Summary
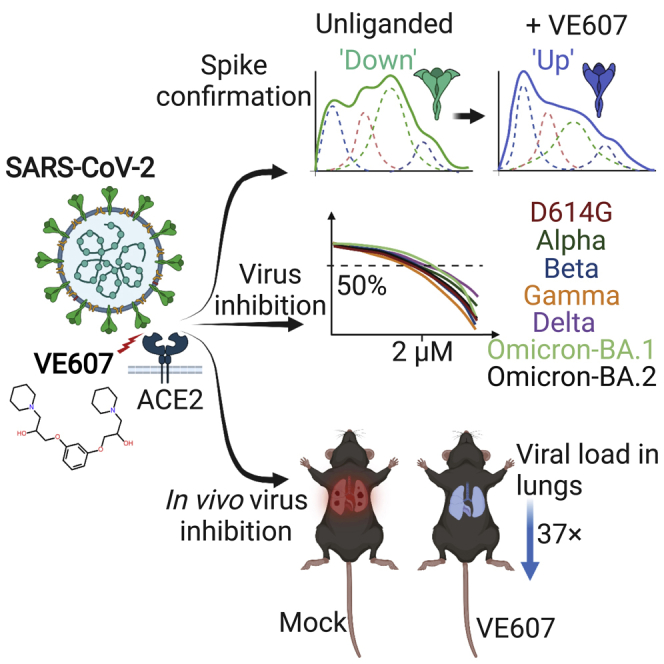
SARS-CoV-2 infection of host cells starts by binding the Spike glycoprotein (S) to the ACE2 receptor. The S-ACE2 interaction is a potential target for therapies against COVID-19 as demonstrated by the development of immunotherapies blocking this interaction. VE607 — a commercially available compound composed of three stereoisomers — was described as an inhibitor of SARS-CoV-1. Here, we show that VE607 broadly inhibits pseudoviral particles bearing the Spike from major VOCs (D614G, Alpha, Beta, Gamma, Delta, Omicron – BA.1, and BA.2) as well as authentic SARS-CoV-2 at low micromolar concentrations. In silico docking, mutational analysis, and smFRET revealed that VE607 binds to the receptor binding domain (RBD)-ACE2 interface and stabilizes RBD in its “up” conformation. Prophylactic treatment with VE607 did not prevent SARS-CoV-2-induced mortality in K18-hACE2 mice, but it did reduce viral replication in the lungs by 37-fold. Thus, VE607 is an interesting lead for drug development for the treatment of SARS-CoV-2 infection.
Subject areas: Drugs, Virology
Graphical abstract
Highlights
-
•
VE607 stabilizes RBD in its “up” conformation
-
•
VE607 inhibits SARS-CoV-1 and SARS-CoV-2 variants of concern
-
•
VE607 reduces SARS-CoV-2 replication in the lung of infected hACE2-K18 mice
Drugs; Virology
Introduction
The COVID-19 pandemic continues to cause widespread morbidity and mortality (Wu et al., 2020; Zhu et al., 2020a). This is largely because of insufficient vaccination levels as vaccines offer good protection against infection and severe disease (Ball, 2021). The currently used vaccines exploit modified versions of the Spike (S) glycoprotein that is exposed on the surface of viral particles (Krammer, 2020) and infected cells (Ding et al., 2022). S is processed by cellular proteases furin and TMPRSS2 on host cells. After binding to ACE2 via its receptor binding domain (RBD), S undergoes significant conformational changes that ultimately lead to fusion of the viral membrane with human cells. Fusion allows translocation of the RNA genome and associated replicase proteins into mammalian cells, leading to viral replication (Harrison et al., 2020; Hoffmann et al., 2020a, 2020b; Yang and Rao, 2021). S is a trimeric glycoprotein that is present in multiple conformations that have been resolved primarily by cryo-electron microscopy (Cai et al., 2020; Lan et al., 2020; Shang et al., 2020; Wrapp et al., 2020; Yan et al., 2020). Its conformational dynamics can be monitored by single molecule FRET (Li et al., 2021; Lu et al., 2020; Ullah et al., 2021; Yang et al., 2021). Vaccine-elicited antibodies act in several ways including neutralizing viral particles but also through Fc-mediated effector functions (Tauzin et al., 2021, 2022). The selective pressure during the pandemic has led to a growing list of variants carrying mutations in the S-glycoprotein (Gong et al., 2021; Li et al., 2021; Mannar et al., 2021; Nabel et al., 2021; Prevost and Finzi, 2021; Yang and Rao, 2021), resulting in different degrees of resistance to previous infection and vaccine-elicited antibody neutralization.
Despite the efficacy of currently used vaccines and ongoing work to generate broadly protective pan-coronavirus vaccines (Cohen, 2021; Nabel et al., 2021; Rappazzo et al., 2021), there is an urgent need for efficient and specific treatments for infected patients. The viral replication machinery offers different possible drug targets (Yang and Rao, 2021) and several small molecule inhibitors targeting the SARS-CoV-2 protease (Dai et al., 2020; Zhang et al., 2020) or replicase (Kokic et al., 2021; Yin et al., 2021) have been published with some recently showing promise in clinical trials (Owen et al., 2021). In contrast, relatively little attention has been given to the S-ACE2 interaction as a potential target for small molecule inhibitors (Tong, 2009; Wang et al., 2021; Zhu et al., 2020b). Research on SARS-CoV-1 and Middle East respiratory syndrome (MERS) has inspired work on potential drug targets, and some previous studies explored the isolation of small molecule inhibitors against various potential targets. Some of these molecules were described as potential inhibitors of the SARS-CoV-1 RBD interaction with ACE2 (Adedeji et al., 2013; Kao et al., 2004), but the binding was not demonstrated directly and there was no biological follow-up to characterize their mode of action.
Here, we employed differential scanning fluorimetry (DSF) to identify the capacity of the small molecule inhibitor VE607 (Kao et al., 2004), composed of three stereoisomers [(S,S)-VE607, (R,R)-VE607, and (R,S)-VE607], to bind the SARS-CoV-2 RBD. We found that this VE607 mixture of isomers (hereafter referred to as “VE607”) is capable of specific inhibition of infection of human cells with pseudoviral particles that express the SARS-CoV-1 or SARS-CoV-2 S-glycoproteins. VE607 was also able to inhibit the infection with authentic SARS-CoV-2 viruses. We found that VE607 inhibits the Spike by stabilizing the “up” conformation of the RBD. The mode of binding to RBD was elucidated by in silico docking experiments followed by validation of critical residues through mutagenesis and functional studies. Moreover, VE607 remained potent against current variants of concern (VOCs) of SARS-CoV-2. In vivo efficacy analyses revealed that prophylaxis with VE607 did not prevent SARS-CoV-2 induced mortality in the highly susceptible K18-hACE2 mice. However, VE607 reduced infection in the lungs by 37-fold. These data suggest that VE607 is an interesting lead for the development of drugs for the prevention or treatment of COVID-19 infections.
