Abstract
Small-subunit rRNA sequences were obtained for two saturated fatty acid-β-oxidizing syntrophic bacteria, Syntrophomonas sapovorans and Syntrophomonas wolfei LYB, and sequence analysis confirmed their classification as members of the family Syntrophomonadaceae. S. wolfei LYB was closely related to S. wolfei subsp. wolfei, but S. sapovorans did not cluster with the other members of the genus Syntrophomonas. Five oligonucleotide probes targeting the small-subunit rRNA of different groups within the family Syntrophomonadaceae, which contains all currently known saturated fatty acid-β-oxidizing syntrophic bacteria, were developed and characterized. The probes were designed to be specific at the family, genus, and species levels and were characterized by temperature-of-dissociation and specificity studies. To demonstrate the usefulness of the probes for the detection and quantification of saturated fatty acid-β-oxidizing syntrophic bacteria in methanogenic environments, the microbial community structure of a sample from a full-scale biogas plant was determined. Hybridization results with probes for syntrophic bacteria and methanogens were compared to specific methanogenic activities and microbial numbers determined with most-probable-number estimates. Most of the methanogenic rRNA was comprised of Methanomicrobiales rRNA, suggesting that members of this order served as the main hydrogen-utilizing microorganisms. Between 0.2 and 1% of the rRNA was attributed to the Syntrophomonadaceae, of which the majority was accounted for by the genus Syntrophomonas.
In full-scale biogas plants in Denmark, manure is digested along with industrial organic waste (IOW) (2). Since digestion of lipids results in a higher methane yield than digestion of carbohydrates and proteins, biogas plants often use IOW that is rich in lipids (2). The degradation of lipids results in the production of fatty acids through the hydrolysis of triglycerides. Subsequently, saturated fatty acids are degraded by proton-reducing acetogens in syntrophic association with hydrogen-utilizing methanogens and aceticlastic methanogens (3, 8, 31, 36). Thus, sufficient levels of saturated fatty acid-β-oxidizing syntrophs should be present in biogas reactors to maximize the consumption of butyrate and higher fatty acids.
Only a limited number of saturated fatty acid-degrading syntrophs (SFAS) have been isolated (either in pure culture or in defined binary or ternary mixed cultures) and characterized (31). They all group into a separate cluster within the phylum consisting of gram-positive organisms and have been placed in the family Syntrophomonadaceae based on comparative small-subunit (SSU) rRNA sequence analysis (42, 43). This family currently contains three genera, Syntrophomonas, Syntrophospora, and Thermosyntropha, as well as two closely related isolates, strains FSM2 and FSS7 (19, 36, 42, 43). Even though SSU rRNA sequences are available for strains FSM2 and FSS7, these isolates were lost before they were deposited in a culture collection (43). SFAS are extremely fastidious organisms (31), with doubling times ranging from 40 h for Syntrophomonas sapovorans (29) to 90 h for Syntrophomonas wolfei subsp. wolfei (21) grown under optimal conditions with butyrate as a substrate. Their low growth rates and syntrophic associations make the characterization of anaerobic microbial consortia particularly challenging.
Previously, oligonucleotide probes targeting the SSU rRNA of phylogenetic groups of methanogens (26), which are also fastidious and sometimes difficult to culture, were used to determine the abundance of and to study the population dynamics of these microorganisms in anaerobic digesters (16, 27). With laboratory-scale digesters, it was demonstrated that the combined application of oligonucleotide probes and traditional performance measurements can result in an improved understanding of the operation of engineered reactor systems (16). In this study, we obtained SSU rRNA sequences for S. wolfei LYB and S. sapovorans, and we designed and characterized oligonucleotide probes for different phylogenetic groups of the mesophilic members of the family Syntrophomonadaceae. The new probes, as well as previously designed probes, were used to characterize the microbial community in an anaerobic biogas reactor. These hybridization results were compared with specific methanogenic activities and the numbers of various microbial groups determined by traditional approaches (most-probable-number [MPN] estimates).
MATERIALS AND METHODS
Organisms, culture techniques, and nucleic acid extractions.
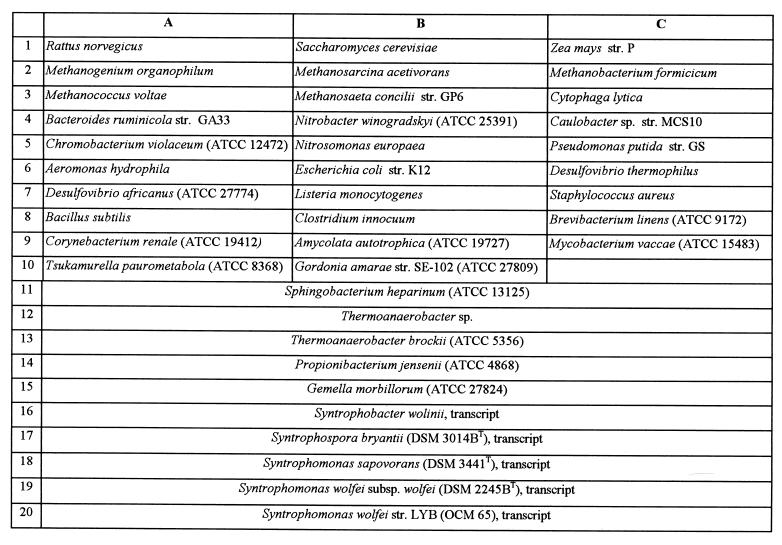
The organisms used in this study are listed in Fig. 1. Most organisms were obtained from the American Type Culture Collection (Manassas, Va.) and the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany). Additional strains were obtained from various collections at the University of Illinois at Urbana-Champaign and the Oregon Collection of Methanogens.
