Abstract
Despite the importance of Thiobacillus ferrooxidans in bioremediation and bioleaching, little is known about the genes encoding electron transfer proteins implicated in its energetic metabolism. This paper reports the sequences of the four cox genes encoding the subunits of an aa3-type cytochrome c oxidase. These genes are in a locus containing four other genes: cyc2, which encodes a high-molecular-weight cytochrome c; cyc1, which encodes a c4-type cytochrome (c552); open reading frame 1, which encodes a putative periplasmic protein of unknown function; and rus, which encodes rusticyanin. The results of Northern and reverse transcription-PCR analyses indicated that these eight genes are cotranscribed. Two transcriptional start sites were identified for this operon. Upstream from each of the start sites was a ς70-type promoter recognized in Escherichia coli. While transcription in sulfur-grown T. ferrooxidans cells was detected from the two promoters, transcription in ferrous-iron-grown T. ferrooxidans cells was detected only from the downstream promoter. The cotranscription of seven genes encoding redox proteins suggests that all these proteins are involved in the same electron transfer chain; a model taking into account the biochemistry and the genetic data is discussed.
The gram-negative eubacterium Thiobacillus ferrooxidans is important for industry and ecology because (i) this microorganism is able to solubilize metals from ores, such as copper, uranium, and cobalt, and to decompose recalcitrant gold-containing ores (39) and (ii) it is able to remove heavy metals from contaminated industrial effluents or soils and to desulfurize fossil fuels to avoid corrosion and atmospheric acid depositions (7, 21, 22). In addition to its industrial importance, T. ferrooxidans is of fundamental interest since its way of life is one of the “most primitive extant” (8): for growth, this microorganism requires only air, which provides carbon from carbon dioxide, nitrogen, and oxygen, and ores containing ferrous iron (Fe2+) or reduced sulfur compounds, from which it derives its energy. Furthermore, it thrives at extremely low pHs (between 4 and 1.5). T. ferrooxidans is one of the most studied bioleaching microorganisms, but little is known about its physiology and, more particularly, its energy metabolism. Because its energy metabolism is responsible for its bioleaching and bioremediation abilities, any attempt to improve these properties is dependent on an understanding of the respiratory mechanisms. Although several redox proteins have been identified (49), the electron pathways from Fe2+ to oxygen (O2) and from reduced sulfur compounds to O2 are not established. Several models for the iron respiratory chain which differ with regard to the electron carriers and the side of the cytoplasmic membrane on which oxygen reduction takes place have been proposed (2, 5, 13, 17, 18, 50).
As an approach for elucidating the T. ferrooxidans respiratory chains, we are studying the genes encoding electron transfer proteins. We have previously cloned and sequenced the rus, cyc1, and cyc2 genes, which encode, respectively, the rusticyanin (4) and two cytochromes c (2) from strain ATCC 33020. The sequences of internal fragments of the rus genes from strain ATCC 19859 (38) and from strain ATCC 23270 (15) and the sequence of the iro gene from strain Fe-1 (26) have also been reported. We have also shown that in strain ATCC 33020 the cyc1 and cyc2 genes are cotranscribed with at least two downstream open reading frames (ORFs) (2) and that the rus gene is cotranscribed with at least three upstream ORFs (4). In this paper, we demonstrate that all these genes together with the genes encoding the four subunits of a cytochrome c oxidase belong to the same operon.
MATERIALS AND METHODS
Strains, plasmids, growth conditions, and β-galactosidase assays.
T. ferrooxidans ATCC 33020 was obtained from the American Type Culture Collection. Escherichia coli MC4100 [araD139 Δ(lacIPOZYA-argF)U169 rpsL thi] was used when β-galactosidase activities had to be determined.
Phagemid Bluescript SK was purchased from Stratagene. Plasmid pGE593 is an operon fusion vector containing the lacZ gene as the reporter gene (10).
E. coli strains were grown in Luria-Bertani medium (31). Conditions for growth of T. ferrooxidans on iron or sulfur medium have been described previously (4).
β-Galactosidase activities were determined according to the method of Miller (31) in whole cells grown on Luria-Bertani medium containing ampicillin.
DNA manipulations.
General molecular biology techniques were carried out by standard procedures (3) or as recommended by the manufacturer. T. ferrooxidans genomic DNA preparation has been described previously (4). Ultrapure plasmid DNA was obtained with a Wizard plus Spin or vacuum minipreps DNA purification system plasmid kit from Promega. Ligations were generally carried out in the presence of the restriction enzyme used to cleave the vector to prevent its recircularization.
Routine PCRs were performed with Boehringer Mannheim Taq DNA polymerase according to the manufacturer’s recommendations in a Mini-cycler (MG Research). For cloning purposes, the Pwo polymerase was preferred. Amplification of flanking sequences by inverse PCR was as described by Ochman et al. (36). All synthetic oligonucleotides for PCR and sequencing were purchased from Genset.
Sequences were determined with a Thermo Sequenase II dye terminator cycle sequencing premix kit from Amersham. The DNA sequences were compiled and analyzed through the Worldwide Web Netscape facilities. The predicted proteins were compared with EMBL, GenBank, and SwissProt database entries by using the BLASTP program (1) with the BLOSUM62 scoring matrix or, when specified, with the BLOSUM30 matrix.
Plasmid constructions.
