The authors show that tumor-specific T cells upregulate and require the transcriptional regulator BHLHE40 for tumor microenvironment remodeling and effective immune checkpoint therapy. BHLHE40 is a potential biomarker for immune checkpoint therapy efficacy and possible therapeutic target.
Abstract
Immune checkpoint therapy (ICT) using antibody blockade of programmed cell death protein 1 (PD-1) or cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) can provoke T cell–dependent antitumor activity that generates durable clinical responses in some patients. The epigenetic and transcriptional features that T cells require for efficacious ICT remain to be fully elucidated. Herein, we report that anti–PD-1 and anti–CTLA-4 ICT induce upregulation of the transcription factor BHLHE40 in tumor antigen–specific CD8+ and CD4+ T cells and that T cells require BHLHE40 for effective ICT in mice bearing immune-edited tumors. Single-cell RNA sequencing of intratumoral immune cells in BHLHE40-deficient mice revealed differential ICT-induced immune cell remodeling. The BHLHE40-dependent gene expression changes indicated dysregulated metabolism, NF-κB signaling, and IFNγ response within certain subpopulations of CD4+ and CD8+ T cells. Intratumoral CD4+ and CD8+ T cells from BHLHE40-deficient mice exhibited higher expression of the inhibitory receptor gene Tigit and displayed alterations in expression of genes encoding chemokines/chemokine receptors and granzyme family members. Mice lacking BHLHE40 had reduced ICT-driven IFNγ production by CD4+ and CD8+ T cells and defects in ICT-induced remodeling of macrophages from a CX3CR1+CD206+ subpopulation to an iNOS+ subpopulation that is typically observed during effective ICT. Although both anti–PD-1 and anti–CTLA-4 ICT in BHLHE40-deficient mice led to the same outcome—tumor outgrowth—several BHLHE40-dependent alterations were specific to the ICT that was used. Our results reveal a crucial role for BHLHE40 in effective ICT and suggest that BHLHE40 may be a predictive or prognostic biomarker for ICT efficacy and a potential therapeutic target.
Introduction
Antitumor immunity induced by anti–CTLA-4 and/or anti–PD-1 immune checkpoint therapy (ICT) requires effector T cells (Teff) directed against tumor antigens (1–5). T cells undergo epigenetic, transcriptional, and post-transcriptional modifications to become Teff (6–8). Basic Helix-Loop-Helix Family Member E40 (BHLHE40, also called DEC1) is a transcription factor expressed in both non-hematopoietic and hematopoietic cells that can promote or repress transcription (9, 10). BHLHE40 is upregulated in T cells by select cytokines (11, 12) and by activation, with CD28-signaling further enhancing BHLHE40 expression (13). In models of infectious disease and autoimmunity, BHLHE40 regulates Th1, Th2, and Th17 cytokine production, with a key role in promoting GM-CSF and repressing IL10 production (11, 13–19). The mechanisms by which BHLHE40 promotes or suppresses transcription are incompletely understood. In mouse CD4+ T cells, BHLHE40 can suppress Il10 transcription, possibly by binding to an enhancer region in Il10 that is also the target of transcriptional activators (10, 16, 20). In GM-CSF–expressing human memory CD4+ T cells, BHLHE40 facilitates inflammatory gene expression by directly dampening expression of ZC3H12D, which encodes an RNase capable of degrading inflammatory transcripts, and indirectly inhibiting miR-146a, a negative regulator of NF-κB signaling (21). BHLHE40 can also interact with other transcription factors such as TBET and RUNX1 to mediate transcriptional activation of target genes (22, 23). Specifically, activation of Ifng transcription by BHLHE40 can be mediated by TBET-dependent and -independent mechanisms (15, 23).
In models of melanoma, BHLHE40 has been found to be required for IFNγ production by invariant natural killer T (iNKT) cells activated with α-galactosylceramide (23). It also has been shown that BHLHE40 regulates the mitochondrial metabolism required for production of sufficient acetyl-CoA for histone acetylation and maintenance of transcriptionally active chromatin domains in tissue resident memory T cells (Trm) and tumor-infiltrating lymphocytes (TIL) (12). This study also revealed that BHLHE40 is required for reinvigoration of TIL upon anti–PD-L1 ICT, however, the role of BHLHE40 in CD4+, as well as CD8+, T cells, during anti–PD-1 or anti–CTLA-4 ICT has yet to be fully defined.
Herein, we report that ICT upregulates Bhlhe40 in both CD4+ and CD8+ T cells and that T cells require BHLHE40 for antitumor effector function and response to anti–PD-1 or anti–CTLA-4. Single-cell RNA sequencing (scRNAseq) analysis revealed that ICT-treated tumor-bearing BHLHE40-deficient mice have altered remodeling of intratumoral lymphoid and myeloid cells, including a profound absence of a shift from a CX3CR1+CD206+ macrophage subpopulation to an iNOS+ subpopulation that is typically observed in wild-type (WT) mice during effective ICT. These differences observed in the absence of BHLHE40 were associated with gene expression changes implicating dysregulated T-cell metabolism, NF-κB signaling, and IFNγ response, along with defects in IFNγ production by BHLHE40-deficient CD4+ and CD8+ T cells.
Materials and Methods
Mice
All mice used were on a C57BL/6 background. WT C57BL/6J, CD4-Cre (B6.Cg-Tg(Cd4-cre)1Cwi/BfluJ), LysM-Cre (B6N.129P2(B6)-Lyz2tm1(cre)Ifo/J), Bhlhe40 knockout (KO) (B6.129S1(Cg)-Bhlhe40tm1.1Rhli/MpmJ), OT-I T-cell receptor (TCR) transgenic (Tg(TcraTcrb)1100Mjb/J) and OT-II TCR transgenic (B6.Cg-Tg(TcraTcrb)425Cbn/J) mice were purchased from Jackson Labs. All in vivo experiments used 8- to 12-week-old female or male mice (to match the sex and strain of the tumors). All mice were housed in a specific pathogen-free animal facility. Bhlhe40–/– (ten generations backcrossed to the C57BL/6 background) and Bhlhe40f/f mice have been previously reported (11, 16, 24). For experiments using conditional Bhlhe40 KO mice, Bhlhe40f/f littermates were used as controls. All animal studies were performed in accordance with, and with the approval of the Institutional Animal Care and Use Committee (IACUC) of Washington University (St. Louis) and The University of Texas MD Anderson Cancer Center (Houston, TX).
Tumor cell lines
The MCA-induced sarcomas used in this study were generated in female C57BL/6 WT (the edited, progressor 1956 MCA sarcoma cell line) or Rag2−/− (the unedited, highly immunogenic 1969 MCA sarcoma cell line) mice and were banked as low-passage tumor cells as previously described (25). Both sarcoma cell lines were obtained from Robert Schreiber (Washington University School of Medicine, St. Louis, MO) in January 2020. The B16 murine melanoma cell line expressing the full-length chicken ovalbumin (herein referred to as B16-OVA) was obtained from Stephanie Watowich (The University of Texas MD Anderson Cancer Center, Houston, TX) in October 2020. Tumor cell lines were found to be free of common mouse pathogens and Mycoplasma as assessed by IDEXX IMPACT I mouse pathogen testing [PCR evaluation for: Corynebacterium bovis, Corynebacterium sp. (HAC2), Ectromelia, EDIM, Hantaan, K virus, LCMV, LDEV, MAV1, MAV2, mCMV, MHV, MNV, MPV, MTV, MVM, Mycoplasma pulmonis, Mycoplasma sp., Polyoma, PVM, REO3, Sendai, TMEV] in December 2020. Tumor cell lines from the same cryopreserved stocks that were used in this study tested negative for Mycoplasma and were authenticated and found to be free of nonmouse cells as assessed by mouse cell STR profiling in December 2021 (IDEXX CellCheck mouse 19 plus Mycoplasma spp. testing).
Tumor transplantation
MCA sarcoma cell lines and B16-OVA melanoma cells were propagated in R-10 plus BME media [RPMI media (HyClone/Cytiva: catalog no. SH30096.02) supplemented with 1% l-glutamine, 1% penicillin–streptomycin, 1% sodium pyruvate, 0.5% sodium bicarbonate, 0.1% 2-mercaptoethanol, and 10% FCS (HyClone/Cytiva: catalog no. SH30070.03HI)]. Upon thawing, tumor lines were passaged 3 to 6 times before experimental use. Prior to injection, cells were washed extensively, resuspended at a concentration of 6.67×106 cells per mL in endotoxin-free PBS for MCA sarcoma cell lines and at a concentration of 2.67×106 cells per mL in endotoxin-free PBS for B16-OVA cells and then 150 µL (1×106 cells per mouse for MCA sarcomas and 0.4×106 cells per mouse for B16-OVA) injected subcutaneously into the flanks of recipient mice. Tumor cells were >90% viable at the time of injection as assessed by Trypan blue exclusion. Tumor growth was quantified by caliper measurements and expressed as the average of two perpendicular diameters. Lack of survival was defined as mouse death or mean tumor diameter size of 20 mm.
In vivo antibody treatments
For ICT, tumor-bearing mice were treated intraperitoneally with 200 μg of anti–PD-1 or anti–CTLA-4, used alone or in combination on days 3, 6, 9, and 12 post tumor transplant. For experiments where tumors were harvested on day 9, mice were treated on days 3 and 6 post tumor transplant; and for tumors harvested on day 11, mice were treated on days 3, 6, and 9 post tumor transplant. For controls, mice were injected with 200 μg of IgG2a isotype control antibodies. For antibody depletion studies, 250 μg of control mAb, anti-CD4, or anti-CD8a was injected intraperitoneally into mice at day −1 and every 7 days thereafter until day 20. For in vivo experiments, “In vivo Platinum”-grade antibodies that were verified to be free of mouse pathogens (IDEXX IMPACT I mouse pathogen testing) were purchased from Leinco Technologies: anti–PD-1 (rat IgG2a clone RMP1–14), anti–CTLA-4 (murine IgG2b clone 9D9), anti-CD4 (rat IgG2b clone GK1.5), anti-CD8a (rat IgG2b clone YTS169.4), and isotype controls (rat IgG2a clone 1–1, mouse IgG2a clone OKT3, and rat IgG2b clone 1–2).
Tetramers
Peptide–MHC class I monomers refolded with an ultraviolet light (UV)-cleavable conditional ligand were prepared in-house as previously described (7). Briefly, recombinant H-2Kb heavy chain and human β2 microglobulin light chain were produced in Escherichia coli as inclusion bodies and refolded in the presence of a UV-cleavable peptide (SIINFE-J-L, where J represents 3-amino-3-(2-nitro) phenyl-propionic acid (Peptide 2.0). Monomers were captured by anion exchange (HiTrap Q HP, GE), biotinylated, and purified by gel filtration FPLC. UV-induced ligand exchange with mutant Lama4 (mLama4) peptide was performed as described previously (7). OVA-I (SIINFEKL)-H-2Kb-PE and mutant Lama4-H-2Kb-PE tetramers were additionally obtained from the Baylor College of Medicine MHC Tetramer Production Facility.
Tumor and spleen harvest
Established tumors were excised from mice, minced and treated with 1 mg/mL type IA collagenase (Sigma: catalog no. C9891) in HBSS (Hyclone/Cytiva: catalog no. SH30588.02) for 45 minutes at 37°C. Cells were washed thrice. Red blood cells were lysed using ACK lysis buffer (Gibco: catalog no. A1049201). To remove aggregates and clumps, cells were passed through a 40-μm strainer. Spleens were harvested, crushed, and vigorously resuspended to make single-cell suspensions. To remove aggregates and clumps, cells were passed through a 70-μm strainer and subsequently through a 40-μm strainer.