FIG. 1.
Membrane template used for the specificity study (Fig. 4). OCM, Oregon Collection of Methanogens; DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH; ATCC, American Type Culture Collection.
Cultures were grown as recommended by the respective culture collections and were harvested while the cells were in the exponential growth phase (25 ml of culture was centrifuged [2,040 × g, 10 min, 4°C] and the supernatant was discarded). Gram-negative cells were lysed with lysozyme and guanidine isothiocyanate lysing buffer, and DNA was extracted by a phenol-chloroform extraction procedure (30). DNA was extracted from gram-positive cells by a microwave protocol involving a phenol-chloroform-isoamyl alcohol extraction step (11). Total RNA was extracted from pure cultures and from a full-scale biogas reactor sample by a low-pH hot-phenol extraction protocol similar to the one described by Stahl et al. (34), except that samples were “bead-beaten” in intervals of 2 min instead of 3 min. The quality of the extracted RNA was evaluated by polyacrylamide gel electrophoresis (4) and quantified with BIT image analysis software (Bioimaging Technologies, Elburn, Ill.).
Cloning, sequencing, and in vitro transcription.
SSU rRNA gene amplifications were performed in 50-μl reaction mixtures with 1× PCR buffer (Gibco BRL, Gaithersburg, Md.), 1 to 100 ng of DNA, 10 pmol of each primer, 2 μl of 50 mM MgCl2, 2 μl of each deoxynucleoside triphosphate at 1.25 mM (Promega Corp., Madison, Wis.), and 1.25 U of Taq polymerase (Gibco BRL). The PCR was carried out in a PTC-200 thermocycler (MJ Research Inc., Watertown, Mass.) with an initial cycle of 4 min at 95°C and then 30 cycles of 1 min at 92°C, 1 min at 50°C, and 1 min at 72°C. After the last cycle, the mixtures were kept at 72°C for an additional 5 min before termination of the reaction. The following primer pair was used: S-D-Bact-0011-a-S-17 (GTTTGATCCTGGCTCAG) and S-D-Bact-1492-a-A-21 (ACGGYTACCTTGTTACGACTT) (5, 17) or S-D-Bact-0008-a-S-21 (CAGAGTTTGATCCTGGCTCAG) and S-∗-Univ-1508-a-A-21 (ACGGCTACCTTGTTACGACTT) (40). The purity of the PCR product was evaluated by 0.7% agarose gel electrophoresis. Pure PCR product was ligated into a TA cloning vector (pCR2.1) with a TA cloning kit (Invitrogen Corporation, San Diego, Calif.). Cell pellets were prepared from an overnight growth of transformed Escherichia coli cells. One each of the clones containing the SSU rRNA genes of S. sapovorans and S. wolfei LYB was sequenced with four primers specific for the bacterial domain, S-∗-Bact-0343-a-S-15 (TACGGGAGGCAGCAG), S-∗-Bact-0519-a-A-18 (GTATTACCGCGGCTGCTG), S-∗-Bact-0907-a-S-20 (AAACTCAAATGAATTGACGG), and S-∗-Bact-1100-a-A-16 (AGGGTTGCGCTCGTTG) (12), and the M13(−20) forward and M13 reverse primers (Invitrogen Corporation). An additional clone each for S. sapovorans and S. wolfei LYB was sequenced with the M13 primer set, S-∗-Bact-0343-a-S-15, and S-∗-Bact-1100-a-A-16. Sequencing was performed by the University of Illinois Biotechnology Center, Genetic Engineering Facility (Urbana).
Clones of S. wolfei subsp. wolfei and Syntrophospora bryantii were partially sequenced [with only the M13(−20) forward primer] to ensure that the gene was inserted in the right direction for subsequent in vitro transcription. SSU rRNA transcripts were produced in vitro with purified linearized plasmid and T7 RNA polymerase from the Ampliscribe transcription kit (Epicentre Technologies, Madison, Wis.) (22).
Phylogenetic analyses.
Alignment analyses were performed with the sequences available for the members of the Syntrophomonadaceae with the CLUSTAL W program (version 1.6) (38). The sequences were further aligned by hand and gaps and unknown bases were not considered, resulting in 1,081 nucleotides per sequence. A phylogenetic analysis was performed with DNAML, a maximum-likelihood program available in the PHYLIP package, with a transition/transversion rate set at 2.000 (13).
Design and characterization of oligonucleotide probes.
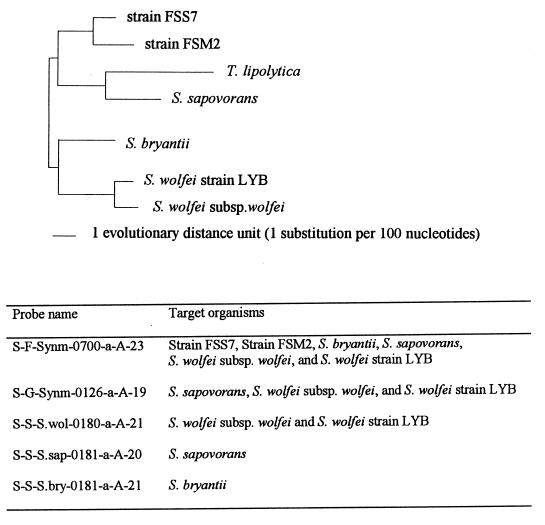
The SSU rRNA sequences of S. wolfei LYB and S. sapovorans (obtained in this study), of S. bryantii and S. wolfei subsp. wolfei (available in the Ribosomal Database Project [20]), and of strains FSM2 and FSS7 (kindly provided by Carl R. Woese) were used for oligonucleotide probe design. Five probes were designed to target the mesophilic members of the family Syntrophomonadaceae at different levels of specificity. The probes and their target groups are shown in Fig. 2 and 3. The probes were synthesized with a DNA synthesizer (Applied Biosystems, Foster City, Calif.) at the University of Illinois Biotechnology Center, Genetic Engineering Facility, and purified with an oligonucleotide purification cartridge (Applied Biosystems), and the 5′ ends were labeled with [γ-32P]ATP (ICN Radiochemicals, Irvine, Calif.) by bacteriophage T4 polynucleotide kinase (Promega Corp.) (26). A universal base analogue, 5-nitroindole (N5) (Glen Research, Sterling, Va.) (18), was incorporated during the synthesis of one of the probes (S-F-Synm-0700-a-A-23).
