Abstract
A bacterial strain, PM1, which is able to utilize methyl tert-butyl ether (MTBE) as its sole carbon and energy source, was isolated from a mixed microbial consortium in a compost biofilter capable of degrading MTBE. Initial linear rates of MTBE degradation by 2 × 106 cells ml−1 were 0.07, 1.17, and 3.56 μg ml−1 h−1 for initial concentrations of 5, 50, and 500 μg MTBE ml−1, respectively. When incubated with 20 μg of uniformly labeled [14C]MTBE ml−1, strain PM1 converted 46% to 14CO2 and 19% to 14C-labeled cells within 120 h. This yield is consistent with the measurement of protein accumulation at different MTBE concentrations from which was estimated a biomass yield of 0.18 mg of cells mg MTBE−1. Strain PM1 was inoculated into sediment core material collected from a contaminated groundwater plume at Port Hueneme, California, in which there was no evidence of MTBE degradation. Strain PM1 readily degraded 20 μg of MTBE ml−1 added to the core material. The rate of MTBE removal increased with additional inputs of 20 μg of MTBE ml−1. These results suggest that PM1 has potential for use in the remediation of MTBE-contaminated environments.
Since its initial use as a gasoline oxygenate in the 1980s, methyl tert-butyl ether (MTBE) production has risen to 7.7 billion kilograms per year and currently comprises up to 15% (vol/vol) of some reformulated gasolines (9). This increased usage coupled with high incidences of leaking underground storage tanks and recreational watercraft operation has led to MTBE contamination of surface waters, groundwater, soils, and sediments. The compound is extremely water soluble and moderately volatile; thus, it is highly mobile in both groundwater and surface waters and can volatilize to contaminate the vadose zone, surface soils, and sediments. Currently, the U.S. Environmental Protection Agency (EPA) lists MTBE as a possible carcinogen; however, toxicity limits are a subject of debate. Since MTBE can be detected by both taste and odor at concentrations as low as 35 μg liter−1, the EPA has recommended keeping concentrations in drinking water below a nuisance limit of 40 μg liter−1 (1).
Unlike other gasoline components, including benzene (14, 16) and toluene (6, 14), there are few reports of microorganisms in either pure or mixed cultures capable of biodegrading MTBE. Salanitro et al. were the first to report the bacterial degradation of MTBE (17). In that study, a mixed microbial consortium was found to degrade 120 μg of MTBE ml−1 at a rate of 34 mg g of cells−1 h−1. The metabolic intermediate tert-butyl alcohol (TBA) was observed; however, further analysis of the metabolic pathway was complicated by the presence of more than one bacterial species in the degrading culture. Eweis et al. have reported the enrichment of a second microbial consortium capable of degrading MTBE (3). In that study, a mixed bacterial culture was obtained by subculturing the solid support material from a compost biofilter located at the Los Angeles County Joint Water Pollution Control Plant (Carson, Calif.) that began removing MTBE after a 1-year acclimation period. The microbial consortium was used to inoculate a bench-scale biofilter established for treatment of MTBE-contaminated airstreams (4). The compost-derived consortium was the source of the bacterial isolate described in our study.
To date, there has been a single study describing pure bacterial cultures capable of using MTBE as a sole carbon and energy source. Mo et al. described three bacterial strains (an Arthrobacter, a Rhodococcus, and a Methylobacterium strain) that degraded up to 29% of an initial concentration of 200 μg of MTBE ml−1 in 2 weeks; however, complete MTBE degradation by these cultures was not observed (11). In cometabolism experiments researchers have reported that propane-enriched environmental isolates are capable of mineralizing MTBE, but these organisms cannot grow on MTBE without prior induction by another compound (20). There is also one report of a strain of the fungal genus Graphium capable of cometabolizing MTBE in the presence of n-butane (7).