Quantitative RT-PCR
RNA was extracted from sorted T cells using RNAeasy Plus Mini Kit (Qiagen: catalog no. 74034). One-hundred micrograms of RNA was reverse-transcribed and subjected to qPCR using the SuperScript III Platinum Two-Step qRT-PCR Kit with SYBR Green (Invitrogen: catalog no. 11744500). qPCR was performed on the StepOne Real-Time PCR System (Applied Biosystems). Each sample was run in triplicate for each gene and the cDNA from each sample was divided equally per reaction in a 20 µL volume. The qPCR conditions were as follows: 50°C for 2 minutes and 95°C for 2 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 30 seconds. Melting curve analysis was performed to confirm a single amplicon. Differences in gene expression were determined using the equation 2–ΔΔCt, where the Ct value of Bhlhe40 was subtracted from the Ct value of the Gapdh control to yield the ΔCt value. For each sample, the ΔCt value of Bhlhe40 done in triplicate was averaged and compared to give one ΔΔCt value per sample. Mouse quantitative RT-PCR primers for Bhlhe40 were as follows: forward primer-5′ ACGGAGACCTGTCAGGGATG3′ and reverse primer-5′GGCAGTTTGTAAGTTTCCTTGC3′. Mouse quantitative RT-PCR primers for Gapdh were as follows: forward primer-5′AGGTCGGTGTGAACGGATTTG3′ and reverse primer-5′TGTAGACCATGTAGTTGAGGTCA3′ (Integrated DNA Technologies, custom order).
Reanalysis of published scRNAseq
Published scRNAseq data [Gene Expression Omnibus (GEO): GSE119352; ref. 26] of intratumoral CD45+ cells from mice bearing T3 murine MCA-induced sarcomas treated with control mAb, anti–PD-1, anti–CTLA-4, or anti–PD-1 and anti–CTLA-4 were reanalyzed using the same pipelines and parameters as previously described (26). Single-cell explorer [an open-source project for processing and visualization of scRNAseq data developed by the Artyomov Lab (Washington University; https://artyomovlab.wustl.edu/shiny/single_cell_explorer/)] was used to visualize and generate plots in Fig. 1 and Supplementary Fig. S1.
Figure 1.
Subsets of intratumoral myeloid and lymphoid cells express Bhlhe40. A,Bhlhe40 mRNA expression in intratumoral mLama4-specific CD8+ T cells sorted from T3 sarcoma–bearing WT mice treated with anti–CTLA-4, anti–PD-1, or both anti–CTLA-4 and anti–PD-1. B, tSNE plot from merged treatment data of exclusively intratumoral lymphocytes showing Bhlhe40 expression within indicated lymphoid subpopulations identified by scRNAseq in the T3 MCA sarcoma. C, Violin plots showing Bhlhe40 expression in T3 intratumoral lymphoid cells by cluster and treatment. D, Heatmap displaying normalized expression of select genes in T3 intratumoral lymphoid cells by cluster and treatment. E, Tumor growth in Bhlhe40–/– or Bhlhe40+/+ mice transplanted with 1956 sarcoma cells and subsequently treated with control, anti–CTLA-4, anti–PD-1, or both anti–CTLA-4 and anti–PD-1. F, Cumulative Kaplan–Meier survival curves for the 1956 tumor–bearing Bhlhe40–/– or Bhlhe40+/+ mice treated as in E. For A, each dot represents 5 pooled mice harvested on day 11 posttransplant and assessed independently (N = 3; *, P < 0.05, unpaired t test). B–D, scRNAseq data generated in Gubin et al (39) were reanalyzed for Bhlhe40 expression. Data in E are presented as average tumor diameter ± SEM of 5 mice per group and are representative of at least four independent experiments. Data in (F) are cumulative survival curves from 4 independent experiments of 3–5 mice per group [***, P < 0.001 (log-rank (Mantel–Cox) test)].
scRNAseq
scRNAseq library generation
Droplet-based 5′ end massively parallel scRNAseq was performed by encapsulating sorted live CD45+ tumor-infiltrating cells into droplets and libraries were prepared using Chromium Next GEM Single-cell 5′ Reagent Kit v2 (10x Genomics: catalog no. 100263) according to manufacturer's protocol. The generated scRNAseq libraries were sequenced using an Illumina NovaSeq6000 S2 flow cell.
scRNAseq alignment, barcode assignment, and unique molecular identifier counting
The Cell Ranger Single-Cell Software Suite available at https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome was used to perform sample demultiplexing, barcode processing, and single-cell 5′ counting. Cellranger mkfastq was used to demultiplex raw base call files from the NovaSeq6000 sequencer, into sample-specific fastq files. Files were demultiplexed with 81.9% to 97.1% perfect barcode match, and 90%+ q30 reads. Afterward, fastq files for each sample were processed with Cellranger count, which was used to align samples to mm10 genome, filtered, and quantified. For each sample, the recovered cells’ parameter was specified as 10,000 cells that we expected to recover for each individual library.
Preprocessing analysis with Seurat package
The Seurat pipeline was applied to each dataset following tutorial specifications from https://satijalab.org/seurat/articles/archive; version 3.2 and https://hbctraining.github.io/scRNA-seq_online/. For each experiment, data from all groups were merged into a single Seurat object, and integration was performed using the reciprocal principal component analysis (PCA) workflow to identify integration anchors. After integration, genes that were expressed in fewer than 3 cells and cells that contained fewer than 500 transcripts (unique molecular identifiers; UMI) were excluded. In addition, cells that had fewer than 250 or more than 5,000 genes (for the day 9 experiment) or fewer than 250 or more than 4,000 genes (for the day 11 experiment) were excluded. Cells with high content of mitochondrial transcripts (more than 20% for the day 9 experiment and more than 10% for the day 11 experiment) were also excluded from analysis. The cutoffs used were set based on the characteristics of the cell population in each dataset. Data were normalized using LogNormalize method (counts for each cell divided by the total counts for that cell, multiplied by the scale factor of 104 and natural-log transformed using log1p). PCA was performed on about 4,000 genes with PCA function. A uniform manifold approximation and projection (UMAP) dimensional reduction was performed on the scaled matrix (with most variable genes only) using the first 30 PCA components to obtain a two-dimensional representation of the cell states. For clustering, we used the function FindClusters that implements SNN (shared nearest neighbor) modularity optimization–based clustering algorithm on 30 PCA components, leading to 19–23 clusters.
Identification of cluster-specific genes and marker-based classification
To identify marker genes, the FindAllMarkers function was used with likelihood-ratio test for single-cell gene expression. To characterize clusters, we used ImmGen database. For heatmap representation, mean expression of markers inside each cluster was used. To compare gene expression for the clusters inside cohorts (e.g., T cells, macrophages) we used FindMarkers function to calculate average log2 fold change and identify differentially expressed genes between each pair of experimental conditions using a Wilcoxon rank-sum test for calculating P values and Bonferroni correction for Padj values.
T-cell population analysis
To examine T cells, clusters expressing Cd8b1 and/or Cd4 were extracted from aggregated samples. Clusters extracted for day 9 scRNAseq analysis were T_15.d9 and Treg_16.d9 and for day 11 scRNAseq analysis were γδ_15.d11, CD8_16.d11, CD8_17.d11, CD8_18.d11, CD4_19.d11, Treg_20.d11, and Mki67hi_21.d11. Identification of most variable genes, PCA, UMAP, clustering, and marker selection analysis were performed as described above. Pearson correlation (PC) coefficients were used as the measure of relationship between expression of Bhlhe40 and other genes in reclustered T cells from day 9 posttransplant and day 11 posttransplant, from both Bhlhe40+/+ and Bhlhe40–/– mice treated with ICT or control antibody.
Ex vivo T-cell stimulation
For PMA/ionomycin T-cell stimulation, cells from tumors, isolated as described above (in Tumor and spleen harvest section), were enriched for CD4+ and CD8+ cells using the Miltenyi Biotec Mouse CD4+ Enrichment Kit (catalog no. 130–104–454) and Mouse CD8+ T Cell Enrichment Kit (catalog no. 130–104–075), respectively, according to the manufacturer's protocols. Cells were then incubated with 10 ng/mL of PMA (Millipore: catalog no. 5.00582.0001) and 1 μg/mL of ionomycin (Fisher: catalog no. BP25271) in the presence of BD GolgiPlug (BD Biosciences: catalog no. 555028) for 6 hours at 37°C. Cells were then stained for IFNγ (as described below in Flow cytometry section).
For OVA peptide restimulation of intratumoral T cells, CD4+ and CD8+ cells were enriched using the Miltenyi Biotec Mouse CD4+ Enrichment Kit (catalog no. 130–104–454) and Mouse CD8+ T Cell Enrichment Kit (catalog no. 130–104–075), respectively, according to the manufacturer's protocols. T cells were stimulated with naïve irradiated splenocytes from WT mice (isolated as described above in Tumor and spleen harvest section) pulsed with 1 μmol/L of OVA-SIINFEKL (Peptide 2.0, custom order), or OVA-II (ISQAVHAAHAEINEAGR) peptide (Peptide 2.0, custom order), respectively, in the presence of BD GolgiPlug for 6 hours at 37°C. Cells were then stained for IFNγ (as described below in Flow cytometry section).
For longitudinal monitoring of Bhlhe40 expression in TCR transgenic CD4+ and CD8+ splenic T cells upon antigen-specific activation, CD8+ T cells from spleens of naïve 8-week-old female OT-I mice and CD4+ T cells from spleens of naïve 8-week-old female OT-II mice were isolated as described above (in the Tumor and spleen harvest section) and enriched using the Miltenyi Biotec Mouse CD8+ Enrichment Kit and Mouse CD4+ T Cell Enrichment Kit, respectively, according to the manufacturer's protocols. Splenocytes from naïve 8-week-old female C57BL/6J mice were pulsed for 2 hours at 37°C with 1 μmol/L of OVA-I-SIINFEKL or OVA-II (ISQAVHAAHAEINEAGR) peptide (Peptide 2.0, custom order) and then washed twice with R10 + BME media. Enriched CD8+ OT-I or CD4+ OT-II T cells were cocultured with OVA-I-SIINFEKL or OVA-II–pulsed splenocytes, respectively. Individual wells were harvested at 24, 48, 72, and 96 hours poststimulation for mRNA analysis. For anti-CD3 and anti-CD28 T-cell stimulation experiments, CD4+ or CD8+ splenic T cells were isolated from naïve male C57BL/6J mice and stimulated with plate-bound anti-CD3 (1 μg/mL) alone or with soluble anti-CD28 (5 μg/mL) for 72 hours in R-10 plus BME media. Purified anti-mouse CD3ε (clone 145–2C11) and purified anti-mouse CD28 (clone 37.51) were purchased from BioLegend.
Flow cytometry
For flow cytometry, cells were stained for 5 minutes at room temperature with rat anti-mouse CD16/32 (mouse BD Fc Block; clone 2.4G2, BD Biosciences) at 1 μg/million cells and then surface stained for 20 minutes at 4°C. Surface antibodies were diluted in FACS staining buffer (PBS with 2% FCS, 2 mmol/L EDTA, and 0.05% NaN3; Sigma). Anti-mouse CD64-BV421 (clone X54–5/7.1, BioLegend; 1:200 dilution), anti-mouse Ly6G-Alexa Fluor 700 (clone 1A8, BD Biosciences; 1:400 dilution), anti-mouse CD4-BV711 (clone RM4–5, BioLegend; 1:200 dilution), anti-mouse CD8a-BV786 (clone 53–6.7, BioLegend; 1:200 dilution), anti-mouse CX3CR1-FITC (clone SA011F11, BioLegend; 1:1,000 dilution), anti-mouse CD90.2/Thy1.2-PE/Cy7 (clone 30-H12, BioLegend, 1:500 dilution), anti-mouse CD8b-PerCP/Cy5.5 (clone YTS156.7.7, BioLegend; 1:200 dilution), anti-mouse PD-1-BV421 (clone 29F.1A12, BioLegend; 1:200), anti-mouse LAG-3-PerCP/Cy5.5 (clone C9B7W, BioLegend; 1:200), anti-mouse CD3e-APC (clone 145–2C11, BioLegend; 1:200 dilution), anti-mouse I-A/I-E-BV650 (clone M5/114.15.2, BioLegend; 1:3,000 dilution), anti-mouse CD24-BV711 (clone M1/69, BD Biosciences; 1:500 dilution), anti-mouse CD11c-BV786 (clone HL3, BD Biosciences; 1:400 dilution), anti-mouse F4/80-BUV395 (clone T45–2342, BD Biosciences; 1:400 dilution), anti-mouse CD64-APC (clone X54–5/7.1, BioLegend; 1:400 dilution), anti-mouse CD45 BV605 (clone 30-F11, BioLegend; 1:800 dilution), anti-mouse CD25-PC (clone 3C7, BioLegend; 1:200 dilution), and anti-mouse CD11b-APC (clone M1/70, BioLegend; 1:400 dilution) were used for surface staining at the indicated dilutions. Zombie NIR Viability dye (BioLegend: catalog no. 423105) was added at 1:500 during surface staining.