FIG. 2.
Unrooted phylogenetic tree for the family Syntrophomonadaceae, inferred from comparisons of SSU rRNA sequences with a maximum-likelihood algorithm. The tree was constructed with the evolutionary distances shown in Table 2.
FIG. 3.
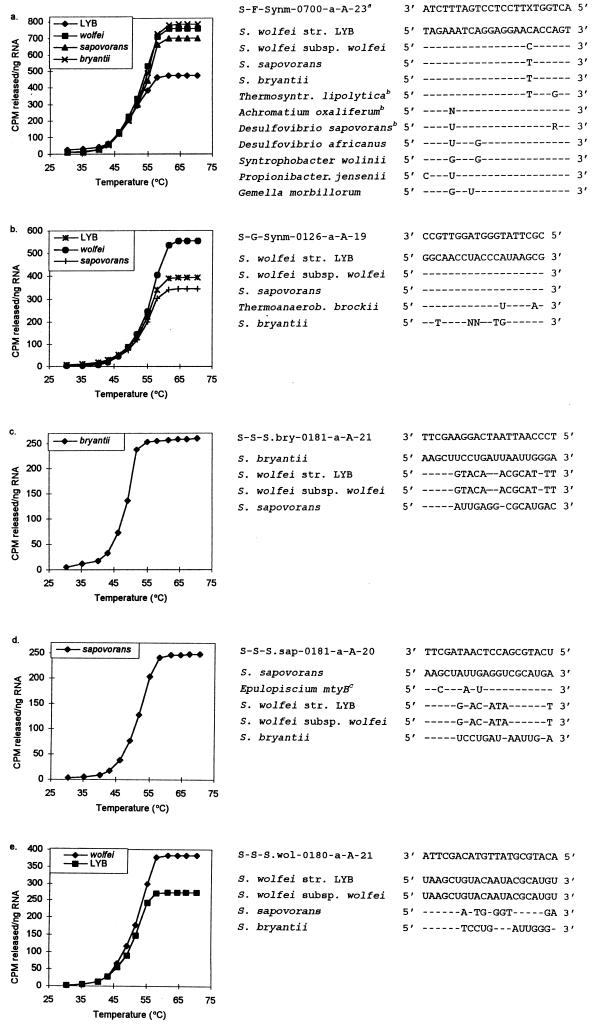
Td studies for probes S-F-Synm-0700-a-A-23, S-G-Synm-0126-a-A-19, S-S-S.bry-0181-a-A-21, S-S-S.sap-0181-a-A-20, and S-S-S.wol-0180-a-A-21. Adjacent to the probe dissociation results are SSU rRNA sequences of target and nontarget species and probe sequences. The top SSU rRNA sequences for each list of organisms are those of the target organisms. Dashes in the succeeding sequences signify identical nucleotides. A superscript a indicates that an unlabeled version of the competitive probe was used with the labeled probe; X represents the universal base analogue N5. A superscript b indicates organisms that were not included in the experimental evaluation of probe specificity. A superscript c indicates that the organism was not included in the experimental evaluation of probe specificity due to three significant mismatches.
Temperature-of-dissociation (Td) curves were determined for the five oligonucleotide probes with various target and nontarget SSU rRNAs, according to the elution method described by Zheng et al. (44). Subsequently, the posthybridization wash temperature (Tw) was determined by the replicate-slot Td method (44) for four temperatures around the previously determined Tds.
The specificities of the five oligonucleotide probes and universal probe S-∗-Univ-1390-a-A-18 (44) were evaluated by applying approximately 30 ng of total RNA extracted from 39 organisms, representing a wide phylogenetic diversity (Fig. 1), to six Magna Charge membranes (Micron Separations, Inc., Westboro, Mass.). Prehybridization, hybridization, wash, and imaging procedures were described previously (25, 44). A Tw of 44°C was used for the universal probe (44), and the Tws used for the five probes designed in this study are listed in Table 1.
TABLE 1.
Experimentally determined Tds and posthybridization Tws used in quantitative membrane hybridizations
| Probe | Tw (°C) | Trial no.a |
Td (°C)
|
|||
|---|---|---|---|---|---|---|
| S. wolfei LYB | S. wolfei subsp. wolfei | S. sapovorans | S. bryantii | |||
| S-F-Synm-0700-a-A-23 | 54 | 1 | 51.0 | 52.0 | NDc | ND |
| 2 | 51.2 | 52.0 | 52.9 | 52.9 | ||
| 3 | 50.1 | 52.5 | 53.0 | 53.4 | ||
| S-G-Synm-0126-a-A-19 | 53 | 1 | 55.2 | ND | ND | NTb |
| 2 | 54.7 | 54.1 | 54.6 | NT | ||
| 3 | 55.4 | 55.8 | 54.1 | NT | ||
| S-S-S.bry-0181-a-A-21 | 48 | 1 | NT | NT | NT | 53.0 |
| 2 | NT | NT | NT | 49.4 | ||
| 3 | NT | NT | NT | 48.8 | ||
| S-S-S.sap-0181-a-A-20 | 50 | 1 | NT | NT | ND | NT |
| 2 | NT | NT | 51.6 | NT | ||
| 3 | NT | NT | 51.0 | NT | ||
| S-S-S.wol-0180-a-A-21 | 50 | 1 | 50.5 | ND | NT | NT |
| 2 | 52.5 | 52.5 | NT | NT | ||
| 3 | 52.1 | 51.5 | NT | NT | ||
Most Td studies were performed three times.
