Abstract
Artificial light has been increasingly in use for the past 70 years. The aberrant light exposure and round‐the‐clock nature of work lead to the disruption of biological clock. Circadian rhythm disruption (CRD) contributes to multiple metabolic and neurodegenerative diseases. However, its effect on vision is not understood. Moreover, the mammalian retina possesses an autonomous clock that could be reset with light exposure. We evaluated the impact of CRD on retinal morphology, physiology, and vision after housing mice in a disruption inducing shorter light/dark cycle (L10:D10). Interestingly, the mice under L10:D10 exhibited three different entrainment behaviors; “entrained,” “free‐running,” and “zigzagging.” These behavior groups under CRD exhibited reduced visual acuity, retinal thinning, and a decrease in the number of photoreceptors. Intriguingly, the electroretinogram response was decreased only in the mice exhibiting “entrained” behavior. The retinal proteome showed distinct changes with each entrainment behavior, and there was a dysfunctional oxidative stress‐antioxidant mechanism. These results demonstrate that CRD alters entrainment behavior and leads to visual dysfunction in mice. Our studies uniquely show the effect of entrainment behavior on retinal physiology. Our data have broader implications in understanding and mitigating the impact of CRD on vision and its potential role in the etiology of retinal diseases.
Keywords: biological clock, circadian rhythm disruption, entrainment, retina, vision
Abbreviations
- ANOVA
analysis of variance
- BCA
bicinchoninic acid
- BMAL
brain and muscle ARNT‐like 1
- CRD
circadian rhythm disruption
- EIF2
eukaryotic initiation factor
- ERG
electroretinogram
- GPX
glutathione peroxidase
- HPLC
high‐performance liquid chromatography
- IDH3B
isocitrate dehydrogenase (NAD(+)) 3 non‐catalytic subunit beta
- IPA
ingenuity pathway analysis
- L10:D10
10‐hour light: 10‐hour dark
- MT‐CO2
mitochondrially encoded cytochrome C oxidase II
- MT‐CYB
mitochondrially encoded cytochrome B
- MT‐ND6
mitochondrially encoded NADH: ubiquinone oxidoreductase core subunit
- OP
oscillatory potential
- PER
period 1
- PPM1A
protein phosphatase, Mg2+/Mn2+ dependent 1A
- RIPA
radio immuno precipitation assay
- SCN
suprachiasmatic nucleus
- SD‐OCT
spectral domain optical coherent tomography
- SOD1
superoxide dismutase 1
- SOD2
superoxide dismutase 2
- TCA
trichloroacetic acid
- TMT
tandem mass tag
- ZT
zeitgeber time
1. INTRODUCTION
Circadian rhythms are the near 24‐h patterns in physiology and behavior that persist without external stimuli. In mammals, a neuronal network in the suprachiasmatic nucleus (SCN) generates endogenous circadian rhythm and orchestrates a complex system of circadian oscillators throughout the body via behavioral, hormonal, and neuronal signals. 1 The SCN maintains its phase coherence with the environmental light‐dark cycles through neuronal projections from the retina via the retina‐hypothalamic tract. 1 Within the cells of SCN and every other tissue, circadian oscillation is driven by a cell‐autonomous autoregulatory feedback loop of clock genes. 2 Environmental and systemic inputs could feed into this core clock mechanism, 2 possibly modulating its precision and flexibility while adapting to perturbations.
Disruption of this fine‐tuned machinery is widespread in the modern world with the extensive use of artificial lighting, the round‐the‐clock nature of work, and trans meridian travel. Chronic circadian rhythm disruption (CRD) has been identified as a risk factor for multiple metabolic, cardiovascular, and neurodegenerative diseases. 3 , 4 Animal models with clock gene mutations also showed metabolic and neurodegenerative disorders. 3 Moreover, exposure to shorter light‐dark cycles resulted in metabolic and neuropsychiatric alterations in genetically intact mice. 5 , 6
The mammalian retina possesses an autonomous circadian clock that can be entrained by light, 7 , 8 and the retinal clock is also exposed to systemic circadian time cues. Thus, retinal gene expression and function are tightly controlled by the circadian clock and exposure to light. 7 , 9 Moreover, ablation of the clock gene Bmal in mice resulted in alterations of ocular gene expression, reduced electroretinogram (ERG) amplitude, and cone photoreceptor viability. 9 , 10 Furthermore, mutant mice for Bmal or Per2 exhibited changes in their retinal vasculature, suggesting the role of clock genes in the pathogenesis of vascular eye diseases such as diabetic retinopathy. 11 , 12 However, the effect of altered light exposure on the visual system has not been evaluated. Therefore, in this study, we exposed genetically intact mice to a shorter light‐dark cycle (L10:D10) to assess the effect of pronounced CRD on visual acuity and retinal structure, function, and proteomes.
2. METHODS
2.1. Animals
C57BL/6J male (5 weeks old) mice were purchased from Jackson Laboratory. After a week of acclimatization, mice were housed individually in cages with access to running wheels in sound‐attenuated and ventilated isolation cabinets (Phenome Technologies, Chicago, IL, USA) for 10 weeks. The light schedules were either 12‐h light: 12‐h dark (L12:D12) for control or 10‐h light: 10‐h dark (L10:D10) for circadian disruption with 300 lux light intensity during the light phase, from a cool white LED source. Food and water were provided ad libitum. All experiments were conducted between 10 and 14 weeks of L10: D10 exposure, at Zeitgeber Time (ZT) 3 to 6, where ZT 0 is the “lights on” time, unless otherwise specified. “Free‐running” and “zigzagging” mice were monitored for pinpointing the day at which activity onset was at lights‐off, and the experimentation was performed on the following day. This enabled behavior rhythm coincided with the light cycle on the day of experimentation. All animal experimentations were carried out in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals and the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research and approved by the Institutional Animal Care and Use Committee of Indiana University School of Medicine. The same set of mice was used for all the subsequent studies.
2.2. Wheel running activity
Wheel running activity was recorded every minute for 10 weeks using Actimetrics (Actimetrics, Chicago, IL, USA) hardware and monitored and analyzed using Clocklab (Actimetrics, Chicago, IL, USA). The actogram data from 15 to 70 days were used to determine the period and quantitate wheel‐running activity using Clocklab. C57BL/6J mice exposed to L10:D10 exhibited three distinct entrainment behaviors in their wheel‐running activity: (i) “entrained” (E), (ii) “free‐running” (F), and (iii) “zigzagging” (Z); the data were analyzed based on these distinct behaviors. The period was determined either by periodogram analysis (L12:D12 and L10:D10E) or by fitting manual regression lines on 3 to 4 consecutive onsets on the actograms in Clocklab (L10:D10 F and L10:D10 Z). Wheel running activity counts were determined by averaging the total activity during light and dark phases and were expressed for 24 h to make them comparable between groups.
2.3. Electroretinography
ERG recordings were performed with an LKC Technologies UTAS system (LKC Technologies, Inc, Gaithersburg, MD, USA) under dark and light‐adapted conditions. Before testing, all mice were dark‐adapted for 24 h to nullify the effect of light history. 13 We used the previous day's light cycle and activity onset to predict ZT‐10 on the day of experimentation, and the measurements were made between ZT 10 and 13, the reported peak time for ERG response. 14 The mice were anesthetized with an i.p. injection of ketamine (100 mg/kg) and xylazine (5 mg/kg). Topical 0.5% proparacaine‐HCl eye drops were applied, and the pupils were dilated by topical application of 1% tropicamide and 2.5% phenylephrine (Alcon laboratories). The eyes were kept moist using a 2.5% hypromellose ophthalmic demulcent solution (Akorn). The core body temperature was maintained using a heating pad at 37.0°C. The ground needle electrode was placed on the base of the tail and the reference electrode was placed subdermally between the eyes. The gold loop electrodes (LKC Technologies, Inc, Gaithersburg, MD, USA) placed over the cornea were used for recording ERG response. The stimulus flash intensities of 0.025, 0.25, and 2.5 cd.s/m2 for scotopic conditions and of 1.25, 4.99, 9.95, and 25 cd.s/m2 for photopic conditions were presented in an UTAS ganzfeld illuminator (LKC Technologies). For photopic ERG recordings, the mice were light‐adapted for 10 min inside the ganzfeld prior to testing. The values for a‐wave and b‐wave amplitudes, their implicit times, individual oscillatory potential (OP), and average oscillatory potential amplitude were obtained from an inbuilt analysis tool by LKC Technologies.