For intracellular staining, surface-stained cells were fixed and permeabilized with BD Fixation and Permeabilization Kit (BD Bioscience: catalog no. 555028). Fixed and permeabilized cells were then stained with anti-mouse Mrc1 (CD206)-PE-Cy7 (clone C068C2, BioLegend; 1:400 dilution) and anti-mouse iNOS/NOS2-PE (clone CXNFT, Thermo Fisher; 1:400 dilution) for 30 minutes at 4°C.
For FOXP3 staining, surface-stained cells were fixed and permeabilized using the eBioscience FOXP3/Transcription Factor Staining Buffer Set (eBioscience/Thermo Fisher Scientific: catalog no. 00–5523–00). Fixed and permeabilized cells were then stained with anti-mouse FOXP3-FITC (clone FJK-16s, Thermo Fisher Scientific; 1:100 dilution) for 30 minutes at 4°C.
For intracellular cytokine staining of lymphocytes, cells were isolated, then restimulated and incubated at 37°C for 6 hours with GolgiStop. Cells were then washed and stained for 5 minutes at room temperature with Fc block at 1 μg/million cells and then surface stained for 30 minutes at 4°C, and then fixed and permeabilized with BD Fixation and Permeabilization Kit. Fixed and permeabilized cells were then stained with anti-IFNγ-APC (XMG1.2, BioLegend; 1:200 dilution) for 30 minutes at 4°C.
All flow cytometry was performed on the LSR Fortessa X-20 (BD Biosciences) and analyzed using FlowJo software (v. 10.6.1, TreeStar).
FACS
For sorting of tetramer-positive CD8+ T cells, intratumoral cells were stained for 5 minutes at room temperature with Fc block at 1 μg/million cells and then were stained with mLama4-H-2Kb-PE or OVA-I-257–264-H-2Kb-PE tetramers in FACS buffer for 15 minutes at 37°C. Surface antibodies against CD45 (clone 30-F11, BioLegend; 1:800 dilution), CD90.2/Thy1.2 (clone 30-H12, BioLegend, 1:500 dilution), and CD8b (clone YTS156.7.7, BioLegend; 1:200 dilution) as well as Zombie NIR Viability dye were then added and tetramers plus surface antibodies were incubated for 30 minutes at 4°C. Cells were washed twice with FACS buffer and sorted gating on live CD45+CD90.2/Thy1.2+CD8β+H-2Kb-tetramer+ cells. For sorting of intratumoral CD8+PD-1+LAG-3+T cells and CD4+PD-1+LAG-3+ T cells, intratumoral cells were stained for 5 minutes at room temperature with Fc block at 1 μg/million cells. Surface antibodies against CD45 (clone 30-F11, BioLegend; 1:800 dilution), CD90.2/Thy1.2 (clone 30-H12, BioLegend, 1:500 dilution), CD4 (clone GK1.5, BioLegend, 1:200), CD8b (clone YTS156.7.7, BioLegend; 1:200 dilution), LAG-3 (clone C9B7W, BioLegend, 1:200 dilution), and PD-1 (clone 29F.1A12, BioLegend, 1:200 dilution) as well as Zombie NIR Viability dye were then added. Cells were incubated for 30 minutes at 4°C. Live CD45+Thy1.2+CD8β+PD-1+LAG-3+ and live CD45+Thy1.2+CD4+PD-1 LAG-3+ cells were then sorted on a BD FACSAria II (BD Biosciences). Sorted cells were pelleted and processed for RNA analysis. The purity of the sorted cells for scRNAseq was greater than 97% as assessed during postsort cellular analysis.
Peptides
Mutant Lama4 (VGFNFRTL), OVA-I-257–264 (SIINFEKL) and OVA-II-323–339 (ISQAVHAAHAEINEAGR) peptides were custom ordered from Peptide 2.0. All peptides were HPLC purified to >95% purity.
Statistical analysis
Samples were compared using an unpaired, two-tailed Student t test, two-way ANOVA, or log-rank (Mantel–Cox) test unless specified otherwise.
Data and software availability
Data files for the scRNAseq data reported in this article have been deposited in the GEO database. The accession number is GEO: GSE192546. Software used in this study is available online: current version of Cell Ranger: https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest; Seurat 4.0: https://satijalab.org/seurat/; ggplot2 3.3.3: https://ggplot2.tidyverse.org/index.html; and ImmGen: https://www.immgen.org. All other data generated in this study are available within the article and its Supplementary Data files.
Results
ICT upregulates Bhlhe40 in CD4+ and CD8+ T cells
We previously documented transcriptional and protein changes within mutant neoantigen–specific CD8+ TIL in mice bearing the MCA-induced T3 sarcoma line treated with control mAb or ICT (1, 26, 27). RNAseq revealed a significant upregulation of the transcription factor Bhlhe40 exclusively in tumor-specific, neoantigen-reactive CD8+ TIL in mice treated with anti–CTLA-4, alone or in combination with anti–PD-1. We confirmed Bhlhe40 was upregulated in CD8+ TILs recognizing the T3-specific neoantigen mLama4 (1) in mice treated with anti–CTLA-4, alone or in combination with anti–PD-1 (Fig. 1A). Reanalysis of scRNAseq profiling of the intratumoral immune cells in T3 tumor–bearing mice treated with control mAb or ICT (26) revealed expression of Bhlhe40 in both myeloid and lymphoid populations (Supplementary Fig. S1A–S1C). Further analysis of reclustered lymphocytes identified previously (26) revealed Bhlhe40 expression correlated with the transcriptionally-defined activation/functional T-cell status (Fig. 1B–D). With ICT treatment, Bhlhe40 transcript was highly expressed in CD8_s1, a cluster defined by transcripts upregulated by TCR stimulation such as the negative regulatory molecules Havcr2 (TIM-3), Lag3, and Pdcd1 (PD-1). Within CD4+ Teff cells, CD4_s2 expressed high levels of Bhlhe40 and transcripts consistent with T-cell activation, as well as Ifng. CD4_s1 also expressed activation/functional markers (to a lesser extent than CD4_s2), and expressed Bhlhe40, in particular upon ICT. In addition, two clusters composed of both CD4+ and CD8+ T cells, Mki67hi and T activated (Tact) expressed Bhlhe40. Tact cells displayed relatively low Bhlhe40 expression with control treatment, but much higher expression with ICT treatment, accompanied by a substantial increase in Ifng expression. Altogether, these results indicate that Bhlhe40 is transcriptionally upregulated with ICT in CD4+ and CD8+ TILs that display hallmarks of activation and function.
BHLHE40 is essential for ICT-mediated tumor regression
For further study, we chose a different MCA sarcoma cell line [as opposed to T3 (129S6 background)] to match the genetic background of Bhlhe40–/– mice (C57BL/6), the 1956 cell line. Injection of 1956 cells into syngeneic WT or Bhlhe40–/– mice treated with control mAb resulted in subsequent tumor outgrowth (Fig. 1E). When 1956 tumor–bearing Bhlhe40+/+ WT mice were treated with anti–PD-1 or anti–CTLA-4, nearly all mice rejected tumors and survived 50 or more days (Fig. 1E and F). In contrast, the majority of 1956 tumor–bearing Bhlhe40–/– mice treated with anti–PD-1 and/or anti–CTLA-4 displayed tumor outgrowth and reached endpoint by day 50 (Fig. 1E and F), indicating that BHLHE40 is critical for ICT efficacy.
We then assessed whether Bhlhe40–/– mice could mediate regression of unedited, highly immunogenic tumors that are rejected spontaneously, even in the absence of ICT, by using the 1969 MCA sarcoma line derived from a Rag2–/– mouse (25). When 1969 cells were transplanted into WT mice rendered immunodeficient by anti–CD4 and anti–CD8 depletion, tumors grew progressively (Supplementary Fig. S2A–S2C); whereas all WT immunocompetent mice rejected 1969 tumors. Bhlhe40–/– mice were also able to reject the 1969 tumor, indicating that BHLHE40-deficient mice can spontaneously reject highly immunogenic tumors despite a profound deficiency in ICT efficacy against immune-edited tumors.
T cells require BHLHE40 for ICT-mediated rejection
To delineate which cells require BHLHE40 for effective ICT, we used CD4-Cre+Bhlhe40f/f(Bhlhe40ΔT) mice that lack BHLHE40 in both CD4+ and CD8+ T cells (15, 16), and LysM-Cre+Bhlhe40f/f mice that lack BHLHE40 in monocytes, mature macrophages, and granulocytes (17). 1956 sarcomas grew progressively when injected into Bhlhe40f/f (CD4-Cre–negative Bhlhe40f/f) mice treated with control mAb, whereas they were rejected in most Bhlhe40f/f mice treated with ICT (Fig. 2A and B). In contrast, Bhlhe40ΔT mice displayed outgrowth of 1956 tumors, even when given ICT (Fig. 2A and B). ICT-treated LysM-Cre+Bhlhe40f/fmice, however, were able to reject 1956 tumors, indicating BHLHE40 is dispensable in LysM+ myeloid cells during ICT (Supplementary Fig. S2D–S2F). Together, these results reveal that BHLHE40 is selectively required in T cells for effective ICT.
Figure 2.
Bhlhe40 is selectively required in T cells for anti–PD-1 or anti–CTLA-4 ICT-mediated tumor rejection. A, Tumor growth in Bhlhe40ΔT or Bhlhe40f/f mice transplanted with 1956 sarcoma cells and subsequently treated with control mAb, anti–CTLA-4, or anti–PD-1. B, Cumulative Kaplan–Meier survival curves of 1956 tumor–bearing Bhlhe40ΔT or Bhlhe40f/f mice treated as in A. Bhlhe40 mRNA expression in intratumoral PD-1+LAG-3+CD8+ T cells (C) and PD-1+LAG-3+CD4+ T cells (D) sorted on day 11 posttransplant from 1956 sarcoma–bearing WT mice treated with control mAb, anti–CTLA-4, or anti–PD-1. Bhlhe40 mRNA expression in CD8+ OT-I T cells (E) stimulated with 1 μmol/L OVA-I peptide and CD4+ OT-II T cells (F) stimulated with 1 μmol/L OVA-II peptide for the indicated time. Data in A are presented as average tumor diameter ± SEM of 5–6 mice per group and are representative of 3 independent experiments. Data in B are cumulative survival curves from 3 independent experiments of 5–6 mice per group [***, P < 0.001; NS, not significant, (log-rank (Mantel–Cox) test)]. For C and D, each dot represents mice harvested and assessed independently (N = 5; *, P < 0.05; **, P < 0.01, unpaired t test). Data in E, and F are presented as mean mRNA fold change. Bar indicates mean ± SEM (*, P < 0.05; **, P < 0.01, unpaired t test) and are representative of 3 independent experiments.