NT, nontarget organism.
ND, not determined.
Anaerobic biogas reactor analyses.
A 2-liter sample was collected from the full-scale mesophilic biogas reactor in Hashøj, Denmark (37). The biogas plant consists of a single continuously stirred tank reactor of 3,000 m3 and is operated at a retention time of approximately 30 days. The plant treats a mixture of swine and cattle manure and a variety of IOW streams. Analyses of volatile solids (VS) and total solids were performed according to standard methods (15). Volatile fatty acids (VFAs) were determined with a gas chromatograph (model 5890; Hewlett-Packard, Palo Alto, Calif.) equipped with a flame ionization detector. Specific methanogenic activities (SMA) were determined with BA medium (9) containing acetate, hydrogen and carbon dioxide, butyrate, and propionate as substrates (32, 33). The SMA tests were initiated less than 5 h after sampling.
Triplicate MPN determinations were performed in BA medium (9) with glucose, butyrate, propionate, acetate, and hydrogen as substrates. Furthermore, sulfate-reducing bacteria were enumerated by using Postgate medium, which contains lactate as the substrate (24).
Quantitative membrane hybridizations were performed by triplicate applications of 30 ng of denatured RNA, extracted from the sample obtained from the biogas reactor, to Magna Charge membranes (Micron Separations, Inc.) in combination with dilution series of pure-culture RNA samples (28). Hybridizations with 32P-labeled oligonucleotide probes were performed as previously described (25), and hybridization results were analyzed with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) (23, 44). In addition to the five probes designed in this study, the following SSU rRNA-targeted probes were used, with the Tws given in parentheses: S-∗-Univ-1390-a-A-18 (44°C), S-D-Euca-0502-a-A-16 (58°C), S-D-Bact-0338-a-A-18 (54°C), S-D-Arch-0915-a-A-20 (58°C), S-O-Msar-0860-a-A-21 (60°C), S-G-Msar-0821-a-A-21 (60°C), S-F-Mcoc-1109-a-A-20 (55°C), S-F-Mbac-310-a-A-22 (57°C), S-O-Mmic-1200-a-A-21 (53°C), and S-S-M.con-0381-a-A-22 (56°C [45]). Original citations for the first nine probes can be found in the Oligonucleotide Probe Database (5). The Tws for the five probes designed in this study are listed in Table 1.
Nucleotide sequence accession numbers.
Nucleotide sequences have been deposited in the GenBank database under the accession no. AF022248 for S. wolfei LYB and AF022249 for S. sapovorans.
RESULTS AND DISCUSSION
Sequence analysis.
The phylogenetic analysis of the SSU rRNA sequences confirmed that S. sapovorans and S. wolfei LYB, the sequences of which were obtained in this study, are members of the family Syntrophomonadaceae (Fig. 2 and Table 2). As expected, S. wolfei LYB is closely related to S. wolfei subsp. wolfei (evolutionary distance, 1.88 [Table 2]). However, the alignment of the rRNA sequence for S. sapovorans indicates that this strain does not cluster with the other members of the genus Syntrophomonas (Fig. 2) and that the evolutionary distance between S. sapovorans and the two other members of the Syntrophomonas genus is about 9 (Table 2). S. sapovorans is somewhat more closely related to one (strain FSS7) of the two uncharacterized members of the family Syntrophomonadaceae and to the recently characterized thermophilic syntrophic species Thermosyntropha lipolytica. Based on our phylogenetic analysis, we suggest that S. sapovorans be considered a representative of a new genus.
TABLE 2.
Evolutionary distances based on SSU rRNA sequences of Syntrophomonadaceae speciesa
| Organism | Evolutionary distance from organism
|
|||||
|---|---|---|---|---|---|---|
| S. wolfei subsp. wolfei | S. wolfei LYB | S. bryantii | S. sapovorans | T. lipolytica | Strain FSS7 | |
| S. wolfei LYB | 1.88 | |||||
| S. bryantii | 6.37 | 6.17 | ||||
| S. sapovorans | 9.23 | 9.03 | 8.33 | |||
| T. lipolytica | 11.6 | 11.4 | 10.7 | 7.47 | ||
| Strain FSS7 | 7.12 | 6.92 | 6.22 | 7.38 | 9.75 | |
| Strain FSM2 | 9.16 | 8.96 | 8.26 | 9.42 | 11.8 | 4.11 |
The distances were generated by a maximum-likelihood algorithm, with a perfect alignment of 1,081 nucleotides.
Design of oligonucleotide probes.
Probe design was based on the previously published phylogeny of the Syntrophomonadaceae (43) and Syntrophomonas genus characterization (19). Probes targeting each of the three mesophilic Syntrophomonadaceae species were designed: S-S-S.bry-0181-a-A-21 (S. bryantii), S-S-S.sap-0181-a-A-20 (S. sapovorans), and S-S-S.wol-0180-a-A-21 (S. wolfei) (Fig. 2). The specificities of the oligonucleotide probes were determined with CHECK_PROBE software provided through the Ribosomal Database Project (20) and the Oligonucleotide Probe Database (5) and with BLAST software available from the National Center for Biotechnology Information (6). Figure 3 shows sequence alignments of the probes with target organisms and a few nontarget organisms. The probes were found to have at least three mismatches with the nearest nontarget SSU rRNA. The three probes target a region of the SSU rRNA (positions 180 to 201, according to E. coli numbering) which corresponds to the target region of a number of probes that were previously used successfully for whole-cell hybridizations (7), indicating that the species-specific probes designed in this study will likely perform well for whole-cell hybridizations.