Several studies have investigated the potential for natural attenuation of MTBE in soils and sediment. Yeh and Novak measured the anaerobic biodegradation of MTBE in soil microcosms and found that MTBE was degraded only in the low organic matter soils (22). Mormille et al. (12) detected anaerobic degradation of MTBE in one replicate of a fuel-contaminated river sediment after a 152-day acclimation period. Most recently, Bradley et al. (2) have reported mineralization of both MTBE and TBA in streambed sediments under aerobic conditions. In addition to soil and sediment studies, the potential for natural attenuation of MTBE has been evaluated through modeling studies, for example, in the Borden aquifer (19). Laboratory confirmation of biodegradation potential in samples from sites where natural attenuation has been hypothesized is not part of these studies.
Given the increasing incidence of MTBE in the environment and the apparently low rates of MTBE natural attenuation, it is important to find bacterial cultures capable of rapid MTBE degradation and survival outside of the laboratory. Our objectives in this study were to isolate and characterize a bacterial culture capable of using MTBE as its sole carbon and energy source. We then evaluated this organism, designated strain PM1, for the ability to degrade MTBE when inoculated into groundwater core material. Our results indicate that strain PM1 may be effective for use in the bioaugmentation of MTBE-contaminated environments.
MATERIALS AND METHODS
Isolation, culturing, and identification of the organism.
A mixed bacterial culture capable of degrading MTBE was obtained from the University of California, Davis, Department of Civil and Environmental Engineering (3). This bacterial consortium was originally enriched from a compost biofilter at the Los Angeles County Joint Water Pollution Control Plant. The mixed culture was plated onto 0.1× tryptic soy agar (TSA). Isolated colonies were tested for the ability to grow in mineral salts medium (MSM) (13) with 25 μg of MTBE (high-pressure liquid chromatography grade, >99.9% pure; Fisher Scientific, New Brunswick, N.J.) ml−1 as the sole added carbon and energy source. Cultures were incubated in 250-ml bottles sealed with Teflon-lined Mini-Nert caps (Alltech, Deerfield, Ill.) at 25°C in the dark on an orbital shaker (rotation speed of 150 rpm). Cultures were passed from TSA to MTBE-containing MSM a minimum of four times to establish cell line purity. Detection of MTBE degradation in microcosms was performed by gas chromatography by using a Shimadzu GC-14A equipped with a photonionization detector. Then, 50 μl of microcosm headspace was injected and analyzed by using a 15-m 0.53-mm DB1 column (J & W Scientific, Folsom, Calif.). Gas chromatography (GC) analyses were performed isothermally at 90°C, with He flow rates of 45 cm s−1. Once MTBE-degrading cultures were obtained, they were maintained in MSM amended with MTBE (100 μg ml−1).
Measurement of MTBE mineralization and disappearance.
MTBE mineralization was determined by using uniformly labeled [14C]MTBE (NEN Life Science Products, Boston, Mass.) with a specific activity of 5.0 mCi/mmol and a radiochemical purity of 99% as determined by GC coupled with an online radioactivity monitor. For inoculum preparation, cells were grown in MSM with 100 μg of MTBE ml−1. When cultures reached an optical density at 550 nm (OD550) of ≥0.5, the cells were centrifuged, washed in MSM, and resuspended to achieve an OD550 of ≥1.0. A total of 4.5 × 108 cells, estimated from total protein analysis (see below) and assuming 1 pg of carbon per cell, were resuspended in 25 ml of MSM in 250-ml biometer flasks. Unlabeled MTBE was added to yield a final concentration of 20 μg of MTBE ml−1, with [14C]MTBE added at a concentration of 6,000 dpm ml−1. Then, 1 ml of 0.5 M NaOH was added to flask side arms to serve as a trap for 14CO2. Biometer flasks with killed cells (1% sodium azide added) and uninoculated flasks were used as controls. All treatments were performed in triplicate. Flasks were incubated at 25°C on a rotary shaker in the dark. At various time points, samples of base were withdrawn, and the radioactivity was measured by using a liquid scintillation counter (Beckman Model LS 6000IC). In a preliminary MTBE mineralization experiment, the [14C]HCO3− in the withdrawn base was precipitated by using Ba(OH)2, followed by washing, to determine the amount of radioactivity attributable to the partitioning of [14C]MTBE into the base traps, as described previously (5). For the volumes described, there was no radioactivity above background levels in the Ba(OH)2 supernatants, indicating that all of the radioactivity was precipitated as BaCO3. For subsequent experiments, the base precipitation step was eliminated. This experiment was conducted twice.