Because the tumor–antigen specificities of the T cells infiltrating 1956 tumors are unknown, we flow sorted intratumoral CD4+ and CD8+ T cells that coexpressed PD-1 and LAG-3 because these surface proteins are known to be associated with activated, antigen-specific T cells (1, 26, 27). Expression of Bhlhe40 mRNA was significantly higher in PD-1+LAG-3+CD8+ and PD-1+LAG-3+CD4+ TIL from mice treated with anti–PD-1 and/or anti–CTLA-4 when compared with those from control-treated mice (Fig. 2C and D). Longitudinal monitoring of Bhlhe40 expression in CD8+ OT-I TCR transgenic T cells activated by OVA257–264 SIINFEKL (OVA-I) peptide revealed Bhlhe40 upregulation as early as 24 hours poststimulation and an increase of almost 6-fold from baseline by 72 hours (Fig. 2E). In OVA323–339 (OVA-II) peptide–stimulated CD4+ OT-II cells, Bhlhe40 was upregulated by 24 hours poststimulation, with expression further increased by 48 hours (Fig. 2F). To assess whether CD28 costimulation of T cells amplified Bhlhe40 expression, we stimulated splenic T cells from naïve WT mice with plate-bound anti-CD3 in the presence or absence of soluble anti-CD28. Bhlhe40 mRNA was upregulated by plate-bound anti-CD3 in both CD4+ and CD8+ T cells; the addition of anti-CD28 further increased Bhlhe40 transcript expression (Supplementary Fig. S3A).
Because both CD4+ and CD8+ T cells express Bhlhe40, we assessed whether both T-cell subsets were necessary for ICT-mediated tumor rejection in the 1956 sarcoma model. Although control-treated mice experienced tumor outgrowth, most ICT-treated mice experienced complete tumor rejection (Supplementary Fig. S3B and S3C). Depletion of either CD4+ or CD8+ T cells led to tumor outgrowth despite treatment with ICT, indicating both CD4+ and CD8+ T cells are required for ICT-mediated 1956 tumor regression.
Defective ICT-induced remodeling of intratumoral immune cells in Bhlhe40–/– mice
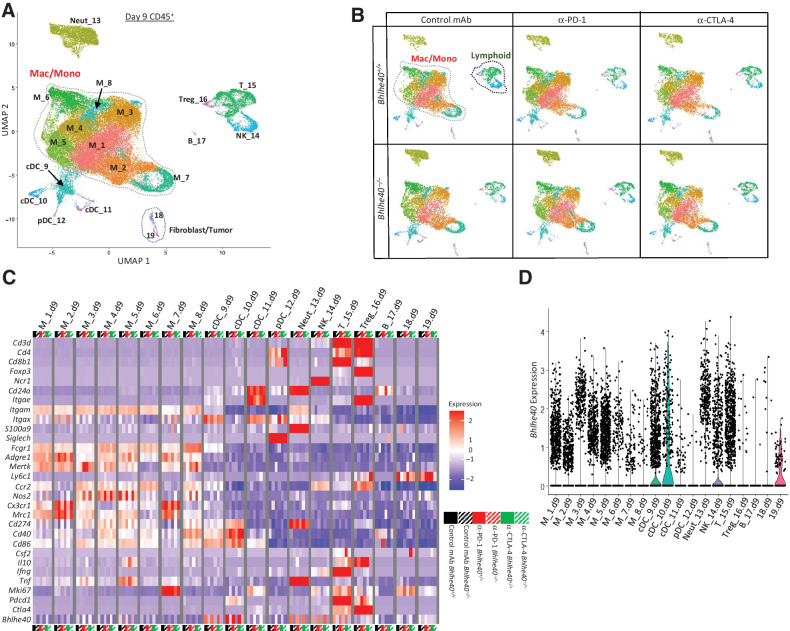
To analyze the deficiencies underlying insensitivity to ICT in the absence of BHLHE40, we performed scRNAseq on intratumoral CD45+ cells isolated on day 9 and 11 posttransplant from 1956 tumor–bearing Bhlhe40+/+ or Bhlhe40–/– mice treated with control, anti–PD-1, or anti–CTLA-4 mAb (Supplementary Fig. S4A). Graph-based clustering revealed distinct subpopulations of myeloid and lymphoid cells on day 9 (Fig. 3A and B) and day 11 posttransplant (Supplementary Fig. S4B and S4C), with cluster cell type identified by expression of lineage markers (Fig. 3C; Supplementary Fig. S4D). Bhlhe40 was expressed in myeloid and lymphoid populations, with increased Bhlhe40 expression in certain lymphoid subpopulations from tumor-bearing WT mice treated with ICT (Fig. 3C and D; Supplementary Fig. S4D and S4E).
Figure 3.
Bhlhe40 is required for ICT-induced remodeling of intratumoral immune cells. A, UMAP plot from scRNAseq of intratumoral CD45+ cells harvested on day 9 posttumor transplant from 1956 tumor–bearing Bhlhe40+/+ or Bhlhe40–/– mice treated with control mAb, anti–CTLA-4, or anti–PD-1. Cluster cell types identified via known cellular subset marker expression and comparison with Immgen database. B, UMAP plots from scRNAseq of CD45+ intratumoral cells. C, Heatmap displaying normalized expression of select genes in each cell cluster by group. D, Violin plot showing Bhlhe40 expression by cluster.
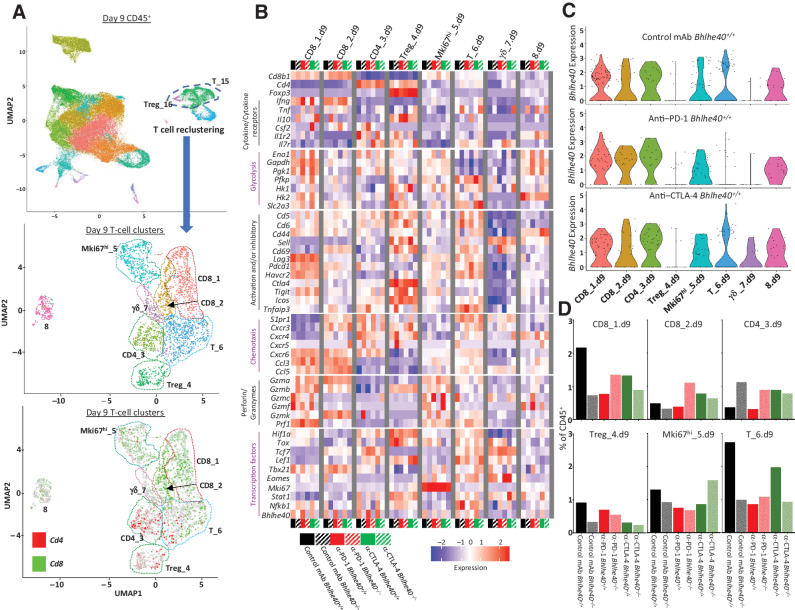
To increase the resolution of our T-cell analysis, we reclustered T cell–containing clusters (Fig. 4A; Supplementary Figs. S4A and S5). T-cell reclustering on day 9 (Fig. 4B–D; Supplementary Fig. S6) and day 11 posttransplant (Supplementary Figs. S5B–S5D and S7) revealed 8 and 10 clusters, respectively, whose proportions and gene expression patterns were altered by ICT in both a BHLHE40-dependent and -independent manner. Although the zenith of TIL presence occurs on day 11 posttransplant in this tumor model, the main focus of our analysis was on day 9 posttransplant, as this timepoint ensures tumor regression has not occurred under any of the experimental conditions.
Figure 4.
Bhlhe40 regulates CD4+ and CD8+ T-cell function. Intratumoral CD45+ cells were harvested on day 9 posttumor transplant from 1956 tumor–bearing Bhlhe40+/+ or Bhlhe40–/– mice treated with control mAb, anti–CTLA-4, or anti–PD-1, and analyzed by scRNAseq. A, UMAP displaying reclustering of T cell–containing clusters (middle plot) and Cd4 and Cd8 expression (bottom plot) of all experimental conditions computationally combined. B, Heatmap displaying normalized expression of select genes in each T-cell cluster. C, Violin plot showing Bhlhe40 expression in Bhlhe40+/+ mice by cluster and treatment. D, Percentage of cells in individual T-cell clusters by condition and treatment represented as percent of CD45+ cells.
CD8+ T cells
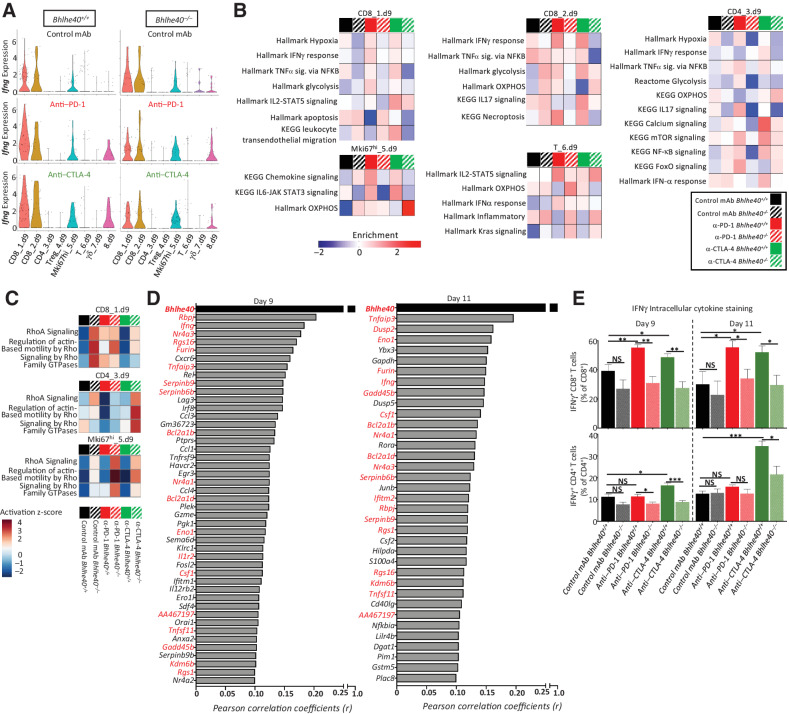
Two CD8+ TIL clusters (CD8_1.d9 and CD8_2.d9) were among the reclustered T cells, with anti–PD-1 ICT inducing Bhlhe40 upregulation in both clusters in WT mice. CD8_1.d9 expressed Bhlhe40 and transcripts associated with T-cell activation and dysfunction/exhaustion [Lag3, Pdcd1 (PD-1), Havcr2 (TIM-3), and Rgs16; Fig. 4B; Supplementary Fig. S8]. In addition, CD8_1.d9 expressed chemokine transcripts Ccl3 and Ccl4, along with Nkg7 and Ifng (Fig. 4B; Supplementary Fig. S8). Overall, CD8_1.d9 cells from Bhlhe40–/– mice expressed higher levels of Tigit (negative T-cell regulator), Cxcr3, Ccl5, Klrc2, and Pfn1 [actin-binding negative regulator of T-cell cytotoxicity (28)] and two related genes: Cd5, which can negatively regulate TCR signaling (29), and Cd6 (Fig. 4B; Supplementary Fig. S6). In contrast, reduced expression of Lag3, Ybx3, Ndufb1-ps, and the transcription factor–encoding genes Hif1a and Tbx21 (TBET) was observed in Bhlhe40–/– CD8_1.d9 (Fig. 4B; Supplementary Fig. S6). A divergent transcript expression pattern within granzyme family members was observed, whereby Gzmb was more highly expressed and Gzmc and Gzmf expression was dampened in Bhlhe40–/– mice (Fig. 4B). ICT induced an increase in Ifng expression that was noticeably blunted in Bhlhe40–/– mice (Figs. 4B and 5A). ICT also induced BHLHE40-dependent upregulation of glycolysis-related transcripts (Pgk1, Pfkp, Hk1, Eno1, Slc2a3, and Gapdh), and two TCR signaling–induced transcription factor genes Nr4a1 (Nur77) and Nr4a3 in WT mice (Fig. 4B; Supplementary Fig. S6; ref. 30). Gene set enrichment analysis (GSEA) revealed that CD8_1.d9 displayed BHLHE40-dependent enrichment in glycolysis, IFNγ response, TNFα signaling via NF-κB, and hypoxia-related transcripts. Specific to the ICT used, anti–PD-1 induced BHLHE40-dependent upregulation of the perforin gene (Prf1), whereas anti–CTLA-4 upregulated Csf2 (GM-CSF) and displayed enrichment for IL2–STAT5 signaling and leukocyte transendothelial migration that was severely diminished in Bhlhe40–/– mice (Figs. 4B and 5B). In addition, high Il10 expression was observed in CD8_1.d9 from anti–CTLA-4–treated Bhlhe40–/– mice (Fig. 4B). Both Csf2 and Il10 are known to be differentially regulated by BHLHE40 (11, 16). Although anti–CTLA-4 reduced the apoptosis gene signature in both WT and Bhlhe40–/– mice, it was more drastically diminished by anti–PD-1 in WT mice, and it was lower than in anti–PD-1–treated Bhlhe40–/– mice (Fig. 5B). Ingenuity Pathway Analysis (IPA) indicated actin-based motility by Rho and RhoA signaling upregulation within all Bhlhe40–/– treatment groups (Fig. 5C).
