Summary
The recent characterization of RNA-targeting CRISPR nucleases has enabled diverse transcriptome engineering and screening applications that depend crucially on prediction and selection of optimized CRISPR guide RNAs (gRNAs). Previously, we developed a computational model to predict RfxCas13d gRNA activity for all human protein-coding genes. Here, we extend this framework to six model organisms (human, mouse, zebrafish, fly, nematode, and flowering plants) for protein-coding genes and noncoding RNAs (ncRNAs) and also to four RNA virus families (severe acute respiratory syndrome coronavirus 2 [SARS-CoV-2], HIV-1, H1N1 influenza, and Middle East respiratory syndrome [MERS]). We include experimental validation of predictions by testing knockdown of multiple ncRNAs (MALAT1, HOTAIRM1, Gas5, and Pvt1) in human and mouse cells. We developed a freely available web-based platform (cas13design) with pre-scored gRNAs for transcriptome-wide targeting in several organisms and an interactive design tool to predict optimal gRNAs for custom RNA targets entered by the user. This resource will facilitate CRISPR-Cas13 RNA targeting in model organisms, emerging viral threats to human health.
Keywords: Cas13, CRISPR, model organisms, RNA viruses, on-target efficiency prediction
Graphical abstract
Highlights
Optimized CRISPR-Cas13d guide RNAs for mRNA and noncoding RNA knockdown
Pre-designed guide RNAs for 6 model organisms and 4 RNA virus families
Top-scoring guide RNAs improve knockdown of human and mouse noncoding RNAs
Web-based interface enables guide RNA design for custom RNA targets
To accelerate CRISPR-based targeting of RNA, Guo et al. present a resource with optimized RfxCas13d guide RNAs (gRNAs) to target messenger RNAs and noncoding RNAs in six common model organisms and four RNA virus families. An accompanying open access web-based platform and design tool enable optimal gRNA design for any RNA target.
Introduction
CRISPR-Cas13 mediates robust transcript knockdown in human cells through direct RNA targeting.1, 2, 3, 4 Compared with DNA-targeting CRISPR enzymes like Cas9, RNA targeting by Cas13 is transcript and strand specific; it can distinguish and specifically knock down processed transcripts, alternatively spliced isoforms, and overlapping genes, all of which frequently serve different functions. Several recent studies targeting different types of transcripts in diverse organisms have demonstrated the wide applicability of CRISPR-Cas13 RNA knockdown. In mammalian systems, CRISPR-Cas13 targeting has been used to select specific isoforms in cellular models of neurodegeneration,5 to identify noncoding transcripts that modulate cancer phenotypes like chemotherapy resistance6 and tumor proliferation,7 and to block infection by RNA viruses via targeted cleavage of viral RNA.8,9 Cas13 transcriptome modification has also been applied in vivo in diverse organisms, including Drosophila,10,11 zebrafish embryos,12 mouse embryos,12 and plants.13 Although there is a growing interest in targeting different types of transcripts across organisms, the biomedical community lacks resources to facilitate easy design of optimized Cas13d guide RNAs (gRNAs) for noncoding RNAs (ncRNAs),6,7 viral RNAs,8,9 and protein-coding transcripts in other commonly used organisms.5,10, 11, 12, 13
Previously, we used a massively parallel screening approach to identify a set of optimal design rules for RfxCas13d gRNAs and developed a computational model to predict gRNA efficacy for all human protein-coding genes.14 Here, we extended this framework to predict optimized Cas13 gRNAs for messenger RNAs and ncRNAs in six model organisms (human, mouse, zebrafish, fly, nematode, and flowering plants) and four abundant RNA virus families (severe acute respiratory syndrome coronavirus 2 [SARS-CoV-2], HIV-1, H1N1 influenza, and Middle East respiratory syndrome [MERS]). For four ncRNAs, we experimentally validated these predictions by comparing Cas13 knockdown of predicted high- and low-efficacy gRNAs in human and mouse cell lines. To allow more flexible gRNA design, we also developed an open access web-based application to enable prediction of optimal Cas13d gRNAs for any RNA target entered by the user.
