low carbon–nitrogen ratio of 3 (CODCr/TN ratio of 2.6), indicating that this strain could be a promising candidate for treating the oligotrophic wastewater.
Figure 5.
Influence of C/N ratio on -N removal characteristic of strain JM10B5aT. Values are mean ± SE (standard error) for four replicates.
Description of Pseudomonas oligotrophica sp. nov.
Pseudomonas oligotrophica (o.li.go.tro'phi.ca. Gr. adj. oligos, few; Gr. adj. trophikos, nursing, tending, or feeding; N.L. fem. adj. oligotrophica, eating little, referring to a bacterium living on low-nutrient media).
Cells are rods (1.4–1.5 μm in length and 0.6–0.7 μm in width), gram-stain-negative, facultative anaerobic, and motile by the polar flagellum. Colonies are irregular, filmy sheet, non-transparent, and ecru white after 48 h of incubation on NA agar at 30°C. Growth was determined at 10–45°C (optimum, 25–30°C), at pH 5.5–11.0 (optimum, 6.0), and in 0–4.0% (w/v) NaCl (optimum, 0.5–2.0%) and is positive for catalase and oxidase, and hydrolysis of Tween 20 and 60. In the API ZYM and 20NE system, it is positive for esterase (C4), esterase lipase (C8), lipase (C14), leucine arylamidase and naphtol-AS-B1-phosphoamidase, nitrate and nitrite reduction, β-glucosidase, D-glucose, D-maltose, potassium gluconate, capric acid, adipic acid, malic acid, and trisodium citrate. In the Biolog GNE III MicroPlate system, it is positive for gentiobiose, α-D-glucose, D-fructose, glycerol, L-alanine, L-arginine, L-aspartic acid, L-glutamic acid, D-galacturonic acid, L-galactonic acid lactone, D-gluconic acid, D-glucuronic acid, glucuronamide, mucic acid, quinic acid, D-saccharic acid, L-lactic acid, citric acid, α-ketoglutaric acid, D-malic acid, L-malic acid, bromosuccinic acid, γ-aminobutyric acid, α-hydroxybutyric acid, β-hydroxy-D, L-butyric acid, α-ketobutyric acid, propionic acid, and acetic acid. The major fatty acids are C16:0, summed feature 3 (C16:1 ω6c and/or C16:1 ω7c), and summed feature 8 (C18:1 ω6c and/or C18:1 ω7c). The major polar lipids are PE, PG, and DPG, and the predominant respiratory quinone is Q-9. The DNA G + C content of strain JM10B05aT is 67.2%. Accession numbers of the complete 16S rRNA gene sequence and draft genome in DDBJ/ENA/GenBank are OM341414 and JAKJRU000000000, respectively. The type strain, JM10B5aT (= GDMCC 1.2828T = JCM 35033T), was isolated from pond water for juvenile Litopenaeus vannamei collected from Jiangmen city Guangdong Province, China.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
MZ, AL, and QY were involved in conceptualization and project administration. MZ involved in data curation, software, visualization, and writing—original draft. MZ and BX designed the formal analysis and methodology. AL, HZ, and BX were involved in funding acquisition. MZ, AL, QY, BX, and HZ investigated the study. HZ collected the resources and supervised the study. MZ and AL validated the study. MZ, QY, and BX were involved in writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the Key-Area Research and Development Program of Guangdong Province (2020B0202080005), the National Natural Science Foundation of China (32070115, 11772133), and the GDAS' Special Project of Science and Technology Development (2020GDASYL-20200301003).
Conflict of Interest
AL was employed by Guangdong BOWOTE BioSciTech, Co. Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are very grateful to Mr. Fan Yang at Guangdong Microbial Culture Collection Center (Guangzhou, China) for purchasing the related type strains as references.