Detection of nonradiolabeled MTBE degradation in microcosms was performed by GC as described above. In an experiment measuring the disappearance of MTBE at different initial concentrations, the inoculum density was 2 × 106 cells ml−1. All treatments were performed in duplicate, and samples were compared to uninoculated controls. Initial degradation rates were estimated by calculating the total amount of MTBE degraded per ml during the period of linear MTBE disappearance for each initial concentration.
Cell densities were established by measurement of the total cellular protein. Cells were harvested by centrifugation, washed, and resuspended in 500 μl of 0.85% NaCl. Cell lysis was accomplished by adding a 0.5 volume of acid-washed glass beads and vortexing the samples three times for 1-min bursts. Total protein was measured in 50-μl subsamples by using the Micro Protein Determination Kit (Sigma Diagnostics, St. Louis, Mo.) according to the manufacturer’s instructions. All assays were performed in triplicate. For yield determination, cells were grown in MSM at various MTBE concentrations, and protein levels were measured before and after MTBE degradation. Cells were assumed to be 55% protein by mass (dry weight), and the conversion factor 155 × 10−15 g of protein cell−1 was used to convert the protein measurements to cell densities (15).
Groundwater matrix inoculation experiments.
Groundwater matrix inoculation experiments were performed to measure the ability of strain PM1 to degrade MTBE in environmental samples. Groundwater studies were conducted in samples collected from an MTBE plume at Port Hueneme Naval Base (Oxnard, Calif.). MTBE contamination at the site is the result of a gasoline spill which occurred in 1984. The sandy aquifer is shallow (10 feet below the land surface) and perched on a clay lens (20 feet from the land surface). Groundwater flow at the site is approximately 0.1 to 0.3 feet day−1. The aquifer temperature in February 1999 was 23°C, and concentrations were as follows: oxygen, 2 mg liter−1; phosphate, 2.2 to 3.5 g liter−1; and nitrate, 0.2 to 0.63 g liter−1 (10). Core samples containing approximately 8 μg of MTBE ml−1 were obtained from a depth of 15 to 18 feet below the surface by using a geoprobe fitted with plastic coring inserts. Samples were stored at 4°C for less than 1 week prior to inoculation. The gravimetric moisture content of the samples was 22%.
Groundwater matrix material (20 g [dry weight]) was placed in 250-ml bottles sealed with Teflon-lined Mini-Nert caps. Microcosms were inoculated with 107 cells g of matrix material−1, and MTBE was added at a concentration of 20 μg of MTBE ml of matrix solution−1. Uninoculated core material and core material treated with 1% sodium azide were used as controls. All treatments were performed in triplicate. MTBE disappearance was monitored by GC as described above. When MTBE was no longer detectable in the microcosms, the pollutant was added again at a concentration of 20 μg ml−1. This was repeated for a total of three MTBE additions.
RESULTS AND DISCUSSION
Characterization of isolate.
A mixed microbial culture originally enriched from a compost biofilter at the Los Angeles Joint Water Treatment Plant was subcultured in MSM containing MTBE as the sole carbon source. When streaked onto 0.1× TSA, six different colony morphologies were observed. Of these, 53 colonies were purified and tested for the ability to use MTBE as their sole source of carbon and energy. Two strains were able to degrade MTBE as their sole carbon and energy source, one forming large yellow colonies and the other forming white pinpoint colonies on 0.1× TSA. The white-colony-forming strain, which had the faster rate of MTBE removal, was selected for further study.
Microscopic analysis of the isolated MTBE-degrading strain, designated PM1, shows that this organism is a gram-negative, uniflagellated rod that produces an extracellular matrix. This exuded material causes the cells to form flocs when grown in liquid media, making accurate determinations of the cell numbers difficult, particularly at low cell densities. Strain PM1 was found to be a member of the β1 subgroup of Proteobacteria by 16S rDNA analysis (2a).
Degradation and mineralization of MTBE.