A probe targeting all currently known species of the genus Syntrophomonas, including S. sapovorans, was also designed (S-G-Synm-0126-a-A-19) (Fig. 2). This probe perfectly matched the SSU rRNA of all target organisms and had at least two mismatches with the nearest nontarget organisms (Fig. 3).
Finally, a probe targeting all mesophilic members of the family Syntrophomonadaceae (S-F-Synm-0700-a-A-23) was designed (Fig. 2). At position 716 (according to E. coli numbering), the SSU rRNA of the target organisms contains a C or a T (Fig. 3). In order to ensure equal specificity of the probe to all target organisms, a universal base analogue, N5, was used in this location (18). This approach was previously successful for the design of an SSU rRNA-targeted probe for mycolic acid-containing actinomycetes (10). Probe S-F-Synm-0700-a-A-23 has one T-G mismatch (position 720, according to E. coli numbering) with the recently described thermophilic member of the Syntrophomonadaceae T. lipolytica. Since a T-G mismatch is considered to have little effect on hybridization (39), it is likely that the probe also binds to T. lipolytica (this was not evaluated experimentally as part of this study). Consequently, probe S-F-Synm-0700-a-A-23 may be a family-specific probe for all presently described members of the Syntrophomonadaceae. Furthermore, probe S-F-Synm-0700-a-A-23 may have only one mismatch with the nontarget organism Achromatium oxaliferum (Fig. 3). This organism has been isolated from freshwater sediments and appears to have a role in the oxidation of reduced sulfur species to sulfate (14). Therefore, it is unlikely that A. oxaliferum plays a role in anaerobic reactors. The SSU rRNA of Desulfovibrio sapovorans has one or two mismatches with the family-specific probe (Fig. 3). In order to eliminate the potential binding of this probe to nontarget species, a competitive probe targeting D. sapovorans was designed (S-F-Synm-0700-p-A-23 [ACTGGTN5TTCCTCCTGATATCTA]). An unlabeled version of this probe was always used with the labeled probe S-F-Synm-0700-a-A-23 in a 1:1 ratio.
Optimization of Tws.
Since the oligonucleotide probes designed in this study were to be used to quantify the SSU rRNA extracted from complex microbial systems, it was necessary to determine the optimal Tw experimentally. The Td, or the temperature at which 50% of the oligonucleotide remains bound to a perfect target (39), is generally used as the posthybridization Tw to ensure the dissociation of duplexes with one or more mismatches (35). Figure 3 shows representative Td curves for the five probes and sequence alignments of the probes with the target and nontarget organisms used in the Td studies. The experimentally determined Tds are summarized in Table 1. The Tds of the five oligonucleotides ranged from 49 to 55°C. Probe S-F-Synm-0700-a-A-23 demonstrated the largest variation in Td among different target organisms (50 to 53°C) (Table 1). This variation can be explained by small differences in the duplex stability for the various target SSU rRNAs due to the incorporation of N5 into this probe (18).
Tds determined by the elution method (which involves a 10-min wash) are typically slightly higher than those obtained with a Td experiment involving a 30-min wash (44). Since the posthybridization wash step during routine quantitative hybridizations lasts 30 min (44), a hybridization experiment was performed to determine the optimal Tws. This experiment involved the use of four different posthybridization Tws close to the experimentally determined Tds. Tws were selected such that the probes bound only to the desired targets while maintaining as much of the hybridization signals as possible. The resulting Tws (Table 1) were generally 1 to 2°C below the Tds, except for that of probe S-F-Synm-0700-a-A-23.
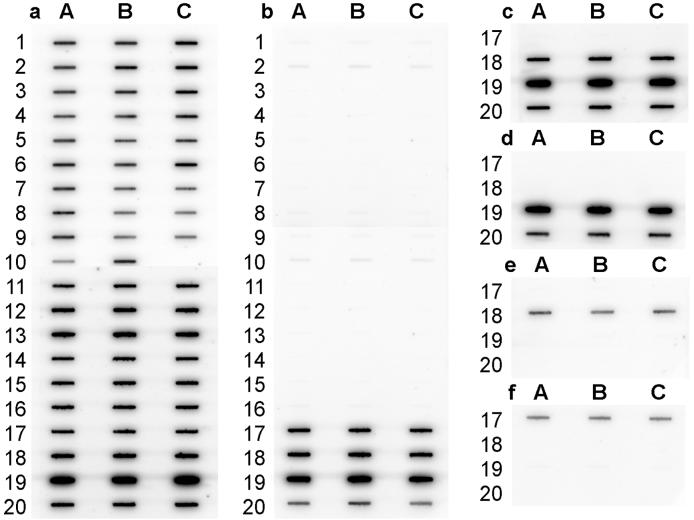
Probe specificity studies.
RNA samples obtained from a diverse selection of 39 target and nontarget organisms (Fig. 1) were hybridized with a universal probe and the five newly designed probes to assess probe specificities. Figure 4 shows the hybridization membranes for this specificity study. The hybridization result obtained with the universal probe, S-∗-Univ-1390-a-A-18 (Fig. 4a), indicates that target and nontarget RNAs were applied to the membranes at approximately the same levels. Hybridization results for the five newly designed probes show that they bound only to their target RNA (Fig. 4b to f), as expected from database searches. The hybridization signal of probe S-F-Synm-0700-a-A-23 for the SSU rRNA of S. wolfei LYB was slightly lower than the response obtained for the SSU rRNA of the other members of the family Syntrophomonadaceae (Fig. 4b). This observation was in accordance with the experimentally determined Tds (Table 1).