Different DNA fragments containing the putative promoters of the cyc operon were amplified by PCR with oligonucleotides d to h, f to h, d to k, or c to l (Table 1 and see Fig. 1A and 3A) from T. ferrooxidans genomic DNA and cloned into the SmaI site of pGE593 to yield plasmids 1, 2, 4, and 5, respectively. The insert of plasmid 3 is the larger fragment obtained after digestion with SspI of the PCR fragment obtained with oligonucleotides d to h.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence | Positions (orientation)a |
|---|---|---|
| a | 5′-CGTATTCATCTATGTGCTGGTCGG-3′ | 3472–3495 (+) |
| b | 5′-CACCATCCCGTCTGAAACCCG-3′ | 4757–4787 (−) |
| c | 5′-TTGGCATGTCGATTTTTGGACC-3′ | 1–22 (+) |
| d | 5′-ATGGTTAACATGATAAAATAACG-3′ | 53–75 (+) |
| e | 5′-TGTTCGTTATTTACTTTATTGC-3′ | 98–119 (+) |
| f | 5′-GTTTGTATTAAATAGAACGTGTGG-3′ | 269–292 (+) |
| g | 5′-GGTTATGGTGTCATCGTCCGTTGG-3′ | 446–469 (+) |
| h | 5′-CCAACGGACGATGACACCATAACC-3′ | 446–469 (−) |
| i | 5′-CGCAAAGGATGGCAGTGCCCAGG-3′ | 535–557 (−) |
| j | 5′-GAACATTATTGTTGGGAGAAGC-3′ | 810–831 (−) |
| k | 5′-ACACGTTCTATTTAATACAAACCG-3′ | 267–290 (−) |
| l | 5′-ATGGTCATAACTATAATGCTTTTA-3′ | 167–190 (−) |
| m | 5′-GCTCATGCGCCCGGTCTTCCTGCC-3′ | 374–397 (−) |
| n | 5′-GACCTTTAACCAATGTTGTTGCG-3′ | 306–328 (−) |
| o | 5′-TGCCGTTTAATTATAGGCCG-3′ | 34–53 (−) |
| p | 5′-ATGTCTCGTTGCCACAAATCAAGG-3′ | 3963–3985 (−) |
The positions refer to the nucleotide sequences submitted to the EMBL data bank. + corresponds to the coding strand, and − corresponds to the noncoding strand.
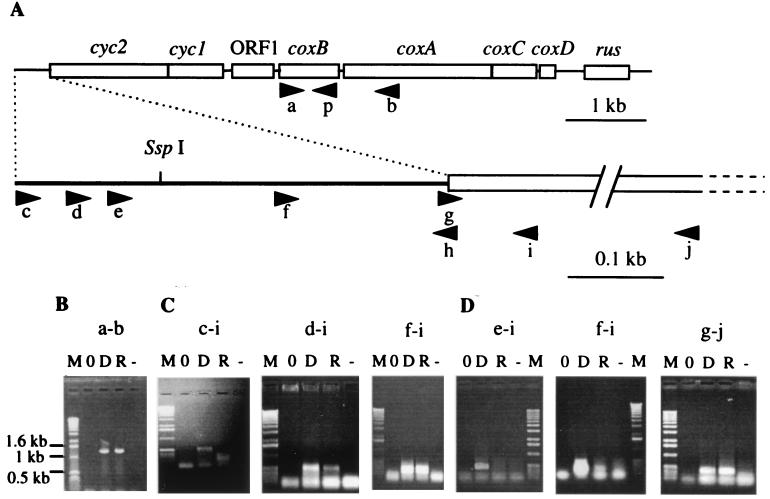
FIG. 1.
RT-PCR on T. ferrooxidans total RNA. (A) Localizations of the oligonucleotides used for RT-PCR experiments; (B) RT-PCR experiments with oligonucleotides a and b on no template (lane 0), T. ferrooxidans genomic DNA (lane D), total RNA from T. ferrooxidans cells (lane R), total RNA from T. ferrooxidans but without reverse transcriptase (lane −); (C) RT-PCR experiments with oligonucleotides c and i, d and i, and f and i on RNA extracted from sulfur-grown cells (lanes R and −); (D) RT-PCR experiments with oligonucleotides e and i, f and i, and g and j on RNA extracted from ferrous-iron-grown cells (lanes R and −). Lanes M, 1-kb molecular weight ladder from Boehringer Mannheim.
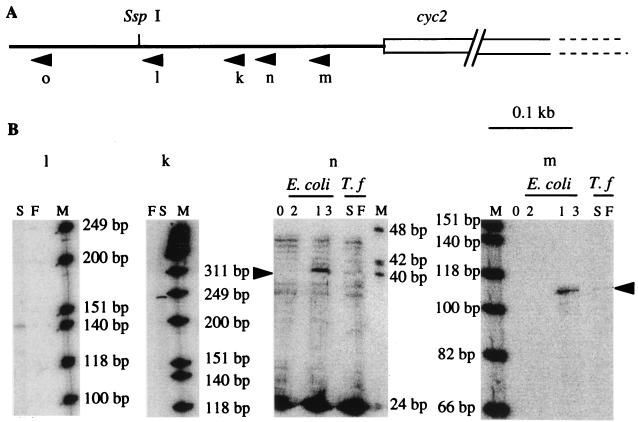
FIG. 3.
Primer extension experiments. (A) Localizations of the oligonucleotides used. (B) RT experiments with the k, l, m, and n oligonucleotides on no template (lanes 0); E. coli carrying plasmid 1, 2, or 3 (lanes 1 to 3, respectively); total RNA from ferrous-iron (lanes F)- or sulfur (lanes S)-grown T. ferrooxidans (T.f) cells. Lanes M, [γ-32P]ATP-labeled φX174 HinfI markers (from Promega).
Plasmid SK/coxB was obtained by cloning the DNA PCR fragment amplified between oligonucleotides a and p (Table 1; see Fig. 1A) into the EcoRV site of the Bluescript SK vector. Screening was performed by PCR with the oligonucleotides used to amplify the DNA. In all cases, the sequence of the cloned fragment was checked.
RNA manipulations.
T. ferrooxidans total RNA was prepared as described previously (14). RNA electrophoresis was performed with agarose-formaldehyde gels (3). RNA was transferred to a positively charged nylon membrane from Boehringer Mannheim by capillary blotting. The digoxigenin (DIG)-labeled RNA probe was obtained by in vitro transcription with T7 RNA polymerase from the EcoRI-linearized SK/coxB plasmid described above with DIG-UTP as the substrate according to the instructions of the Boehringer Mannheim DIG RNA labeling kit. Prehybridization and hybridization with a DIG-labeled probe were performed under stringent conditions according to the recommendations of Boehringer Mannheim. RNA was detected by a chemiluminescent reaction with disodium 3-(4-methoxyspuro{1,2-dioxelane-3,2′-(5′-chloro)tricyclo [3.3.1.13.7] decan}-4-yl) (CSPD) as recommended by Boehringer Mannheim.