Results
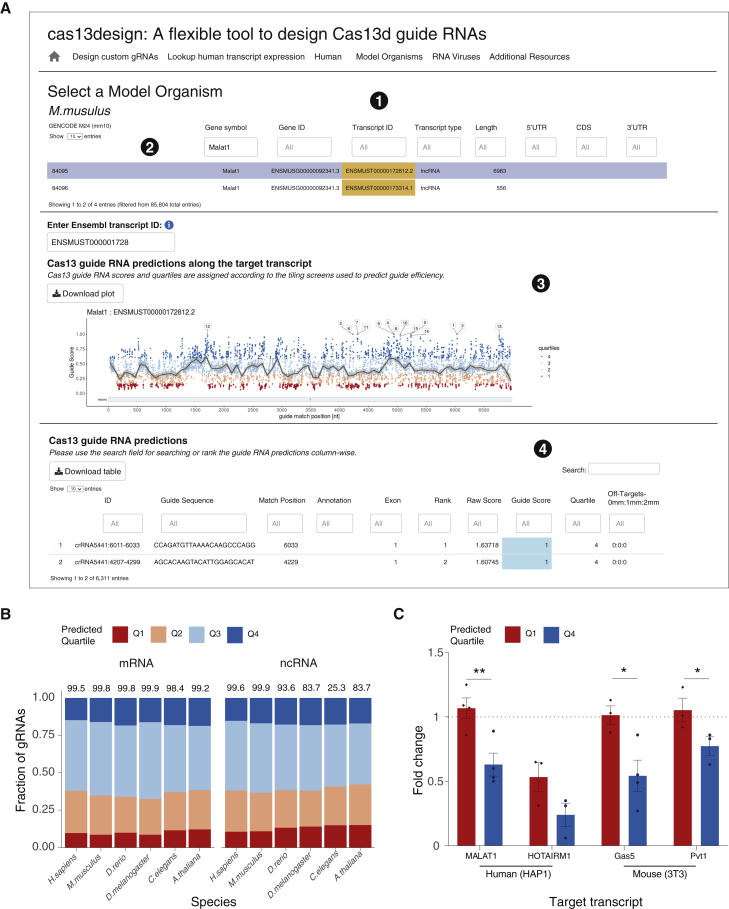
To select optimal gRNAs for transcripts produced from the reference genomes of human, mouse, zebrafish, fly, nematode, and flowering plants, we created the cas13design online platform (https://cas13design.nygenome.org/; Figure 1A). We previously found that optimal Cas13 gRNAs depend on specific sequence and structural features, including position-based nucleotide preferences in the gRNA and the predicted folding energy (secondary structure) of the combined direct repeat plus gRNA.14 Using this algorithm, we pre-computed gRNA efficacies, where possible, for all mRNAs and ncRNAs with varying transcript lengths for the six model organisms (Figure 1B). For the scored gRNAs for each organism, we found that approximately 20% are ranked in the top quartile (Q4 gRNAs) for both mRNAs and ncRNAs. Remarkably, even though the nucleotide composition can vary between RNAs from different species,15, 16, 17 we find a similar proportion of optimal RfxCas13d gRNAs across all six species.
Figure 1.
A graphical interface for optimized CRISPR-Cas13d gRNA design for messenger RNAs (mRNAs) and noncoding RNAs (ncRNAs) from six common model organisms
(A) Example output of the cas13design webtool. (1) Selection of model organisms. (2) Searches by gene symbol or transcript ID for gRNA design, with options to download generated plots and data tables. (3) Interactive display of gRNAs along the target transcript, color coded by the predicted targeting efficacy scores separated into four quartiles. Q4 gRNAs correspond to those with the highest predicted efficacy, and Q1 gRNAs correspond to those with the lowest predicted efficacy. (4) Display of gRNA options with on-target score predictions and potential off-targets by number of mismatches (number of sequences in the transcriptome with 0, 1, or 2 mismatches).
(B) The predicted guide efficacy quartiles for mRNAs and ncRNAs across six model organisms. The percentage of scored transcripts that meet the minimal length requirement for target RNAs (80 nt) is indicated above each bar.
(C) Average lncRNA knockdown for Q4 and Q1 gRNAs (∗p < 0.05, ∗∗p < 0.01, two-tailed Student’s t test; mean ± SEM, n = 3–4 different gRNAs from the specified prediction quartile, each transduced with three biological replicates).
See also Figure S3.
Next, we examined how many predicted high-efficacy gRNAs are present, on average, in different transcripts. To do this, we determined what fraction of the transcripts in each organism include n top-scoring (Q4) gRNAs for values of n between 1 and 25 (Figure S1). We found that coding sequences contained a higher number of top-scoring gRNA per transcript across all organisms, whereas targeting the noncoding transcriptome and UTRs (3′ UTRs and 5′ UTRs) was more challenging (Figure S2). This reduction in the number of top-scoring gRNAs was most pronounced in C. elegans, possibly because of its noncoding transcriptome containing many short ncRNAs. On average, we were able to find at least 25 Q4 gRNAs for more than 99% of coding exons in mRNAs but only 80% of ncRNAs.
Previously, we demonstrated that Q4 gRNAs result in better knockdown for protein-coding genes than Q1 gRNAs.14 To validate gRNA predictions for ncRNA knockdown, we targeted four long ncRNAs (lncRNAs) in the human and mouse transcriptome (human, MALAT1 and HOTAIRM1; mouse, Gas5 and Pvt1). Using RNA sequencing, we first confirmed that the selected lncRNAs were expressed in HAP1 (human) or NIH 3T3 (mouse) cells. For each lncRNA, we cloned and lentivirally transduced at least three gRNAs predicted as Q4 gRNAs and at least three gRNAs predicted as Q1 gRNAs. In total, each lncRNA was targeted with 6–8 distinct gRNAs. After 3 days, we extracted RNA and measured lncRNA knockdown by qPCR. We found that, for all targeted lncRNAs, Q4 gRNAs resulted in greater transcript knockdown than Q1 gRNAs (Figure 1C; Figure S3). The highest knockdown achieved for an individual Q4 gRNA in our dataset was 99% when targeting the lncRNA HOTAIRM1. For 3 of 4 targeted lncRNAs, we observed no statistically significant knockdown with the Q1 gRNAs, further reinforcing the importance of gRNA prediction for effective transcript knockdown.