Figure 5.
Bhlhe40 deficiency alters effector phenotype of intratumoral T cells during ICT. A, Violin plot showing Ifng expression by T-cell cluster and treatment determined using scRNAseq data from intratumoral CD45+ cells harvested on day 9 posttumor transplant from 1956 tumor–bearing Bhlhe40+/+ (left) or Bhlhe40−/− (right) mice treated with control mAb, anti–CTLA-4, or anti–PD-1. B, Heatmap of select enriched gene sets by T-cell cluster and condition as determined by GSEA. C, Heatmap of select IPA pathways by T-cell cluster and condition. D, Correlation between gene expression and Bhlhe40 expression. Genes displayed are those with a Pearson correlation coefficient (r) greater than or equal to 0.1 from day 9 (left) or day 11 (right). Genes in red indicate overlap between day 9 and day 11 scRNAseq (P ≤ 1.5e-7 for all genes listed). E, Percent of intratumoral IFNγ+CD8+ cells and IFNγ+CD4+ cells as assessed by intracellular cytokine staining. Data in E are representative of 5 individual mice per condition and represent 3 independent experiments. Bar indicates mean percent ± SEM as assessed by flow cytometry (*, P < 0.05; **, P < 0.01; ***, P < 0.005; NS, not significant, unpaired t test).
CD8_2.d9 was distinguished from CD8_1.d9 by overall higher expression of Cd69, Cd7, Ly6c2, S1pr1, Gzmk, and Tcf7 (TCF-1), along with overall lower expression of Lag3 and Havcr2 (Fig. 4B; Supplementary Fig. S6 and S8). Based on transcript expression, this population likely represents early-activated CD8+ T cells with features of effector/effector-memory T cells (31). CD8_2.d9 also shares some qualities with the Tcf7+CD8+ T cells that have been linked to exhausted T-cell precursors/progenitors in chronic viral infection and cancer (6, 32–34), whereas other transcriptional features point to Tcf7+Pdcd1+CD8+ T cells that were previously described to have stem-like properties (35), although T_6.d9 more closely aligns with this population. As seen in CD8_1.d9, expression of Tigit, Cxcr3, Ccl5, and Gzmb was higher in Bhlhe40–/– mice, whereas expression of Cx3cr1 (36), Nr4a1, Nr4a3, and Slc2a3 was reduced in Bhlhe40–/– mice under like treatment conditions (Fig. 4B; Supplementary Fig. S6). Bhlhe40–/– CD8_2.d9 displayed reduced expression of the apoptosis inhibitor transcript Api5 and Cd44 (Fig. 4B; Supplementary Fig. S6). In WT mice only, ICT induced upregulation of Eno1 and Tbx21. As observed in CD8_1.d9, CD8_2.d9 also displayed enrichment, but to a greater extent, of IFNγ response and glycolysis genes that was dampened in Bhlhe40–/– CD8_2.d9 (Fig. 5B).
CD4+ T cells
CD4_3.d9 was the major CD4+ Teff cluster. In WT mice, anti–PD-1 and anti–CTLA-4 increased Bhlhe40 expression to an equal or greater extent than was observed in CD8+ TILs (Fig. 4B and C). CD4_3.d9 from Bhlhe40–/– mice was characterized by higher transcript expression of two actin-binding proteins involved in cytoskeletal rearrangement (Cnn2 and Pfn1), the latter of which was also more highly expressed in Bhlhe40–/– CD8+ clusters. Bhlhe40–/– CD4_3.d9 displayed reduced expression of Hif1a, Eno1, and Gapdh, paralleling observations in Bhlhe40–/– CD8+ clusters, along with reduced expression of the IFN-induced transmembrane genes Ifitm1, Ifitm2, and Ifitm3. A divergent expression pattern between specific chemokine receptor transcripts was also detected, whereby Cxcr3 and Cxcr5 were expressed higher in Bhlhe40–/– mice, whereas Cxcr6 was expressed lower. CD4_3.d9 from Bhlhe40–/– mice treated with ICT showed upregulation of Tigit and Cd5 (a pattern also detected in CD8+ TIL) and the proapoptotic Bak1 gene (Fig. 4B; Supplementary Fig. S6). Bhlhe40–/– CD4_3.d9 displayed downregulation of glycolysis, IL17 signaling, and hypoxia, with IPA revealing upregulation of RhoA signaling (Fig. 5B and C). In contrast, CD4_3 from ICT-treated WT mice displayed BHLHE40-dependent upregulation of Lef1, S1pr1, Cox6a1, and mt-Nd3 and leukocyte transendothelial migration, TNFα signaling via NF-κB, and mTOR signaling gene set enrichment (Fig. 5B and C). Specific to the ICT used, anti–PD-1 increased expression of Api5 (similar to CD8_2.d9), as well as Csf2, Tnfaip3, and Tnfsf8 (CD153) that was notably absent in Bhlhe40–/– mice (Fig. 4B; Supplementary Fig. S6). Exclusive to anti–CTLA-4, Bhlhe40+/+ CD4_3.d9 displayed higher Icos and lower Ctla4 as compared with Bhlhe40–/– CD4_3.d9. CD4_3.d9 from WT mice treated with anti–PD-1 exhibited enriched FoxO signaling, whereas WT mice treated with anti–CTLA-4 had enrichment in calcium and chemokine signaling and IFNα response.
Tregs
Cluster Treg_4.d9 containing Cd4+Foxp3+ Tregs displayed the lowest Bhlhe40 transcript expression when compared to the other T-cell clusters. Treg_4.d9 represented 0.91% and 0.33% of reclustered cells in control mAb-treated Bhlhe40+/+ and Bhlhe40–/– mice, respectively. In anti–PD-1–treated WT mice, the percentage decreased to 0.70%, whereas WT mice treated with anti–CTLA-4 experienced a more dramatic decrease (Fig. 4D). This is consistent with the previous observation that the anti-CTLA-4 mAb used (9D9 mouse IgG2b) depletes populations of CD4+FOXP3+ Tregs, which express high levels of surface CTLA-4 (37, 38).
Additional changes occurring in the T-cell compartment
Cluster Mki67hi_5.d9 highly expressed the proliferation marker gene Mki67 (Fig. 4A; Supplementary Fig. S8) and contained proliferating CD8+ T cells with a small number of proliferating CD4+ T cells. Similar to the other T-cell clusters, Pfn1 and Tigit were more highly expressed in Bhlhe40–/– cells under all corresponding conditions, along with Gimap4, Tox, Cd6, and Gzmb, whereas Bcl2 was expressed at lower levels in Bhlhe40–/– mice. This cluster exhibited pathway enrichment in regulation of RhoA signaling, signaling by Rho family GTPases, and actin-based motility by Rho in Bhlhe40–/– mice (Fig. 5C). Bhlhe40-dependent ICT-induced changes were also observed, with increased Eno1, Hk1, Tbx21, and Gzmc expression exclusively in WT mice treated with ICT (Fig. 4B). Anti–PD-1 also induced upregulation of Ifng, whereas anti–CTLA-4 induced upregulation of Gzmf, Cx3cr1, and Cd7 in WT mice. Exclusively in anti–CTLA-4–treated Bhlhe40–/– mice, Mki67hi_5.d9 displayed enhanced OXPHOS as indicated by GSEA (Fig. 5B).
T_6.d9 contained CD8+ T cells with a small fraction of CD4+ T cells. This cluster expressed Tcf7 and several other transcripts previously associated with a naïve, central memory, or stem-like phenotype that has been observed to be important for tumor control by ICT (Fig. 4B; Supplementary Figs. S6 and S8; refs. 31, 34, 35). This cluster was also distinguished by expression of Gm26917 and Bcl2 (Supplementary Fig. S8). Within T_6.d9, expression of Cd5, Cd6, and S1p1r was higher in Bhlhe40–/– mice, whereas Stat1, Tox, and Tcf7 were expressed at lower levels as compared with WT mice under all respective treatment conditions (Fig. 4B). ICT-treated Bhlhe40–/– mice also displayed higher Pdcd1, Havcr2, and Il10 expression. T_6.d9 from WT mice treated with anti–PD-1 displayed enrichment in IFNα response and IL2–STAT5 signaling (Fig. 5B). Conversely, anti–CTLA-4 upregulated IL2–STAT5 signaling but only in Bhlhe40–/– mice. OXPHOS gene set enrichment also diverged with each ICT treatment, with anti–PD-1–treated Bhlhe40–/– mice displaying enrichment of this gene set and anti–CTLA-4–treated Bhlhe40–/– mice downregulating transcripts in this gene set.
Although the number of T cells was higher on day 11, T cells had overall similar alterations between Bhlhe40+/+ and Bhlhe40–/– mice treated with control mAb or ICT on both days assessed (Fig. 5E; Supplementary Fig. S5D). However, in contrast to day 9 where anti–PD-1 induced the most robust upregulation of Bhlhe40 in subsets of T cells, anti–CTLA-4 induced the strongest upregulation on day 11, with the highest expression occurring in CD8+ T-cell clusters expressing cytokine genes Ifng, Tnf, and Csf2, as well as in CD4+ T cells (CD4_6.d11; Supplementary Fig. S5B and S5C).
Bhlhe40 correlation analysis (Pearson) on reclustered T cells from day 9 and 11 revealed multiple statistically significant gene expression correlations, including transcripts known to be regulated by BHLHE40 (e.g., Ifng and Csf2; Fig. 5D). Consistent with observations in individual T-cell clusters, glucose metabolism transcripts (Eno1, Gapdh, and Pgk1) and several genes that are upregulated by T-cell activation [Tnfaip3, Nr4a3, Lag3, Havcr2, and Tnfrsf9 (CD137/4–1BB)] were correlated with Bhlhe40 expression.