Results
Differential scanning fluorimetry and docking suggest that VE607 may bind the RBD
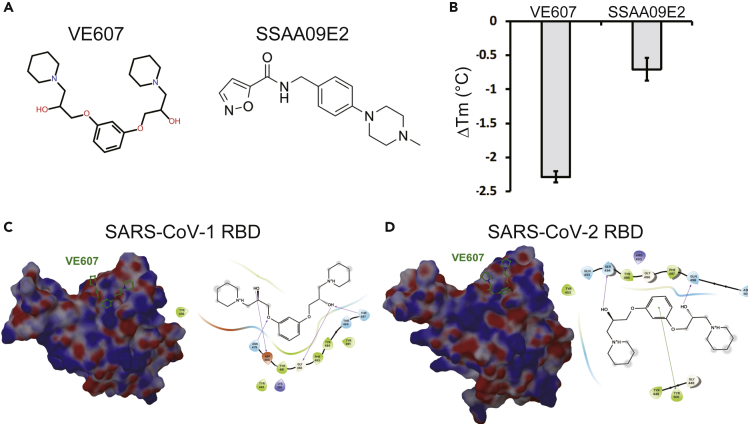
We tested the ability of previously described SARS-CoV-1 inhibitors VE607 (Kao et al., 2004) and SSAA09E2 (Adedeji et al., 2013) to bind the SARS-CoV-2 RBD (Figure 1A). We used differential scanning fluorimetry (DSF) that measures the effect of small molecules on the melting temperature of proteins (Mashalidis et al., 2013). Incubation with VE607 led to a significant decrease of the melting temperature (ΔTm, −2.3°C), whereas SSAA09E2 had a smaller yet measurable effect (ΔTm, −0.7°C) (Figure 1B). Because this result suggested binding of VE607 to RBD, we next performed in silico docking against RBD using Glide (Schrödinger, 2020). We identified moderately favorable potential VE607 binding sites overlapping the ACE2 epitopes in both SARS-CoV-1 and SARS-CoV-2 RBDs (Figures 1C and 1D).
Figure 1.
Potential interactions of SARS-CoV-1 inhibitors with the RBD
(A) Chemical structures of VE607 and SSAA09E2.
(B) Differential scanning fluorimetry of the SARS-CoV-2 RBD in the presence of SARS-CoV-1 inhibitors, results from two experiments (eight replicates total) are shown.
(C) Virtual docking of VE607 to SARS-CoV-1 and (D) SARS-CoV-2 RBD. Left panels, the electrostatic potential is displayed over the molecular surface of the RBD and colored red and blue for negative and positive potential, respectively. Right panels, scheme showing a docking model of VE607 to the RBD. The presumable RBD contact residues are shown as spheres.
VE607 inhibits infection of pseudoviral particles and authentic SARS-CoV-2
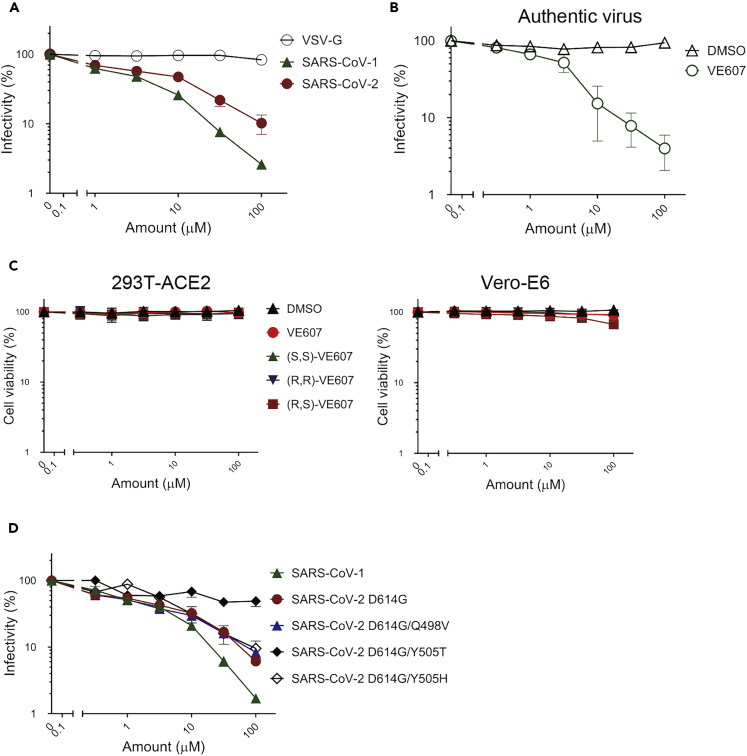
To assess the effects of VE607 and SSAA09E2 on infection, we expressed the SARS-CoV-1 and SARS-CoV-2 S glycoproteins on the surface of pseudoviral particles carrying a luciferase reporter gene. Pseudoviral particles carrying the VSV-G glycoprotein served as control. Infection was measured using ACE2-expressing 293T (293T-ACE2) cells (Prevost et al., 2020) in the presence of increasing concentrations of VE607 and SSAA09E2. VE607 specifically inhibited pseudoviral particles bearing the SARS-CoV-1 Spike (IC50 = 1.47 μM, Figure 2A) in agreement with previous findings (Kao et al., 2004). Interestingly, VE607 also inhibited pseudoviral particles expressing the Spike from SARS-CoV-2 (IC50 = 3.06 μM, Figure 2A), albeit slightly less efficiently than for SARS-CoV-1. No inhibition was observed for pseudoviral particles bearing the VSV-G (IC50 > 100 μM, Figure 2A). To ensure that the inhibitory capacity of VE607 against SARS-CoV-2 was not limited to pseudoviral particles, we evaluated whether the inhibitory capacity of VE607 was maintained against authentic viruses. As shown in Figure 2B, VE607 inhibited authentic SARS-CoV-2 D614G with an IC50 of 2.42 μM. No cell toxicity of VE607 or the three different enantiomers was observed with concentrations up to 100 μM on 293T-ACE2 cells or Vero-E6 cells (Figure 2C). In contrast, SSAA09E2 at concentrations up to 100 μM did not inhibit infection of pseudoviral particles (data not shown), and we did not further pursue work with this small molecule.
Figure 2.
VE607 inhibits infection of SARS-CoV-1 and SARS-CoV-2 pseudoviral particles and of authentic SARS-CoV-2
(A) VE607 inhibition of SARS-CoV-1, SARS-CoV-2, or VSV-G (specificity control) pseudovirus.
(B) VE607 inhibition of authentic live SARS-CoV-2 virus.
(C) VE607 and the three different enantiomers are not toxic on 293T-ACE2 (left) or Vero-E6 (right) cells, as measured by CellTiter-Glo One Solution Assay for the quantitation of ATP presented in live cells.
(D) Pseudovirus neutralization of SARS-CoV-2 S mutants predicted by our in silico analysis to modulate the inhibition by VE607. Data represents the average of at least four independent experiments ± SEM.
As stated before, commercially available VE607 is a mixture of three stereochemical isomers, comprised of the (S,S)-VE607, (R,R)-VE607, and the meso (R,S)-VE607 (Data S1), which are mixed at a ratio of 1:1:2 (not shown). We observed no differences in the SARS-CoV-2 pseudoviral inhibition among these enantiomers obtained by synthesis and the commercially available mixture of all three isomers (Figure S1).