Recently, several groups have proposed using CRISPR-Cas13 nucleases to directly target viral RNAs8,18 for viral diagnostics and treatment, which has become an area of rapid technology development because of the recent coronavirus disease 2019 (COVID-19) pandemic.19 However, these approaches do not use optimized Cas13 gRNAs. Previously, we showed that optimal gRNAs targeting an EGFP transgene can result in an ∼10-fold increase in knockdown efficacy compared with other gRNAs.14 Therefore, to facilitate functional studies of viral genetic elements, we applied our design algorithm to target SARS-CoV-2 and other pathogenic RNA viruses using Cas13d.
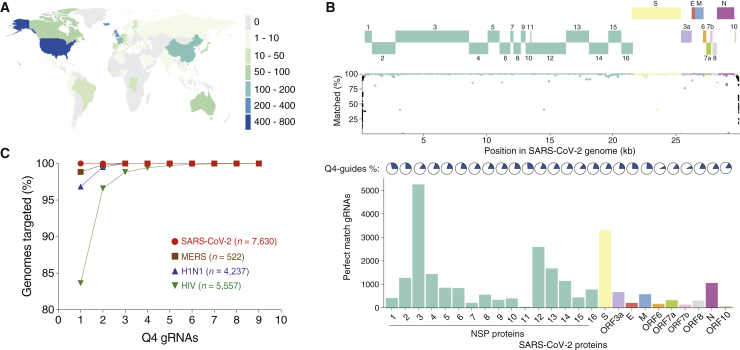
To ensure coverage of diverse isolates from affected individuals, we collected 7,630 sequenced SARS-CoV-2 genomes submitted to the Global Initiative on Sharing All Influenza Data (GISAID) database from 58 countries/regions (Figure 2A).20 Using the first sequenced SARS-CoV-2 isolate from New York City (USA/NY1-PV08001/2020) as a reference,21 we evaluated how many individual SARS-CoV-2 genomes each reference gRNA can target (Figure 2B). gRNAs targeting protein-coding regions are mostly well conserved across all genomes, with lower conservation in more variable regions, such as non-structural protein 14 (NSP14) and spike (S) protein. We found that gRNAs targeting in the 5′ and 3′ UTRs tended to be poorly conserved, as might be expected given the lack of coding function of these regions (Figure S4). Upon examination of each of the 26 SARS-CoV-2 genes, we found that all gene transcripts could be targeted with Q4 gRNAs.
Figure 2.
Optimal CRISPR-Cas13d gRNAs to target common human pathogenic RNA viruses
(A) World map of analyzed SARS-CoV-2 isolates (data from GISAID, April 17, 2020). Numbers in the legend denote isolate counts.
(B) gRNA design for each SARS-CoV-2 gene. Top: SARS-CoV-2 gene annotations. Center: percentage of SARS-CoV-2 genomes targeted by each NY1 reference gRNA. Bottom: fraction of gRNAs in Q4 per gene (pies) and total number of Q4 gRNAs per gene that targets at least 99% of the total genomes (bars).
(C) Predicted minimum number of Q4 gRNAs to target all analyzed SARS-CoV-2, MERS-CoV, H1N1, and HIV-1 genomes (n = 7,630, 522, 4,237, and 5,557 viral genomes, respectively).
Similarly, we designed and scored all gRNAs for the MERS coronavirus and two other RNA viruses: HIV-1, which drives acquired immunodeficiency syndrome (AIDS), and H1N1 pandemic influenza. Unlike SARS-CoV-2, where a single high-efficacy (Q4) gRNA can target all analyzed genomes, we found that at least two gRNAs are needed to target nearly all available genomes. For the highly mutagenic virus HIV-1,22 we found that nine gRNAs are needed to target all available genomes (Figure 2C). Given the tremendous current interest in viral RNA targeting using Cas13 enzymes, this dataset of optimized gRNAs will be useful as a platform for broad targeting of viral populations from diverse isolates from affected individuals. All designed gRNAs for model organism and viral transcripts can be browsed interactively or downloaded in bulk on the design tool website. Finally, to target transcripts from non-model organisms, synthetic RNAs, and transcripts carrying genetic variants not found in reference genomes, we developed a web-based interactive design mode where the user can enter a custom RNA sequence for selection and scoring of gRNAs.