Primer extension was performed with the Superscript II RNase H− reverse transcriptase from Gibco BRL. The k, l, m, n, and o primers used (Table 1; see Fig. 3A) were labeled with [γ-32P]ATP. Coupled reverse transcription and PCR amplification (RT-PCR) was performed with the Promega Access RT-PCR system as described previously (14) with the a, b, c, d, e, f, g, h, i, and j oligonucleotides (Table 1; see Fig. 1A). For each RT-PCR experiment, three control experiments were performed: one without template to detect any contamination, one with genomic DNA as a control for PCR amplification, and one with RNA but without the reverse transcriptase to ensure that there were no DNA traces in the RNA preparation.
Nucleotide sequence accession number.
The EMBL accession number of the 8,007-bp DNA nucleotide sequence containing the cyc2, cyc1, ORF1, coxB, coxA, coxC, coxD, and rus genes is AJ006456.
RESULTS AND DISCUSSION
Characterization of the cytochrome oxidase genes.
The cyc2 and cyc1 genes encoding a high-molecular-weight cytochrome c and a c4-type cytochrome are cotranscribed with at least one other gene (ORF1) encoding a putative periplasmic protein of unknown function (2). We have determined the nucleotide sequence downstream from ORF1 by chromosome walking using PCR and inverse-PCR approaches. Four putative open reading frames, each preceded by a correctly positioned putative ribosome binding site, were found on the same DNA strand between positions 3296 and 4060, 4417 and 6000, 6019 and 6570, and 6611 and 6805. The rus gene, which we have already characterized (4), lies immediately downstream. Downstream from ORF1, the first and second ORFs encode proteins presenting significant similarities to subunits II and I, respectively, of an aa3-type cytochrome c oxidase and will be referred to herein as the coxB and coxA genes. The two ORFs downstream from the coxA gene are those found upstream from rus (ORF1 and ORF2 in reference 4) and will be referred to herein as coxC and coxD (see below). Thus, the gene order in this locus is cyc2-cyc1-ORF1-coxB-coxA-coxC-coxD-rus.
Analysis of the coxB-encoded polypeptide.
The coxB gene encodes a putative 254-amino-acid polypeptide (CoxBTf) with a calculated molecular weight of 28,240. The first 51 amino acids may constitute a long but standard signal sequence. The mature protein has a higher similarity to subunit II of aa3-type cytochrome c oxidases (and more particularly to those of Synechococcus vulcanus, Anabaena sp. strain PCC7120, and Synechocystis sp. strain PCC6803) than to subunit II of quinol oxidases. Similar to the cytochrome oxidases, the mature CoxBTf has two putative N-transmembrane segments that serve as membrane anchors and a large periplasmic carboxy-terminal domain (for extensive references, see references 40 and 41). In the periplasmic domain, the aromatic amino-acid-rich region (145-WKWTFSY-151) involved in the electron transfer between subunits I and II is present (20, 45). Furthermore, the residues binding the dinuclear copper center (CuA) (H181, C222, C226, H230, and M233) (20, 25, 40), the residues stabilizing CuA (W145 and D178), and three of the four highly conserved residues interacting with cytochrome c (Q144, D178, and D193) (20, 27) are also present, suggesting that CoxBTf belongs to a c-type cytochrome oxidase.
In spite of the acidic pH of the T. ferrooxidans periplasm, the periplasmic domain of CoxBTf is well conserved. Acid-stable proteins generally contain a relatively low number of charged residues (29), but this is not the case in the CoxBTf periplasmic domain (11.6% R+H+K; 8% D+E). It is noteworthy that two T. ferrooxidans periplasmic redox proteins, the high-potential iron sulfur protein (HiPIP) encoded by iro and rusticyanin, have already been noted as exceptions to this rule (29). A possibility is that these electron transfer proteins are inaccessible to the periplasm medium because they are buried in a supercomplex.
Analysis of the coxA-encoded polypeptide.
The coxA gene encodes a putative 627-amino-acid polypeptide (CoxATf) with a calculated molecular weight of 69,090. This protein is related to subunit I of both quinol and cytochrome c oxidases from archaea, eucarya, and bacteria but more particularly to subunit I of the aa3-type cytochrome c oxidase from Synechocystis sp. strain PCC6803, Synechococcus vulcanus, and Anabaena sp. strain PCC7120. CoxATf contains the 12-transmembrane segment core common to all cytochrome oxidase subunits I (41). In this core region are the residues binding and stabilizing the low- and high-spin hemes (a and a3) and the copper atom (CuB): H159, H333, H382, H383, H467, H469 (Cu and heme binding), W329, Y337 (CuB stabilization), W145, R529, and R530 (heme stabilization) (20, 40, 44). The invariant phenylalanine residue (F468) involved in electron transfer between the two hemes is also present (41). Based on the predicted topology of CoxATf, all the residues binding hemes and copper are within the membrane bilayer but near the periplasm. This may explain why Kai et al. (23, 24) found that the pH optimum for T. ferrooxidans cytochrome oxidase was pH 3.5, a value corresponding to that of the periplasm. From the crystal structure of the Paracoccus denitrificans cytochrome c oxidase, two proton transfer pathways have been proposed: the K and the D channels (16, 20, 37). Although not strictly invariant, most of the residues involved in these channels are conserved in the CoxATf subunit. Five of the seven residues constituting the K channel and six of the nine residues constituting the D channel are present. Altogether, these data strongly suggest that the cox genes encode an aa3-type cytochrome c oxidase.
In addition to the core region, CoxATf contains an extended N terminus with two hydrophobic regions that have no similarity to sequences in the protein data banks. Extra hydrophobic regions have been also described for the N termini of subunits I of the cbb3-type cytochrome oxidases from Bradyrhizobium japonicum and Sinorhizobium meliloti (41).