Initial in silico docking identified RBD residues Y505 and Q498 as potential specific contact sites for VE607 (Figure 1D). We mutated these residues in the full-length SARS-CoV-2 D614G Spike and prepared pseudoviral particles to test whether they affect VE607 inhibition. Although the Q498V mutation had only a minor effect (IC50 = 1.8 μM), the Y505T mutant was resistant to VE607 inhibition (IC50 > 40 μM, Figure 2D). But when Y505 was replaced with Histidine (H), which is a naturally occurring mutation in Omicron VOCs, the pseudoviral particles harboring Y505H spike became sensitive to VE607 (IC50 = 4.5 μM) (Figure 2D). These results are in agreement with the in silico analysis, where a strong π-π interaction between Y/H505’s aromatic side-chain and the central aromatic ring of VE607 is predicted. Because it is solvent-exposed in our model, the hydroxyl of tyrosine does not engage VE607, and neither would we expect a threonine hydroxyl to do so. Alignment of sACE2 on the known ACE2 epitope of the VE607-bound model of RBD displayed significant steric clashes between ACE2 and VE607, suggesting some direct competition for the ACE2 epitope. Alignment of the VE607-bound RBD model onto the RBD of the closed S glycoprotein spike complex (Xu et al., 2021) showed that VE607 also clashes with the neighboring S1 protomer, suggesting some possible destabilization of the closed state of the S glycoprotein.
VE607 stabilizes the “up” conformation of the S glycoprotein
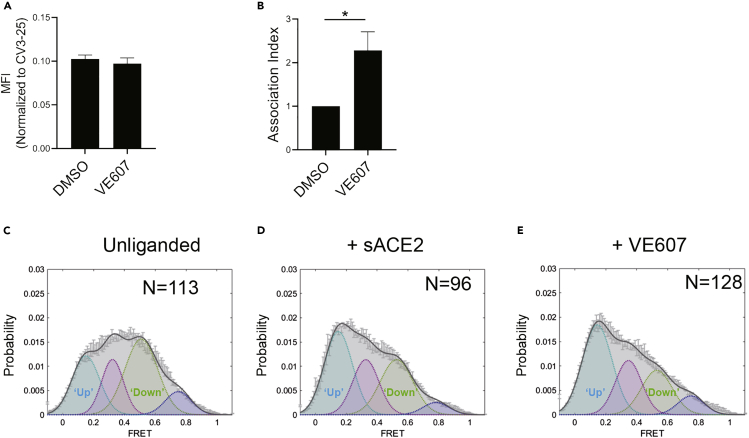
We next assessed whether VE607 affects the RBD-ACE2 interaction. Briefly, using flow cytometry, we measured the capacity of VE607 to compete with soluble ACE2 (sACE2) for interaction with the full SARS-CoV-2 Spike, expressed at the cell surface, as described previously (Anand et al., 2020). We observed no competition between VE607 (100 μM) and sACE2 (Figure 3A). Because the mode of action of some neutralizing antibodies such as CV3-1 involve S1 shedding (Li et al., 2022), we tested if VE607 acted in a similar manner. To this effect, Spike-expressing 293T cells were radioactively labeled followed by the immunoprecipitation of cell lysates and supernatant as described (Li et al., 2022). In contrast to CV3-1, VE607 decreased shedding, resulting in a significantly more stable trimer (Figure 3B).
Figure 3.
VE607 stabilizes SARS-CoV-2 S in the “up” conformation
(A) VE607 does not compete for sACE2 interaction as measured by flow cytometry. The values represent the median fluorescence intensities (MFI) normalized to binding signals obtained with the conformationally independent CV3-25 Ab. Five experiments are represented as mean ± SEM and statistical significance was tested using unpaired t-test.
(B) SARS-CoV-2 Spike stability was measured by radioactive labeling of 293T Spike expressing cells followed by immunoprecipitation of cell lysates and supernatants. At least four experiments are represented as mean ± SEM and statistical significance was tested using unpaired t-test, ∗p < 0.05.
(C–E) Single molecule FRET analysis of SARS-CoV-2 S unliganded (C), in presence of sACE2 (D) or VE607 (E).
To evaluate whether the enhanced stability observed in presence of VE607 altered the conformational landscape of the Spike, we performed single-molecule FRET (smFRET) analysis using viral pseudoparticles carrying modified S glycoproteins labeled with FRET donor and acceptor dyes, enabling us to distinguish the “up” and “down” conformations (Lu et al., 2020) (Li et al., 2022). In agreement with previous observations, the unliganded Spike predominantly sampled the “down” conformation (Figure 3C). As expected, the addition of sACE2 shifted the conformational landscape of the Spike to the “up” conformation reflecting the receptor bound state (Figure 3D). Interestingly, we observed that VE607 stabilized the “up” conformation mimicking sACE2 (Figure 3E).
VE607 inhibits infection of some SARS-CoV-2 variants of concern
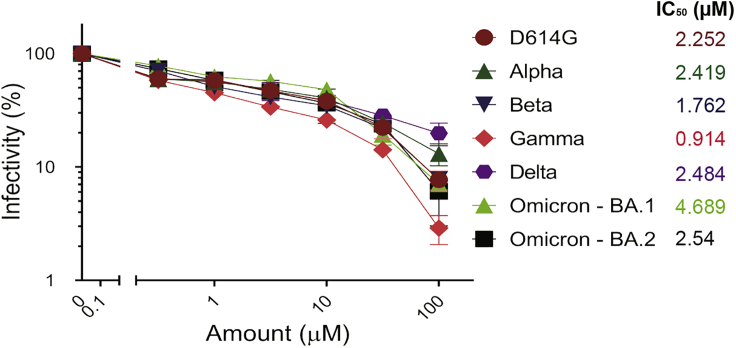
SARS-CoV-2 is in constant evolution as VOCs keep emerging. VE607 was identified as an inhibitor of SARS-CoV-1 — a related Beta-coronavirus — suggesting some inhibitory breadth. Therefore, we tested whether it inhibits pseudoviral particles bearing the Spike glycoproteins from the major VOCs (Alpha, Beta, Gamma, Delta, Omicron - BA.1, and Omicron - BA.2). In agreement with its broad SARS coronavirus activity, VE607 inhibited all VOCs with similar potency with IC50 values in the low micromolar range (Figure 4). These results demonstrate that the various amino acid changes in the S-glycoprotein of these variants do not impact the inhibitory potential of VE607 and show promise for the development of a new generation of anti-SARS-CoV-2 small molecule inhibitors blocking viral entry.
Figure 4.
VE607 inhibits infection of SARS-CoV-2 variants Alpha, Beta, Gamma, Delta, and Omicron pseudovirus particles
VE607 inhibits SARS-CoV-2 pseudoviral particles infection of 293T-ACE2 cells. IC50 values are shown next to the Spikes of different VOCs. Data represents the average of at least four independent experiments ± SEM.