Flow cytometry analysis revealed fewer CD45+ cells as a percent of live intratumoral cells between control mAb–treated Bhlhe40+/+ and Bhlhe40–/– mice on day 9 posttransplant with the same trend observed under all treatment conditions on day 11 posttransplant (Supplementary Figs. S9 and S10A). No major differences in the overall intratumoral CD8+ or CD4+ Teff cells as a percent of CD45+ cells were observed when comparing ICT-treated Bhlhe40–/– and Bhlhe40+/+ mice (Supplementary Figs. S9, S10B, and S10C). We did not detect significant quantitative differences in CD4+FOXP3+ Tregs between Bhlhe40+/+ and Bhlhe40–/– mice treated with control mAb or anti–PD-1 (Supplementary Figs. S9 and S10D). However, anti–CTLA-4 ICT reduced the percent of Tregs in both Bhlhe40–/– and Bhlhe40+/+ tumor–bearing mice, consistent with scRNAseq results. Assessment of IFNγ by intracellular cytokine staining revealed the percent of intratumoral IFNγ+CD8+ T cells was increased significantly by ICT in a BHLHE40-dependent manner (Fig. 5E). Whereas both anti–PD-1 and anti–CTLA-4 induced more IFNγ-producing CD8+ T cells in WT, but not Bhlhe40–/– mice, only anti–CTLA-4 induced more IFNγ-producing CD4+ T cells on both day 9 and day 11 posttransplant in Bhlhe40+/+ mice, with Bhlhe40–/– mice again having significantly less IFNγ-expressing CD4+ T cells (Fig. 5E). Although anti–PD-1 treatment did not increase the percentage of IFNγ+CD4+ T cells at either timepoint assessed, anti–PD-1–treated WT mice had a significantly greater percentage of IFNγ+CD4+ T cells as compared with Bhlhe40–/– mice at the day 9 timepoint. Together, these results indicate both CD4+ and CD8+ T cells require BHLHE40 for normal IFNγ production and ICT-induced changes to the tumor microenvironment that are associated with tumor regression.
BHLHE40 is required for ICT-induced remodeling of the myeloid cell compartment
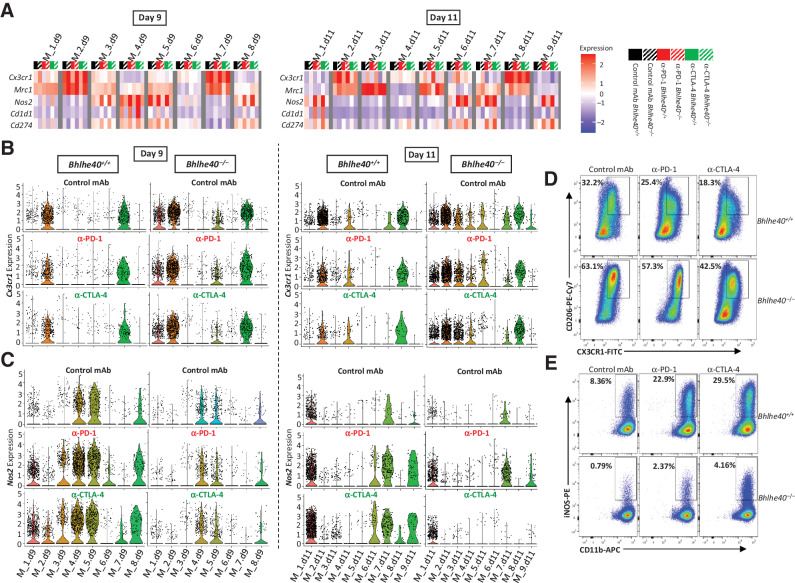
Similar to previous observations in the T3 MCA sarcoma model (26), ICT led to a shift in Bhlhe40+/+ mice from Cx3cr1+Mrc1+ (CD206+) macrophages to Nos2+ (iNOS+) macrophages including Nos2+Cd1d1+ macrophages, as revealed by scRNAseq (Fig. 6A–C; Supplementary Fig. S11A and S11B). These changes were completely dependent upon BHLHE40, as subsets of intratumoral macrophages in ICT-treated Bhlhe40–/– mice displayed high expression of Cx3cr1 with few Nos2+ macrophages present. Furthermore, clusters containing Nos2+ macrophages also expressed high levels of Cd274 (PD-L1) in WT, but not Bhlhe40–/–, mice treated with ICT (Fig. 6A; Supplementary Fig. S11A and S11B).
Figure 6.
ICT-induced remodeling of intratumoral monocyte/macrophages requires Bhlhe40. A, Heatmap displaying normalized expression of select genes in each monocyte/macrophage cluster by group determined using scRNAseq data from 1956 intratumoral CD45+ cells harvested on day 9 (left) or day 11 (right) posttumor transplant from 1956 tumor–bearing Bhlhe40+/+ or Bhlhe40–/– mice treated with control mAb, anti–CTLA-4, or anti–PD-1. B, Violin plots showing Cx3cr1 expression in intratumoral monocytes/macrophages on day 9 (left) and day 11 (right) by cluster and treatment. C, Violin plots showing Nos2 expression in intratumoral monocytes/macrophages on day 9 (left) and day 11 (right) by cluster and treatment. Flow cytometry plots of 1956 intratumoral macrophages from day 11 posttransplant showing percent of CX3CR1+CD206+ macrophages (D) and iNOS+macrophages (E) in Bhlhe40–/– or Bhlhe40+/+ mice treated with control mAb, anti–PD-1, or anti–CTLA-4. Flow cytometry plots in D and E are gated on macrophages as described in the study by Noguchi and colleagues (39).
To analyze different monocyte/macrophage populations by protein expression, we used a myeloid cell flow cytometry panel (39). Consistent with the scRNAseq data, macrophages constituted a plurality of CD45+ cells (Supplementary Figs. S9 and S10E). ICT significantly reduced the percentage of CX3CR1+CD206+ macrophages in Bhlhe40+/+ mice (Fig. 6D; Supplementary Fig. S10F). However, this decrease was absent in Bhlhe40–/– mice treated with ICT, consistent with scRNAseq data. Furthermore, a sharp increase in intratumoral iNOS+ macrophages was observed in tumor-bearing Bhlhe40+/+ mice treated with ICT (Fig. 6E; Supplementary Fig. S10G). In contrast, control-treated tumor-bearing Bhlhe40–/– mice contained a paucity of intratumoral iNOS+ macrophages that increased with ICT but represented only a fraction of what was observed in ICT-treated Bhlhe40+/+ mice. In sum, BHLHE40 is required for ICT-driven remodeling of intratumoral macrophages from a CX3CR1+CD206+ phenotype to an iNOS+ phenotype.
Tumor antigen–specific CD4+ and CD8+ T cells require BHLHE40 for effector function
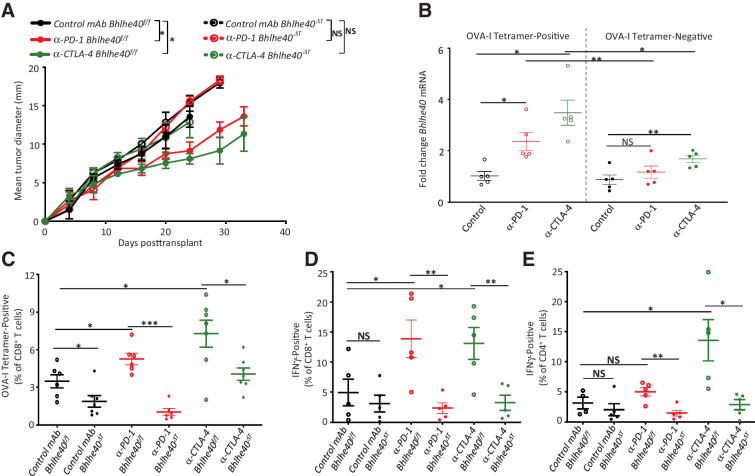
To extend our study to a non-MCA sarcoma line, we injected B16-OVA melanoma cells into Bhlhe40f/f and Bhlhe40ΔT mice. Whereas B16-OVA–bearing Bhlhe40f/f mice treated with either ICT displayed a decrease in tumor growth when compared with control mAb–treated mice, tumor-bearing T cell–deficient Bhlhe40ΔT mice showed no significant reduction in tumor growth after ICT (Fig. 7A; Supplementary Fig. S12A and S12B). We harvested B16-OVA tumors from Bhlhe40f/f and Bhlhe40ΔT mice treated with control or ICT at day 15 posttransplant and performed peptide–MHC-I tetramer staining to monitor tumor antigen–specific CD8+ T-cell responses. Sorted intratumoral CD8+ T cells recognizing the immunodominant OVA-I epitope (SIINFEKL) presented on H-2Kb (OVA-I tetramer–positive) upregulated Bhlhe40 in Bhlhe40f/f mice treated with ICT and displayed increased expression of Bhlhe40 mRNA when compared with tetramer-negative sorted intratumoral CD8+ T cells (Fig. 7B). Although OVA-I tetramer–positive T cells displayed the more robust upregulation of Bhlhe40 upon ICT, an increase in Bhlhe40 expression was also observed in tetramer-negative T cells with anti–CTLA-4 treatment. These tetramer-negative T cells are likely composed of not only bystander T cells, but also subpopulations of T cells recognizing shared, non-mutant antigens and other subdominant antigens (40, 41), which may account for the increase in Bhlhe40 expression we observed in OVA-I tetramer–negative populations with anti–CTLA-4 treatment. It is also known that signals other than TCR ligation contribute to Bhlhe40 upregulation in T cells (11, 12). Next, we observed a decreased percent of intratumoral CD8+ T cells specific for OVA-I in Bhlhe40ΔT mice as compared with treatment-matched Bhlhe40f/f (Fig. 7C). Finally, we observed a reduced percentage of IFNγ+ intratumoral CD8+ T cells from ICT-treated Bhlhe40ΔT mice upon restimulation with the OVA-I peptide when compared with those from ICT-treated Bhlhe40f/f mice (Fig. 7D). In addition, intratumoral CD4+ T cells from ICT-treated Bhlhe40ΔT mice restimulated with OVA-II peptide–loaded splenocytes displayed a significant reduction by percent in IFNγ+ cells (Fig. 7E), indicating defects in both CD4+, as well as CD8+, T cells in the absence of BHLHE40.
Figure 7.
Bhlhe40 is required for generation of functional tumor antigen–specific T cells and ICT efficacy against B16-OVA melanoma. A, B16-OVA tumor growth in Bhlhe40ΔT and Bhlhe40f/f mice treated with control mAb, anti–CTLA-4, or anti–PD-1. B,Bhlhe40 mRNA expression in intratumoral OVA-I tetramer–positive or –negative CD8+ T cells sorted from B16-OVA melanoma–bearing WT mice treated with control mAb, anti–CTLA-4, or anti–PD-1. C, Percent of intratumoral OVA tetramer–positive CD8+ T cells in B16-OVA melanoma–bearing Bhlhe40ΔT and Bhlhe40f/f mice treated as in B. Percent of intratumoral IFNγ-positive CD8+ T cells (stimulated ex vivo with OVA-I peptide; D) or CD4+ T cells (E; stimulated ex vivo with OVA-II peptide) in B16-OVA melanoma–bearing Bhlhe40ΔTor Bhlhe40f/f mice treated as in B. Data in A are presented as average tumor diameter ± SEM of 4–5 mice per group and are representative of at least 3 independent experiments (*, P < 0.05; **, P < 0.01, two-way ANOVA). Data in B are presented as mean ± SEM Bhlhe40 mRNA fold change. Each dot represents a Bhlhe40 mRNA data point from sorted OVA-I tetramer–positive and –negative CD8+ T cells isolated from 4–5 individual mice per group and are representative of at least 3 independent experiments. Data in D and E are presented as mean ± SEM of IFNγ+ cells expressed as a percent of CD8+ T cells or CD4+ T cells as assessed by flow cytometry. For C–E, cells were gated on live CD45+Thy1.2+ and CD8+ or CD4+ T cells. For B–E, cells were isolated from 4–5 individual mice per group on day 15 posttransplant (*, P < 0.05; **, P < 0.01; NS, not significant, unpaired t test).
Discussion
Here we have demonstrated that BHLHE40 expression in T cells is vital for anti–PD-1 or anti–CTLA-4 ICT efficacy against B16-OVA melanoma and the 1956 edited progressively growing MCA sarcoma, but is dispensable for spontaneous rejection of a highly immunogenic unedited MCA sarcoma line (1969). Interrogation of the immune tumor microenvironment by both scRNAseq and flow cytometry demonstrated that ICT upregulated Bhlhe40 in both CD4+ and CD8+ TIL subsets and was associated with expression of activation/effector transcripts and proteins, with BHLHE40-deficient T cells displaying reduced expression of the critical cytokine IFNγ. Defects in IFNγ production by BHLHE40-deficient CD8+ T cells may be attributed to not only BHLHE40’s known role in enhancing IFNγ expression (10, 23), but also may reflect poor priming, differentiation, trafficking, or survival (12) of BHLHE40-deficient tumor antigen–specific CD8+ T cells, as we observed fewer OVA-I–specific CD8+ TILs in Bhlhe40ΔT tumor-bearing mice (Fig. 7C).