2.4. Optomotor response recording
Quantification of mouse spatial vision was performed by detecting the spatial frequency threshold of optomotor response behavior using an OptoMotry device (CerebralMechanics, Inc.) as described earlier 15 between ZT3 and ZT6. Tracking head movements in response to rotating sine‐wave gratings (100% contrast) were recorded in free‐moving mice. Spatial frequency was systematically increased in a staircase method until the animal did not respond, and the highest spatial frequency the animal could track was identified as the threshold. The threshold obtained for each eye was reported.
2.5. Spectral‐domain optical coherence tomography (SD‐OCT)
The mice were anesthetized and pupils were dilated and kept moist as described for electroretinogram. SD‐OCT images were obtained using a Bioptigen SD‐OCT system between ZT3 and ZT6. The optic nerve head was located manually and was used to center the scan. Retinal thickness was measured using manual calipers, 2 each at 0.35 mm and 0.45 mm away from the optic nerve head on each B‐scan's nasal and temporal axis. Three B scans from each eye were used for measurement, and the average of all the calipers of each eye is reported.
2.6. Immunohistochemistry
Eyes for immunohistochemistry were isolated between ZT‐5 and ZT‐8 and fixed in paraformaldehyde (4%) in PBS overnight at 4°C. The eyes were immersed in 30% ethanol (v/v) in PBS and paraffin‐embedded. Paraffin sections (5 µm) were cleared by 2 × 5 min incubation in xylene before rehydration through a graded series of ethanol [100%, 95% and 80% (v/v) in PBS]. Epitope retrieval was performed by incubating in pre‐warmed citrate buffer (pH 6), overnight at 56°C. The sections were washed in 0.3% Triton X‐100 in PBS and blocked with 10% goat serum for 2 h at room temperature. Sections were incubated overnight at 4°C with one of the following primary antibodies: (i) anti‐rhodopsin (Abcam ab5417, 1:300), (ii) anti‐cone arrestin (Millipore Sigma AB15282, 1:450), (iii) anti‐PRDX6 (ThermoFisher 13585‐1‐AP, 1:50), and (iv) anti‐protein kinase Cα (Sigma‐Aldrich P4334, 1:100) diluted in 10% goat serum, washed in PBS, followed by incubation with appropriate secondary antibodies (Alexa Fluor 488 and Alexa Fluor 555; ThermoFisher) for 2 h at room temperature. After washing in PBS, slides were mounted with Vectashield containing DAPI for nuclei staining. Retinal sections were imaged with a confocal microscope (Zeiss LSM700), and images were analyzed with Zeiss ZEN image processing software. Sections with optic nerve head visible were used for immunohistochemistry, and imaging was performed between 350 and 650 µm on both sides of the optic nerve head. The density of photoreceptors was determined by counting DAPI positive nuclei in the outer nuclear layer (ONL) in 6 random fields on each side of the optic nerve and the ONL thickness was determined using ImageJ 16 by placing calipers at three different places, such two slides for each mouse were assessed. The number of cones was enumerated as arrestin positive cells. The investigator was blinded for these measurements.
2.7. Proteomics
The total proteins from the isolated retina were extracted using RIPA buffer and quantitated using BCA assay. The solubilized proteins were precipitated using trichloroacetic acid and dissolved in 8 M Urea in 100 mM Tris.HCl. An equal amount of protein starting material from each sample was reduced and alkylated with Tris (2‐carboxyethyl) phosphine and chloroacetamide and digested with trypsin overnight. Samples were labeled with tandem mass tag (TMT) reagents (Thermo Fisher Scientific). The labeled sample was fractionated using the Pierce High pH Reversed‐Phase Peptide Fractionation Kit (Thermo Fisher Scientific) and subjected to liquid chromatography–MS/MS analysis using Orbitrap Fusion Lumos mass spectrometer (Thermo Scientific) coupled to an EASY‐nLC HPLC system (Thermo Scientific). Data were acquired with the complete MS acquisition with an Orbitrap resolution of 60,000, and MS/MS analysis was performed at a resolution of 50,000 and with a collision energy of 36. MS/MS database search was performed against in silico tryptic digest of mouse proteins UniProt FASTA database using SEQUEST HT within Proteome Discoverer 2.2 (PD 2.2, Thermo Fisher Scientific) with precursor mass tolerance of 10 ppm and fragment mass tolerance of 0.02 Da. Percolator False Discovery Rate was set to 1%, and peptide abundance values were normalized in Proteome Discoverer 2.2. The experiments were performed in two separate sets by comparing the following groups: (1) E versus C and (2) F and Z versus C.
Ingenuity Pathway Analysis (IPA) (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity‐pathway‐analysis) was used to identify pathways associated with differentially expressed proteins. The complete dataset was uploaded to IPA. Comparative analysis was done on the z‐scored pathways as well as diseases and conditions to compare between entrainment groups.
2.8. Real‐time qRT‐PCR
The retina was homogenized in Trizol solution (ThermoFisher Scientific) followed by separation in chloroform and ethanol precipitation. The isolated RNA was further purified using the RNEasy MiniElute cleanup kit (Qiagen). One microgram of RNA was used to make cDNA using the SuperScript Vilo cDNA synthesis kit (ThermoFisher Scientific). The mRNA expression was determined using the following gene‐specific Taqman Gene Expression Assays (ThermoFisher Scientific), Mdh2 (Mm01208232_m1), GPx1 (Mm04207457_g1), Sod1 (Mm01344233_g1), Arntl (Mm00500226_m1), Anxa6 (Mm00478966_m1), Espn (Mm07303610_m1), Idh3b (Mm00504589_m1), Ppm1a (Mm00725963_s1), Sod2 (Mm01313000_m1), MT‐Co2 (Mm03294838_g1), MT‐Cyb (Mm04225271_g1), MT‐Nd6 (Mm04225325_g1), and Taqman Master Mix (ThermoFisher Scientific) using a ViiA7 real‐time PCR system. The mRNA expression was expressed as a 2−ΔCt value after normalization of respective gene expression to a housekeeping gene Tbp or 18S.
2.9. Statistical analysis
The data were analyzed either using Welch one‐way ANOVA followed by Dunnett's test to account for heteroscedasticity or two‐way ANOVA followed by post hoc multiple comparisons with Tukey's or by linear mixed models EM means followed by a comparison of individual groups with least square design (LSD). The following software programs were used GraphPad Prism version 9.0.0 for Windows, GraphPad Software, San Diego, CA, USA, www.graphpad.com and IBM SPSS Statistics, New York, USA, www.ibm.com/products/spss‐statistics. Data are expressed as mean ± SEM, the Welch F statistic (W) or Fisher F statistic, and p‐values are reported in the results section. The significance level was set at p ≤ 0.05.
2.9.1. IPA analysis
A threshold filter of p = 0.05 in IPA was applied, and only experimentally observed interactions were selected for analysis. The significance in the canonical pathway function was defined by a p‐value calculated using Fisher's exact test that determines if the probability of association between proteins in the dataset and in the pathway is due to chance. Prediction activation scores (z‐score) were also used as a statistical measure of the match between an expected relationship direction in a pathway and the observed protein expression. The p‐value adjustment of proteomics data and IPA was performed as described previously 17 and using The R Project for Statistical Computing www.r‐project.org/. The significance level was set at p ≤ 0.05.