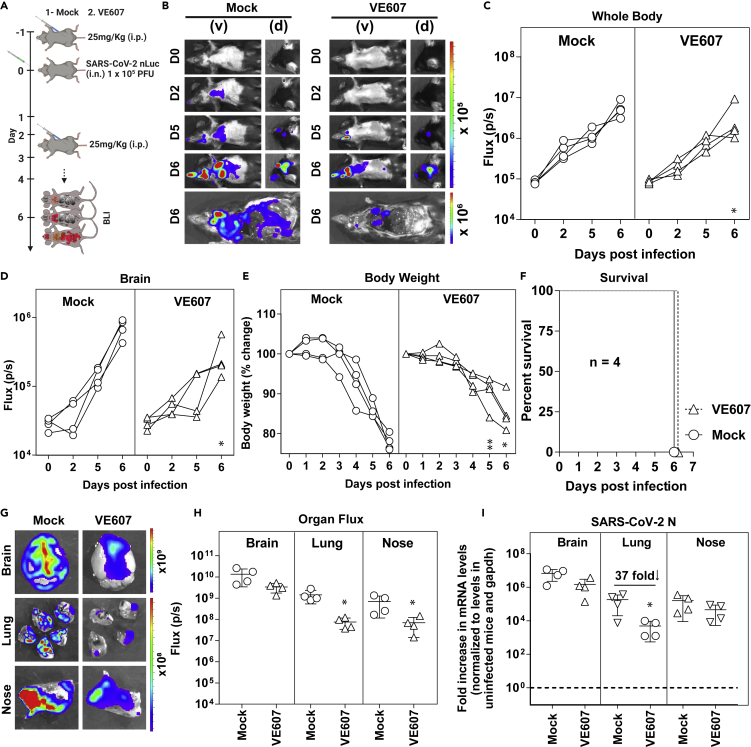
Prophylaxis with VE607 reduces infection in the lungs of SARS-CoV-2-challenged K18-hACE2 mice
We carried out pharmacokinetic (PK) analysis of VE607 before evaluating in vivo efficacy against SARS-CoV-2. K18-hACE2 mice were administered VE607 or vehicle (DMSO) intraperitoneally at doses of 12.5, 25, and 50 mg/kg followed by blood collection at intervals of 0.5, 2, 10, and 24 h. We estimated VE607 concentrations in plasma as well as organs after necropsy at 24 h. We achieved a concentration of 2.6 μM close to the IC50 (2.42 μM) of SARS-CoV-2 D614G in the lungs at 25 mg/kg (Figure S2). For testing the ability of VE607 to inhibit SARS-CoV-2-nLuc replication in K18-hACE2 mice, we administered this dosage intraperitoneally 24 h before challenge and each day thereafter for 4 days. SARS-CoV-2-nLuc replication was followed noninvasively using bioluminescence imaging. VE607 reduced viral replication in the lungs as shown by the nLuc signal but not in the brain, where concentrations of VE607 were more than a log lower than the IC50 (Figures 5A–5D). Although body weight loss in VE607-treated mice was significantly lower than in mock treated controls at 5 and 6 dpi, all the mice in both groups succumbed to SARS-CoV-2-induced mortality (Figure 5F). Reimaging target organs including lungs after necropsy confirmed that VE607 significantly reduced replication in the lungs (Figures 5G and 5H). An analysis of nucleocapsid (N) mRNA copy numbers corroborated the imaging data and revealed that VE607 was able to significantly reduce SARS-CoV-2 replication in the lungs by ∼37 fold, but the reduction was expectedly low (3-4 fold) in the brain and nasal cavity based on PK analyses (Figure 5I). Thus, our in vivo efficacy analyses suggest that VE607 is a promising small molecule inhibitor suited for the development of new anti-SARS-CoV-2 agents that inhibit viral entry.
Figure 5.
VE607 reduces SARS-CoV-2 replication in lungs of K18-hACE2 mice
(A) Experimental design to test the efficacy of VE607 in K18-hACE2 mice challenged with SARS-CoV-2-nLuc (WA1, 1 × 105 FFU, i.n.) and treated intraperitoneally (i.p.) from 1 to 4 dpi, (25 mg/kg) with VE607. Vehicle (DMSO)-treated mice were used as controls (Mock). Animals were followed by noninvasive BLI every 2 days as indicated.
(B) Representative BLI images of SARS-CoV-2-nLuc-infected mice in ventral (v) and dorsal (d) positions. Scale bars denote radiance (photons/sec/cm2/steradian).
(C and D) Temporal quantification of nLuc signal as flux (photons/sec) computed noninvasively.
(E) Temporal changes in mouse body weight with initial body weight set to 100% for an experiment shown in A.
(F) Kaplan-Meier survival curves of mice (n = 4 per group) statistically compared by log-rank (Mantel-Cox) test for experiment as in A.
(G and H) Ex vivo imaging of indicated organs and quantification of nLuc signal as flux (photons/sec) after necropsy.
(I) Fold change in SARS-CoV-2 nucleocapsid (N gene) expression in brain, lung, and nose tissues. The data were normalized to Gapdh mRNA in the same sample and that in noninfected mice after necropsy. Viral loads (I) were determined after necropsy at 6 dpi.
Each curve in (C–E) and each data point in (C–E) represents an individual mouse. Data in panels are from two independent experiments and n = 2 mice per group. The data in (C–E), (H–I) were analyzed by Mann Whitney nonparametric test. ∗, p < 0.05; ∗∗, p < 0.01; Mean values ± SD are depicted.
Discussion
Here, we present data suggesting that small molecule inhibitors of SARS-CoV-2 entry, such as VE607, have potential for the development of drugs for the prevention and/or treatment of COVID-19. VE607 was originally described as an inhibitor of the S glycoprotein-ACE2-mediated SARS-CoV-1 entry (Kao et al., 2004), and we confirmed these results using a pseudovirus infection assay. The inhibition of SARS-CoV-1 carrying pseudoviruses was stronger than that of SARS-CoV-2 pseudoviruses, but the IC50 values remained in the low μM range in both cases. In silico docking identified a potential binding site at the RBD-ACE2 interface in both cases (Lan et al., 2020; Shang et al., 2020). The results of mutational analysis are consistent with the predicted binding site suggesting a critical role of Y505 for the activity of VE607. Therefore, VE607 may inhibit viral entry by blocking ACE2-mediated Spike conformational changes required for fusion.
Single molecule FRET data indicate that VE607 alone stabilized the Spike in the preferred “up” conformation for sACE2 binding. However, in contrast to RBD-targeting antibodies such as CV3-1 (Li et al., 2022), VE607 did not induce shedding of S1. These results suggest a unique mechanistic basis for the inhibition of infection by VE607. We hypothesize that VE607 stabilizes one ACE2-bound conformation of Spike but is an allosteric inhibitor of downstream ACE2-triggered conformational changes required for fusion. The binding site predicted by in silico docking was confirmed by mutational analysis suggesting that VE607 binds at the S-ACE2 interface where it may block the protein-protein interaction required to activate the Spike for fusion. The emergence of variants over the course of the pandemic is a continuing concern and many of them carry mutations in the S glycoprotein including the RBD domain that contributes to increased infectivity and immune escape. To validate the binding site predicted by in silico docking, we introduced the mutations Q498V and Y505T; although the first change had only a modest effect, the change Y505 T led to an S glycoprotein that is resistant to VE607. These results are consistent with the fact that pseudoviruses carrying S from the variants Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.167.2), and Omicron (B.1.1.529) are inhibited by VE607 at low μM IC50 values, suggesting that common mutations in the vicinity of the binding site such as N501Y do not impact the inhibitory potential of VE607. Changes of Q498 are rarely observed in the Beta variant (less than 0.1% of all sequences), but the mutation Q498R is very frequent in Omicron (90% of all sequences), which is rapidly spreading and leading to the next phase of the pandemic (https://outbreak.info/compare-lineages). Interestingly, the Y505H mutation is also frequent in Omicron (90% of all sequences), but it is rarely present in other variants such as Alpha (less than 0.1% of all sequences). Nevertheless, the inhibitory effect of VE607 on pseudoviral particles carrying Spike from Omicron was comparable to other VOCs. Of note, the single Y505H mutation did not affect the inhibitory activity of VE607 against Omicron, whereas the Y505T change rendered pseudoviral particles resistant to the molecule, showing the importance of the aromatic motif on the side chain of residue 505 in a direct π−π interaction with the central phenylene of VE607, an interaction unavailable to Y505T. It is also possible that the role of Y505 in VE607 binding depends on the overall conformation of the Spike, which might differ in Omicron, because it accumulated more than 30 mutations in the Spike. Indeed, the likelihood that VE607 binding appears inconsistent with protein-protein interfaces between S1 subunits in the closed S glycoprotein complex suggests that VE607 prefers an open spike configuration.