Glossary
Abbreviations
- ANI
average nucleotide identity
- CCUG
Culture Collection University of Gothenburg
- CDS
protein-coding sequences
- CGMCC
China General Microbiological Culture Collection Center
- dDDH
digital DNA–DNA hybridization
- DPG
diphosphatidylglycerol
- DSM
denitrification screening medium
- GDMCC
Guangdong Microbial Culture Collection Center
- JCM
Japan Collection of Microorganisms
- ML
maximum likelihood
- NA
nutrient broth agar medium
- NB
nutrient broth medium
- OGRIs
overall genome relatedness indices
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- RAST
Rapid Annotation using Subsystem Technology
- TLC
thin-layer chromatography
- UBCG
up-to-date bacterial core gene.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.882890/full#supplementary-material
References
- Association A. P. H., Association A. W. W., Federation W. E. (2005). Standard Methods for the Examination of Water and Wastewater. Washington, DC: American Public Health Association Press. [Google Scholar]
- Baars O., Zhang X., Morel F. M. M., Seyedsayamdost M. R. (2016). The siderophore metabolome of Azotobacter vinelandii. Appl. Environ. Microbiol. 82, 27–39. 10.1128/AEM.03160-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasar A., RosselloMora R., Lalucat J., Moore E. R. B. (1996). 16S rRNA gene sequence analysis relative to genomovars of Pseudomonas stutzeri and proposal of Pseudomonas balearica sp. nov. Int. J. Syst. Bacteriol. 46, 200–205. 10.1099/00207713-46-1-200 [DOI] [PubMed] [Google Scholar]
- Bernardet J. F., Nakagawa Y., Holmes B. (2002). Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int. J. Syst. Evol. Microbiol. 52, 1049–1070. 10.1099/ijs.0.02136-0 [DOI] [PubMed] [Google Scholar]
- Chen C., Ali A., Su J., Wang Y., Huang T., Gao J. (2021). Pseudomonas stutzeri GF2 augmented the denitrification of low carbon to nitrogen ratio: possibility for sewage wastewater treatment. Bioresour. Technol. 333, 125169. 10.1016/j.biortech.2021.125169 [DOI] [PubMed] [Google Scholar]
- Chun J., Oren A., Ventosa A., Christensen H., Arahal D. R., da Costa M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466. 10.1099/ijsem.0.002516 [DOI] [PubMed] [Google Scholar]
- Collins M. D., Jones D. (1981). A note on the separation of natural mixtures of bacterial ubiquinones using reverse-phase partition thin-layer chromatography and high-performance liquid chromatography. J. Appl. Bacteriol. 51, 129–134. 10.1111/j.1365-2672.1981.tb00916.x [DOI] [PubMed] [Google Scholar]
- Deng L., Ngo H. H., Guo W., Wang J., Zhang H. (2018). Evaluation of a new sponge addition-microbial fuel cell system for removing nutrient from low C/N ratio wastewater. Chem. Eng. J. 338, 166–175. 10.1016/j.cej.2018.01.028 [DOI] [Google Scholar]
- Deng S., Li D., Yang X., Zhu S., Li J. (2016). Process of nitrogen transformation and microbial community structure in the Fe(0)-carbon-based bio-carrier filled in biological aerated filter. Environ. Sci. Pollut. Res. Int. 23, 6621–6630. 10.1007/s11356-015-5892-6 [DOI] [PubMed] [Google Scholar]
- Di Y., Xia H., Jiao Y., Zhang X., Fang Q., Li F., et al. (2019). Biodegradation of polyhydroxybutyrate by Pseudomonas sp. DSDY0501 and purification and characterization of polyhydroxybutyrate depolymerase 3. Biotechnology 9, 359. 10.1007/s13205-019-1871-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep Thi H., Chernomor O., von Haeseler A., Minh B. Q., Le Sy V. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutra F. M., Cidemar Alab J. H., Costa Gomes M. K., Furtado P. S., Valenti W. C., Cupertino Ballester E. L. (2020). Nitrate acute toxicity to post larvae and juveniles of Macrobrachium amazonicum (Heller, 1862). Chemosphere 242, 125229. 10.1016/j.chemosphere.2019.125229 [DOI] [PubMed] [Google Scholar]