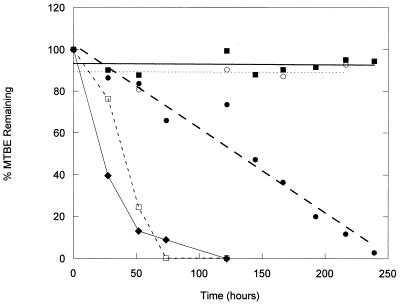
An initial inoculum of 2 × 106 cells of strain PM1 ml−1 was added to flasks containing different concentrations of MTBE in MSM, and the headspace concentrations of MTBE were measured. Strain PM1 was capable of degrading concentrations as great as 500 μg of MTBE ml−1 without an appreciable lag period, whereas concentrations of 5,000 μg of MTBE ml−1 were not degraded (Fig. 1). There was insignificant MTBE disappearance from the headspace in the abiotic controls. The initial linear rates of degradation increased with initial MTBE concentration. Estimated rates were 0.07, 1.17, and 3.56 μg of MTBE degraded ml−1 h−1 for the initial MTBE concentrations of 5, 50, and 500 μg ml−1, respectively.
FIG. 1.
Degradation of increasing MTBE concentrations by strain PM1. Strain PM1 was incubated with MTBE at initial concentrations of 5 (⧫), 50 (□), 500 (●), and 5,000 (■) μg ml−1. A representative abiotic control (50 μg ml−1) is also shown (○); controls at all concentrations were similar.
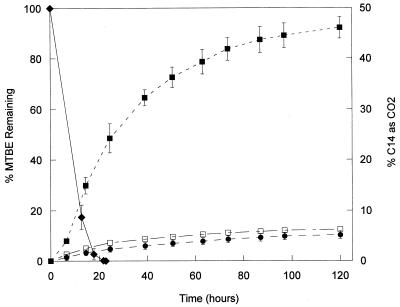
Mineralization of 20 μg of MTBE ml−1 spiked with 6,000 dpm of uniformly labeled [14C]MTBE by 2 × 107 cells of strain PM1 ml−1 in solution culture was measured and compared with the disappearance of GC-detectable MTBE in headspace samples (Fig. 2). MTBE was degraded to below the detection limit of 50 ng ml−1 by 23 h, at which time 24% of the 14C had evolved as 14CO2. By 120 h, 14CO2 accounted for 46% of the initial 14C added. Reprecipitation of the carbonate as barium carbonate, followed by washing, indicated that all of the 14C was associated with carbonate and not MTBE or volatile metabolites. In the uninoculated and killed controls, less than 7% of the initial isotope was recovered in the base trap within the 120-h incubation period. Analysis of the 14C associated with the particulate fraction collected on 0.2-μm (pore size) filters indicated that 19% (±0.65) of the radioisotope was incorporated into cell biomass, whereas less than 1% of initial counts added were found in the particulate fraction of control flasks.
FIG. 2.
Mineralization and degradation of MTBE by strain PM1. Strain PM1 was incubated with an initial concentration of 20 μg of MTBE ml−1 with or without 6,000 dpm of uniformly labeled 14C-MTBE ml−1 as a tracer. Symbols: ⧫, GC-detectable MTBE in headspace; ■, 14CO2 from inoculated microcosms; □, 14CO2 from uninoculated microcosms; ●, 14CO2 from killed microcosms.
In the mineralization study 14CO2 production lagged behind the disappearance of MTBE measured by headspace analysis. This discrepancy may be due to the production of a slowly metabolized intermediate, such as TBA, reported to be an intermediate of MTBE degradation in all MTBE-degrading cultures for which it was assayed (7, 17, 20). Others have found that TBA is more slowly metabolized than MTBE (17, 20).
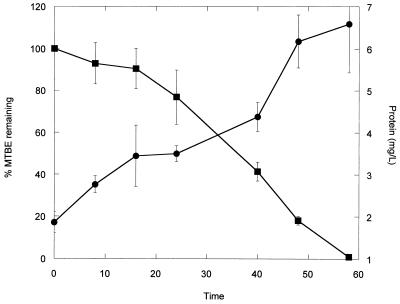
In order to demonstrate growth of strain PM1 on MTBE, 3 × 107 cells ml−1 were inoculated in MSM flasks containing 25 μg of MTBE ml−1. MTBE headspace concentrations and protein contents were measured over time (Fig. 3). As the MTBE concentration in microcosms decreased, the protein concentrations increased, indicating that cells were utilizing MTBE for biomass production. Inoculated treatments in which MTBE was not present exhibited a slight decline in initial protein concentration over the course of the experiment.