PK analyses revealed that VE607 accumulated primarily in the spleen, liver, and kidneys following intraperitoneal administration. We were nevertheless able to reach levels close to the IC50 in the lungs, where SARS-CoV-2 establishes infection. Although this concentration is not enough to achieve complete virologic control, we did observe a promising ∼37 fold reduction in SARS-CoV-2 replication in lungs. Our data suggest that a 5–10 fold higher concentration must be administered to achieve effective virologic control. High-resolution structures followed by structure-activity relationship studies will be required to confirm the mode of action and guide Structure Activity Relationship (SAR) to improve the in vivo efficacy of VE607.
Altogether, our results constitute a proof of concept showing that small molecules targeting the SARS-CoV-2 Spike have potential for the development of drugs that may contribute to the fight against COVID-19.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Polyclonal Goat anti-human ACE2 | R&D Systems | Cat# AF933 |
| AlexaFluor-647-conjugated goat anti-human IgG (H+L) | Invitrogen | Cat# A-21445 |
| AlexaFluor-647-conjugated donkey anti-goat IgG (H+L) | Invitrogen | Cat# A-21447 |
| Polyclonal rabbit antiserum raised against SARS-CoV-2 RBD protein | Finzi Lab, Montreal University | N/A |
| SARS-CoV-2 Nucleocapsid Protein (1C7) Monoclonal Antibody (mouse) | Bioss Antibodies | Cat# bsm-41411M |
| Goat anti-Mouse IgG (H+L) Secondary Antibody, HRP | ThermoFisher | Cat# 62-6520 |
| LIVE-DEAD Fixable AquaVivid Cell Stain | Thermo Fischer Scientific | Cat# P34957 |
| CV3-25 | (Jennewein et al., 2021) | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Dulbecco’s Modified Eagle’s medium (DMEM) | Wisent | Cat# 319-005-CL |
| EasyTag™ EXPRESS35 S Protein Labeling Mix | Perkin-Elmer | Cat# NEG772007MC |
| Penicillin/Streptomycin | Wisent | Cat# 450-201-EL |
| Bovine Serum Albumin (BSA) | Sigma-Aldrich | Cat# A9647-100G |
| Fetal Bovine Serum (FBS) | VWR | Cat# 97068-085 |
| Dulbecco's modified Eagle's medium lacking methionine and cysteine | Millipore Sigma | Cat# D0422-100ML |
| GlutaMAX | Gibco | Cat# 35050061 |
| Phosphate Buffered Saline (PBS) | ThermoFischer Scientific | Cat# 10010023 |
| Dialyzed fetal bovine serum | Gibco | Cat# 26400-044 |
| Puromycin Dihydrochloride | Millipore Sigma | Cat# P8833 |
| Passive Lysis Buffer | Promega | Cat# E1941 |
| Freestyle 293F expression medium | Thermo Fischer Scientific | Cat# A14525 |
| D-Luciferin Potassium Salt | Thermo Fischer Scientific | Cat# L2916 |
| Formaldehyde 37% | Thermo Fischer Scientific | Cat# F79-500 |
| QuikChange II XL Site-Directed Mutagenesis Kit | Agilent | Cat# 200521 |
| ExpiFectamine 293 transfection reagent | ThermoFisher Scientific | Cat# A14525 |
| Western Lightning Plus-ECL, Enhanced Chemiluminescence Substrate | Perkin Elmer Life Sciences | Cat# NEL105001EA |
| Protein A Sepharose CL-4B | Cytiva | Cat # 17096303 |
| Ni-NTA agarose | Invitrogen | Cat # R90110 |
| The PEG-it Virus precipitation solution (5X) | System Bioscience | Cat # LV810A-1 |
| Triton X-100 | Sigma | Cat# 9036-19-5 |
| Vector TrueView Autofluorescence Quenching Kit | Vector Laboratories | SP-8400 |
| 2x Laemmli Sample Buffer | BIO-RAD | Cat# 1610737EDU |
| 2-Mercaptoethanol | Sigma-Aldrich | Cat# M3148 |
| CellTiter-Glo® One Solution Assay | Promega | Cat# |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat # D2650-5X5ML CAS: 67-68-5 |
| PepMix™ SARS-CoV-2 (Spike Glycoprotein) | JPT | Cat# PM-WCPV-S-1 |
| SYPRO Orange | ThermoFisher | Cat# S6650 |
| Avicel (Pharma Grade) | FMC | Cat # RC-581 NF |
| RNeasy plus Mini kit | Qiagen | Cat # 74136 |
| iScript advanced cDNA kit | Bio-Rad | Cat #1725036 |
| SSAA009E2 | This paper | N/A |
| VE607 | This paper | N/A |
| (S,S)-VE607 | This paper | N/A |
| (R,R)-VE607 | This paper | N/A |
| (R,S)-VE607 | This paper | N/A |
| Experimental models: cell lines | ||
| HEK293 T cells (female, Homo sapiens) | ATCC | Cat# CRL-3216; RRID: CVCL_0063 |
| 293 T-ACE2 cells (female, Homo sapiens) | (Prévost et al., 2020) | N/A |
| FreeStyle 293F cells (female, Homo sapiens) | ThermoFischer Scientific | Cat# R79007; RRID: CVCL_D603 |
| Vero E6 cells (female, Chlorocebus sabaeus) | ATCC | Cat# ARP-4376; RRID: CRL-1586™ |
| Vero E6-TMPRSS2 (female, Chlorocebus sabaeus) | Craig B. Wilen, Yale University | N/A |
| Recombinant DNA | ||
| pNL4.3 R-E- Luc | NIH AIDS reagent program | Cat# 3418 |
| pCAGGS-SARS-CoV-2 D614G-Spike | (Beaudoin-Bussières et al., 2020) | N/A |
| pCG1-SARS-CoV-1-Spike | (Hoffmann et al., 2013) | N/A |
| pCAGGS-SARS-CoV-2-B.1.1.7 Spike | (Tauzin et al., 2021) | N/A |
| pcDNA3.1-SARS-CoV-2-B.1.351 Spike | (Gong et al., 2021) | N/A |
| pCAGGS-SARS-CoV-2-B.1.617.2 Spike | (Gong et al., 2021) | N/A |
| pcDNA3.1-SARS-CoV-2-P.1 Spike | (Gong et al., 2021) | N/A |
| pcDNA3.1-SARS-CoV-2-Omicron-BA.1 Spike | (Gong et al., 2021) | N/A |
| pcDNA3.1-SARS-CoV-2-Omicron-BA.2 Spike | (Tauzin et al., 2022) | N/A |
| pCAGGS-SARS-CoV-2 D614G/Q498V-Spike | This paper | N/A |
| pCAGGS-SARS-CoV-2 D614G/Y505H-Spike | This paper | N/A |
| pCAGGS-SARS-CoV-2 D614G/Y505T-Spike | (Gong et al., 2021) | N/A |
| pSVCMV-IN-VSV-G | (Emi et al., 1991) | N/A |
| pIRES-GFP | Clontech | Cat# 6029-1 |
| pcDNA3.1-SARS-CoV-2 RBD | (Beaudoin-Bussières et al., 2020) | N/A |
| Soluble ACE2 (residues 1-615 of human ACE2) | (Wrapp et al. 2020) | N/A |
| Bacterial and virus strains | ||
| Authentic SARS-CoV-2 D614G virus (LSPQ/231457/2020) | (Prévost et al., 2021) | N/A |