- Elkarrach K., Merzouki M., Atia F., Laidi O., Benlemlih M. (2021). Aerobic denitrification using Bacillus pumilus, Arthrobacter sp., and Streptomyces lusitanus: novel aerobic denitrifying bacteria. Bioresour. Technol. Rep. 14, 100663. 10.1016/j.biteb.2021.100663 [DOI] [Google Scholar]
- Fan Y., Su J., Zheng Z., Gao J., Ali A. (2021). Denitrification performance and mechanism of a novel isolated Acinetobacter sp. FYF8 in oligotrophic ecosystem. Bioresour. Technol. 320 (Pt A), 124280. 10.1016/j.biortech.2020.124280 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376. 10.1007/BF01734359 [DOI] [PubMed] [Google Scholar]
- Feng L., Yang J., Ma F., Xing L., Pi S., Cui D., et al. (2021). Biological stimulation with Fe(III) promotes the growth and aerobic denitrification of Pseudomonas stutzeri T13. Sci. Total. Environ. 776, 145939. 10.1016/j.scitotenv.2021.145939 [DOI] [PubMed] [Google Scholar]
- Gao L., Han F., Zhang X., Liu B., Fan D., Sun X., et al. (2020). Simultaneous nitrate and dissolved organic matter removal from wastewater treatment plant effluent in a solid-phase denitrification biofilm reactor. Bioresour. Technol. 314, 123714. 10.1016/j.biortech.2020.123714 [DOI] [PubMed] [Google Scholar]
- Guo L. J., Zhao B., An Q., Tian M. (2016). Characteristics of a novel aerobic denitrifying bacterium, Enterobacter cloacae strain HNR. Appl. Biochem. Biotechnol. 178, 947–959. 10.1007/s12010-015-1920-8 [DOI] [PubMed] [Google Scholar]
- He T., Ye Q., Sun Q., Cai X., Ni J., Li Z., et al. (2018). Removal of nitrate in simulated water at low temperature by a novel psychrotrophic and aerobic bacterium, Pseudomonas taiwanensis strain. J. Biomed Res. Int. 2018, 4984087. 10.1155/2018/4984087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Huang G., An C., Song P., Xin X., Chen X., et al. (2019). Enhanced nitrogen removal in the treatment of rural domestic sewage using vertical-flow multi-soil-layering systems: experimental and modeling insights. J. Environ. Manage. 240, 273–284. 10.1016/j.jenvman.2019.03.097 [DOI] [PubMed] [Google Scholar]
- Huang L., Ye J., Xiang H., Jiang J., Wang Y., Li Y. (2020). Enhanced nitrogen removal from low C/N wastewater using biodegradable and inert carriers: performance and microbial shift. Bioresour. Technol. 300, 122658. 10.1016/j.biortech.2019.122658 [DOI] [PubMed] [Google Scholar]
- Jain C., Rodriguez -R. L. M, Phillippy A. M., Konstantinidis K. T., Aluru S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9, 5114. 10.1038/s41467-018-07641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Feng L., Zheng X., Chen Y. (2020). Bio-denitrification performance enhanced by graphene-facilitated iron acquisition. Water Res. 180, 115916. 10.1016/j.watres.2020.115916 [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S., Minh B. Q, Wong T. K. F., von Haeseler A., Jermiin L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2014). MAFFT: iterative refinement and additional methods. Methods Mol. Biol. 1079, 131–146. 10.1007/978-1-62703-646-7_8 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuypers M. M. M., Marchant H. K., Kartal B. (2018). The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 16, 263–276. 10.1038/nrmicro.2018.9 [DOI] [PubMed] [Google Scholar]
- Lalucat J., Gomila M., Mulet M., Zaruma A., Garcia-Valdes E. (2021). Past, present and future of the boundaries of the Pseudomonas genus: proposal of Stutzerimonas gen. Nov. Syst. Appl. Microbiol. 45, 126289. 10.1016/j.syapm.2021.126289 [DOI] [PubMed] [Google Scholar]
- Lanyi B. (1987). Classical and rapid identification methods for medically important bacteria. Method. Microbiol. 19, 1–67. 10.1016/S0580-9517(08)70407-0 [DOI] [Google Scholar]