FIG. 3.
MTBE degradation and cellular protein production by strain PM1. Strain PM1 was incubated with an initial concentration of 25 μg of MTBE ml−1. MTBE disappearance (■) and protein concentration (●) were measured over time.
Protein analysis was also used to estimate biomass yield for strain PM1 grown on MTBE. At initial MTBE concentrations of 0, 5, and 50 μg of MTBE ml−1, the mass of protein produced was 2.7, 4.5, and 57 mg ml−1, respectively. Thus, approximately 0.1 mg of protein was produced per mg of MTBE consumed and, assuming cells are 55% protein as in Escherichia coli (15), this corresponds to a yield of 0.18 mg of cells mg of MTBE−1 (±0.06). The estimated yields were substantially less than cell yields typical of growth on aromatics, sugars, and aliphatics. The low cell yields found in this study are consistent with those reported for other MTBE-degrading cultures (Table 1). One possible explanation for the low yields may be the high energetic cost required to cleave the ether linkage in the MTBE molecule (21). Others have also suggested that MTBE may act as an uncoupler to ATP synthesis or that intermediates may be toxic to cells (17).
TABLE 1.
Comparison of mixed and pure cultures capable of MTBE biodegradation
| Strain | Organism | Type of metabolism | MTBE degradation rate | Notes | Source or reference |
|---|---|---|---|---|---|
| BC-1 | Mixed culture | Aerobic heterotrophic | 34 mg g of cells−1 h−1 | 17 | |
| UCD culture | Mixed culture | Aerobic heterotrophic | 0.061 mg of MTBE liter−1 in 2 days | 3, 4 | |
| ENV421 | Similar to nocardioform bacteria | Cometabolic with propane | 9.2 nmol min−1 mg of protein−1 | Αfter growth on propane | 20 |
| JOB5 | Mycobacterium vaccae | Cometabolic with propane | Not reported | 20 | |
| ENV425 | Most closely related to Nocardia sp. | Cometabolic with propane | 4.6 nmol min−1 mg of cell protein−1 | After growth on propane | 20 |
| Isolate 24 | Methylobacterium mesophilicum | Aerobic heterotrophic | 29% of 200 μg ml−1 in 2 weeks | Incomplete MTBE degradation | 11 |
| Isolate 33 | Rhodococcus sp. | Aerobic heterotrophic | 28% of 200 μg ml−1 in 2 weeks | Incomplete MTBE biodegradation | 11 |
| Isolate 41 | Arthrobacter ilicis | Aerobic heterotrophic | 28% of 200 μg ml−1 in 2 weeks | Incomplete MTBE biodegradation | 11 |
| ATCC 58400 | Graphium (fungus) | Cometabolic with n-butane | 0.6 nmol h−1 mg−1 in the presence of n-butane and 1.9 nmol h−1 mg−1 in the absence of n-butane | Induction by n-butane required for MTBE biodegradation | 7 |
| PM1 | Member of the β1 subgroup of Proteobacteria | Aerobic heterotrophic | 0.07, 1.17, and 3.56 g ml−1 h−1 for MTBE concentrations of 5, 50, and 500 g ml−1 | Inoculation density of 2 × 106 cells ml−1 | This work |
Bioaugmentation potential in groundwater samples.