FIG. 4.
Results of the probe specificity study. Membrane hybridization results were analyzed with a PhosphorImager and were scanned and printed with Adobe Photoshop 3.0 (Adobe, Seattle, Wash.). (a) Probe S-∗-Univ-1390-a-A-18; (b) probe S-F-Synm-0700-a-A-23; (c) probe S-G-Synm-0126-a-A-19; (d) probe S-S-S.bry-0181-a-A-21; (e) probe S-S-S.sap-0181-a-A-20; (f) probe S-S-S.wol-0180-a-A-21.
Anaerobic biogas reactor analyses.
Influent and effluent grab samples were obtained from the full-scale mesophilic biogas reactor in Hashøj, Denmark. The reactor influent, which consisted of a mixture of swine and cattle manure and a variety of IOW streams, had a VS content of 79 kg/m3. The VS level of the reactor effluent was 53 kg/m3, corresponding to a 33% removal of VS. The plant produced 0.38 m3 of biogas per kg of VS, with a methane content of 70%. Acetate, propionate, butyrate, and isobutyrate levels in the effluent were low (Table 3), indicating that the reactor exhibited stable performance.
TABLE 3.
Concentrations of VFAs in samples of influent and effluent of the full-scale mesophilic biogas reactor in Hashøj, Denmark
| Source of sample | Concn (mg/liter) of:
|
|||
|---|---|---|---|---|
| Acetate | Propionate | Butyrate | Isobutyrate | |
| Influent | 101 | 50 | 4 | 35 |
| Effluent | 53 | 9 | 1 | 5 |
Fifteen different oligonucleotide probes targeting the SSU rRNA of the three domains, phylogenetically defined groups of methanogens, and the SFAS were used in quantitative membrane hybridizations with RNA extracted from a sample obtained from the biogas reactor. Only seven of the probes resulted in a hybridization response above the detection limit. Detection limits varied with probe labeling efficiency, the amount of nucleic acid applied to the hybridization membrane, and the length of exposure of the hybridization membranes and are therefore different for each sample and probe. In general, the detection limits for our standard hybridization protocol varied between 0.1 and 0.2%. The values obtained with the domain-specific probes correspond with previous studies of anaerobic digester microbial communities. For example, the archaeal (methanogen) rRNA level of 13% ± 2% (mean ± standard deviation, determined with probe S-D-Arch-0915-a-A-20) corresponds well with the archaeal rRNA concentration of 8 to 12% for stable mesophilic sewage sludge digesters (27). The levels of SSU rRNA were low for the Eucarya (0.3% ± 0.2%) and high for the Bacteria (79% ± 3%) (determined with probes S-D-Euca-0502-a-A-16 and S-D-Bact-0338-a-A-18, respectively), which is consistent with values obtained in other digester studies (16, 27). The combined use of the domain-specific probes targeted most of the rRNA present in the biogas reactor sample. The sum of the percentages obtained with the three domains was 92% ± 4%, which is close to the expected value of 100%.
The results obtained with the various methanogenic probes indicate that most of the methanogenic SSU rRNA was comprised of Methanomicrobiales SSU rRNA (probe S-O-Mmic-1200-a-A-21 detected 10% ± 1% of the total SSU rRNA). Thus, members of the Methanomicrobiales were the dominant hydrogenotrophic methanogens and may have served as the hydrogen-utilizing partners of the Syntrophomonadaceae in the biogas reactor. A small but significant number of Syntrophomonadaceae was present (probe S-F-Synm-0700-a-A-23 targeted 0.2 to 1% of the SSU rRNA). The only specific probe that resulted in a detectable signal was probe S-G-Synm-0126-a-A-19 (0.2 to 1%), indicating that species of the genus Syntrophomonas were likely responsible for butyrate degradation in the biogas reactor. Methanosarcinales were the only other methanogens that were present at detectable levels (0.6% ± 0.5%). A probe specific for Methanosaeta concilii (S-S-M.con-0381-a-A-22) (45) did not result in a detectable signal. Even though it is possible that other acetate scavengers (e.g., Methanosaeta spp. not detected by probe S-S-M.con-0381-a-A-22) were present in the biogas reactor, it is likely that Methanosarcina spp. served as the aceticlastic methanogens in the biogas reactor. This result is consistent with previous studies, which demonstrated that Methanosarcina spp. were almost always found to be the main acetate utilizers in Danish biogas plants (1). The low acetate level in the reactor (Table 3) was close to the threshold level of mesophilic Methanosarcina spp. (41), indicating that this group performed well in this reactor. The low overall levels of aceticlastic methanogens (Methanosarcinales) and relatively high levels of hydrogenotrophic methanogens (Methanomicrobiales) may also indicate that a syntrophic relationship between an acetate-oxidizing organism and a hydrogenotrophic methanogen was the major route of acetate degradation in this biogas reactor.