The estimated molecular masses of subunits I of the aa3-type cytochrome oxidases purified from the T. ferrooxidans Fe-1, AP19-3, and OK1-50 strains (53, 53 and 55 kDa, respectively) (19, 24) do not correspond with the molecular mass deduced from the coxA gene sequence reported here (69 kDa). This discrepancy may be due to an aberrant migration of subunit I on sodium dodecyl sulfate gels, as was previously observed with other integral membrane proteins. Another possibility is that the cytochrome oxidase encoded by the coxBACD gene cluster described in this paper is distinct from the oxidases which have been purified.
Analysis of the coxC- and coxD-encoded polypeptides.
coxC and coxD genes encode putative polypeptides of 183 and 64 amino acids with calculated molecular weights of 20,202 and 7,211, respectively. These proteins are integral membrane proteins, with five transmembrane helices for CoxCTf and one for CoxDTf (4).
By specifying alternate scoring matrices, CoxCTf exhibits some similarity with the Mycobacterium tuberculosis, Mycobacterium leprae, Synechocystis sp. strain PCC6803, Synechococcus vulcanus, and Anabaena sp. strain PCC7120 cytochrome c oxidase subunits III. Although slight, this similarity is significant. Furthermore, the carboxy-terminal region is the best conserved, as has been observed for other cytochrome oxidase subunits III (40).
As a general rule, subunit III is the second most conserved subunit in the cytochrome oxidase family after subunit I. Surprisingly, CoxCTf is less conserved than subunit II, suggesting that it has evolved more quickly because of an interaction with another protein partner(s) specific to T. ferrooxidans.
Whatever the scoring matrices used, no similarity was detected for CoxDTf. Because the bacterial cytochrome c oxidase structural genes are always in the order coxB-coxA-coxC-coxD if they are clustered in the same locus (11, 46) and because cytochrome c oxidase subunit IV is generally a small protein which has one transmembrane helix and a sequence which is not always conserved (48), we have inferred that the coxD gene encodes cytochrome oxidase subunit IV.
Interestingly, even though T. ferrooxidans belongs to the phylogenetic β subdivision of the Proteobacteria, CoxATf, CoxBTf, and CoxCTf amino acid sequences are most closely related to those of cyanobacteria (Synechococcus vulcanus, Synechocystis sp., and Anabaena sp). However, no significative similarities were detected at the nucleotide level, dismissing the hypothesis of a lateral gene transfer. On the other hand, the ancestors of the cyanobacteria were the first to introduce oxygen into an anaerobic environment and the T. ferrooxidans way of life has been described as one of the “most primitive extant” by Cairns-Smith et al. (8), arguing rather for a convergent evolution.
Transcription of the coxB, coxA, coxC, and coxD genes in T. ferrooxidans.
If the genes encoding the different cytochrome oxidase subunits are clustered in the same locus, they are always cotranscribed (11, 46). To determine if the cox genes of T. ferrooxidans ATCC 33020 are also organized in an operon, we used the RT-PCR approach, which combines RNA RT and cDNA amplification (PCR). An amplification product of the expected size was obtained between oligonucleotides a and b, corresponding to the coxB and coxA genes, respectively (Fig. 1A), indicating that these two genes are cotranscribed (Fig. 1B). We have shown previously that (i) coxB (ORF2 in reference 2) is cotranscribed with cyc2, cyc1, and ORF1 and (ii) coxA (ORFA in reference 4) is cotranscribed with coxC, coxD (ORF1 and ORF2, respectively, in reference 4), and rus, the rus gene being the last gene of this operon. All these results suggest that cyc2, cyc1, ORF1, coxB, coxA, coxC, coxD, and rus constitute an operon.
To confirm this hypothesis, the transcription of the cox genes was studied by Northern hybridization with total RNAs from ferrous-iron-grown cells. The RNA probe was chosen to hybridize to the coxB transcript. As predicted, one major transcript of approximately 7.4 kb was detected (Fig. 2). The size of this transcript is in agreement with cyc2-cyc1-ORF1-coxB-coxA-coxC-coxD-rus cotranscription. Larger minor transcripts were also observed. These minor transcripts can be due either to mRNA processing, to transcription initiation from several promoters (see below), or to the presence of more than one cytochrome c oxidase in T. ferrooxidans. The last hypothesis is supported by the facts that (i) different cytochrome oxidases have been detected in several T. ferrooxidans strains (9, 17, 28) and (ii) cytochrome c oxidases, in which subunit I has a lower apparent molecular weight than that predicted for CoxATf, have been previously purified in T. ferrooxidans (19, 24).
FIG. 2.
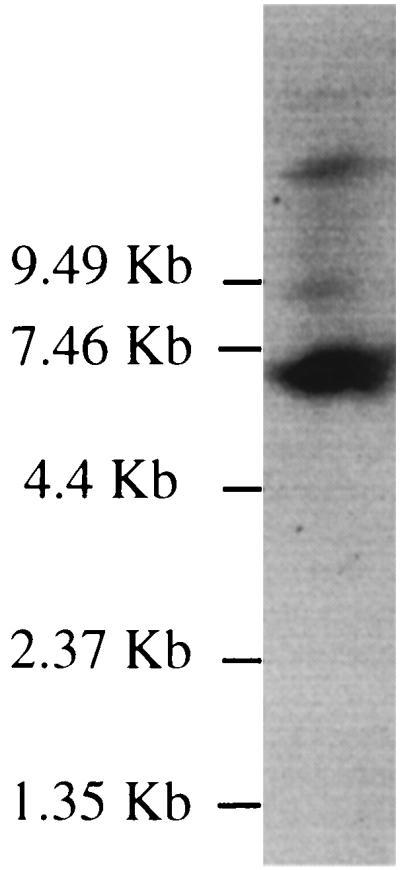
Northern blot of total RNA from ferrous-iron-grown T. ferrooxidans cells (1.1 μg) probed with DIG-UTP-labeled coxB RNA. The positions and the sizes of the RNA ladder from Gibco BRL are indicated on the left.