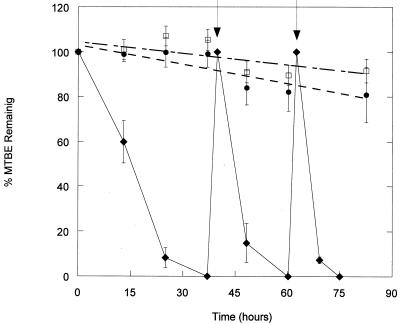
To assess the potential for MTBE biodegradation in groundwater samples inoculated with strain PM1, 107 cells g of matrix material−1 were added to groundwater core material amended with MTBE. Strain PM1 degraded an initial addition of 20 μg of MTBE ml−1 within 40 h (Fig. 4). The rate of MTBE degradation increased substantially with each subsequent MTBE addition. The second addition of 20 μg of MTBE ml−1 degraded within 20.5 h, and the third degraded within 12.5 h.
FIG. 4.
MTBE degradation in groundwater matrix cores inoculated with strain PM1. Groundwater cores were inoculated with 107 cells g of matrix material−1 and amended with 20 μg of MTBE ml−1. Arrows indicate times of subsequent MTBE additions of 20 μg ml−1. Treatments include inoculated microcosms (⧫), uninoculated microcosms (□), and killed microcosms (●).
MTBE is becoming a widespread groundwater contaminant with an estimated 3,000 plumes in California alone (8). Because of the compound’s high solubility in water, standard remediation technologies, such as air stripping, are energy intensive and thus may prove economically unfavorable for use in field situations. An alternative to abiotic cleanup methods is the use of microorganisms for both in situ and ex situ groundwater treatment. Salanitro et al. (18) provide evidence that inoculation of the MTBE-contaminated plume at Port Hueneme with a mixed culture of bacteria, combined with sparging the plume with oxygen, has resulted in a >90% reduction in MTBE concentration in the immediate area. We found that strain PM1 inoculated into groundwater core samples readily degraded 20 μg of MTBE ml−1, and the organism remained active on MTBE for up to 83 h after its inoculation. From these data it appears that strain PM1 may be a promising candidate for use in bioaugmentation of MTBE-contaminated groundwater sites. Field trials with PM1 to inoculate an MTBE-contaminated groundwater plume at Port Hueneme are currently underway.
ACKNOWLEDGMENTS
We thank Marc Deshusses for supplying the radiolabelled MTBE used in this study, and we thank Ernie Lowry at Port Hueneme for providing groundwater sediment samples. We also thank the anonymous reviewers for their helpful comments.
This study was supported, in part, by the University of California Toxic Substances Research and Teaching Program, with special funds allocated under SB-521 legislation. Additional funding was provided by National Institutes of Environmental Health Science Superfund Basic Research Program (2P42 ES04699) and the EPA (R819658) Center for Ecological Health Research at the University of California, Davis.
REFERENCES
- 1.Anonymous. Health advisory set for fuel oxygenate MTBE. Environ Sci Technol. 1988;32:83A. doi: 10.1021/es9833671. [DOI] [PubMed] [Google Scholar]
- 2.Bradley P, Landmeyer J, Chapelle F. Aerobic mineralization of MTBE and tert-butyl alcohol by stream-bed sediment microorganisms. Environ Sci Technol. 1999;33:1877–1879. [Google Scholar]
- 2a.Bruns, M. A., J. R. Hanson, J. Mefford, M. Aswath, and K. M. Scow. DNA fingerprinting confirms that isolate PM1 is a dominant member of an MTB-degrading consortium. Submitted for publication.
- 3.Eweis, J. B., D. P. Chang, E. D. Schroeder, K. M. Scow, R. L. Morton, and R. C. Caballero. 1997. Presented at the Air and Waste Management Association 90th Annual Meeting and Exhibition, Toronto, Ontario, Canada. AWMA, Washington, D.C.