The MPN estimates and SMA measurements are summarized in Table 4. In addition, the levels of SSU rRNA obtained for different groups that generally correspond to groups utilizing the different substrates used in the MPN and activity tests are also given in Table 4. The MPN estimates, SMA measurements, and oligonucleotide probe hybridization results correlated well. For example, the hydrogen and carbon dioxide utilizers (hydrogenotrophic methanogens) were found to be the most abundant (or most active) microorganisms by all three methods. The SMA values obtained with butyrate and propionate as the substrates were similar to the SMA measurement obtained with the control system, to which no external substrate was added. Since butyrate- and propionate-degrading organisms have low affinities for their substrates, it is likely that the butyrate and propionate concentrations in the control system were close to their respective substrate saturation constants (32). The relatively low numbers of butyrate-degrading organisms further imply that these bacteria have high metabolic rates, which is in accordance with the low energy yields that can be obtained from the oxidation of VFAs. Finally, it is also possible that bacteria other than the ones detected by our probes are responsible for the transformation of butyrate in this biogas reactor.
TABLE 4.
Results obtained with three different activity tests (MPN estimates, SMA measurements, and quantitative membrane hybridizations) for grab samples taken from the full-scale mesophilic biogas reactor in Hashøj, Denmark
| Substrate(s) or control for MPN and SMA tests and hybridization probesa | MPN (cells/ml) | SMA (mM CH4/h · ml) | % SSU rRNA |
|---|---|---|---|
| Control | 7 ± 2 | ||
| H2, CO2, and Mmic-1200 | 2.4 × 106 | 39 ± 3 | 10 ± 1 |
| Acetate and Msar-0860 | 1.1 × 103 | 14 | 0.6 ± 0.5 |
| Propionate | 1.1 × 104 | 5 ± 1 | NDb |
| Butyrate and Synm-0700 | 3.9 × 104 | 7 ± 1 | 0.2–1.0 |
| Sulfatec | 2.4 × 101 | ND | ND |
Mmic-1200 represents S-O-Mmic-1200-a-A-21, which targets Methanomicrobiales. Msar-0860 represents S-O-Msar-0860-a-A-21, which targets Methanosarcinales. Synm-0700 represents S-F-Synm-0700-a-A-23, which targets Syntrophomonadaceae.
ND, not determined.
MPN determinations were performed in Postgate medium.
ACKNOWLEDGMENTS
We thank Carl Woese for providing the SSU rRNA sequences of strains FSM2 and FSS7, Katherine D. McMahon for assistance with the generation of transcripts, and Daniel B. Oerther and Dandan Zheng for helpful discussions.
This research was funded by grants from the Office of Solid Waste Research (University of Illinois) to L.R. and from the Danish Science Committee, Biotechnology Program, to K.H.H. and B.K.A.
REFERENCES
- 1.Ahring B K. Methanogenesis in thermophilic biogas reactors. Antonie Leeuwenhoek. 1995;67:91–102. doi: 10.1007/BF00872197. [DOI] [PubMed] [Google Scholar]
- 2.Ahring B K, Angelidaki I, Johansen K. Anaerobic treatment of manure together with industrial waste. Water Sci Technol. 1992;25:311–318. [Google Scholar]
- 3.Ahring B K, Westermann P. Thermophilic anaerobic degradation of butyrate by a butyrate-utilizing bacterium in coculture and triculture with methanogenic bacteria. Appl Environ Microbiol. 1987;53:429–433. doi: 10.1128/aem.53.2.429-433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alm, E. W., and D. Stahl. Critical factors influencing the recovery and integrity of rRNA extracted from environmental samples: use of an optimized protocol to measure depth-related biomass distribution in freshwater sediments. Submitted for publication. [DOI] [PubMed]
- 5.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 7.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angelidaki I, Ahring B K. Establishment and characterization of an anaerobic thermophilic (55°C) enrichment culture degrading long-chain fatty acids. Appl Environ Microbiol. 1995;61:2442–2445. doi: 10.1128/aem.61.6.2442-2445.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angelidaki I, Petersen S P, Ahring B K. Effects of lipids on thermophilic anaerobic digestion and reduction of lipid inhibition upon addition of bentonite. Appl Microbiol Biotechnol. 1990;33:469–472. doi: 10.1007/BF00176668. [DOI] [PubMed] [Google Scholar]
- 10.de los Reyes F L, Ritter W, Raskin L. Group-specific small-subunit rRNA hybridization probes to characterize filamentous foaming in activated sludge systems. Appl Environ Microbiol. 1997;63:1107–1117. doi: 10.1128/aem.63.3.1107-1117.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de los Reyes M F, de los Reyes F L, III, Hernandez M, Raskin L. Quantification of Gordona amarae strains in foaming activated sludge and anaerobic digester systems with oligonucleotide hybridization probes. Appl Environ Microbiol. 1998;64:2503–2512. doi: 10.1128/aem.64.7.2503-2512.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorsch M, Stackebrandt E. Some modifications in the procedure of direct sequencing of PCR amplified 16S rDNA. J Microbiol Methods. 1992;16:271–279. [Google Scholar]
- 13.Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- 14.Gray N D, Pickup R W, Jones J G, Head I M. Ecophysiological evidence that Achromatium oxaliferum is responsible for the oxidation of reduced sulfur species to sulfate in a freshwater sediment. Appl Environ Microbiol. 1997;63:1905–1910. doi: 10.1128/aem.63.5.1905-1910.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenberg A E, Clesceri L S, Eaton A D, editors. Standard methods for the examination of water and wastewater. 18th ed. Washington, D.C.: American Public Health Association; 1992. [Google Scholar]