3. RESULTS
3.1. CRD resulted in different entrainment behaviors
C57BL/6J mice exposed to L10:D10 exhibited three distinct entrainment behaviors in their wheel‐running activity: (i) “entrained” (E), (ii) “free‐running” (F), and (iii) “zigzagging” (Z). Mice that “entrained” to L10:D10 restricted their activity only in the dark phase with a period of ~20 h and a phase of entrainment of ≤1 h to lights off (Figure 1B and E; Figure S1). The mice which exhibited “free‐running” did not entrain to the external light/dark cycle and showed a period of 23.09 ± 0.24 h (Figure 1C and E; Figure S2). The “zigzagging” mice lacked a stable phase of entrainment and switched their period every 3–5 days between 21.48 ± 0.12 h and 18.47 ± 0.21 h (Figure 1D and E; Figure S3). Lomb‐Scargle periodogram analysis revealed 20 h and a >20‐h periodicities for “free‐running” mice (Figure S2) and 20‐h period for “zigzagging” mice (Figure S3). Since these periods did not match the period calculated by fitting regression lines on activity onsets, we also report the latter (Figure 1E). Three of the F mice initially showed unstable periodicities of 22.31 ± 0.26, which shortened when light exposure was at the end of the active phase and lengthened when light exposure was at the beginning of the active phase (Figure S2D, E, H). These three mice settled at stable longer periodicities between 8–12 weeks. Next, we quantified wheel revolution activity to evaluate if it has any protective effect on retinal physiology. 18 Total wheel‐running activity was not significantly different between control and L10:D10 behavior groups (Figure 1F; W3,15 = 2.45, p = 0.10). However, wheel revolutions differed significantly between behaviors when separated as that of the light phase (Figure 1G; W3,15 = 9.24, p = 0.001) and dark phase (Figure 1H; W3,15 = 3.83, p = 0.03). The “free‐running” mice had higher light phase activity (Figure 1G, p = 0.02) and lower dark phase activity (Figure 1H, p = 0.04) compared to “entrained” mice, since their activity was distributed across all phases of the L10:D10 light/dark cycle.
FIGURE 1.
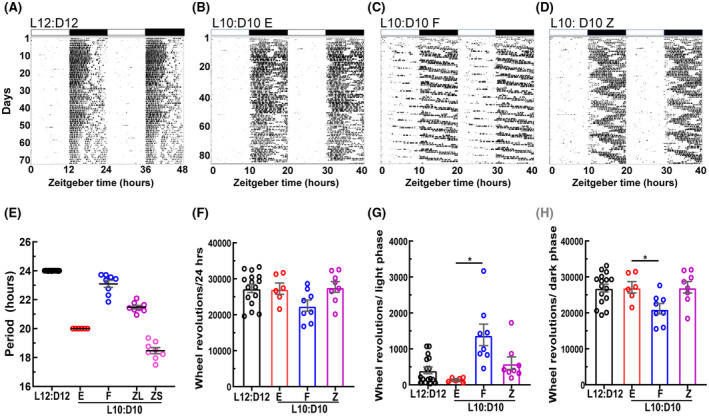
Mice under L10:D10 conditions display different entrainment behaviors. Representative double plotted actograms of wheel‐running activity under control and L10:D10 conditions showing different behavior patterns; control (A), “entrained” (B), “free‐running” (C), and “zigzagging” (D). Periods exhibited in each entrainment behavior, obtained by fitting regression lines on activity onsets on the actograms using Clocklab (E). Quantification of wheel‐running activity (F) and its distribution to light (G) and dark phases (H). n = 17 (L12:D12), 6 (L10:D10 E), 8 (L10:D10 F), 8 (L10:D10 Z). Shown are mean ± SEM. *p ≤ 0.05 (ANOVA with Dunnett's post hoc test)
3.2. CRD resulted in reduced visual acuity
Optomotor response tracking was performed to determine whether the visual function was affected by CRD, and visual acuity defined by spatial frequency threshold was measured. L10:D10 groups demonstrated significantly reduced visual acuity irrespective of their entrainment behavior (Figure 2; W3,30 = 39.15, p < 0.0001).
FIGURE 2.
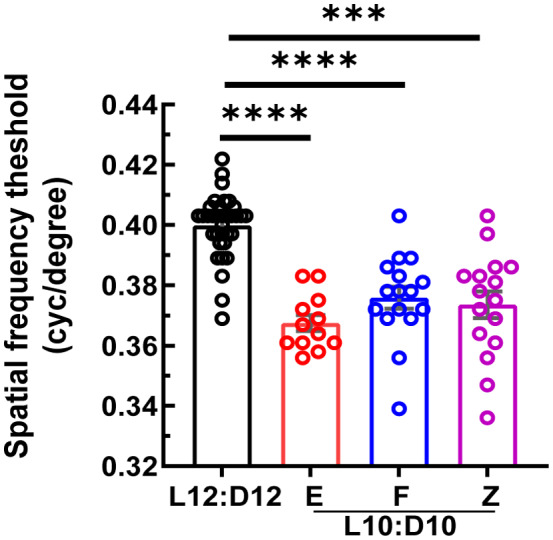
Decrease in visual acuity under L10:D10 conditions: L10:D10 mice showed reduced visual acuity when measured using an optomotor response tracking system. Data from each eye is plotted. n = 17 (L12:D12), 6 (L10:D10 E), 8 (L10:D10 F), 8 (L10:D10 Z). Shown are mean ± SEM. ***p ≤ 0.001, **** p ≤ 0.0001 (ANOVA with Dunnett's post hoc test)
3.3. CRD results in reduced retinal thickness and loss of photoreceptor cells
To investigate the effect of CRD on retinal structure, SD‐OCT and immunohistochemistry were performed. The overall retinal thickness measured from SD‐OCT b‐scans was significantly reduced in all L10:D10 behavior groups (Figure 3A and B; W3,23.72 = 15.35, p < 0.0001). In addition, immunohistochemistry using rhodopsin and cone arrestin antibodies was performed, the DAPI positive nuclei were enumerated to quantify photoreceptors (Figure 4A). The number of photoreceptors was reduced significantly in the entrained group (Figure 4B, p < 0.05) when compared to control mice. Also, the zigzagging group differed significantly in photoreceptors when compared to entrained (p < 0.01) group. While the cone photoreceptors tended to be reduced in all L10: D10 groups, the difference was not statistically significant (Figure 4C). There was an overall decrease in ONL thickness due to CRD, and this difference was significant (p < 0.001) for an entrained group when compared to the control (Figure 4D). The entrained group was also significantly different (p < 0.05) from a zigzagging group.
FIGURE 3.
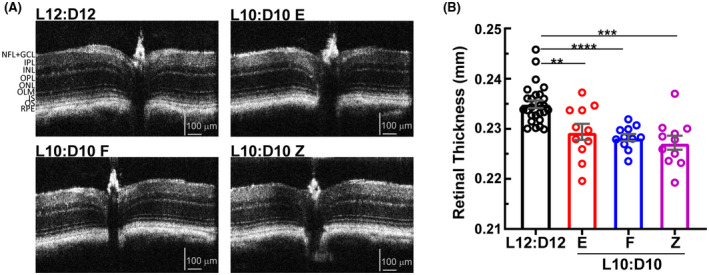
Retinal thinning under L10:D10 conditions. Representative Spectral Domain‐Optical Coherence Tomography (SD‐OCT) B‐scans of the mouse retina (A), showing central optic nerve head and retinal sublayers. L10:D10 mice showed reduced total retinal thickness (B). n = 26 (L12:D12), 11 (L10:D10 E), 11 (L10:D10 F), (L10:D10 Z). Shown are mean ± SEM. **p ≤ 0.01, *** p ≤ 0.001, ****p ≤ 0.0001 (ANOVA with Dunnett's post hoc test)
FIGURE 4.