Discussion
RNA-targeting CRISPR-Cas13 has great potential for transcriptome perturbation and antiviral therapy. In this study, we designed and scored Cas13d gRNAs for mRNAs and ncRNAs in six common model organisms and identified optimized gRNAs to target nearly all sequenced viral RNAs for SARS-CoV-2, HIV-1, H1N1 influenza, and MERS. We expanded our web-based platform to make the Cas13 gRNA design readily accessible for model organisms and created a new application to enable gRNA predictions for custom target RNA sequences. This unique resource provides an advance over existing Cas13 guide design tools23,24 as the first to use on-target efficiencies in gRNA predictions and focus on Cas13 orthologs (e.g., Cas13a) that have significant non-specific cleavage (Table S1).25 To facilitate potential high-throughput design and development of CRISPR-Cas13 libraries for functional transcriptomics screens, we also have made all pre-scored gRNAs available for batch download. We anticipate that this resource will greatly facilitate CRISPR-Cas13 RNA targeting in model organisms, emerging viral threats to human health.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| NEB Stable Cells | New England Biolabs | Cat#C3040I |
| Oligonucleotides | ||
| lncRNA-targeting gRNA oligo sequences, see Table S2 | This paper | N/A |
| qPCR primers for gene expression quantification, see Table S3 | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Polyethyleneimine | Polysciences | Cat#23966 |
| Critical commercial assays | ||
| Direct-zol RNA MicroPrep | Zymo Research | Cat# R2061 |
| RevertAid RT Reverse Transcription Kit | Thermo Fisher Scientific | Cat# K1691 |
| Luna Universal qPCR Master Mix | New England Biolabs | Cat#M3003E |
| Deposited data | ||
| Reference transcriptome (H. sapiens: GENCODE v19, GRCh37) | ENSEMBL | ftp://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_19/gencode.v19.pc_transcripts.fa.gz |
| Reference annotations (H. sapiens: GENCODE v19, GRCh37) | ENSEMBL | ftp://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_human/release_19/gencode.v19.annotation.gtf.gz |
| Reference transcriptome (M. musculus: GENCODE M24, mm10) | ENSEMBL | ftp://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_mouse/release_M24/gencode.vM24.pc_transcripts.fa.gz |
| Reference annotations (M. musculus: GENCODE M24, mm10) | ENSEMBL | ftp://ftp.ebi.ac.uk/pub/databases/gencode/Gencode_mouse/release_M24/gencode.vM24.annotation.gtf.gz |
| Reference transcriptome - mRNA (D. rerio: Ensembl v99, GRCz11) | ENSEMBL | ftp://ftp.ensembl.org/pub/release-99/fasta/danio_rerio/cdna/Danio_rerio.GRCz11.cdna.all.fa.gz |
| Reference transcriptome - ncRNA (D. rerio: Ensembl v99, GRCz11) | ENSEMBL | ftp://ftp.ensembl.org/pub/release-99/fasta/danio_rerio/ncrna/Danio_rerio.GRCz11.ncrna.fa.gz |
| Reference annotations (D. rerio: Ensembl v99, GRCz11) | ENSEMBL | ftp://ftp.ensembl.org/pub/release-99/gtf/danio_rerio/Danio_rerio.GRCz11.99.gtf.gz |
| Reference transcriptome - mRNA (D. melanogaster: Ensembl v99, BDGP6) | ENSEMBL | ftp://ftp.ensembl.org/pub/release-100/fasta/drosophila_melanogaster/cdna/Drosophila_melanogaster.BDGP6.28.cdna.all.fa.gz |
| Reference transcriptome - ncRNA (D. melanogaster: Ensembl v99, BDGP6) | ENSEMBL | ftp://ftp.ensembl.org/pub/release-99/fasta/drosophila_melanogaster/ncrna/Drosophila_melanogaster.BDGP6.28.ncrna.fa.gz |
| Reference annotations (D. melanogaster: Ensembl v99, BDGP6) | ENSEMBL | ftp://ftp.ensembl.org/pub/release-100/gtf/drosophila_melanogaster/Drosophila_melanogaster.BDGP6.28.100.gtf.gz |
| Reference transcriptome - mRNA (C. elegans: Ensembl Plants v46, WBcel235) | ENSEMBL | ftp://ftp.ensembl.org/pub/release-100/fasta/caenorhabditis_elegans/cdna/Caenorhabditis_elegans.WBcel235.cdna.all.fa.gz |
| Reference transcriptome - ncRNA (C. elegans: Ensembl Plants v46, WBcel235) | ENSEMBL | ftp://ftp.ensembl.org/pub/release-99/fasta/caenorhabditis_elegans/ncrna/Caenorhabditis_elegans.WBcel235.ncrna.fa.gz |
| Reference annotations (C. elegans: Ensembl Plants v46, WBcel235) | ENSEMBL | ftp://ftp.ensembl.org/pub/release-100/gtf/caenorhabditis_elegans/Caenorhabditis_elegans.WBcel235.100.gtf.gz |
| Reference transcriptome - mRNA (A. thaliana: Ensembl v99, TAIR10) | ENSEMBL | ftp://ftp.ensemblgenomes.org/pub/plants/release-46/fasta/arabidopsis_thaliana/cdna/Arabidopsis_thaliana.TAIR10.cdna.all.fa.gz |
| Reference transcriptome - ncRNA (A. thaliana: Ensembl v99, TAIR10) | ENSEMBL | ftp://ftp.ensemblgenomes.org/pub/plants/release-46/fasta/arabidopsis_thaliana/ncrna/Arabidopsis_thaliana.TAIR10.ncrna.fa.gz |
| Reference annotations (A. thaliana: Ensembl v99, TAIR10) | ENSEMBL | ftp://ftp.ensemblgenomes.org/pub/plants/release-46/gff3/arabidopsis_thaliana/Arabidopsis_thaliana.TAIR10.46.gff3.gz |