- 4.Eweis J B, Schroeder E D, Chang D P Y, Scow K M. Biodegradation of MTBE in a pilot-scale biofilter. In: Wickramanayake G B, Hinchee R E, editors. Natural attenuation: chlorinated and recalcitrant compounds. Columbus, Ohio: Battelle Press; 1998. pp. 341–346. [Google Scholar]
- 5.Fan S, Scow K M. Biodegradation of trichloroethylene and toluene by indigenous microbial populations in soil. Appl Environ Microbiol. 1993;59:1911–1918. doi: 10.1128/aem.59.6.1911-1918.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harayama S. Aerobic biodegradation of aromatic hydrocarbons by bacteria. Metal Ions Biol Syst. 1992;28:99–156. [Google Scholar]
- 7.Hardison L K, Curry S S, Ciuffetti L M, Hyman M R. Metabolism of diethyl ether and cometabolism of methyl tert-butyl ether by a filamentous fungus, a Graphium sp. Appl Environ Microbiol. 1997;63:3059–3067. doi: 10.1128/aem.63.8.3059-3067.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keller A, Froines J, Koshland C, Reuter J, Suffet I, Last J. Health and environmental assessment of MTBE: report to the governor and legislature of the state of California, sponsored by SB 521. Calif: University of California, Davis; 1998. [Google Scholar]
- 9.Kirschner E M. Production of top 50 chemicals increased substantially in 1994. Chem Eng News. 1995;73:16–20. [Google Scholar]
- 10.Lory, E. 1999. Personal communication.
- 11.Mo K, Lora C O, Wanken A E, Javanmardian M, Yang X, Kulpa C F. Biodegradation of methyl t-butyl ether by pure bacterial cultures. Appl Microbiol Biotechnol. 1997;47:69–72. doi: 10.1007/s002530050890. [DOI] [PubMed] [Google Scholar]
- 12.Mormille M R, Liu S, Suflita J M. Anaerobic biodegradation of gasoline oxygenates: extrapolation of information to multiple sites and redox conditions. Environ Sci Technol. 1994;28:1727–1732. doi: 10.1021/es00058a026. [DOI] [PubMed] [Google Scholar]
- 13.Mu D Y, Scow K M. Effect of trichloroethylene (TCE) and toluene concentrations on TCE and toluene biodegradation and the population density of TCE and toluene degraders in soil. Appl Environ Microbiol. 1994;60:2661–2665. doi: 10.1128/aem.60.7.2661-2665.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Natarajan M R, Lu A, Oriel P. Cloning and expression of a pathway for benzene and toluene from Bacillus stearothermophilus. Biodegradation. 1994;5:77–82. doi: 10.1007/BF00700632. [DOI] [PubMed] [Google Scholar]
- 15.Neidhardt F C, Ingraham J L, Schaecter M. The physiology of the bacterial cell. Sunderland, Mass: Sinauer Associates, Inc.; 1990. [Google Scholar]
- 16.Paje M L F, Neilan B A, Couperwhite I. A Rhodoccus species that thrives on medium saturated with liquid benzene. Microbiology. 1997;143:2975–2981. doi: 10.1099/00221287-143-9-2975. [DOI] [PubMed] [Google Scholar]
- 17.Salanitro J P, Diaz L A, Williams M P, Wisniewski H L. Isolation of a bacterial culture that degrades methyl t-butyl ether. Appl Environ Microbiol. 1994;60:2593–2596. doi: 10.1128/aem.60.7.2593-2596.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salanitro J P, Spinnler G E, Neaville C C, Maner P M, Stearns S M, Johnson P C, Bruce C. Demonstration of the enhanced MTBE bioremediation (EMB) in situ process. In: Alleman B C, Leeson A, editors. In situ bioremediation of petroleum hydrocarbon and other organic compounds. Columbus, Ohio: Battelle Press; 1999. pp. 37–46. [Google Scholar]
- 19.Schirmer M, Barker J F. A study of long-term MTBE attenuation in the Borden Aquifer, Ontario, Canada. Groundwater Monitor Res. 1998;18:113–122. [Google Scholar]
- 20.Steffan R J, McClay K, Vainberg S, Condee C W, Zhang D. Biodegradation of the gasoline oxygenates methyl tert-butyl ether, ethyl tert-butyl ether, and tert-amyl methyl ether by propane-oxidizing bacteria. Appl Environ Microbiol. 1997;63:4216–4222. doi: 10.1128/aem.63.11.4216-4222.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White G F, Russell N J, Tidswell E C. Bacterial scission of ether bonds. Microbiol Rev. 1996;60:216–232. doi: 10.1128/mr.60.1.216-232.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeh C K, Novak J T. Anaerobic biodegradation of gasoline oxygenates in soils. Water Environ Res. 1994;66:744–752. [Google Scholar]