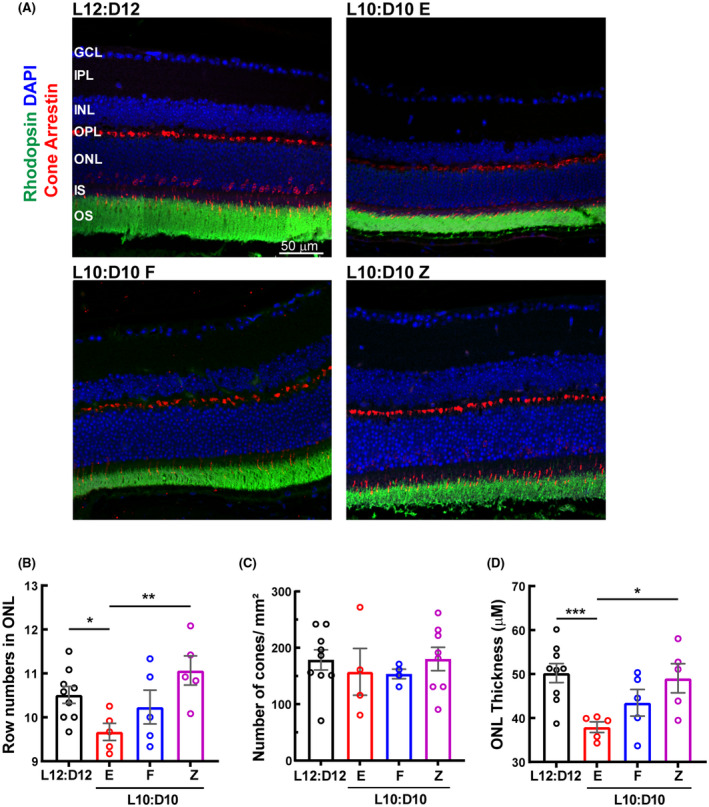
A decrease in the number of photoreceptor cells in L10:D10 treatment. (A) Retinal sections were immunostained for photoreceptors. Cell densities of photoreceptors (row numbers in ONL) (B) and cones (C) per mm2 and (D) ONL thickness. n = 9 (L12:D12), 5 (L10:D10 E), 5 (L10:D10 F), 5 (L10:D10 Z). Each data point in the bar chart represents an average count in 6 fields on each side of the optic nerve. The raw data were analyzed using Linear Mixed Models EM Means followed by a comparison of individual groups with Least Square design (LSD) *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.01
3.4. Scotopic electroretinogram a‐wave and b‐wave amplitudes were reduced after CRD depending on the entrainment behavior
To assess the retinal function, ERG response to flashes of light under scotopic (rod driven) and photopic (cone isolated) conditions were recorded. Under scotopic conditions both a‐wave that originates from photoreceptor cells and b‐wave that originates from bipolar cells were significantly reduced in the “entrained” mice while those of “free‐running” and “zigzagging” mice were unaffected (Figure 5A; flash intensity—F2,216 = 312, p < 0.0001, behavior—F3,216 = 9.29, p < 0.0001, interaction—F3,216 = 9.29, p = 0.34), (Figure 5B; flash intensity—F2,216 = 9.8, p < 0.0001, behavior—F6,216 = 24, p < 0.0001, interaction—F6,216 = 0.31, p = 0.93). The average oscillatory potential was also different between the behaviors (Figure 5C; flash intensity—F2,216 = 39.2, p < 0.0001, behavior—F3,216 = 14.7, p < 0.0001, interaction—F6,216 = 1.3, p = 0.27). The individual oscillatory potential was further separated based on different oscillations and for each flash intensity. The oscillatory potential at 0.25 cd.s/m2 showed the greatest effect with a statistically significant difference between L12:D12 vs. L10:D10 E (p < 0.05) and L10:D10E vs. L10:D10F (p < 0.05) or LD10:D10Z (p < 0.0001; Figure S4). The peak latency of oscillatory potential at 0.025 cd.s/m2 exhibited the greatest difference showing a significant decrease when the L12:D12 group was compared with L10:D10 E (Figure S5). L10:D10 entrainment behavior showed an overall increase on scotopic a‐wave peak latency (Figure S6A; flash intensity—F2,216 = 335, p < 0.0001, behavior—F3,216 = 2.86, p = 0.04, interaction—F6,216 = 0.43, p = 0.86) however, there was no change on b‐wave peak latency (Figure S6B; flash intensity—F2,216 = 384, p < 0.0001, behavior—F3,216 = 2.58, p = 0.06, interaction—F6,216 = 0.76, p = 0.6) at all the intensities that were tested. These results further emphasize the loss of rod photoreceptor function in the “entrained” mice.
FIGURE 5.
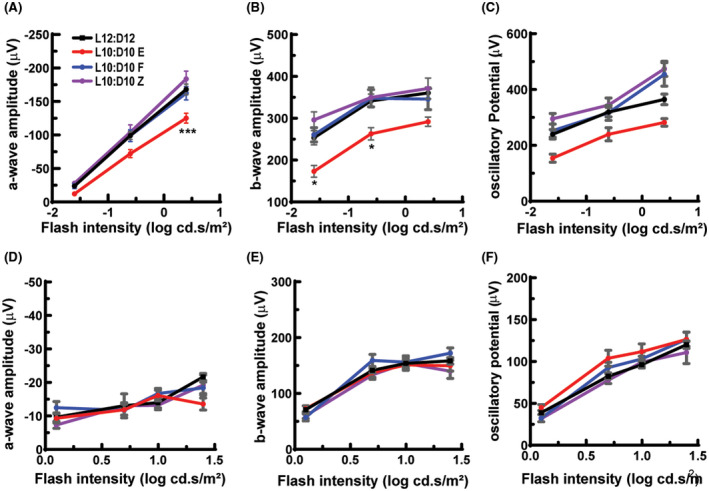
Altered ERG responses in mice under L10: D10 conditions. Entrained mice showed reduced amplitudes for scotopic a‐wave (A) and b‐wave (B) and average oscillatory potential (C), while “free‐running” and “zigzagging” mice did not show any difference. Amplitudes of photopic a‐wave (D) and b‐wave (E) and average oscillatory potential (F) were not significantly different between the groups. n = 17 (L12:D12), 6 (L10:D10 E), 8 (L10:D10 F), 8(L10:D10 Z). Shown are mean ± SEM. *p ≤ 0.05, ***p ≤ 0.001 (Two‐way ANOVA with Tukey's post hoc test)
Under photopic conditions a‐wave and b‐wave amplitudes were not significantly different between control and L10:D10 behavior groups (Figure 5D; flash intensity—F3,288 = 17.4, p < 0.0001, behavior—F3,288 = 1.35, p = 0.26, interaction—F9,288 = 1.71, p = 0.09, Figure 5E; flash intensity—F3,288 = 111.9, p < 0.0001, behavior—F3,288 = 2.05, p = 0.11, interaction—F9,288 = 1.14, p = 0.34). Entrainment behavior did show an increase in an average photopic oscillatory potential amplitude, (Figure 5F; flash intensity—F3,275 = 138, p < 0.0001, behavior—F3,275 = 4.47, p = 0.004, interaction—F9,275 = 0.74, p = 0.09). The analysis of individual oscillatory potentials exhibited a greatest change at 4.99 cd.s/m2 with a significant increase in entrained group when compared to control mice (p < 0.05), however, for zigzagging groups there was a decrease in comparison of entrained animals (p < 0.05), Figure S4. The latency of oscillatory potentials showed greatest difference at 9.95 cd.s/m2 with a statistically significant difference between zigzagging animals and free‐running or control animals (p < 0.05), the free‐running group was also reduced significantly (p < 0.05) in comparison to entrained mice (Figure S5). There was a mixed response for a‐wave peak latency depending on the frequency, (Figure S6C; flash intensity—F3,288 = 20.7, p < 0.0001, behavior—F3,288 = 4.4, p = 0.005, interaction—F9,288 = 2.1, p = 0.03) and to the b‐wave peak latency (Figure S6D; flash intensity—F3,288 = 6.2, p = 0.0004, behavior—F3,288 = 9.8, p < 0.0001, interaction—F9,288 = 4.2, p < 0.0001). The free‐running group showed significantly shortened peak latencies for a‐ wave and b‐wave at a flash intensity of 1.25 cd.s/m2 (Figure S6C, D).
3.5. CRD affects retinal circuitry
Because we observed a decrease in bipolar cell response to scotopic stimulation in ERG studies of entrained mice, we decided to study structural changes in the outer nuclear layer. The retinal sections were stained for anti‐PKCα to label bipolar cells as described earlier. 10 , 19 The dendritic processes of rod bipolar cells in entrained mice appeared to be stunted compared to control animals. While there was some defect in arborization in the free‐running group, the zig‐zagging group appeared to be similar to the L12:D12 animals (Figure 6).
FIGURE 6.