| Reference genome (SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2) | NCBI | https://www.ncbi.nlm.nih.gov/nuccore/1834374999 |
| Reference annotation (SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2) | NCBI | https://www.ncbi.nlm.nih.gov/nuccore/NC_045512.2 |
| Reference genome (MERS: Middle East respiratory syndrome coronavirus, complete genome) | NCBI | https://www.ncbi.nlm.nih.gov/nuccore/NC_019843.3 |
| Reference annotation (MERS: Middle East respiratory syndrome coronavirus, complete genome) | NCBI | https://www.ncbi.nlm.nih.gov/nuccore/NC_019843.3 |
| Reference genome (HIV1: Human immunodeficiency virus 1, complete genome) | NCBI | https://www.ncbi.nlm.nih.gov/nuccore/NC_001802.1 |
| Reference annotation (HIV1: Human immunodeficiency virus 1, complete genome) | NCBI | https://www.ncbi.nlm.nih.gov/nuccore/NC_001802.1 |
| Reference genome (H1N1: Influenza A virus) | NCBI | https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=641809 |
| Reference annotation (H1N1: Influenza A virus) | NCBI | https://www.ncbi.nlm.nih.gov/nuccore/?term=Influenza+A+virus+(A%2FCalifornia%2F07%2F2009(H1N1)) |
| Analyses and summary statistics of designed guide RNAs | This paper | https://gitlab.com/sanjanalab/cas13_webtool |
| Experimental models: Cell lines | ||
| HAP1 | Landau lab | N/A |
| NIH 3T3 | ATCC | CRL-1658 |
| HAP1-Cas13d | This study | N/A |
| NIH 3T3-Cas13d | This study | N/A |
| HEK293FT | Thermo Fisher Scientific | Cat#R70007 |
| Recombinant DNA | ||
| pLentiRNACRISPR_007 - TetO-NLS-RfxCas13d-NLS-WPRE-EFS-rtTA3-2A-Blast | Wessels et al.14 | Addgene 138149 |
| pLentiRNAGuide_001 - hU6-RfxCas13d-DR1-BsmBI-EFS-Puro-WPRE | Wessels et al.14 | Addgene 138150 |
| pMD2.G | Trono Lab packaging and envelope plasmids | Addgene 12259 |
| psPAX2 | Trono Lab packaging and envelope plasmids | Addgene 12260 |
| Software and algorithms | ||
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com/ |
| RStudio | RStudio | https://www.rstudio.com/ |
| Python version 2.7.8 | Python Software Foundation | https://www.python.org |
| Cas13 guide design algorithm | Wessels et al.14 | https://gitlab.com/sanjanalab/cas13 |
| Cas13 design tool | This paper | https://cas13design.nygenome.org/ |
Resource availability
Lead contact
Further information requests should be directed to the Lead Contact, Neville Sanjana (neville@sanjanalab.org).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human and mouse cell culture
HAP1 cells (male) were obtained from Horizons. NIH 3T3 (male) and HEK293FT (female) cells were obtained from ATCC. HAP1 cells were maintained at 37°C with 5% CO2 in I10 media: Iscove’s Modified Dulbecco’s Medium (Thermo Fisher) supplemented with 10% Serum Plus II (Sigma-Aldrich). NIH 3T3 cells were maintained at 37°C with 5% CO2 in D10 media: Dulbecco’s Modified Eagle’s Medium with high glucose and stabilized L-glutamine (Caisson Labs) supplemented with 10% Serum Plus II (Sigma-Aldrich). HEK293FT cells were maintained at 37°C with 5% CO2 in D10 media.
Method details
gRNA design for model organisms
Reference transcriptomes and corresponding annotations were obtained for each model organism: H. sapiens (GENCODE v19, GRCh37), M. musculus (GENCODE M24, mm10),26 D. rerio (Ensembl v99, GRCz11), D. melanogaster (Ensembl v99, BDGP6), C. elegans (Ensembl v99, WBcel235) and A. thaliana (Ensembl Plants v46, TAIR10).27 For each organism, we performed the on-target efficiency predictions for both mRNAs and ncRNAs using command-line RfxCas13d designer version 0.2 as previously described.14 We scored gRNAs for all RNA targets with a length of at least 80 nucleotides.
Prediction of gRNA off-targets
Each gRNA designed was aligned against the corresponding transcriptome with bowtie (v1.1.2)28 using the following command: bowtie–nofw -a–threads 20 -n 2 -f %s %s -S–sam-nohead %s–un %s. The process outputs all valid alignments with no greater than 2 mismatches, and refrains any mapping against the forward-reference strand. We then determined from each SAM file the number of unique off-target gene that individual gRNA sequences mapped to at varying mismatch thresholds (perfect match, one mismatch, or two mismatches).
Knock-down of lncRNAs with Cas13d
We first established doxycycline-inducible Cas13d cell lines for HAP1 cells and NIH 3T3 by transducing cells with an inducible RfxCas13d lentivirus (Addgene 138149). Transduced HAP1 and NIH 3T3 are maintained in I10 media with 10 μg/ml of blasticidin S (Thermo Fisher), and D10 with 5 μg/ml of blasticidin S, respectively. To produce lentivirus, we transfected HEK293FT cells with 1 μg of the transfer plasmid together with viral packaging plasmids (0.8 μg of psPAX2: Addgene 12260; and 0.55 μg pMD2.G: Addgene 12259) using 5.5 μL of 1 mg/mL polyethylenimine (PEI, Polysciences).