Characterization of the operon promoter(s).
Because no ORF has been detected in the 450 bp upstream from cyc2 (2), this gene is likely the first cistron of the operon. To determine approximately where the transcription of this operon is initiated, RT-PCR experiments were performed with a set of oligonucleotides hybridizing in the 5′ untranslated region of cyc2 (c, d, e, and f) and convergent oligonucleotides hybridizing at the beginning of cyc2 (h and i) (Fig. 1A) on total RNA extracted from sulfur- or Fe2+-grown cells. With RNAs extracted from sulfur-grown cells, cDNAs of the expected sizes were obtained between all the different pairs of oligonucleotides except with oligonucleotide c (Fig. 1C), suggesting that the mRNA starts between oligonucleotides c and d, which are between positions 1 and 75. With RNA extracted from Fe2+-grown cells, however, no amplification product was obtained with the different pairs of oligonucleotides except a faint band with oligonucleotide f (Fig. 1D), suggesting that under Fe2+ growth conditions, the mRNA starts in the region where oligonucleotide f hybridizes. Because an amplification product was obtained with two cyc2 internal oligonucleotides, g and j (Fig. 1D), the inhibition of avian myeloblastosis virus reverse transcriptase or Tfl polymerase in RNA preparation from Fe2+-grown cells is excluded. From these RT-PCR experiments, we conclude that cyc2 is the first gene of the operon and that this operon is transcribed from at least two promoters, the upstream promoter being nonfunctional under Fe2+ growth conditions. According to these results, the eight genes of the operon are transcribed in sulfur- as well as in ferrous-iron-grown ATCC 33020 cells, confirming that rusticyanin is synthesized when thiosulfate, sulfur, or ferrous iron is present in the growth medium (4) and suggesting that the proteins encoded by the operon play a role not only in ferrous-iron- but also in sulfur-grown cells.
The 5′ ends of these transcripts were determined more precisely by primer extension analysis with RNA samples prepared from T. ferrooxidans cells grown with ferrous iron or sulfur as the energy source. No signal was obtained with oligonucleotide o (data not shown), indicating that the transcription of the cyc operon initiates downstream from this oligonucleotide (position 22). A band was obtained with oligonucleotides k and l (Fig. 3) from sulfur- but not from Fe2+-grown cells. This signal corresponds to a G (position 51) located 398 bp upstream from the translational initiation site of cyc2. From Fe2+- as well as from sulfur-grown cells, a weak band was obtained with oligonucleotide m (Fig. 3, lanes S and F). This signal corresponds to a G (position 289) located 161 bp upstream from the cyc2 translational initiation site. This signal was confirmed with oligonucleotide n, even though other bands of unknown origin were observed with this oligonucleotide without any template (Fig. 3B, lanes 0). Correctly positioned upstream from the operon transcriptional initiation sites identified are two E. coli ς70-type promoters: TTGGAC(17 bp)TATAAT for the upstream promoter and TTGCAA(17 bp)TAAATA for the downstream promoter.
To determine if these promoters are functional in E. coli, different regions of the 5′ untranslated region of the operon have been cloned in the operon fusion vector pGE593 (see Materials and Methods). β-Galactosidase activities of strain MC4100 (Δlac) carrying the resulting plasmids were determined. The results unambiguously show that a sequence functioning as a promoter in E. coli is present between the SspI site and oligonucleotide k (positions 159 and 290) and that there is a second such sequence between oligonucleotides c and l (positions 1 to 190) (data not shown). Primer extension experiments with RNA samples prepared from E. coli MC4100 carrying plasmids 1, 2, and 3 (Fig. 3B, oligonucleotides m and n) and plasmid 5 (data not shown) have confirmed these results and have shown that the same transcriptional start sites are used in both T. ferrooxidans and E. coli.
The transcription of the genes of the operon appears to be complex because at least three promoters have been characterized: two upstream from cyc2 (this paper) and one between coxD and rus (4). The function of the internal promoter may be to uncouple expression of the rus gene from that of the other genes of the operon under certain growth conditions. Two of these promoters are active only under sulfur growth conditions: the most upstream promoter from cyc2 (this paper) and the internal promoter (4). The requirement for multiple promoters suggests that the expression of the different genes of the operon must be tightly regulated depending on the growth conditions to allow the cells to adapt quickly to the environmental changes. Further work will focus on (i) looking for environmental signals involved in the transcriptional regulation of this operon and (ii) determining how these signals are transmitted to the transcriptional machinery.
Electron transfer pathway between ferrous iron and molecular oxygen in T. ferrooxidans ATCC 33020.