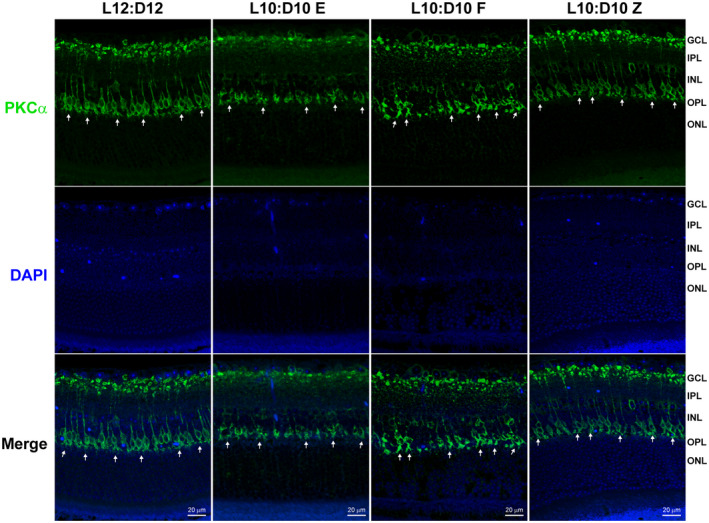
Changes in rod bipolar cell dendrites under L10:D10 conditions. Retinal sections were immunostained for bipolar cells (anti‐PKCα, green). The white arrows show the presence of dendritic arborization in L12:D12 mice, which were stunted in LD10:D10 entrained and free‐running group; however, LD10:10 zigzagging group remained unchanged (n = 3 each behavior)
3.6. CRD resulted in distinct changes in the retinal proteome with each entrainment behavior
To begin to identify the molecular alterations underlying the observed changes in retinal structure and physiology, we performed proteomic analysis of the retinas from mice with all entrainment behaviors. Overall, a total of 7620 proteins were identified. Interestingly, the quantitation of protein abundance revealed distinct proteomic changes in the retina with each entrainment behavior, as demonstrated by the volcano plots (Figure S7A, C, E). The “entrained” retina showed 317 proteins upregulated and 174 proteins downregulated. The “free‐running” retina showed 149 proteins upregulated and 165 proteins downregulated. Surprisingly, the most significant number of proteins were differentially expressed in the “zigzagging” behavior; 441 upregulated and 538 downregulated. A complete list of differentially expressed proteins is provided in the Table S1.
We used IPA to gain more information about the molecular pathways associated with altered protein expression. This analysis predicted activation or inhibition of specific canonical pathways related to each entrainment behavior (Figure S7B, D, F). In the “entrained” retina, pathways associated with mitochondrial function were affected. In the “free‐running” group, multiple protective pathways were activated, including the TCA cycle, long‐term synaptic potentiation, and AMPK signaling. 20 , 21 In the “zigzagging” retina, EIF2 signaling, which was activated in endoplasmic reticulum stress, was found to be inhibited 22 while mTOR and Ephrin signaling was activated.
Further statistical analysis was performed to adjust the p values for protein targets in Figure S7A, C, E and false discovery rate (FDR) of IPA analysis in Figure S7B, D, F. For the comparison E vs. C and F vs. C, none of the proteins survived the FDR and adjusted p‐values greater than 0.05. However, for the comparison Z vs. C, 33 out of 49 proteins survived adjusted p‐value cut‐off 0.05/0.2. Similarly, p‐value adjustment for the enrichment analysis for comparison E vs. C and F vs. C, none of the pathways passed the FDR and adjusted p‐values were greater than 0.05; for the comparison Z vs. C, one pathway (EIF2 signaling) stayed under adjusted p‐value cutoff 0.05 or 0.2. These data are provided in the supplemental information (Table S1).
A comparative analysis of differentially regulated pathways (based on p‐value cut‐offs) and protein targets revealed distinct changes between the behavior groups (Figure S8) and the identification of 15 unique targets common to these three behaviors (Figure 7A); these targets were further separated by overlaying IPA data set over “ophthalmic diseases” and “small molecule biochemistry” under disease and function category. This resulted in the mapping of 7 targets with overlapping functions (Figure 7B); the remaining proteins were unmapped. The insulin secretion signaling pathway was predicted to be activated in all L10:D10 groups. Interestingly TCA cycle pathway was predicted to be inhibited in the “entrained” retina, while it was activated in the “free‐running” and “zigzagging” retina, suggesting reduced retinal energy production after L10:D10 entrainment (Figure S8).
FIGURE 7.
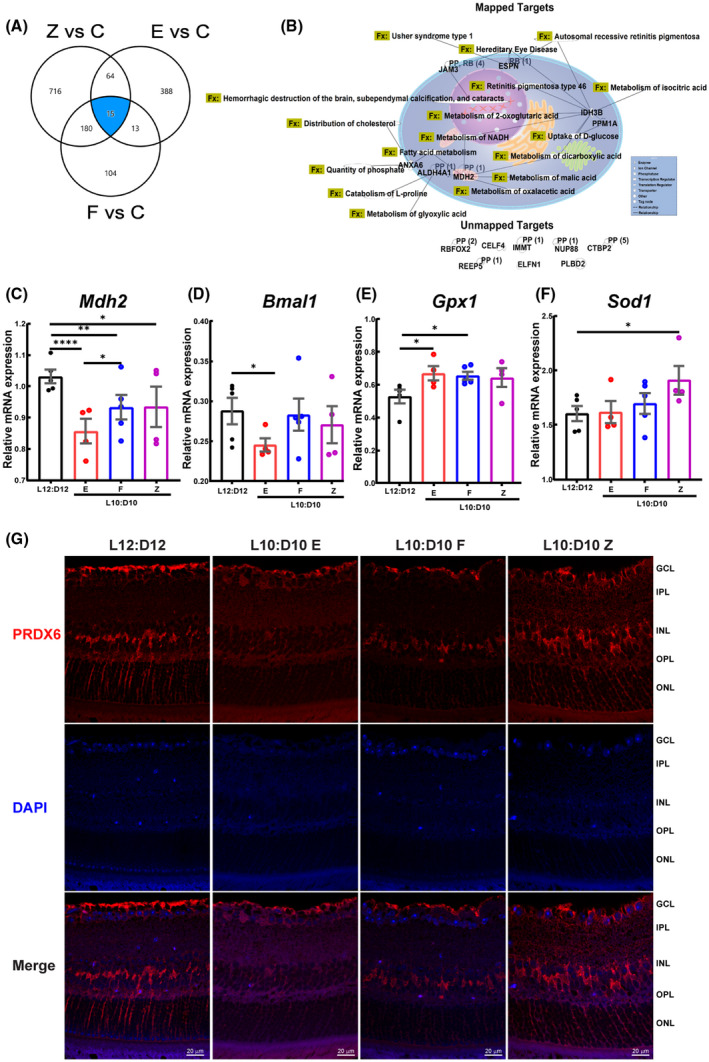
Comparative analysis of retinal proteome and Prdx6 expression in retinal sections. A comparison analysis was performed using IPA (A) Venn diagram showing common dysregulated proteins shared by different behaviors. (B) The protein targets common to these three behaviors were mapped to show their connectivity and function (Fx) and PP‐protein‐protein interactions. RB‐regulation of binding, RBFOX2‐RNA binding fox‐1 homolog 2, ELFN1‐extracellular leucine‐rich repeat and fibronectin type III domain containing 1, JAM3‐junctional adhesion molecule 3, NUPP88‐ nucleoporin 88, CTBP2‐c‐terminal binding protein 2, PLBD2‐ phospholipase B domain containing 2, CELF4‐ CUGBP elav‐like family member 4, REEP5‐receptor accessory protein 5, IMMT‐inner membrane mitochondrial protein. Target validation using qRT‐PCR showing mRNA expression of (C) Mdh2 (D) Bmal (ARNTL), (E) Gpx, and (F) Sod1. mRNA expression was determined in triplicate for each mouse and represented as an average data point for each animal. The raw data were analyzed by Linear Mixed Models EM Means followed by a comparison of individual groups with Least Square design (LSD), *p ≤ 0.05, **p ≤ 0.01, ****p ≤ 0.0001. (G) Retinal sections were immunostained for peroxiredoxin 6 (PRDX6), representative photomicrographs showing the staining for Prdx6 in the cells of INL, putative Müller cell end feet, and nerve fiber layer
To validate targets in Figure 7B, mRNA analysis was performed on mapped targets, sharing functions, and having a role either in TCA cycle (Mdh2, Idh3b), glucose uptake (Ppm1a), mitochondrial function (Anxa6 23 ), or photoreceptor function (Espn 24 ). In mRNA validation, none of the above targets exhibited a statistical significance (Figure S9) except Mdh2, a vital member of the TCA cycle and malate aspartate shuttle 25 , 26 ; thus, a critical enzyme of mitochondrial respiration showed a statistically significant decrease between different behaviors and the control group (Figure 7C), suggesting a crucial role of TCA cycle in our studies. Because Bmal1 ablation in other tissues such as the heart results in mitochondrial defects and a decrease in expression of genes for fatty acid oxidative pathway, TCA cycle, and mitochondrial respiratory chain, including MDH2, 27 we reasoned to test the mRNA expression of clock gene Bmal1. We observed the downregulation of Bmal1 in all the behaviors compared to control mice; however, only the entrained group showed a statistically significant decrease (Figure 7D). Since Bmal1 loss is known to increase oxidative stress 28 and TCA cycle intermediates are protective against oxidative stress, 29 we next evaluated oxidative stress genes Sod1, Sod2, Gpx1, MT‐Cytb, MT‐Co2, and MT‐Nd6, which are known to be involved with oxidative stress in the retina. 30 Of these genes, Gpx1 was significantly upregulated in the entrained and free‐running group (Figure 7E), while Sod1 was significantly up in the zigzagging group (Figure 7F), none of the remaining genes showed a statistical significance (Figure S9).