Candidate lncRNAs were characterized in the past in either mouse or human models.29,30 We first acquired RNA-seq data for human HAP1 cells (accession: GSE80793) and mouse NIH 3T3 cells (accession: GSM2897262) from NCBI Gene Expression Omnibus (GEO), and confirmed that the selected lncRNAs were expressed in respective cell lines. For each gene, we designed at least three predicted Quartile 4 gRNAs (Q4, or predicted high efficacy) and at least three predicted Quartile 1 gRNAs (Q1, or predicted low efficacy) with our cas13design webtool. The gRNA sequences and predicted scores can be found in Table S2. We synthesized oligonucleotides with these sequences (IDT) and cloned them into a U6-driven RfxCas13d gRNA lentiviral vector (Addgene 138150). We annealed and phosphorylated the oligos before ligation into the backbone using T7 ligase (NEB). All constructs were sequence confirmed with Sanger sequencing. For each gRNA construct, we produced lentivirus as described above. At day 3 post-transfection, viral supernatant was collected and stored at −80°C until use.
All lentiviral gRNA transduction experiments were performed in biological triplicate. At day 1 post transduction, we treated cells with 1 μg/mL puromycin and 1 μg/mL doxycycline for transduction selection and Cas13 expression induction and then cultured for 2 additional days before RNA extraction. We extracted total RNA from each sample using Direct-zol RNA MicroPrep (Zymo). For each sample, we reverse-transcribed 830 ng of total RNA into cDNA with RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher). We performed SYBR Green quantitative PCR (qPCR) with Luna Universal qPCR Master Mix (NEB). The qPCR primers were designed using Primer-BLAST and wherever possible we selected amplicons that spanned an intron to minimize the possibility of genomic DNA amplification (Table S3). We quantified qPCR changes using the ΔΔCt method: For each biological sample, we first normalized for input using GAPDH gene expression31 and then computed fold-change relative to the non-targeting gRNA control.
RNA virus genome collection
All full-length RNA virus genomes were downloaded on April 17th, 2020, from the GISAID20 and NCBI Virus32 databases. We downloaded 7,630 complete SARS-CoV-2 viral genomes classified as high coverage and 4,237 Influenza A H1N1 viral genomes with a complete set of eight genomic segments. SARS-CoV-2 and H1N1 genomes were obtained from GISAID (https://www.gisaid.org/). We also analyzed 522 MERS-CoV and 5,557 full length HIV-1 viral genomes, which were downloaded from NCBI Virus (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/).
gRNA design to target SARS-CoV-2
We split multi-FASTA files into single-entry FASTA files using the UCSC tool faSplit.33 All possible 23-mer gRNAs targeting individual genomes were scored with the RfxCas13 on-target model described previously.14 All scored guide RNAs were classified into four quartiles. Quartile 4 guide RNAs (or Q4) are designated to be the predicted best-performing guide RNAs. We used USA/NY1-PV08001/2020 (NY1 isolate) for the SARS-CoV-2 reference gRNA design. Compared to the earlier (Wuhan) isolate, NY1 contains 3 nucleotide substitutions (G3243A, C25214T, G29027T) resulting in two amino acid mutations (N: A252S, ORF1a: G993S). The SARS-CoV-2 transcript annotation was obtained from NCBI (GenBank: NC_045512.2).
Prediction of minimal numbers of gRNAs to target RNA viruses
For each RNA virus, we identified a minimal set of high-scoring Q4 gRNAs that could target all genomes collected. We used a greedy algorithm as described previously:8 For each iteration, the gRNAs with the highest number of targeting genomes are added to the set. During each iteration, if multiple gRNAs target the same highest number of genomes, we will pick one for the minimal set and start the next iteration.
Quantification and statistical analysis
Data analysis was performed in GraphPad Prism 8 and RStudio (R v3.5.1). All transduction experiments show the mean of three replicates, with error bars representing the standard error of mean, see each figure legend for specific replicate details. Significant tests were performed in GraphPad Prism 8 using two-tailed Student’s t test (∗ denotes p < 0.05, ∗∗ denotes p < 0.01).
Additional resources
The webtool described in the paper contains designed Cas13 guide RNAs for model organisms and RNA viruses with an interactive design application, as well as a web application for custom RNA input: https://cas13design.nygenome.org/.
Acknowledgments
We thank the entire Sanjana laboratory for support and advice. We thank M. Zaran and S. Brock for assistance with the web tool server. N.E.S. is supported by New York University and New York Genome Center startup funds, the National Institutes of Health (NIH)/National Human Genome Research Institute (DP2HG010099), the NIH/National Cancer Institute (R01CA218668), the Defense Advanced Research Projects Agency (D18AP00053), the Cancer Research Institute, and the Brain and Behavior Foundation.
Author contributions
N.E.S. and H.-H.W. conceived the project. N.E.S., H.-H.W., X.G., and A.M.-M. designed the study. A.M.-M. and D.H. designed Cas13d gRNAs for all model organisms. H.-H.W. performed analyses for model organisms. X.G. designed Cas13d gRNAs and performed analyses for viruses. H.-H.W. and J.A.R. built the web tool and computed off-target counts per guide, X.G., A.M.-M., and X.C. designed and tested guide efficiency in human and mouse models. X.G., H.-H.W., and D.H. produced the figures. N.E.S. supervised the work. X.G. and N.E.S. wrote the manuscript with input from all authors.