In most cases, the cytochrome oxidase subunits are encoded within an operon with the order coxB-coxA-coxC-coxD (11, 46). In some cases, other genes required in the biogenesis of the oxidase (biosynthesis of heme A or heme O) (11, 46) or encoding other redox proteins belong to this operon. This is the case for cbb3-type cytochrome c oxidases (46) and for Sulfolobus acidocaldarius terminal oxidase complexes (41, 42). The cytochrome oxidase subunit I gene of the cbb3-type oxidases is cotranscribed with the genes encoding a mono- and a bihemic membrane-bound cytochrome c and a small membrane protein of unknown function (46). In Sulfolobus acidocaldarius, the genes encoding subunits I and II of one cytochrome oxidase are cotranscribed with the genes encoding an a-type cytochrome and a small hydrophobic subunit (41, 42). In a second cytochrome oxidase, the gene encoding subunit II and the gene encoding a subunit I-subunit III fusion protein are cotranscribed with the genes encoding sulfocyanin, an iron-sulfur protein, and a cytochrome b (41, 42). In these three cases, the redox proteins encoded by the operon constitute a respiratory chain organized as a supercomplex. Because the T. ferrooxidans aa3-type cytochrome c oxidase genes are cotranscribed with the genes encoding two cytochromes c (cyc2 and cyc1) and rusticyanin (rus), we infer that all these proteins belong to the same electron transfer chain and constitute a respiratory supercomplex. We have previously proposed the following electron pathway (1): Fe2+ → X → rusticyanin → cytochrome c4 → cytochrome oxidase → O2, in which the carrier which transfers electrons from ferrous iron to rusticyanin (X) is unknown. The HiPIP encoded by the iro gene has been proposed to receive the electrons from ferrous iron and thus to be the first electron carrier in the respiratory chain (13, 50). However, several observations appear to disagree with this hypothesis: (i) HiPIPs are generally found in photosynthetic bacteria, where they are assumed to transfer electrons between two integral transmembrane complexes, from the bc1 complex to the photosynthetic reaction center or to the terminal oxidase (6, 30); (ii) no HiPIP protein has been detected in the Tf-3 and F424 T. ferrooxidans strains (9, 17, 28); and (iii) the iro gene is monocistronic in strain Fe-1 (26), which suggests that the HiPIP is not synthesized concomitantly with the electron carriers encoded by the cyc2, cyc1, coxBACD, and rus genes. The HiPIP would rather be a nonobligatory carrier in ferrous-iron oxidation or an intermediate carrier between the bc1 complex and the terminal oxidase. We propose that the high-molecular-weight cytochrome would be a better candidate for the role of the first electron acceptor because (i) the cyc2 gene encoding this cytochrome is the first gene of the operon, (ii) this cytochrome is indeed translocated to the periplasm because it has a signal sequence that is cleaved (1), (iii) this cytochrome is likely to be acid stable because it has a low number of charged amino acids, and (iv) from its amino-acid sequence, the Psort program (35) predicts that this cytochrome c is an outer membrane protein. Some organisms, such as Desulfovibrio gigas (47), Geobacter sulfurreducens (12, 43), and Shewanella putrefaciens (32–34), have outer membrane cytochromes c. Interestingly, Geobacter sulfurreducens and Shewanella putrefaciens are able to reduce ferric iron and this ferric iron reductase activity is associated with an outer membrane cytochrome c which makes direct contact with the solid substrate.
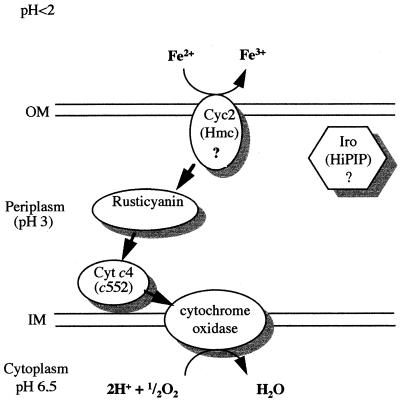
Based on genetic and biochemical evidence, we propose the pathway shown in Fig. 4 for electron transfer between ferrous iron and oxygen in T. ferrooxidans ATCC 33020; the high-molecular-weight cytochrome c encoded by the cyc2 gene transfers electrons from ferrous iron to rusticyanin, which passes them to the cytochrome c4 and from there to the cytochrome oxidase. To confirm this model, subcellular localization of the high-molecular-weight cytochrome c will be determined and the interaction between the cytochrome c4 and the cytochrome oxidase subunit II will be studied.
FIG. 4.
Proposed electron transfer pathway between ferrous iron and oxygen in T. ferrooxidans ATCC 33020. OM, outer membrane; IM, inner membrane; Cyt c4, cytochrome c4; Hmc, high-molecular-weight cytochrome c.
ACKNOWLEDGMENTS
We owe special thanks to L. Thöny-Meyer and Y. Quentin for helpful discussions. We thank M. Chippaux and more particularly J. DeMoss for critical readings of the manuscript. We are grateful to the Centre de Séquençage d’ADN (I.B.S.M., Marseille, France). We thank Patrice Brucella for skillful technical assistance.
This work was supported by grants from the CNRS, the Agence de l’Environnement et de la Maîtrise de l’Energie (A.D.E.M.E.), the Bureau de Recherche Géologique et Minière, and the Compagnie Générale des Matériaux (CO.GE.MA). N.G. acknowledges the support of a graduate scholarship from A.D.E.M.E., and C.A.-A. acknowledges support from M.E.S.R.
REFERENCES
- 1.Altschul S F, Gish W. Local alignment statistics. Methods Enzymol. 1996;266:460–480. doi: 10.1016/s0076-6879(96)66029-7. [DOI] [PubMed] [Google Scholar]
- 2.Appia-Ayme C, Bengrine A, Cavazza C, Giudici-Orticoni M-T, Bruschi M, Chippaux M, Bonnefoy V. Characterization and expression of the cotranscribed cyc1 and cyc2 genes encoding the cytochrome c4 (c552) and a high molecular weight cytochrome c from Thiobacillus ferrooxidans ATCC33020. FEMS Microbiol Lett. 1998;167:171–177. doi: 10.1111/j.1574-6968.1998.tb13224.x. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing; 1992. [Google Scholar]