- Li B., Jing F., Wu D., Xiao B., Hu Z. (2021). Simultaneous removal of nitrogen and phosphorus by a novel aerobic denitrifying phosphorus-accumulating bacterium, Pseudomonas stutzeri ADP-19. Bioresour. Technol. 321, 124445. 10.1016/j.biortech.2020.124445 [DOI] [PubMed] [Google Scholar]
- Li J., Wang L. H., Xiang F. G., Ding W. L., Xi L. J., Wang M. Q., et al. (2020). Pseudomonas phragmitis sp. nov., isolated from petroleum polluted river sediment. Int. J. Syst. Evol. Microbiol. 70, 364–372. 10.1099/ijsem.0.003763 [DOI] [PubMed] [Google Scholar]
- Ling Y., Yan G., Wang H., Dong W., Wang H., Chang Y., et al. (2021). Release mechanism, secondary pollutants and denitrification performance comparison of six kinds of agricultural wastes as solid carbon sources for nitrate removal. Int. J. Environ. Res. Public Health 18, 1232. 10.3390/ijerph18031232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Chen Q., Ma T., Wang M., Ni J. (2018). Genomic insights into metabolic potentials of two simultaneous aerobic denitrification and phosphorus removal bacteria, Achromobacter sp. GAD3 and Agrobacterium sp. LAD9. FEMS Microbiol. Ecol. 94, 4. 10.1093/femsec/fiy020 [DOI] [PubMed] [Google Scholar]
- Liu X., Huang M., Bao S., Tang W., Fang T. (2020). Nitrate removal from low carbon-to-nitrogen ratio wastewater by combining iron-based chemical reduction and autotrophic denitrification. Bioresour. Technol. 301, 122731. 10.1016/j.biortech.2019.122731 [DOI] [PubMed] [Google Scholar]
- Mamtimin T., Anwar N., Abdurahman M., Kurban M., Rozahon M., Mamtimin H., et al. (2021). Pseudomonas lopnurensis sp. nov., an endophytic bacterium isolated from Populus euphratica at the ancient Ugan river. Antonie Van Leeuwenhoek 114, 399–410. 10.1007/s10482-021-01524-8 [DOI] [PubMed] [Google Scholar]
- Meier-Kolthoff J. P., Auch A. F., Klenk H. P., Goeker M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformat. 14, 60. 10.1186/1471-2105-14-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migula W. (1894). Über ein neues system der bakterien. Arb. Bakteriol. Inst. Karlsruhe. 1, 235–238. [Google Scholar]
- Minh B. Q., Schmidt H. A., Chernomor O., Schrempf D., Woodhams M. D., von Haeseler A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 2461. 10.1093/molbev/msaa131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnikin D. E., Odonnell A. G., Goodfellow M., Alderson G., Athalye M., Schaal A., et al. (1984). An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods. 2, 233–241. 10.1016/0167-7012(84)90018-6 [DOI] [Google Scholar]
- Moreno-Vivian C., Cabello P., Martinez-Luque M., Blasco R., Castillo F. (1999). Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J. Bacteriol. 181, 6573–6584. 10.1128/JB.181.21.6573-6584.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na S. I., Kim Y. O., Yoon S. H., Ha S. M., Baek I., Chun J. (2018). UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 280–285. 10.1007/s12275-018-8014-6 [DOI] [PubMed] [Google Scholar]
- Overbeek R., Olson R., Pusch G. D., Olsen G. J., Davis J. J., Disz T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y., Wang J. (2021). Various electron donors for biological nitrate removal: a review. Sci. Total. Environ. 794, 148699. 10.1016/j.scitotenv.2021.148699 [DOI] [PubMed] [Google Scholar]
- Parks D. H., Imelfort M., Skennerton C. T., Hugenholtz P., Tyson G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. 10.1101/gr.186072.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajta A., Bhatia R., Setia H., Pathania P. (2020). Role of heterotrophic aerobic denitrifying bacteria in nitrate removal from wastewater. J. Appl. Microbiol. 128, 1261–1278. 10.1111/jam.14476 [DOI] [PubMed] [Google Scholar]