Declaration of interests
The New York Genome Center and New York University have applied for patents relating to the work in this article. N.E.S. is an adviser to Vertex and Qiagen.
Published: August 30, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xgen.2021.100001.
Supplemental information
Data and code availability
All designed Cas13 guide RNAs (for model organisms and RNA viruses) and the interactive cas13design tool are available here: https://cas13design.nygenome.org/. For additional reproducibility, we provide shell scripts, R code, Python scripts and summary statistics to count gRNA off-targets and reproduce the figures here: https://gitlab.com/sanjanalab/cas13_webtool. The guide design algorithm used in the cas13design tool is available here: https://gitlab.com/sanjanalab/cas13.
The following reference transcriptomes/genes were used: H. sapiens (GENCODE v19, GRCh37), M. musculus (GENCODE M24, mm10), D. rerio (Ensembl v99, GRCz11), D. melanogaster (Ensembl v99, BDGP6), C. elegans (Ensembl v99, WBcel235), A. thaliana (Ensembl Plants v46, TAIR10), SARS-CoV-2 (MT370904, NC_045512), MERS (NC_019843), HIV1 (NC_001802) and H1N1 (NC_026431 to NC_026438). The following gene expression datasets were downloaded from NCBI Gene Expression Omnibus (GEO): HAP1 (GEO: GSE80793) and NIH 3T3 (GEO: GSM2897262).
References
- 1.Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., Verdine V., Cox D.B.T., Kellner M.J., Regev A., et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smargon A.A., Cox D.B.T., Pyzocha N.K., Zheng K., Slaymaker I.M., Gootenberg J.S., Abudayyeh O.A., Essletzbichler P., Shmakov S., Makarova K.S., et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol. Cell. 2017;65:618–630.e7. doi: 10.1016/j.molcel.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Konermann S., Lotfy P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell. 2018;173:665–676.e14. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan W.X., Chong S., Zhang H., Makarova K.S., Koonin E.V., Cheng D.R., Scott D.A. Cas13d Is a Compact RNA-Targeting Type VI CRISPR Effector Positively Modulated by a WYL-Domain-Containing Accessory Protein. Mol. Cell. 2018;70:327–339.e5. doi: 10.1016/j.molcel.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou H., Su J., Hu X., Zhou C., Li H., Chen Z., Xiao Q., Wang B., Wu W., Sun Y., et al. Glia-to-Neuron Conversion by CRISPR-CasRx Alleviates Symptoms of Neurological Disease in Mice. Cell. 2020;181:590–603.e16. doi: 10.1016/j.cell.2020.03.024. [DOI] [PubMed] [Google Scholar]
- 6.Xu D., Cai Y., Tang L., Han X., Gao F., Cao H., Qi F., Kapranov P. A CRISPR/Cas13-based approach demonstrates biological relevance of vlinc class of long non-coding RNAs in anticancer drug response. Sci. Rep. 2020;10:1794. doi: 10.1038/s41598-020-58104-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li S., Li X., Xue W., Zhang L., Yang L.-Z., Cao S.-M., Lei Y.-N., Liu C.-X., Guo S.-K., Shan L., et al. Screening for functional circular RNAs using the CRISPR–Cas13 system. Nat. Methods. 2021;18:51–59. doi: 10.1038/s41592-020-01011-4. [DOI] [PubMed] [Google Scholar]
- 8.Abbott T.R., Dhamdhere G., Liu Y., Lin X., Goudy L., Zeng L., Chemparathy A., Chmura S., Heaton N.S., Debs R., et al. Development of CRISPR as an Antiviral Strategy to Combat SARS-CoV-2 and Influenza. Cell. 2020;181:865–876.e12. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui J., Techakriengkrai N., Nedumpun T., Suradhat S. Abrogation of PRRSV infectivity by CRISPR-Cas13b-mediated viral RNA cleavage in mammalian cells. Sci. Rep. 2020;10:9617. doi: 10.1038/s41598-020-66775-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchman A.B., Brogan D.J., Sun R., Yang T., Hsu P.D., Akbari O.S. Programmable RNA Targeting Using CasRx in Flies. CRISPR J. 2020;3:164–176. doi: 10.1089/crispr.2020.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huynh N., Depner N., Larson R., King-Jones K. A versatile toolkit for CRISPR-Cas13-based RNA manipulation in Drosophila. Genome Biol. 2020;21:279. doi: 10.1186/s13059-020-02193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kushawah G., Hernandez-Huertas L., Abugattas-Nuñez Del Prado J., Martinez-Morales J.R., DeVore M.L., Hassan H., Moreno-Sanchez I., Tomas-Gallardo L., Diaz-Moscoso A., Monges D.E., et al. CRISPR-Cas13d Induces Efficient mRNA Knockdown in Animal Embryos. Dev. Cell. 2020;54:805–817.e7. doi: 10.1016/j.devcel.2020.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Mahas A., Aman R., Mahfouz M. CRISPR-Cas13d mediates robust RNA virus interference in plants. Genome Biol. 2019;20:263. doi: 10.1186/s13059-019-1881-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wessels H.-H., Méndez-Mancilla A., Guo X., Legut M., Daniloski Z., Sanjana N.E. Massively parallel Cas13 screens reveal principles for guide RNA design. Nat. Biotechnol. 2020;38:722–727. doi: 10.1038/s41587-020-0456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boyle A.P., Araya C.L., Brdlik C., Cayting P., Cheng C., Cheng Y., Gardner K., Hillier L.W., Janette J., Jiang L., et al. Comparative analysis of regulatory information and circuits across distant species. Nature. 