The mechanism by which ICT induces upregulation of BHLHE40 in CD4+ and CD8+ T cells may diverge between anti–PD-1 and anti–CTLA-4, as PD-1 and CTLA-4 can exert their suppression through differing means (42). Consistent with previous findings (12, 13), we found that CD28 costimulation enhanced upregulation of Bhlhe40 in both CD4+ and CD8+ T cells (Supplementary Fig. S3A). Therefore, it is possible that anti–CTLA-4 induces upregulation of BHLHE40 by enhancing costimulation, as CTLA-4 outcompetes CD28 for B7 owing to its higher avidity (43). In contrast, PD-1 signaling blockade with anti–PD-1 might be directly responsible for BHLHE40 upregulation, as PD-1 signaling has been previously shown to suppress Bhlhe40 expression (12). Other ICT-induced cues in the tumor microenvironment may also contribute to Bhlhe40 expression (11).
In the absence of BHLHE40, subsets of intratumoral CD4+ and CD8+ Teff displayed altered transcript and/or protein expression of key molecules involved in T-cell function: cytokines/cytokine receptors (e.g., decreased IFNγ and Csf2); effector/cytolytic molecules (e.g., increased Gzmb and decreased Gmzc and Gmzf); chemotaxis molecules (e.g., increased Cxcr3); transcripts affecting the cytoskeleton (e.g., increased Cnn2 and Pfn1); inhibitory receptors (e.g., increased Cd5 and Tigit); and aerobic metabolism–related transcripts (e.g., decreased Cox6a1, Ndufb1-ps, and mt-Nd3); and glycolytic enzyme transcripts (e.g., decreased Pgk1, Pfkp, Hk1, Eno1 and Gapdh). In Bhlhe40+/+ mice, several T-cell clusters showed marked upregulation of multiple glycolytic enzyme transcripts (Fig. 4B; Supplementary Fig. S5B) and a GSEA signature indicative of active glycolysis (Fig. 5B) upon ICT that was blunted in Bhlhe40–/– mice. It was particularly characteristic of CD8_1.d9, CD8_1.d11, and CD8_5.d11, all of which displayed high activation and/or dysfunction/exhaustion marker expression and the highest transcription levels of Ifng in WT mice. CD4+ Teff, but not Tregs, displayed a similar glycolysis transcriptional profile. While Tregs and naïve and memory T cells rely on OXPHOS as the primary energy source, Teff cells additionally require aerobic glycolysis (44, 45). The expression levels of Eno1 and Gapdh showed the most consistent correlation with Bhlhe40 in the different Teff clusters. Higher expression of glycolytic enzymes may be driven by HIF1α, as we observed more abundant Hif1a transcript in Bhlhe40+/+ T cells (Figs. 4B and 5B; Supplementary Fig. S5B). Interestingly, HIF1α is known to directly activate Bhlhe40 transcription (46). Therefore, a consequence of the loss of BHLHE40 expression may be impaired metabolic reprogramming of the Teff cells via hypoxia-related signaling. It should also be noted that in addition to metabolic function, glycolytic enzymes play important regulatory roles independent of their catalytic activity (47, 48).
Although we observed reduced expression of certain mitochondrial complex genes (Cox6a1, mt-Nd3, and Ndufb1-ps) in the absence of BHLHE40, we did not detect as strong of a gene signature as Li and colleagues (12) observed in Bhlhe40–/– CD8+ TIL and Trm cells in a lung infection model. These differences may be connected to the different experimental models used between the studies and the level of available oxygen for aerobic mitochondrial respiration. In addition, the rate of glucose consumption by the tumor cells themselves affects T-cell metabolism and function, and different tumors have unalike rates of glucose consumption (49). Because we did not directly measure T-cell metabolic profiles, our findings at the transcriptional level will need to be further validated by proteomic and metabolomic profiling for more extensive interpretation.
BHLHE40 has been shown to be important in CD4+ T cells in infection and autoimmunity models (10). Furthermore, expression of BHLHE40 has also been detected in a Th1-like CD4+ T-cell population enriched in human colorectal tumors with microsatellite instability (MSI), which generally have a more favorable response to ICT (50). Because ICT efficacy was lost in both 1956- and B16-OVA–bearing Bhlhe40ΔT mice, where both CD4+ and CD8+ T cells display Bhlhe40 deletion (15, 16), it is possible that both CD4+ and CD8+ T cells require BHLHE40 for effective ICT, as it is now clear that CD4+ T cells play a critical role in antitumor immunity (1, 2).
We observed ICT-induced, BHLHE40-dependent macrophage remodeling from “M2-like” CXCR3+CD206+ macrophages to “M1-like” iNOS+ macrophages, consistent with the remodeling that we previously saw associated with tumor regression (26). In the absence of BHLHE40, the percent of CX3CR1+CD206+ macrophages were significantly higher than observed in WT mice under like treatment conditions. This coincided with a strong reduction by percentage of ICT-induced intratumoral iNOS+ macrophages as compared with WT mice. Because iNOS induction in macrophages is known to require IFNγ plus an inducer of NF-κB signaling, it is possible that reduced IFNγ production by T cells in the absence of BHLHE40 or altered NF-κB–inducing stimulus is responsible for the defects in iNOS+ macrophages observed.
Our results reveal that BHLHE40 is essential in T cells for effective ICT. However, it remains to be seen whether this effect is broadly applicable to different forms of immunotherapy, as our study suggests BHLHE40 may be a potential biomarker for ICT efficacy or a therapeutic target for manipulation in cell-based therapies.
Supplementary Material
Acknowledgments
M.M. Gubin is a Cancer Prevention and Research Institute of Texas (CPRIT) Scholar in Cancer Research. This work was supported by CPRIT (Recruitment of First-Time Tenure-Track Faculty Members; RR190017), The Parker Institute for Cancer Immunotherapy Bridge Scholar Award, University of Texas (UT) Rising Stars Award, and the University of Texas MD Anderson Cancer Center Support Grant (CCSG) New Faculty Award supported by the NIH/NCI (P30CA016672 to M.M. Gubin; NIH R01AI113118 and R01AI132653 to B.T. Edelson; and NIH/NCI U01CA247760 to K. Chen). J.P. Ward is supported by the NCI/NIH Paul Calabresi Career Development Award in Clinical Oncology (K12CA167540). N.N. Jarjour is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2427-21). S. Keshari is a Balzan Postdoctoral Research Fellow supported by The International Balzan Prize Foundation. The University of Texas MD Anderson Advanced Cytometry & Sorting Facility at South Campus (ACSF) is partially funded by the NCI Cancer Center Support Grant P30CA16672. scRNAseq was performed by The University of Texas MD Anderson Cancer Center Advanced Technology Genomics Core (ATGC) Facility funded by an NCI grant [CA016672 (ATGC)] and an NIH 1S10OD024977-01 award to the ATGC. Aspects of studies were performed with assistance by the Washington University School of Medicine Immunomonitoring Laboratory (IML), which is supported by the Andrew M. and Jane M. Bursky Center for Human Immunology and Immunotherapy Programs and the Alvin J. Siteman Comprehensive Cancer Center that, in turn, is supported by NCI/NIH Cancer Center Support Grant under award number P30CA91842. We would like to thank David Pollock at The University of Texas MD Anderson Cancer Center Advanced Technology Genomics Core (ATGC) Facility for assistance with scRNAseq. We would like to thank the Baylor College of Medicine MHC Tetramer Core and thank the core director, X. Lily Wang for production of MHC tetramers used in this study. The authors thank all members of the Edelson and Gubin lab for helpful discussions and technical support. We would also like to thank Stephanie Watowich (The University of Texas MD Anderson Cancer Center), Swetha Anandhan (The University of Texas MD Anderson Cancer Center), and James Mancuso (The University of Texas MD Anderson Cancer Center) for critiques and helpful discussions.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Authors' Disclosures
N.N. Jarjour reports grants from Damon Runyon Cancer Research Foundation outside the submitted work. J.P. Ward reports grants from NCI during the conduct of the study; personal fees and other support from Novocure; other support from Pfizer, Genmab A/S, AstraZeneca, and other support from GlaxoSmithKline/GSK outside the submitted work. B.T. Edelson reports grants from NIH/NIAID during the conduct of the study. K. Chen reports grants from the NCI during the conduct of this study. M.M. Gubin reports grants from Cancer Prevention and Research Institute of Texas (CPRIT), grants from Parker Institute for Cancer Immunotherapy (PICI), and grants from The University of Texas MD Anderson Cancer Center Support Grant (CCSG) New Faculty Award supported by the NIH/NCI (P30CA016672) during the conduct of the study; and a personal honorarium of $1000.00 USD per year from Springer Nature Ltd for his role as an Associate Editor for the journal Nature Precision Oncology. No disclosures were reported by the other authors.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Authors' Contributions
A.J. Salmon: Conceptualization, investigation, visualization, writing–original draft, writing–review and editing. A.S. Shavkunov: Conceptualization, investigation, writing–review and editing. Q. Miao: Conceptualization, data curation, formal analysis, investigation, visualization. N.N. Jarjour: Formal analysis, investigation, writing–review and editing. S. Keshari: Conceptualization, data curation, investigation, visualization, writing–review and editing. E. Esaulova: Formal analysis, investigation, writing–review and editing. C.D. Williams: Investigation, writing–review and editing. J.P. Ward: Writing–review and editing. A.M. Highsmith: Data curation, investigation, writing–original draft, writing–review and editing. J.E. Pineda: Data curation, investigation, writing–review and editing. R. Taneja: Conceptualization, resources. K. Chen: Resources, formal analysis, investigation, visualization, writing–review and editing. B.T. Edelson: Conceptualization, data curation, investigation, writing–review and editing. M.M. Gubin: Conceptualization, resources, data curation, formal analysis, supervision, validation, investigation, methodology, writing–original draft, writing–review and editing.