- 4.Bengrine A, Guiliani N, Appia-Ayme C, Jedlicki E, Holmes D S, Chippaux M, Bonnefoy V. Sequence and expression of the rusticyanin structural gene from Thiobacillus ferrooxidans ATCC33020 strain. Biochim Biophys Acta. 1998;1443:99–112. doi: 10.1016/s0167-4781(98)00199-7. [DOI] [PubMed] [Google Scholar]
- 5.Blake R C, II, Shute E A. Respiratory enzymes of Thiobacillus ferrooxidans. Kinetic properties of an acid-stable iron:rusticyanin oxidoreductase. Biochemistry. 1994;33:9220–9228. doi: 10.1021/bi00197a025. [DOI] [PubMed] [Google Scholar]
- 6.Bonora P, Principi I, Monti B, Ciurli S, Zannoni D, Hochkoeppler A. On the role of high-potential iron-sulfur proteins and cytochromes in the respiratory chain of two facultative phototrophs. Biochim Biophys Acta. 1999;1410:51–60. doi: 10.1016/s0005-2728(98)00173-x. [DOI] [PubMed] [Google Scholar]
- 7.Bos P, Kuenen G. Microbial treatment of coal. In: Ehrlich H L, Brierley C L, editors. Microbial mineral recovery. New York, N.Y: McGraw-Hill Publishing Company; 1990. pp. 343–377. [Google Scholar]
- 8.Cairns-Smith A G, Hall A J, Russell M J. Origins of life and evolution of the biosphere. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1992. Mineral theories of the origin of life and an iron sulfide example; pp. 161–180. [Google Scholar]
- 9.Cobley J G, Haddock B A. The respiratory chain of Thiobacillus ferrooxidans: the reduction of cytochromes by Fe2+ and the preliminary characterization of rusticyanin, a novel “blue” copper protein. FEBS Lett. 1975;60:29–33. doi: 10.1016/0014-5793(75)80411-x. [DOI] [PubMed] [Google Scholar]
- 10.Eraso J-M, Weinstock G M. Anaerobic control of colicin E1 production. J Bacteriol. 1992;174:5101–5109. doi: 10.1128/jb.174.15.5101-5109.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García-Horsman J A, Barquera B, Rumbley J, Ma J, Gennis R B. The super-family of heme-copper respiratory oxidases. J Bacteriol. 1994;176:5587–5600. doi: 10.1128/jb.176.18.5587-5600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaspard S, Vazquez F, Holliger C. Localization and solubilization of the iron(III) reductase of Geobacter sulfurreducens. Appl Environ Microbiol. 1998;64:3188–3194. doi: 10.1128/aem.64.9.3188-3194.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giudici-Orticoni M-T, Nitschke W, Cavazza C, Bruschi M. Characterization and functional role of a cytochrome c4 involved in the iron respiratory electron transport chain of Thiobacillus ferrooxidans. In: Ritchie AIM, Pollard D, editors. Biomine. Glenside, Australia: The Australian Mineral Foundation; 1997. pp. PB4.1–PB4.10. [Google Scholar]
- 14.Guiliani N, Bengrine A, Borne F, Chippaux M, Bonnefoy V. Alanyl tRNA synthetase gene of the extreme acidophilic chemolithotrophic Thiobacillus ferrooxidans is highly homologous to alaS from all living kingdoms but cannot be transcribed from its promoter in Escherichia coli. Microbiology. 1997;143:2179–2187. doi: 10.1099/00221287-143-7-2179. [DOI] [PubMed] [Google Scholar]
- 15.Hall J F, Hasnain S S, Ingledew W J. The structural gene for rusticyanin from Thiobacillus ferrooxidans: cloning and sequencing of the rusticyanin gene. FEMS Microbiol Lett. 1996;137:85–89. doi: 10.1111/j.1574-6968.1996.tb08087.x. [DOI] [PubMed] [Google Scholar]
- 16.Hofacker I, Schulten K. Oxygen and proton pathways in cytochrome c oxidase. Proteins. 1998;30:100–107. [PubMed] [Google Scholar]
- 17.Ingledew W J, Cobley J G. A potentiometric and kinetic study on the respiratory chain of ferrous iron grown Thiobacillus ferrooxidans. Biochim Biophys Acta. 1980;590:141–158. doi: 10.1016/0005-2728(80)90020-1. [DOI] [PubMed] [Google Scholar]
- 18.Ingledew W J, Cox J C, Halling P J. A proposed mechanism for energy conservation during Fe2+ oxidation by Thiobacillus ferrooxidans; chemiosmotic coupling to net H+ influx. FEMS Microbiol Lett. 1977;2:193–197. [Google Scholar]
- 19.Iwahori K, Kamimura K, Sugio T. Isolation and some properties of cytochrome c oxidase purified from a bisulfite ion resistant Thiobacillus ferrooxidans strain, OK1-50. Biosci Biotechnol Biochem. 1998;62:1081–1086. doi: 10.1271/bbb.62.1081. [DOI] [PubMed] [Google Scholar]
- 20.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–668. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 21.Jensen A B, Webb C. Treatment of H2S-containing gases: a review of microbiological alternatives. Enzyme Microb Technol. 1995;17:2–10. [Google Scholar]
- 22.Juszczak A, Domka F, Kolowski M, Wachowska H. Microbial desulfurization of coal with Thiobacillus ferrooxidans bacteria. Fuel. 1995;74:725–728. [Google Scholar]
- 23.Kai M, Yano T, Fukumori Y, Yamanaka T. Cytochrome oxidase of an acidophilic iron-oxidizing bacterium, Thiobacillus ferrooxidans, functions at pH 3.5. Biochem Biophys Res Commun. 1989;2:839–843. doi: 10.1016/0006-291x(89)92510-2. [DOI] [PubMed] [Google Scholar]
- 24.Kai M, Yano T, Tamegai H, Fukumori Y, Yamanaka T. Thiobacillus ferrooxidans cytochrome c oxidase: purification, and molecular and enzymatic features. J Biochem. 1992;112:816–821. doi: 10.1093/oxfordjournals.jbchem.a123982. [DOI] [PubMed] [Google Scholar]
- 25.Kelly M, Lappalainen P, Talbo G, Haltia T, van der Oost J, Saraste M. Two cysteines, two histidines, and one methionine are ligands of a binuclear purple copper center. J Biol Chem. 1993;268:16781–16787. [PubMed] [Google Scholar]
- 26.Kusano T, Takeshima T, Sugawara K, Inoue C, Shiratori T, Yano T, Fukumori Y, Yamanaka T. Molecular cloning of the gene encoding Thiobacillus ferrooxidans Fe(II) oxidase. J Biol Chem. 1992;267:11242–11247. [PubMed] [Google Scholar]