Peroxiredoxin 6 (Prdx6, 1‐cys peroxiredoxin) is a unique member of the Prdx6 family known to regulate oxidative stress 31 and reduce phospholipid hydroperoxides through its Gpx activity. 32 Moreover, Prdx6 is affected by changes in circadian rhythm and Bmal1 28 ; therefore, we reasoned to study the Prdx6 expression in retinal sections. Like previous reports, 33 Prdx6 mainly stained an internal limiting membrane, the population of cells in the inner nuclear layer, and Müller cell endfeet (Figure 7F). The entrained animals showed a decrease in Prdx6 staining compared to control mice; the free‐running and zigzagging behaviors also exhibited a reduction in Prdx6, however not to the extent of an entrained group.
4. DISCUSSION
Light exposure at the wrong time could result in CRD and, as such, be associated with metabolic and neurodegenerative diseases. 4 Our study further demonstrates the negative impact of CRD on vision by showing a decrease in visual acuity, altered retinal structure as exhibited by a reduction in retinal thickness, photoreceptor degeneration, and retinal function in ERG assessments. The above retinal parameters were coupled with the unique retinal proteome changes that mapped pathways integral to normal retinal function and could be related to a decrease in clock gene Bmal1 and dysfunctional oxidative stress‐antioxidant defense mechanism. Furthermore, we distinctly report different entrainment behaviors in C57BL/6J mice in response to the shorter light/dark cycle.
In addition to visual dysfunction in response to CRD, in this study, we report different entrainment behaviors in otherwise healthy C57BL/6J mice in response to the L10:D10 cycle. Previous studies show that most mice entrain to L10:D10 and exhibit a shortened free‐running period when exposed to constant darkness. 34 , 35 , 36 However, some reports suggest that C57BL/6J mice do not entrain to these conditions, and no specific activity patterns were described. 35 , 37 , 38 Davis and Menakar reported that mice that did not entrain to L10:D10 had a higher free‐running period (23.686 ± 0.083) than the average (23.224 ± 0.044). 35 Interestingly, in our study, five out of eight “free‐running” mice showed a higher than average period of 23.55 ± 0.073. Thus, it is possible that mice in the “free‐running” group had higher endogenous free‐running periods, and the observed behavior might be attributed to that.
We also observed an interesting entrainment behavior where the mice were “zigzagging” between two periods. These mice likely exhibit advancement in their circadian phase when light exposure was at the end of their dark or active phase. This phase advancement possibly continued to reduce the period length until the light exposure at the beginning of the dark/active phase delayed the activity onset. Recurrent phase advances and phase delays could lead to switching of period every 3–5 days resulting in a “zigzagging” actogram. The individual variation in the ability to undergo a phase shift could explain the observed zigzagging behavior. Previous studies suggest that the inter‐individual difference in the free‐running period correlates with the extent of phase shift in response to a resetting light stimulus. 39 While mice used in our study were from the same genetic background, individual variations in the organization of retinal and/or SCN neuronal networks could result in interindividual differences in the free‐running period and entrainment behaviors. Further studies are necessary to characterize these behaviors in detail.
The mechanism of circadian disruption among these behavioral groups could be different. The “entrained” mice showed clear activity onsets at lights off, suggesting their brain clock aligned itself to the shorter 20‐h cycle. It is plausible that their retinal clock was also aligned to a 20‐h cycle via local entrainment by light exposure. Adapting into a much shorter period inconsistent with the endogenous ~24 h cycle might have caused disruptions in the circadian regulation of homeostatic processes in these mice's retina. The “free‐running” mice in our study were exposed to light and dark at all phases of their circadian cycle. Light exposure at the wrong (active) time could result in circadian disruption. Also, retinal damage caused by light is found to be greater in the subjective night. 40 , 41 Similarly, “zigzagging” mice were also exposed to light at the end of their active phase. Moreover, switching between two periods by the “zigzagging” mice could be affecting their circadian regulation of normal physiology. In similar experiments, mice that did not entrain their activity to L10:D10 but showed 24‐h patterns of core body temperature exhibited strong resetting of SCN clock after dissection and altered phase relationship between SCN and clocks in peripheral organs. 38 This suggested L10:D10 exposure reduced the circadian network's robustness and amplitude and altered the phasing of coupling signals. Moreover, it could further alter systemic circadian cues such as hormonal levels or body temperature, which could, in turn, be affecting the retinal clock and physiology. Earlier studies have demonstrated accelerated weight gain, obesity, and changes in the metabolic hormones in mice exposed to L10:D10. 5 Thus, in addition to the possible direct negative effect of light at the wrong time on the retinal clock and physiology, different entrainment behaviors could possibly contribute to the observed visual dysfunction through altered systemic cues. It would certainly be an interesting future direction to delineate the metabolic cues involved in the visual dysfunction after CRD exposure.
In our study, CRD led to reduced visual acuity and retinal thinning irrespective of the entrainment behavior. Clearly, our studies indicate that a reduced number of photoreceptors could have contributed to the observed retinal thinning and loss of visual acuity. Alterations in the neuronal connectivity of bipolar cells, retinal ganglion cells, or at the level of visual cortical circuitry could decrease visual acuity. In addition, we observe stunting of rod bipolar cell arbors in CRD exposed mice. Moreover, similar light/dark conditions reduce complexity and loss of dendritic lengths in neurons in the prelimbic prefrontal cortex, and decreased cognitive flexibility 5 further supports the possibility of CRD induced cortical changes. We also observed a concurrent decrease in the expression of clock gene Bmal, clearly suggesting a decreased robustness of the ocular clock, which could possibly affect visual function. Our findings demonstrate that a decrease in rod‐driven ERG function was only observed in the “entrained” behavioral group, suggesting a more significant impact of CRD on this group's retinal physiology. Interestingly, despite retinal thinning, scotopic ERG response was not affected in the “free‐running” and “zigzagging” retina. Many recent studies had reported compensatory dendritic synapse formation in retinal bipolar cells when rod or cone photoreceptors were ablated. 42 These rods or cone bipolar dendrites may extend their arbors and form new synapses to compensate for fewer photoreceptor cells 43 ; we observed a similar response in bipolar arbors of retinas in the zigzagging group. A recent study also reported functional compensation in the retina after 50% of rods ablation, without any observable anatomical changes in the bipolar cells. 44 Another possible explanation would be the involvement of Bmal. While Bmal1 was significantly downregulated in entrained mice, there was no significant change in other behaviors. Bmal has been implicated in photoreceptor degeneration, stunting of rod bipolar cells, and retinal thinning. 10 Moreover, Bmal regulates oxidative stress exercising its effect through Prdx6, 28 and heart‐specific deletion of Bmal results in defects in the TCA cycle, mitochondrial respiratory chain, and fatty acid oxidation pathways. 27 Therefore, we speculate that the changes in ERG response could be attributed to Bmal deficiency in entrained group and subsequent dysfunction of the TCA cycle.