2014;512:453–456. doi: 10.1038/nature13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerstein M.B., Rozowsky J., Yan K.K., Wang D., Cheng C., Brown J.B., Davis C.A., Hillier L., Sisu C., Li J.J., et al. Comparative analysis of the transcriptome across distant species. Nature. 2014;512:445–448. doi: 10.1038/nature13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long H., Sung W., Kucukyildirim S., Williams E., Miller S.F., Guo W., Patterson C., Gregory C., Strauss C., Stone C., et al. Evolutionary determinants of genome-wide nucleotide composition. Nat. Ecol. Evol. 2018;2:237–240. doi: 10.1038/s41559-017-0425-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blanchard E.L., Vanover D., Bawage S.S., Tiwari P.M., Rotolo L., Beyersdorf J., Peck H.E., Bruno N.C., Hincapie R., Michel F., et al. Treatment of influenza and SARS-CoV-2 infections via mRNA-encoded Cas13a in rodents. Nat. Biotechnol. 2021 doi: 10.1038/s41587-021-00822-w. [DOI] [PubMed] [Google Scholar]
- 19.World Health Organization . 2020. Coronavirus disease (COVID-19) pandemic.https://www.who.int/emergencies/diseases/novel-coronavirus-2019 [Google Scholar]
- 20.Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Reiche A.S., Hernandez M.M., Sullivan M.J., Ciferri B., Alshammary H., Obla A., Fabre S., Kleiner G., Polanco J., Khan Z., et al. Introductions and early spread of SARS-CoV-2 in the New York City area. Science. 2020;369:297–301. doi: 10.1126/science.abc1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuevas J.M., Geller R., Garijo R., López-Aldeguer J., Sanjuán R. Extremely High Mutation Rate of HIV-1 In Vivo. PLoS Biol. 2015;13:e1002251. doi: 10.1371/journal.pbio.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labun K., Montague T.G., Krause M., Torres Cleuren Y.N., Tjeldnes H., Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47(W1):W171–W174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu H., Richmond E., Liang C. CRISPR-RT: a web application for designing CRISPR-C2c2 crRNA with improved target specificity. Bioinformatics. 2018;34:117–119. doi: 10.1093/bioinformatics/btx580. [DOI] [PubMed] [Google Scholar]
- 25.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B.T., Shmakov S., Makarova K.S., Semenova E., Minakhin L., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frankish A., Diekhans M., Ferreira A.-M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J., et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howe K.L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., Bhai J., et al. Ensembl 2021. Nucleic Acids Res. 2021;49(D1):D884–D891. doi: 10.1093/nar/gkaa942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mongelli A., Martelli F., Farsetti A., Gaetano C. The Dark That Matters: Long Non-coding RNAs as Master Regulators of Cellular Metabolism in Non-communicable Diseases. Front. Physiol. 2019;10:369. doi: 10.3389/fphys.2019.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rea J., Menci V., Tollis P., Santini T., Armaos A., Garone M.G., Iberite F., Cipriano A., Tartaglia G.G., Rosa A., et al. HOTAIRM1 regulates neuronal differentiation by modulating NEUROGENIN 2 and the downstream neurogenic cascade. Cell Death Dis. 2020;11:527. doi: 10.1038/s41419-020-02738-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berkovits B.D., Mayr C. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature. 2015;522:363–367. doi: 10.1038/nature14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatcher E.L., Zhdanov S.A., Bao Y., Blinkova O., Nawrocki E.P., Ostapchuck Y., Schäffer A.A., Brister J.R. Virus Variation Resource - improved response to emergent viral outbreaks. Nucleic Acids Res. 2017;45(D1):D482–D490. doi: 10.1093/nar/gkw1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhn R.M., Haussler D., Kent W.J. The UCSC genome browser and associated tools. Brief. Bioinform. 2013;14:144–161. doi: 10.1093/bib/bbs038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All designed Cas13 guide RNAs (for model organisms and RNA viruses) and the interactive cas13design tool are available here: https://cas13design.nygenome.org/. For additional reproducibility, we provide shell scripts, R code, Python scripts and summary statistics to count gRNA off-targets and reproduce the figures here: https://gitlab.com/sanjanalab/cas13_webtool. The guide design algorithm used in the cas13design tool is available here: https://gitlab.com/sanjanalab/cas13.
The following reference transcriptomes/genes were used: H. sapiens (GENCODE v19, GRCh37), M. musculus (GENCODE M24, mm10), D. rerio (Ensembl v99, GRCz11), D. melanogaster (Ensembl v99, BDGP6), C. elegans (Ensembl v99, WBcel235), A. thaliana (Ensembl Plants v46, TAIR10), SARS-CoV-2 (MT370904, NC_045512), MERS (NC_019843), HIV1 (NC_001802) and H1N1 (NC_026431 to NC_026438). The following gene expression datasets were downloaded from NCBI Gene Expression Omnibus (GEO): HAP1 (GEO: GSE80793) and NIH 3T3 (GEO: GSM2897262).