- 27.Lappalainen P, Watmough N J, Greenwood C, Saraste M. Electron transfer between cytochrome c and the isolated CuA domain: identification of substrate binding residues in cytochrome c oxidase. Biochemistry. 1995;34:5824–5830. doi: 10.1021/bi00017a014. [DOI] [PubMed] [Google Scholar]
- 28.Mansch R, Sand W. Acid-stable cytochromes in ferrous ion oxidizing cell-free preparations from Thiobacillus ferrooxidans. FEMS Microbiol Lett. 1992;92:83–88. [Google Scholar]
- 29.Matzke J, Schwermann B, Bakker E P. Acidostable and acidophilic proteins: the example of the α-amylase from Alicyclobacillus acidocaldarius. Comp Biochem Physiol. 1997;118:475–479. doi: 10.1016/s0300-9629(97)00008-x. [DOI] [PubMed] [Google Scholar]
- 30.Menin L, Gaillard J, Parot P, Schoepp B, Nitschke W, Vermeglio A. Role of HiPIP as electron donor to the RC-bound cytochrome in photosynthetic purple bacteria. Photosynth Res. 1998;55:343–348. [Google Scholar]
- 31.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 32.Myers C R, Myers J M. Localization of cytochromes to the outer membrane of anaerobically grown Shewanella putrefaciens MR-1. J Bacteriol. 1992;174:3429–3438. doi: 10.1128/jb.174.11.3429-3438.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers C R, Myers J M. Outer membrane cytochromes of Shewanella putrefaciens MR-1: spectral analysis, and purification of the 83-kDa c-type cytochrome. Biochim Biophys Acta. 1997;1326:307–318. doi: 10.1016/s0005-2736(97)00034-5. [DOI] [PubMed] [Google Scholar]
- 34.Myers J M, Myers C R. Isolation and sequence of omcA, a gene encoding a decaheme outer membrane cytochrome c of Shewanella putrefaciens MR-1, and detection of omcA homologs in other strains of S. putrefaciens. Biochim Biophys Acta. 1998;1373:237–251. doi: 10.1016/s0005-2736(98)00111-4. [DOI] [PubMed] [Google Scholar]
- 35.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in Gram-negative bacteria. Proteins Struct Funct Genet. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 36.Ochman H, Medhora M M, Garza D, Hartl D L. Amplification of flanking sequences by inverse PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols, a guide to methods and applications. San Diego, Calif: Academic Press, Inc., Harcourt Brace Jovanovich, Publishers; 1990. pp. 219–227. [Google Scholar]
- 37.Ostermeier C, Iwata S, Michel H. Cytochrome c oxidase. Curr Opin Struct Biol. 1996;6:460–466. doi: 10.1016/s0959-440x(96)80110-2. [DOI] [PubMed] [Google Scholar]
- 38.Pulgar V, Nunez L, Moreno F, Orellana O, Jedlicki E. Expression of rusticyanin gene is regulated by growth condition in Thiobacillus ferrooxidans. In: Torma A E, Apel M L, Brierley C L, editors. Biohydrometallurgical technologies. II. Warrendale, Pa: The Minerals, Metals and Material Society; 1993. pp. 541–548. [Google Scholar]
- 39.Rawlings D E, Silver S. Mining with microbes. Bio/Technology. 1995;13:773–778. [Google Scholar]
- 40.Saraste M. Structural features of cytochrome oxidase. Q Rev Biophys. 1990;23:331–366. doi: 10.1017/s0033583500005588. [DOI] [PubMed] [Google Scholar]
- 41.Saraste M, Castresana J, Higgins D, Lübben M, Wilmanns M. Evolution of cytochrome oxidase. In: Baltscheffsky H, editor. Origin and evolution of biological energy conversion. New York, N.Y: VCH; 1994. pp. 255–289. [Google Scholar]
- 42.Schäfer G. Bioenergetics of the archaebacterium Sulfolobus. Biochim Biophys Acta. 1996;1277:163–200. doi: 10.1016/s0005-2728(96)00104-1. [DOI] [PubMed] [Google Scholar]
- 43.Seeliger S, Cord-Ruwisch R, Schink B. A periplasmic and extracellular c-type cytochrome of Geobacter sulfurreducens acts as a ferric iron reductase and as an electron carrier to other acceptors or to partner bacteria. J Bacteriol. 1998;180:3686–3691. doi: 10.1128/jb.180.14.3686-3691.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shapleigh J P, Hosler J P, Tecklenburg M M J, Kim Y, Babcock G T, Gennis R B, Ferguson-Miller S. Definition of the catalytic site of cytochrome c oxidase: specific ligands of heme a and the heme a3-CuB center. Proc Natl Acad Sci USA. 1992;89:4786–4790. doi: 10.1073/pnas.89.11.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steffens G J, Buse G. Studies on cytochrome c oxidase, IV. Primary structure and function of subunit II. Hoppe-Seyler’s Z Physiol Chem. 1979;360:613–619. [PubMed] [Google Scholar]
- 46.Thöny-Meyer L. Biogenesis of respiratory cytochromes in bacteria. Microbiol Mol Biol Rev. 1997;61:337–376. doi: 10.1128/mmbr.61.3.337-376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Ommen Kloeke F, Bryant R D, Laishley E J. Localization of cytochromes in the outer membrane of Desulfovibrio vulgaris (Hildenborough) and their role in anaerobic corrosion. Anaerobe. 1995;1:351–358. doi: 10.1006/anae.1995.1038. [DOI] [PubMed] [Google Scholar]
- 48.Witt H, Ludwig B. Isolation, analysis, and deletion of the gene coding for subunit IV of cytochrome c oxidase in Paracoccus denitrificans. J Biol Chem. 1997;272:5514–5517. doi: 10.1074/jbc.272.9.5514. [DOI] [PubMed] [Google Scholar]
- 49.Yamanaka T, Fukumori Y. Molecular aspects of the electron transfer system which participates in the oxidation of ferrous ion by Thiobacillus ferrooxidans. FEMS Microbiol Rev. 1995;17:401–413. doi: 10.1111/j.1574-6976.1995.tb00222.x. [DOI] [PubMed] [Google Scholar]
- 50.Yamanaka T, Yano T, Kai M, Tamegai H, Sato A, Fukumori Y. The electron transfer system in an acidophilic iron-oxidizing bacterium. In: Mukohata Y, editor. New era of bioenergetics. Tokyo, Japan: Academic Press; 1991. pp. 223–246. [Google Scholar]