| SARS-CoV-2-nLuc (strain 2019-nCoV/ USA_WA1/2020) | Craig B. Wilen, Yale University | K. Plante and Pei-Yong Shi, World Reference Center for Emerging Viruses and Arboviruses, University of Texas Medical Branch) |
| Experimental models: Organisms/strains | ||
| B6.Cg-Tg(K18-ACE2)2Prlmn/J (males and females) | The Jackson Laboratory | Stock No: 034860 RRID:IMSR_JAX:034860 |
| Software and algorithms | ||
| Flow Jo v10.7.1 | Flow Jo | https://www.flowjo.com |
| GraphPad Prism v8.4.3 | GraphPad | https://www.graphpad.com |
| Microsoft Excel v16 | Microsoft Office | https://www.microsoft.com/en-ca/microsoft-365/excel |
| BioRender | BioRender | BioRender.com |
| LightCycler® 480 Software | Roche | N/A |
| ImageQuant 5.2 | Molecular Dynamics | N/A |
| Thermo Scientific Xcalibur 4.2.47 | Thermo | N/A |
| Living Image v4.7.3 in vivo software package | Perkin Elmer | N/A |
| Others | ||
| BD LSRII Flow Cytometer | BD Biosciences | N/A |
| FACS Symphony A5 Cell Analyzer | BD Biosciences | N/A |
| TriStar LB942 Microplate Reader | Berthold Technologies | N/A |
| LightCycler® 480 instrument | Roche | N/A |
| VG Micromass 70/70H or VG ZAB-E spectrometer | VG Micromass Ltd. | University of Pennsylvania Mass Spectroscopy Service Center |
| Model 583 gel dryer | BIO-RAD | N/A |
| Thermo Scientific TSQ Quantiva triple quadrupole mass spectrometer | Thermo | N/A |
| Perkin Elmer IVIS Spectrum In-Vivo Imaging System | PerkinElmer | Yale University ABSL-3 facility. RRID:SCR_018621 |
| XIC-3 animal isolation chamber | PerkinElmer | N/A |
| RAS-4 Rodent Anesthesia System | PerkinElmer | CLS146737 |
| QUANTIFOIL holey carbon grids | Electron Microscopy Sciences | Cat # Q250-CR1 |
| Synergy LX multi-mode reader | Biotek | N/A |
| Superose 6 10/300 GL | GE Healthcare | Cat # 17517201 |
| Hiload 16/600 Superdex 200pg | GE Healthcare | Cat # 28989335 |
| Biacore 3000 | GE Healthcare | N/A |
| Typhoon Trio Variable Mode Imager | Amersham Biosciences | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact andres.finzi@umontreal.ca.
All unique reagents generated during this study are available from the lead contact andres.finzi@umontreal.ca with a completed Materials Transfer Agreement.
Experimental model and subject details
Plasmids
The plasmids expressing the human coronavirus Spike of SARS-CoV-2 was kindly provided by Stefan Pöhlmann and was previously reported (Hoffmann et al., 2020b). The pNL4.3 R-E− Luc was obtained from NIH AIDS Reagent Program. The codon-optimized RBD sequence (encoding residues 319-541) fused to a C-terminal hexahistidine tag was cloned into the pcDNA3.1(+) expression vector and was reported elsewhere (Beaudoin-Bussieres et al., 2020). The plasmids encoding the SARS-CoV-2 variants Spikes D614G, Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1) were codon-optimized and synthesized by Genscript. Plasmid encoding the Delta (B.1.617.2), Omicron – BA.1 (B.1.1.529) and Omicron – BA.2 Spikes were generated by overlapping PCR using a codon-optimized wild-type SARS-CoV-2 Spike gene (GeneArt, ThermoFisher) that was synthesized (Biobasic) and cloned in pCAGGS as a template (Chatterjee et al., 2021; Gong et al., 2021; Tauzin et al., 2022). The vesicular stomatitis virus G (VSV-G)-encoding plasmid (pSVCMV-IN-VSV-G) was previously described (Emi et al., 1991). Plasmids used to generate SARS-CoV-2 pseudoviral particles for smFRET analysis were described previously (Lu et al., 2020).
Cell lines
293T human embryonic kidney cells (obtained from ATCC) and Vero E6 cells (ATCC CRL-1586™) were maintained at 37°C under 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) (Wisent) containing 5% fetal bovine serum (VWR), 100 UI/mL of penicillin and 100 μg/mL of streptomycin (Wisent). The 293T-ACE2 cell line was previously reported (Prevost et al., 2020).
Virus
Authentic SARS-CoV-2 was isolated, sequenced, and amplified from clinical samples obtained from patients infected with SARS-CoV-2 D614G by the Laboratoire de Santé Publique du Québec (LSPQ) and was previously described (Prevost et al., 2021). The virus was sequenced by MinION technology (Oxford Nanopore technologies, Oxford, UK). All work with the infectious SARS-CoV-2 authentic virus was performed in Biosafety Level 3 (BSL3) facilities at CRCHUM using appropriate positive-pressure air respirators and personal protective equipment. For in vivo experiments, the 2019n-CoV/USA_WA1/2019 isolate of SARS-CoV-2 expressing nanoluciferase was obtained from Craig B Wilen, Yale University and generously provided by K. Plante and Pei-Yong Shi, World Reference Center for Emerging Viruses and Arboviruses, University of Texas Medical Branch) (Xie et al., 2020a, 2020b). The viruses were propagated in Vero E6 TMPRSS2 by infecting them in T150 cm2 flasks at a MOI of 0.1. The culture supernatants were collected after 72 h when cytopathic effects were clearly visible. The cell debris was removed by centrifugation and filtered through 0.45-micron filter to generate virus stocks. Viruses were concentrated by adding one volume of cold (4°C) 4x PEG-it Virus Precipitation Solution (40% (w/v) PEG-8000 and 1.2 M NaCl; System Biosciences) to three volumes of virus-containing supernatant. The solution was mixed by inverting the tubes several times and then incubated at 4°C overnight. The precipitated virus was harvested by centrifugation at 1,500 × g for 60 min at 4°C. The concentrated virus was then resuspended in PBS then aliquoted for storage at −80°C. All work with infectious SARS-CoV-2 was performed in Institutional Biosafety Committee approved BSL3 and A-BSL3 facilities at Yale University School of Medicine using appropriate positive pressure air respirators and protective equipment.