- Rezvani F., Sarrafzadeh M. H., Ebrahimi S., Oh H. M. (2019). Nitrate removal from drinking water with a focus on biological methods: a review. Environ. Sci. Pollut. Res. Int. 26, 1124–1141. 10.1007/s11356-017-9185-0 [DOI] [PubMed] [Google Scholar]
- Richter M., Rossello-Mora R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 106, 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout P. R., Bhunia P., Dash R. R. (2017). Simultaneous removal of nitrogen and phosphorous from domestic wastewater using Bacillus cereus GS-5 strain exhibiting heterotrophic nitrification, aerobic denitrification and denitrifying phosphorous removal. Bioresour. Technol. 244, 484–495. 10.1016/j.biortech.2017.07.186 [DOI] [PubMed] [Google Scholar]
- Rudra B., Gupta R. S. (2021). Phylogenomic and comparative genomic analyses of species of the family Pseudomonadaceae: Proposals for the genera Halopseudomonas gen. nov. and Atopomonas gen. nov., merger of the genus Oblitimonas with the genus Thiopseudomonas, and transfer of some misclassified species of the genus Pseudomonas into other genera. Int. J. Syst. Evol. Microbiol. 71, 005011. 10.1099/ijsem.0.005011 [DOI] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Sood U., Hira P., Kumar R., Bajaj A., Rao D. L. N., Lal R., et al. (2019). Comparative genomic analyses reveal core-genome-wide genes under positive selection and major regulatory hubs in outlier strains of Pseudomonas aeruginosa. Front. Microbiol. 10, 53. 10.3389/fmicb.2019.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E., Goebel B. M. (1994). Taxonomic Note: a place for DNA-DNA reassociation and 16s rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44, 846–849. 10.1099/00207713-44-4-846 [DOI] [Google Scholar]
- Stanborough T., Fegan N., Powell S. M., Tamplin M., Chandry P. S. (2018). Vibrioferrin production by the food spoilage bacterium Pseudomonas fragi. FEMS Microbiol. Lett. 365, 6. 10.1093/femsle/fnx279 [DOI] [PubMed] [Google Scholar]
- Stintzi A., Barnes C., Xu L., Raymond K. N. (2000). Microbial iron transport via a siderophore shuttle: a membrane ion transport paradigm. Proc. Natl. Acad. Sci. U. S. A. 97, 10691–10696. 10.1073/pnas.200318797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Wang Y., Zang T., wei J., Wu H., Wei C., et al. (2019). A biosurfactant-producing Pseudomonas aeruginosa S5 isolated from coking wastewater and its application for bioremediation of polycyclic aromatic hydrocarbons. Bioresour. Technol. 281, 421–428. 10.1016/j.biortech.2019.02.087 [DOI] [PubMed] [Google Scholar]
- Tindall B., Sikorski J., Smibert R., Krieg N. (2007). “Phenotypic characterization and the principles of comparative systematics,” in Methods for General and Molecular Microbiology, eds C. Reddy, T. Beveridge, J. Breznak, G. Marzluf, T. Schmidt, and L. Snyder, (Washington, DC: ASM Press; ), 330–393. [Google Scholar]
- Wang J. W., Cai M., Nie Y., Hu B., Yang Y., Wu X. L. (2020). Pseudomonas jilinensis sp. nov., isolated from oil production water of Jilin Oilfield in China. Curr. Microbiol. 77, 688–694. 10.1007/s00284-019-01798-2 [DOI] [PubMed] [Google Scholar]
- Wang T., Wang H., Chang Y., Chu Z., Zhao Y., Liu R. (2018). Enhanced nutrients removal using reeds straw as carbon source in a laboratory scale constructed wetland. Int. J. Environ. Res. Public Health 15, 1081. 10.3390/ijerph15061081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang W., Zhang Y., Sun Z., Zhang J., Chen G., et al. (2019). Simultaneous nitrification and denitrification by a novel isolated Pseudomonas sp. JQ-H3 using polycaprolactone as carbon source. Bioresour. Technol. 288, 121506. 10.1016/j.biortech.2019.121506 [DOI] [PubMed] [Google Scholar]
- Wu Z., Xu F., Yang C., Su X., Guo F., Xu Q., et al. (2019). Highly efficient nitrate removal in a heterotrophic denitrification system amended with redox-active biochar: a molecular and electrochemical mechanism. Bioresour. Technol. 275, 297–306. 10.1016/j.biortech.2018.12.058 [DOI] [PubMed] [Google Scholar]