References
- 1. Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 2014;515:577–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alspach E, Lussier DM, Miceli AP, Kizhvatov I, DuPage M, Luoma AM, et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 2019;574:696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gubin MM, Artyomov MN, Mardis ER, Schreiber RD. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest 2015;125:3413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol 2013;31:e439–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer 2014;14:135–46. [DOI] [PubMed] [Google Scholar]
- 6. Philip M, Fairchild L, Sun L, Horste EL, Camara S, Shakiba M, et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 2017;545:452–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 2018;175:998–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kurtulus S, Madi A, Escobar G, Klapholz M, Nyman J, Christian E, et al. Checkpoint blockade immunotherapy induces dynamic changes in PD-1(-)CD8(+) tumor-infiltrating T cells. Immunity 2019;50:181–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Azmi S, Sun H, Ozog A, Taneja R. mSharp-1/DEC2, a basic helix-loop-helix protein functions as a transcriptional repressor of E box activity and Stra13 expression. J Biol Chem 2003;278:20098–109. [DOI] [PubMed] [Google Scholar]
- 10. Cook ME, Jarjour NN, Lin CC, Edelson BT. Transcription factor Bhlhe40 in immunity and autoimmunity. Trends Immunol 2020;41:1023–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin CC, Bradstreet TR, Schwarzkopf EA, Sim J, Carrero JA, Chou C, et al. Bhlhe40 controls cytokine production by T cells and is essential for pathogenicity in autoimmune neuroinflammation. Nat Commun 2014;5:3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li C, Zhu B, Son YM, Wang Z, Jiang L, Xiang M, et al. The transcription factor Bhlhe40 programs mitochondrial regulation of resident CD8(+) T cell fitness and functionality. Immunity 2019;51:491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martinez-Llordella M, Esensten JH, Bailey-Bucktrout SL, Lipsky RH, Marini A, Chen J, et al. CD28-inducible transcription factor DEC1 is required for efficient autoreactive CD4+ T cell response. J Exp Med 2013;210:1603–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin CC, Bradstreet TR, Schwarzkopf EA, Jarjour NN, Chou C, Archambault AS, et al. IL-1-induced Bhlhe40 identifies pathogenic T helper cells in a model of autoimmune neuroinflammation. J Exp Med 2016;213:251–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu F, Sharma S, Jankovic D, Gurram RK, Su P, Hu G, et al. The transcription factor Bhlhe40 is a switch of inflammatory versus antiinflammatory Th1 cell fate determination. J Exp Med 2018;215:1813–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huynh JP, Lin CC, Kimmey JM, Jarjour NN, Schwarzkopf EA, Bradstreet TR, et al. Bhlhe40 is an essential repressor of IL-10 during Mycobacterium tuberculosis infection. J Exp Med 2018;215:1823–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jarjour NN, Schwarzkopf EA, Bradstreet TR, Shchukina I, Lin CC, Huang SC, et al. Bhlhe40 mediates tissue-specific control of macrophage proliferation in homeostasis and type 2 immunity. Nat Immunol 2019;20:687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jarjour NN, Bradstreet TR, Schwarzkopf EA, Cook ME, Lai CW, Huang SC, et al. BHLHE40 promotes TH2 cell-mediated antihelminth immunity and reveals cooperative CSF2RB family cytokines. J Immunol 2020;204:923–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Henriksson J, Chen X, Gomes T, Ullah U, Meyer KB, Miragaia R, et al. Genome-wide CRISPR screens in T helper cells reveal pervasive crosstalk between activation and differentiation. Cell 2019;176:882–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones EA, Flavell RA. Distal enhancer elements transcribe intergenic RNA in the IL-10 family gene cluster. J Immunol 2005;175:7437–46. [DOI] [PubMed] [Google Scholar]
- 21. Emming S, Bianchi N, Polletti S, Balestrieri C, Leoni C, Montagner S, et al. A molecular network regulating the proinflammatory phenotype of human memory T lymphocytes. Nat Immunol 2020;21:388–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miyazaki K, Miyazaki M, Guo Y, Yamasaki N, Kanno M, Honda Z, et al. The role of the basic helix-loop-helix transcription factor Dec1 in the regulatory T cells. J Immunol 2010;185:7330–9. [DOI] [PubMed] [Google Scholar]
- 23. Kanda M, Yamanaka H, Kojo S, Usui Y, Honda H, Sotomaru Y, et al. Transcriptional regulator Bhlhe40 works as a cofactor of T-bet in the regulation of IFN-gamma production in iNKT cells. Proc Natl Acad Sci U S A 2016;113:E3394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sun H, Lu B, Li RQ, Flavell RA, Taneja R. Defective T cell activation and autoimmune disorder in Stra13-deficient mice. Nat Immunol 2001;2:1040–7. [DOI] [PubMed] [Google Scholar]
- 25. Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 2001;410:1107–11. [DOI] [PubMed] [Google Scholar]
- 26. Gubin MM, Esaulova E, Ward JP, Malkova ON, Runci D, Wong P, et al. High-dimensional analysis delineates myeloid and lymphoid compartment remodeling during successful immune-checkpoint cancer therapy. Cell 2018;175:1014–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fehlings M, Simoni Y, Penny HL, Becht E, Loh CY, Gubin MM, et al. Checkpoint blockade immunotherapy reshapes the high-dimensional phenotypic heterogeneity of murine intratumoural neoantigen-specific CD8(+) T cells. Nat Commun 2017;8:562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schoppmeyer R, Zhao R, Cheng H, Hamed M, Liu C, Zhou X, et al. Human profilin 1 is a negative regulator of CTL mediated cell-killing and migration. Eur J Immunol 2017;47:1562–72. [DOI] [PubMed] [Google Scholar]
- 29. Azzam HS, DeJarnette JB, Huang K, Emmons R, Park CS, Sommers CL, et al. Fine tuning of TCR signaling by CD5. J Immunol 2001;166:5464–72. [DOI] [PubMed] [Google Scholar]
- 30. Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med 2011;208:1279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Andreatta M, Corria-Osorio J, Muller S, Cubas R, Coukos G, Carmona SJ. Interpretation of T cell states from single-cell transcriptomics data using reference atlases. Nat Commun 2021;12:2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen Z, Ji Z, Ngiow SF, Manne S, Cai Z, Huang AC, et al. TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity 2019;51:840–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 2019;20:326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Philip M, Schietinger A. CD8(+) T cell differentiation and dysfunction in cancer. Nat Rev Immunol 2021Jul 12 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Pais Ferreira D, et al. Intratumoral Tcf1(+)PD-1(+)CD8(+) T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 2019;50:195–211. [DOI] [PubMed] [Google Scholar]
- 36. Nishimura M, Umehara H, Nakayama T, Yoneda O, Hieshima K, Kakizaki M, et al. Dual functions of fractalkine/CX3C ligand 1 in trafficking of perforin+/granzyme B+ cytotoxic effector lymphocytes that are defined by CX3CR1 expression. J Immunol 2002;168:6173–80. [DOI] [PubMed] [Google Scholar]
- 37. Peggs KS, Quezada SA, Chambers CA, Korman AJ, Allison JP. Blockade of CTLA-4 on both effector and regulatory T cell compartments contributes to the antitumor activity of anti-CTLA-4 antibodies. J Exp Med 2009;206:1717–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Selby MJ, Engelhardt JJ, Quigley M, Henning KA, Chen T, Srinivasan M, et al. Anti-CTLA-4 antibodies of IgG2a isotype enhance antitumor activity through reduction of intratumoral regulatory T cells. Cancer Immunol Res 2013;1:32–42. [DOI] [PubMed] [Google Scholar]
- 39. Noguchi T, Ward JP, Gubin MM, Arthur CD, Lee SH, Hundal J, et al. Temporally distinct PD-L1 expression by tumor and host cells contributes to immune escape. Cancer Immunol Res 2017;5:106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhai Y, Yang JC, Spiess P, Nishimura MI, Overwijk WW, Roberts B, et al. Cloning and characterization of the genes encoding the murine homologues of the human melanoma antigens MART1 and gp100. J Immunother 1997;20:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Simoni Y, Becht E, Fehlings M, Loh CY, Koo SL, Teng KWW, et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 2018;557:575–9. [DOI] [PubMed] [Google Scholar]
- 42. Wei SC, Levine JH, Cogdill AP, Zhao Y, Anang NAS, Andrews MC, et al. Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell 2017;170:1120–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Engelhardt JJ, Sullivan TJ, Allison JP. CTLA-4 overexpression inhibits T cell responses through a CD28-B7-dependent mechanism. J Immunol 2006;177:1052–61. [DOI] [PubMed] [Google Scholar]
- 44. Buck MD, O'Sullivan D, Klein Geltink RI, Curtis JD, Chang CH, Sanin DE, et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 2016;166:63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sugiura A, Rathmell JC. Metabolic barriers to T cell function in tumors. J Immunol 2018;200:400–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miyazaki K, Kawamoto T, Tanimoto K, Nishiyama M, Honda H, Kato Y. Identification of functional hypoxia response elements in the promoter region of the DEC1 and DEC2 genes. J Biol Chem 2002;277:47014–21. [DOI] [PubMed] [Google Scholar]
- 47. Shyer JA, Flavell RA, Bailis W. Metabolic signaling in T cells. Cell Res 2020;30:649–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ho PC, Bihuniak JD, Macintyre AN, Staron M, Liu X, Amezquita R, et al. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell 2015;162:1217–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chang CH, Qiu J, O'Sullivan D, Buck MD, Noguchi T, Curtis JD, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 2015;162:1229–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 2018;564:268–72. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data files for the scRNAseq data reported in this article have been deposited in the GEO database. The accession number is GEO: GSE192546. Software used in this study is available online: current version of Cell Ranger: https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest; Seurat 4.0: https://satijalab.org/seurat/; ggplot2 3.3.3: https://ggplot2.tidyverse.org/index.html; and ImmGen: https://www.immgen.org. All other data generated in this study are available within the article and its Supplementary Data files.



![Figure 1. Subsets of intratumoral myeloid and lymphoid cells express Bhlhe40. A, Bhlhe40 mRNA expression in intratumoral mLama4-specific CD8+ T cells sorted from T3 sarcoma–bearing WT mice treated with anti–CTLA-4, anti–PD-1, or both anti–CTLA-4 and anti–PD-1. B, tSNE plot from merged treatment data of exclusively intratumoral lymphocytes showing Bhlhe40 expression within indicated lymphoid subpopulations identified by scRNAseq in the T3 MCA sarcoma. C, Violin plots showing Bhlhe40 expression in T3 intratumoral lymphoid cells by cluster and treatment. D, Heatmap displaying normalized expression of select genes in T3 intratumoral lymphoid cells by cluster and treatment. E, Tumor growth in Bhlhe40–/– or Bhlhe40+/+ mice transplanted with 1956 sarcoma cells and subsequently treated with control, anti–CTLA-4, anti–PD-1, or both anti–CTLA-4 and anti–PD-1 on days 3, 6, and 9 post-transplant. F, Cumulative Kaplan–Meier survival curves for the 1956 tumor–bearing Bhlhe40–/– or Bhlhe40+/+ mice treated as in E. For A, each dot represents 5 pooled mice harvested on day 11 posttransplant and assessed independently (N = 3; *, P < 0.05, unpaired t test). B–D, scRNAseq data generated in Gubin et al (39) were reanalyzed for Bhlhe40 expression. Data in E are presented as average tumor diameter ± SEM of 5 mice per group and are representative of at least four independent experiments. Data in (F) are cumulative survival curves from 4 independent experiments of 3–5 mice per group [***, P < 0.001 (log-rank (Mantel–Cox) test)].](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/3953/9381123/80fbdb6552f9/597fig1.jpg)
![Figure 2. Bhlhe40 is selectively required in T cells for anti–PD-1 or anti–CTLA-4 ICT-mediated tumor rejection. A, Tumor growth in Bhlhe40ΔT or Bhlhe40f/f mice transplanted with 1956 sarcoma cells and subsequently treated with control mAb, anti–CTLA-4, or anti–PD-1 on days 3, 6, and 9 posttransplant. B, Cumulative Kaplan–Meier survival curves of 1956 tumor–bearing Bhlhe40ΔT or Bhlhe40f/f mice treated as in A. Bhlhe40 mRNA expression in intratumoral PD-1+LAG-3+CD8+ T cells (C) and PD-1+LAG-3+CD4+ T cells (D) sorted on day 11 posttransplant from 1956 sarcoma–bearing WT mice treated with control mAb, anti–CTLA-4, or anti–PD-1. Bhlhe40 mRNA expression in CD8+ OT-I T cells (E) stimulated with 1 μmol/L OVA-I peptide and CD4+ OT-II T cells (F) stimulated with 1 μmol/L OVA-II peptide for the indicated time. Data in A are presented as average tumor diameter ± SEM of 5–6 mice per group and are representative of 3 independent experiments. Data in B are cumulative survival curves from 3 independent experiments of 4–6 mice per group [***, P < 0.001; NS, not significant, (log-rank (Mantel–Cox) test)]. For C and D, each dot represents mice harvested and assessed independently (N = 5; *, P < 0.05; **, P < 0.01, unpaired t test). Data in E, and F are presented as mean mRNA fold change. Bar indicates mean ± SEM (*, P < 0.05; **, P < 0.01, unpaired t test) and are representative of 3 independent experiments.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/3953/9381123/476fd44a7722/597fig2.jpg)