- 16.Griffin M E, McMahon K D, Mackie R I, Raskin L. Methanogenic population dynamics during start-up of anaerobic digesters treating municipal solid waste and biosolids. Biotechnol Bioeng. 1998;57:342–355. doi: 10.1002/(sici)1097-0290(19980205)57:3<342::aid-bit11>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 17.Kane M D, Poulsen L K, Stahl D A. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl Environ Microbiol. 1993;59:682–686. doi: 10.1128/aem.59.3.682-686.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loakes D, Brown D M. 5-Nitroindole as an universal base analogue. Nucleic Acids Res. 1994;22:4039–4043. doi: 10.1093/nar/22.20.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorowitz W H, Zhao H, Bryant M P. Syntrophomonas wolfei subsp. saponavida subsp. nov., a long-chain fatty-acid-degrading, anaerobic, syntrophic bacterium; Syntrophomonas wolfei subsp. wolfei subsp. nov.; and emended descriptions of the genus and species. Int J Syst Bacteriol. 1989;39:122–126. [Google Scholar]
- 20.Maidak B L, Larsen N, McCaughey M J, Overbeek R, Olsen G J, Forgel K, Blanby J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McInerney M J, Bryant M P, Hespell R B, Costerton J W. Syntrophomonas wolfei gen. nov. sp. nov., an anaerobic, syntrophic, fatty acid-oxidizing bacterium. Appl Environ Microbiol. 1981;41:1029–1039. doi: 10.1128/aem.41.4.1029-1039.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMahon K D, Stahl D A, Raskin L. A comparison of the use of in vitro-transcribed and native rRNA for the quantification of microorganisms in the environment. Microb Ecol. 1998;36:362–371. doi: 10.1007/s002489900122. [DOI] [PubMed] [Google Scholar]
- 23.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflorae. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 24.Postgate J R. Versatile medium for the enumeration of sulfate-reducing bacteria. Appl Microbiol. 1963;11:265–267. doi: 10.1128/am.11.3.265-267.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raskin L, Poulsen L K, Noguera D R, Rittmann B E, Stahl D A. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl Environ Microbiol. 1994;60:1241–1248. doi: 10.1128/aem.60.4.1241-1248.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raskin L, Zheng D, Griffin M E, Stroot P G, Misra P. Characterization of microbial communities in anaerobic bioreactors using molecular probes. Antonie Leeuwenhoek. 1995;68:297–308. doi: 10.1007/BF00874140. [DOI] [PubMed] [Google Scholar]
- 28.Raskin L, Capman W C, Sharp R, Poulsen L K, Stahl D A. Molecular ecology of gastrointestinal ecosystems. In: Mackie R I, White B A, Isaacson R E, editors. Gastrointestinal microbiology. 2. Gastrointestinal microbes and host interactions. New York, N.Y: Chapman & Hall; 1997. pp. 243–298. [Google Scholar]
- 29.Roy F, Samain E, Dubourguir H C, Albagnac G. Syntrophomonas sapovorans sp. nov., a new obligately proton reducing anaerobe oxidizing saturated and unsaturated long chain fatty acids. Arch Microbiol. 1986;145:142–147. [Google Scholar]
- 30.Saunders N A, Harrison T G, Haththotuwa A, Kachwalla N, Taylor A G. A method for typing strains of Legionella pneumophila serogroup 1 by analysis of restriction fragment length polymorphism. J Med Microbiol. 1990;31:45–55. doi: 10.1099/00222615-31-1-45. [DOI] [PubMed] [Google Scholar]
- 31.Schink B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev. 1997;61:262–280. doi: 10.1128/mmbr.61.2.262-280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sørensen A H, Ahring B K. Measurement of the specific methanogenic activity of anaerobic digestor biomass. Appl Microbiol Biotechnol. 1993;40:427–431. [Google Scholar]
- 33.Soto M, Méndez R, Lema J M. Methanogenic and non-methanogenic activity tests. Theoretical basis and experimental set up. Water Res. 1993;27:1361–1376. [Google Scholar]
- 34.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stahl D A, Amann R. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley and Sons, Ltd.; 1991. pp. 205–248. [Google Scholar]
- 36.Svetlitshnyi V, Rainey F, Wiegel J. Thermosyntropha lipolytica gen. nov., sp. nov., a lipolytic, anaerobic, alkalitolerant, thermophilic bacterium utilizing short- and long-chain fatty acids in syntrophic coculture with a methanogenic archaeum. Int J Syst Bacteriol. 1996;46:1131–1137. doi: 10.1099/00207713-46-4-1131. [DOI] [PubMed] [Google Scholar]
- 37.Tafdrup S. Centralized biogas plants combine agricultural and environmental benefits with energy production. Water Sci Technol. 1994;30:133–141. [Google Scholar]
- 38.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tijssen P. Hybridization of nucleic acid probes, part I. Theory and nucleic acid preparation. Amsterdam, The Netherlands: Elsevier Science Publishers B. V.; 1993. p. 268. [Google Scholar]
- 40.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westermann P, Ahring B K, Mah R A. Threshold acetate concentrations for acetate catabolism by aceticlastic methanogenic bacteria. Appl Environ Microbiol. 1989;55:514–515. doi: 10.1128/aem.55.2.514-515.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao H, Yang D, Woese C R, Bryant M P. Assignment of Clostridium bryantii to Syntrophospora bryantii gen. nov., comb. nov. on the basis of a 16S rRNA sequence analysis of its crotonate-grown pure culture. Int J Syst Bacteriol. 1990;40:40–44. doi: 10.1099/00207713-40-1-40. [DOI] [PubMed] [Google Scholar]
- 43.Zhao H, Yang D, Woese C R, Bryant M P. Assignment of fatty acid-β-oxidizing syntrophic bacteria to Syntrophomonadaceae fam. nov. on the basis of 16S rRNA sequence analyses. Int J Syst Bacteriol. 1993;43:278–286. doi: 10.1099/00207713-43-2-278. [DOI] [PubMed] [Google Scholar]
- 44.Zheng D, Alm E W, Stahl D A, Raskin L. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol. 1996;62:4504–4513. doi: 10.1128/aem.62.12.4504-4513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng, D., and L. Raskin. Unpublished data.