Proteomic changes associated with insulin secretion signaling, TCA cycle, oxidative phosphorylation, mitochondrial dysfunction, and phototransduction pathway could collectively affect ERG response. 45 Validation of critical targets of the proteomic analysis further demonstrates mitochondrial defects and dysfunctional oxidative stress‐antioxidant properties. Our studies highlight that Mdh2 was downregulated irrespective of the behavior. MDH2 is a critical enzyme in a malate‐aspartate shuttle and is involved in maintaining cytosolic NADH/NAD redox balance, mitochondrial respiration, and electron transfer. 25 , 26 The highest concentration of mitochondria is observed in retinal photoreceptors, and its dysfunction is associated with oxidative stress and inflammation in the retina. 30 , 46 While we observed an increase in mitochondrial oxidative stress genes, only Sod1 did reach a statistical significance for the zigzagging group; we speculate there is a potential involvement of mitochondrial dysfunction with implications for future studies in this direction. Moreover, our studies demonstrate that Gpx1 is upregulated in all the behaviors with a significant difference in “entrained” and “free‐running” mice. GPX is a cytosolic selenoprotein that catalyzes the reduction of hydrogen peroxide to water and oxygen and catalyzes the reduction of peroxide radicals to alcohol and oxygen 47 and is known to be induced with increased light exposure to the retina. 48 Our studies also highlight that changes in the Prdx6 antioxidant system are integral to the oxidative stress response observed. The protective role of Prdx6 is extensively studied in ocular conditions such as corneal injury, glaucoma, and diabetic retinopathy. 49 Prdx6 is also known to reduce hydrogen peroxide and blue light‐induced oxidative stress in ARPE19 cells 50 and is known to work through its non‐selenium GPX activity 32 , 51 and anti‐oxidant action. In addition, Prdx6 removes free radicals by neutralizing peroxides, peroxynitrite, and phospholipid hydroperoxides. 31 Thus collectively, our studies indicate that dysfunctional oxidative stress‐antioxidant defense mechanisms could have played a role in ERG response and other visual parameters observed in our studies while not pinpointing a particular target for observed behavior.
How these findings of a change in visual function and differential behavior in response to lighting conditions will translate into humans would be an important area of research. Because a chronically disrupted light/dark cycle could put individuals at risk of developing more severe ocular conditions such as diabetic retinopathy and age‐related macular degeneration. Though the selected light cycle might not reflect real‐life scenarios other than some shift workers, our rationale is based on a series of previous studies which used L10:D10 or L11:D11 lighting conditions to report adverse health outcomes such as weight gain, obesity, changes in metabolic hormone levels, 5 sleep 52 and mood disturbances. 53 The ramifications of different entrainment behaviors reported in this study could provide an excellent platform mimicking real‐life scenarios of CRD. The “entrained” behavior could mimic specific rotational shift work schedules. 3 The free‐running behavior is similar to chronic jetlag schedules with light exposure on every endogenous circadian phase. The zigzagging mice have an unstable phase of entrainment, possibly mimicking social jetlag, where people tend to delay their sleep onset later in the night on weekends and sleep early on weekdays. 54 In a human longitudinal study, retinal thinning was observed preceding the development of diabetic retinopathy. 55 These data suggest that the retinal phenotype observed in this study could be preceding vascular pathology. Therefore, we speculate that chronic CRD could lead to photoreceptor degeneration and ophthalmic diseases. In the future, it would be interesting to study whether the observed defects in visual function are permanent or, after switching to a regular light/dark cycle, whether the vision returns to normalcy.
In conclusion, our data provide substantial evidence for CRD and aberrant light exposure as a causative agent for photoreceptor degeneration and visual dysfunction. We uniquely show the effect of entrainment behavior on retinal physiology. Our data has broader implications in understanding the role of CRD in the pathogenesis of ophthalmic diseases and possibly mitigating these negative impacts with lifestyle changes.
CONFLICT OF INTEREST
DM and QL do not have any conflicts of interest; AB is an ad hoc pharmacist at CVS Health/Aetna, and the content of this manuscript does not represent those of CVS Health/Aetna or its management.
AUTHOR CONTRIBUTION
A.B. conceived the study and supervised the project; A.B. and D.M. designed the experiments; D.M. and Q.L. performed experiments and analyzed data; A.B. and D.M. wrote the manuscript.
Supporting information
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Fig S7
Fig S8
Fig S9
Table S1
ACKNOWLEDGEMENTS
This research is supported by funding support from the National Institute of Health (NIH)—National Eye Institute (NEI) grant R01EY027779 to AB. Mass spectrometry services were provided by the Indiana University School of Medicine’s Proteomics Core Facility. We would like to thank Dr. Timothy W. Corson and Dr. Yoshikazu Imanishi (Department of Ophthalmology), Dr. Sha Cao, and Mr. George Eckert (Department of Biostatistics and Health Data Science) at Indiana University, and Dr. Renu Kowluru (Department of Ophthalmology) at Wayne State University for their helpful discussions and suggestions.
Mathew D, Luo Q, Bhatwadekar AD. Circadian rhythm disruption results in visual dysfunction. FASEB BioAdvances. 2022;4:364‐378. doi: 10.1096/fba.2021-00125
REFERENCES
- 1. Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445‐462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Takahashi JS. Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet. 2017;18:164‐179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vetter C. Circadian disruption: what do we actually mean? Eur J Neurosci. 2020;51:531‐550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sulli G, Manoogian ENC, Taub PR, Panda S. Training the circadian clock, clocking the drugs, and drugging the clock to prevent, manage, and treat chronic diseases. Trends Pharmacol Sci. 2018;39:812‐827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karatsoreos IN, Bhagat S, Bloss EB, Morrison JH, McEwen BS. Disruption of circadian clocks has ramifications for metabolism, brain, and behavior. Proc Natl Acad Sci USA. 2011;108:1657‐1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ben‐Hamo M, Larson T, Duge LS, et al. Circadian forced desynchrony of the master clock leads to phenotypic manifestation of depression in rats. eneuro. 2016;3(6):ENEURO.0237‐16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Besharse JC, McMahon DG. The retina and other light‐sensitive ocular clocks. J Biol Rhythms. 2016;31:223‐243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Felder‐Schmittbuhl M‐P, Buhr ED, Dkhissi‐Benyahya O, et al. Ocular clocks: adapting mechanisms for eye functions and health. Invest Ophthalmol Vis Sci. 2018;59:4856‐4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Storch KF, Paz C, Signorovitch J, et al. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;130:730‐741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baba K, Piano I, Lyuboslavsky P, et al. Removal of clock gene Bmal1 from the retina affects retinal development and accelerates cone photoreceptor degeneration during aging. Proc Natl Acad Sci. 2018;115:13099‐13104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bhatwadekar AD, Beli E, Diao Y, et al. Conditional deletion of Bmal1 accentuates microvascular and macrovascular injury. Am J Pathol. 2017;187:1426‐1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bhatwadekar AD, Yan Y, Qi X, et al. Per2 mutation recapitulates the vascular phenotype of diabetes in the retina and bone marrow. Diabetes. 2013;62:273‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barnard AR, Hattar S, Hankins MW, Lucas RJ. Melanopsin regulates visual processing in the mouse retina. Curr Biol. 2006;16:389‐395. [DOI] [PubMed] [Google Scholar]
- 14. Di R, Luo Q, Mathew D, Bhatwadekar AD. Diabetes alters diurnal rhythm of electroretinogram in db/db mice. Yale J Biol Med. 2019;92:155‐167. [PMC free article] [PubMed] [Google Scholar]