Mouse experiments
All experiments were approved by the Institutional Animal Care and Use Committees (IACUC) of and Institutional Biosafety Committee of Yale University (IBSCYU). All the animals were housed under specific pathogen-free conditions in the facilities provided and supported by Yale Animal Resources Center (YARC). hACE2 transgenic B6 mice (heterozygous) were obtained from Jackson Laboratory. 6–8-week-old male and female mice were used for all the experiments. The heterozygous mice were crossed and genotyped to select heterozygous mice for experiments by using the primer sets recommended by Jackson Laboratory.
Method details
Purification of SARS-CoV-2 RBD
FreeStyle 293F cells (Invitrogen) were grown in FreeStyle 293F medium (Invitrogen) to a density of 1 × 106 cells/mL at 37°C with 8% CO2 with regular agitation (150 rpm). Cells were transfected with a plasmid coding for SARS-CoV-2 S RBD using ExpiFectamine 293 transfection reagent, as directed by the manufacturer (Invitrogen). One week later, cells were pelleted and discarded. Supernatants were filtered using a 0.22 μm filter (Thermo Fisher Scientific). The recombinant RBD proteins were purified by nickel affinity columns, as directed by the manufacturer (Invitrogen). The RBD preparations were dialyzed against phosphate-buffered saline (PBS) and stored in aliquots at −80°C until further use. To assess purity, recombinant proteins were loaded on SDS-PAGE gels and stained with Coomassie Blue.
Differential scanning fluorimetry
DSF experiments were essentially performed as described previously (Sharifahmadian et al., 2017). DSF was conducted using 5 μM of purified RBD, 10x concentration of SYPRO Orange (from 5000x stock solution, ThermoFisher) in 50 mM HEPES, 100 mM NaCl, pH 7.5 and 5% final concentration of DMSO. The small molecules were added to final concentrations of 5 mM. SYPRO Orange fluorescence was monitored over 20–95°C with a LightCycler® 480 instrument (Roche, USA). The LightCycler® 480 Software (Roche) was used to calculate the first derivate of the resulting melting curve, with the steepest point of the slope being the Tm.
Molecular modeling
System preparation, modeling, and docking calculation were performed using the Schrödinger Suite package (Schrödinger, 2020), using default parameters unless otherwise noted. The target structures were taken from SARS-CoV-1 RBD (PDB ID: 6waq) and SARS-CoV-2 RBD (PDB ID: 6w41) prepared using the Protein Preparation Wizard (Sastry et al., 2013). To prepare the structures, force field atom types and bond orders were assigned, missing atoms and side-chains were added, protonation states of ionizable amino acid side-chains were determined using PROPKA (Olsson et al., 2011), water orientations were sampled, and hydrogen bond networks were subsequently optimized by flipping Asn/Gln/His residues and sampling hydroxyl/thiol hydrogen. Constrained energy minimization was then performed using the imperf module from impact (Schrödinger, 2020) to generate the structure to be used in the subsequent modeling calculations. Potential binding sites were explored and characterized using the SiteMap tool (Halgren, 2007, 2009). VE607 compound was structurally preprocessed using LigPrep (Schrödinger, 2020) to generate multiple states for stereoisomers, tautomers, ring conformations, and protonation states at a selected pH range. Then, energy minimization was performed with the OPLS3e force field (Roos et al., 2019). The prepared molecular structures were docked into the putative binding sites using Glide (Friesner et al., 2004; Halgren et al., 2004) with the standard precision (SP) scoring function to evaluate enrichment of the calculated docked models.
Neutralization assay using pseudoviral particles
Target cells were infected with single-round luciferase-expressing lentiviral particles as described previously (Prevost et al., 2020). Briefly, 293T cells were transfected by the calcium phosphate method with the lentiviral vector pNL4.3 R-E- Luc (NIH AIDS Reagent Program) and a plasmid encoding for SARS-CoV-2 Spike at a ratio of 5:4. Two days post-transfection, cell supernatants were harvested and stored at −80°C until use. 293T-ACE2 target cells were seeded at a density of 1×104 cells/well in 96-well luminometer-compatible tissue culture plates (Perkin Elmer) 24 h before infection. Recombinant viruses in a final volume of 100 μL were incubated with the indicated concentrations of small molecules (VE607 or SSAA009E2) up to concentrations of 100 μM for 1 h at 37°C and were then added to the target cells followed by incubation for 48 h at 37°C; cells were lysed by the addition of 30 μL of passive lysis buffer (Promega) followed by one freeze-thaw cycle. An LB941 TriStar luminometer (Berthold Technologies) was used to measure the luciferase activity of each well after the addition of 100 μL of luciferin buffer (15 mM MgSO4, 15 mM KPO4 [pH 7.8], 1 mM ATP, and 1 mM dithiothreitol) and 50 μL of 1 mM d-luciferin potassium salt (Prolume). Relative lighting unit (RLU) for luciferase activity was recorded and the ratio to “no compound” was calculated. Minimum RLU for tested pseudoviral particles was 50,000 (background RLU of uninfected cells is ∼400). The half-maximal inhibitory dilution (IC50) represents the compound concentration required to inhibit 50% of the infection of 293T-ACE2 cells by pseudoviral particles.
Cell surface staining and flow cytometry analysis
Using the standard calcium phosphate method, 10 μg of Spike expressor and 2.5 μg of a green fluorescent protein (GFP) expressor (pIRES-GFP) were transfected into 2 × 106 293T cells. 48 h post-transfection Spike-expressing cells were incubated with 100 μM of VE607 or equivalent volume of vehicle (DMSO) and incubated for 30 min at room temperature. CV3-25 (5 μg/mL) or sACE2 (100 μg/mL) was added to the cells and incubated for 45 min at 37°C and sACE2 binding was detected using a polyclonal Goat anti-human ACE2 (R&D Systems) at 1/100 dilution at room temperature for 30 min. AlexaFluor-647-conjugated goat anti-human IgG (H+L) Ab (Invitrogen) or AlexaFluor-647-conjugated donkey anti-goat IgG (H+L) Ab (Invitrogen) was used as secondary antibodies. The percentage of transfected cells (GFP+ cells) was determined by gating the living cell population based on viability dye staining (Aqua Vivid, Invitrogen). sACE2 binding levels were normalized to signals obtained with the conformationally independent anti-S2 CV3-25 mAb (Li et al., 2022; Prevost et al., 2021; Tauzin et al., 2022). Samples were acquired on a LSRII cytometer (BD Biosciences, Mississauga, ON, Canada) and data analysis was performed using FlowJo vX.0.7 (Tree Star, Ashland, OR, USA).