- Xie E., Ding A., Zheng L., Dou J., Anderson B., Huang X., et al. (2016). Screening and characterizing a denitrifying phosphorus-accumulating bacterium isolated from a circular plug-flow reactor. Environ. Technol. 37, 2823–2829. 10.1080/09593330.2016.1167247 [DOI] [PubMed] [Google Scholar]
- Xie F., Thiri M., Wang H. (2021). Simultaneous heterotrophic nitrification and aerobic denitrification by a novel isolated Pseudomonas mendocina X49. Bioresour. Technol. 319, 124198. 10.1016/j.biortech.2020.124198 [DOI] [PubMed] [Google Scholar]
- Yamamoto S., Okujo N., Yoshida T., Matsuura S., Shinoda S. (1994). Structure and iron transport activity of vibrioferrin, a new siderophore of Vibrio parahaemolyticus. J. Biochem. 115, 868–874. 10.1093/oxfordjournals.jbchem.a124432 [DOI] [PubMed] [Google Scholar]
- Yang Z., Sun H., Zhou Q., Zhao L., Wu W. (2020). Nitrogen removal performance in pilot-scale solid-phase denitrification systems using novel biodegradable blends for treatment of waste water treatment plants effluent. Bioresour. Technol. 305, 122994. 10.1016/j.biortech.2020.122994 [DOI] [PubMed] [Google Scholar]
- Yoon S. H., Ha S. M., Kwon S., Lim J., Kim Y., Seo H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. 10.1099/ijsem.0.001755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Feng J., Chen S., Zhao Z., Li B., Wang Y., et al. (2019). Geographical patterns of nirS gene abundance and nirS-type denitrifying bacterial community associated with activated sludge from different wastewater treatment plants. Microb. Ecol. 77, 304–316. 10.1007/s00248-018-1236-7 [DOI] [PubMed] [Google Scholar]
- Zhang M., Li A., Xu S., Chen M., Yao Q., Xiao B., et al. (2021). Sphingobacterium micropteri sp. nov. and Sphingobacterium litopenaei sp. nov., isolated from aquaculture water. Int. J. Syst. Evol. Microbiol. 71, 005091. 10.1099/ijsem.0.005091 [DOI] [PubMed] [Google Scholar]
- Zhang M., Li A., Yao Q., Wu Q., Zhu H. (2020). Nitrogen removal characteristics of a versatile heterotrophic nitrifying-aerobic denitrifying bacterium, Pseudomonas bauzanensis DN13-1, isolated from deep-sea sediment. Bioresour. Technol. 305, 122626. 10.1016/j.biortech.2019.122626 [DOI] [PubMed] [Google Scholar]
- Zhang R. C., Chen C., Wang W., Shao B., Xu X. J., Zhou X., et al. (2020). The stimulating metabolic mechanisms response to sulfide and oxygen in typical heterotrophic sulfide-oxidizing nitrate-reducing bacteria Pseudomonas C27. Bioresour. Technol. 309, 123451. 10.1016/j.biortech.2020.123451 [DOI] [PubMed] [Google Scholar]
- Zhang S., Su J., Zheng Z., Yang S. (2020). Denitrification strategies of strain YSF15 in response to carbon scarcity: based on organic nitrogen, soluble microbial products and extracellular polymeric substances. Bioresour. Technol. 314, 123733. 10.1016/j.biortech.2020.123733 [DOI] [PubMed] [Google Scholar]
- Zhao B., Cheng D. Y., Tan P., An Q., Guo J. S. (2018). Characterization of an aerobic denitrifier Pseudomonas stutzeri strain XL-2 to achieve efficient nitrate removal. Bioresour. Technol. 250, 564–573. 10.1016/j.biortech.2017.11.038 [DOI] [PubMed] [Google Scholar]
- Zheng Z., Ali A., Su J., Huang T., Wang Y., Zhang S. (2021). Fungal pellets immobilized bacterial bioreactor for efficient nitrate removal at low C/N wastewater. Bioresour. Technol. 332, 125113. 10.1016/j.biortech.2021.125113 [DOI] [PubMed] [Google Scholar]
- Zou Y., He S., Sun Y., Zhang X., Liu Y., Cheng Q. (2019). Pseudomonas urumqiensis sp. nov., isolated from rhizosphere soil of Alhagi sparsifolia. Int. J. Syst. Evol. Microbiol. 69, 1760–1766. 10.1099/ijsem.0.003390 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.