- 15. Prusky GT, Alam NM, Beekman S, Douglas RM. Rapid quantification of adult and developing mouse spatial vision using a virtual optomotor system. Invest Ophthalmol Vis Sci. 2004;45:4611‐4616. [DOI] [PubMed] [Google Scholar]
- 16. Schneider CA, Rasband WS, Eliceiri KW. NIH image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671‐675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979:65‐70. [Google Scholar]
- 18. Zhang X, Girardot PE, Sellers JT, et al. Wheel running exercise protects against retinal degeneration in the I307N rhodopsin mouse model of inducible autosomal dominant retinitis pigmentosa. Mol Vis. 2019;25:462‐476. [PMC free article] [PubMed] [Google Scholar]
- 19. Liets LC, Eliasieh K, Van Der List DA, Chalupa LM. Dendrites of rod bipolar cells sprout in normal aging retina. Proc Natl Acad Sci. 2006;103:12156‐12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Du J‐L, Wei H‐P, Wang Z‐R, Wong ST, Poo M‐M. Long‐range retrograde spread of LTP and LTD from optic tectum to retina. Proc Natl Acad Sci. 2009;106:18890‐18896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu L, Kong L, Wang J, Ash JD. Stimulation of AMPK prevents degeneration of photoreceptors and the retinal pigment epithelium. Proc Natl Acad Sci. 2018;115:10475‐10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Starr CR, Pitale PM, Gorbatyuk M. Translational attenuation and retinal degeneration in mice with an active integrated stress response. Cell Death Dis. 2018;9:484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chlystun M, Campanella M, Law A‐L, et al. Regulation of mitochondrial morphogenesis by annexin A6. PLoS One. 2013;8:e53774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ahmed ZM, Jaworek TJ, Sarangdhar GN, et al. Inframe deletion of human ESPN is associated with deafness, vestibulopathy and vision impairment. J Med Genet. 2018;55:479‐488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Y, Asnani A, Zou L, et al. Visnagin protects against doxorubicin‐induced cardiomyopathy through modulation of mitochondrial malate dehydrogenase. Sci Transl Med. 2014;6:170‐266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Molinié T, Cougouilles E, David C, Cahoreau E, Portais J‐C, Mourier A. MDH2 is a metabolic switch rewiring the fuelling of respiratory chain and TCA cycle. bioRxiv. 2021. [DOI] [PubMed] [Google Scholar]
- 27. Kohsaka A, Das P, Hashimoto I, et al. The circadian clock maintains cardiac function by regulating mitochondrial metabolism in mice. PLoS One. 2014;9:e112811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chhunchha B, Kubo E, Singh DP. Clock protein Bmal1 and Nrf2 cooperatively control aging or oxidative response and redox homeostasis by regulating rhythmic expression of Prdx6. Cells. 2020;9:1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sawa K, Uematsu T, Korenaga Y, et al. Krebs cycle intermediates protective against oxidative stress by modulating the level of reactive oxygen species in neuronal HT22 cells. Antioxidants. 2017;6:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Madsen‐Bouterse SA, Zhong Q, Mohammad G, Ho Y‐S, Kowluru RA. Oxidative damage of mitochondrial DNA in diabetes and its protection by manganese superoxide dismutase. Free Radical Res. 2010;44:313‐321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zha X, Wu G, Zhao X, et al. PRDX6 protects ARPE‐19 cells from oxidative damage via PI3K/AKT signaling. Cell Physiol Biochem. 2015;36:2217‐2228. [DOI] [PubMed] [Google Scholar]
- 32. Arevalo JA, Vázquez‐Medina JP. The role of peroxiredoxin 6 in cell signaling. Antioxidants. 2018;7:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chidlow G, Wood JP, Knoops B, Casson RJ. Expression and distribution of peroxiredoxins in the retina and optic nerve. Brain Struct Funct. 2016;221:3903‐3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aton SJ, Block GD, Tei H, Yamazaki S, Herzog ED. Plasticity of circadian behavior and the suprachiasmatic nucleus following exposure to non‐24‐hour light cycles. J Biol Rhythms. 2004;19:198‐207. [DOI] [PubMed] [Google Scholar]
- 35. Davis FC, Menaker M. Development of the mouse circadian pacemaker: independence from environmental cycles. J Comp Physiol. 1981;143:527‐539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pittendrigh CS, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents. II. The variability of phase response curves. J Comp Physiol. 1976;106:253‐266. [Google Scholar]
- 37. Pittendrigh CS, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents. I. The stability and lability of spontaneous frequency. J Comp Physiol A. 1976;106:223‐252. [Google Scholar]
- 38. Leise TL, Goldberg A, Michael J, et al. Recurring circadian disruption alters circadian clock sensitivity to resetting. Eur J Neurosci. 2020;51:2343‐2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Daan S, Pittendrigh CS. A Functional analysis of circadian pacemakers in nocturnal rodents. J Comp Physiol. 1976;106:253‐266. [Google Scholar]
- 40. Organisciak DT, Darrow RM, Barsalou L, Kutty RK, Wiggert B. Circadian‐dependent retinal light damage in rats. Invest Ophthalmol Vis Sci. 2000;41:3694‐3701. [PubMed] [Google Scholar]
- 41. Wiechmann AF, O'Steen WK. Melatonin increases photoreceptor susceptibility to light‐induced damage. Invest Ophthalmol Vis Sci. 1992;33:1894‐1902. [PubMed] [Google Scholar]
- 42. Leinonen H, Pham NC, Boyd T, Santoso J, Palczewski K, Vinberg F. Homeostatic plasticity in the retina is associated with maintenance of night vision during retinal degenerative disease. Elife. 2020;9:e59422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Beier C, Hovhannisyan A, Weiser S, et al. Deafferented adult rod bipolar cells create new synapses with photoreceptors to restore vision. J Neurosci. 2017;37:4635‐4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Care RA, Anastassov IA, Kastner DB, Kuo Y‐M, Della Santina L, Dunn FA. Mature retina compensates functionally for partial loss of rod photoreceptors. Cell Rep. 2020;31:107730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schrier SA, Falk MJ. Mitochondrial disorders and the eye. Curr Opin Ophthalmol. 2011;22:325‐331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Du Y, Veenstra A, Palczewski K, Kern TS. Photoreceptor cells are major contributors to diabetes‐induced oxidative stress and local inflammation in the retina. Proc Natl Acad Sci. 2013;110:16586‐16591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fanucchi MV. Development of antioxidant and xenobiotic metabolizing enzyme systems. The lung. Elsevier; 2004:177‐185. https://www.sciencedirect.com/science/article/pii/B9780123247513500462?via%3Dihub [Google Scholar]
- 48. Ohira A, Tanito M, Kaidzu S, Kondo T. Glutathione peroxidase induced in rat retinas to counteract photic injury. Invest Ophthalmol Vis Sci. 2003;44:1230‐1236. [DOI] [PubMed] [Google Scholar]
- 49. Wahlig S, Lovatt M, Mehta JS. Functional role of peroxiredoxin 6 in the eye. Free Radic Biol Med. 2018;126:210‐220. [DOI] [PubMed] [Google Scholar]
- 50. Zha X, Wu G, Zhang H, Yang Y, Zhang Y, Ma L. PRDX6 regulates the H2O2 and blue light‐induced APRE‐19 cell apoptosis via down‐regulating and interacting with RARA. Anim Cells Syst. 2019;23:241‐245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fisher AB, Dodia C, Manevich Y, Chen J‐W, Feinstein SI. Phospholipid hydroperoxides are substrates for non‐selenium glutathione peroxidase. J Biol Chem. 1999;274:21326‐21334. [DOI] [PubMed] [Google Scholar]
- 52. Hasan S, Foster RG, Vyazovskiy VV, Peirson SN. Effects of circadian misalignment on sleep in mice. Sci Rep. 2018;8:1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ben‐Hamo M, Larson TA, Duge LS, et al. Circadian forced desynchrony of the master clock leads to phenotypic manifestation of depression in rats. Eneuro. 2016;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Roenneberg T, Allebrandt KV, Merrow M, Vetter C. Social jetlag and obesity. Curr Biol. 2012;22:939‐943. [DOI] [PubMed] [Google Scholar]
- 55. Sohn EH, van Dijk HW, Jiao C, et al. Retinal neurodegeneration may precede microvascular changes characteristic of diabetic retinopathy in diabetes mellitus. Proc Natl Acad Sci USA. 2016;113:E2655‐E2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Fig S4
Fig S5
Fig S6
Fig S7
Fig S8
Fig S9
Table S1