Radioactive labeling and immunoprecipitation
For pulse-labeling experiments, 5 × 105 293T cells were transfected by the calcium phosphate method with SARS-CoV-2 D614G Spike expressor. One day after transfection, cells were metabolically labeled for 16–24 h with 100 μCi/mL [35S]methionine-cysteine ([35S] protein labeling mix; Perkin-Elmer) in Dulbecco’s modified Eagle’s medium lacking methionine and cysteine and supplemented with 10% of dialyzed fetal bovine serum and 1% GlutaMAX™. Simultaneously, cells were treated with or without 100 μM VE607. Cells were subsequently lysed in radio immunoprecipitation assay (RIPA) buffer (140 mM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 1% IGEPAL® CA-630, 0.05% sodium dodecyl sulfate [SDS], 1.2 mM sodium deoxycholate [DOC]). Precipitation of radiolabeled SARS-CoV-2 D614G Spike glycoprotein from cell lysates or supernatant was performed with CV3-25 and polyclonal rabbit antiserum raised against SARS-CoV-2 RBD protein, for 1 h at 4°C in the presence of 45 μL of 10% protein A-Sepharose beads (GE Healthcare). Samples were washed twice with RIPA buffer and then boiled 5 min in Laemmli buffer (BIO-RAD) with β-mercaptoethanol before being separated by SDS-PAGE. After migration, gels were dried with a Model 583 gel dryer (Bio-Rad) and exposed to a storage phosphor screen. Densitometry data were acquired with a Typhoon Trio Variable Mode Imager (Amersham Biosciences) in storage phosphor acquisition mode and analyzed using ImageQuant 5.2 (Molecular Dynamics). Association index was determined by precipitation of radiolabeled cell lysates and supernatants with CV3-25 and polyclonal rabbit antiserum raised against SARS-CoV-2 RBD protein. The association index is a measure of the ability of the VE607 treated S1 subunit to remain associated with the trimeric spike (S) glycoprotein on the expressing cell relative to that of the mock-treated S1 and was calculated with the following formula: association index = ([cell S1]treated/[supernatant S1]treated)/([cell S1]mock-treated/[supernatant S1]mock-treated).
Microneutralization with authentic virus
One day prior to infection, 2 × 104 Vero E6 cells were seeded per well in the 96-well flat bottom plate and incubated overnight to permit Vero E6 cell adherence. Compounds dilutions ranged from 0, 0.316, 1, 3.16, 10, 31.6 and 100 μM were performed in a separate 96 well culture plate using DMEM supplemented with penicillin (100 U/mL), streptomycin (100 μg/mL), HEPES, 0.12% sodium bicarbonate, 2% FBS and 0.24% BSA. 104 TCID50/mL of SARS-CoV-2 virus was prepared in DMEM + 2% FBS and combined with an equivalent volume of diluted compounds for one hour. After this incubation, all media was removed from the 96 well plate seeded with Vero E6 cells and virus: compounds mixture was added to each respective well at a volume corresponding to 600 TCID50 per well and incubated for one hour further at 37°C. Both virus only and media only (MEM + 2% FBS) conditions were included in this assay. All virus-compounds supernatant was removed from wells without disrupting the Vero E6 monolayer. Each diluted compound (100 μL) was added to its respective Vero E6-seeded well in addition to an equivalent volume of MEM + 2% FBS and was then incubated for 48 h. Media was then discarded and replaced with 10% formaldehyde for 24 h to cross-link Vero E6 monolayer. Formaldehyde was removed from wells and subsequently washed with PBS. Cell monolayers were permeabilized for 15 min at room temperature with PBS + 0.1% Triton X-100, washed with PBS and then incubated for one hour at room temperature with PBS + 3% non-fat milk. An anti SARS-CoV-2 nucleocapsid protein (Clone 1C7, Bioss Antibodies) primary antibody solution was prepared at 1 μg/mL in PBS + 1% non-fat milk and added to all wells for one hour at room temperature. Following extensive washing (3×) with PBS, an anti-mouse IgG HRP secondary antibody (ThermoFisher) solution was formulated in PBS + 1% non-fat milk. One hour post-room temperature incubation, wells were washed with 3× PBS, substrate (ECL) was added and an LB941 TriStar luminometer (Berthold Technologies) was used to measure the signal each well.
Cell viability test
To measure the cytotoxicity of VE607 and its stereoisomers on 293T-ACE2 or Vero-E6 cells, a cell viability assay using CellTiter-Glo® One Solution Assay (Promega) was performed. Briefly, 293T-ACE2 or Vero-E6 cells were seeded at a density of 1×104 cells/well in 96-well luminometer-compatible tissue culture plates (Perkin Elmer); After 24 h, indicated concentrations of VE607, (S,S)-VE607, (R,R)-VE607 or (R,S)-VE607 up to concentrations of 100 μM were added to the cells followed by incubation for 48 h at 37°C, same volume of its vehicle, DMSO, was added as control. Then a volume of CellTiter-Glo® One Solution equal to the volume of cell culture medium present in each well was added, followed by 2 min mixing on shaker and 10 min incubation at room temperature. An LB941 TriStar luminometer (Berthold Technologies) was used to measure the luciferase activity of each well.
Chemical synthesis of the three enantiomers of VE607
All reactions were conducted in oven-dried glassware under an inert atmosphere of nitrogen, unless otherwise stated. All solvents were reagent or high-performance liquid chromatography (HPLC) grade. Anhydrous THF was obtained from the Pure SolveTM PS-400 system under an argon atmosphere. All reagents were purchased from commercially available sources and used as received. Reactions were magnetically stirred under a nitrogen atmosphere, unless otherwise noted and were monitored by thin layer chromatography (TLC) was performed on pre-coated silica gel 60 F-254 plates (40–55 micron, 230–400 mesh) and visualized by UV light or staining with KMnO4 and heating. Yields refer to chromatographically and spectroscopically pure compounds. Optical rotations were measured on a JASCO P-200 polarimeter. Proton (1H) and carbon (13C) NMR spectra were recorded on a Bruker Avance III 500-MHz spectrometer. Chemical shifts (δ) are reported in parts per million (ppm) relative to chloroform (δ 7.26) or methanol (δ 3.31) for 1H NMR, and chloroform (δ 77.2) or methanol (δ 49.0). High resolution mass spectra (HRMS) were recorded at the University of Pennsylvania Mass Spectroscopy Service Center on either a VG Micromass 70/70H or VG ZAB-E spectrometer. Lyophilization was performed in a Labconco FreeZone 12 Plus lyophilizer (0.148 mbar). The purity of new compounds were judged by NMR and LCMS (>95%) (Data S1).
smFRET analysis
Pseudoviral particles bearing labeled CoV2WH01 S glycoprotein were prepared, imaged, and analyzed as described previously (Li et al., 2021; Lu et al., 2020). Samples were pre-incubated with sACE2 (200 μg/mL) or VE607 (100 μM) for 90 min at room temperature prior to imaging.
Pharmacokinetics of VE607
VE607 or vehicle (DMSO) was administered intraperitoneally (i.p.) to male and female K18-hACE2 mice aged 6 to 8 weeks at doses of 50 mg/kg, 25 mg/kg, and 12.5 mg/kg, and the concentration in blood was measured 30 min, 2, 10, and 24 h after administration. The mice were sacrificed at 24 h, and the indicated organs (Lung, Brain, Liver, Spleen, Kidney and Nose) were isolated for mass spectrometry. Tissue was cut into small pieces, weighed, and resuspended in serum-free RPMI and homogenized using 1.5 mm Zirconium beads with BeadBug 6 homogenizer (Benchmark Scientific, TEquipment Inc). In order to determine drug concentrations, the homogenized tissue was briefly centrifuged at 13,000 rpm at 4°C and the cell free supernatant containing free drug was analyzed by mass spectrometry and data was extrapolated to reflect the amount of drug per organ based on weight.
Mass spectrometry
VE607 mouse plasma and tissue levels were determined by ultra-high performance liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS). Briefly, VE607 was extracted from mouse plasma and tissue homogenate samples using protein precipitation. One thousand microliters of internal standard (IS) solution (25 ng/mL of candesartan in methanol) were added to an aliquot of 10 μL of sample. The sample was vortexed for approximately 5 s and let stand for a period of 10 min, then centrifuged at 14,000 x g for 10 min. The supernatant was transferred to 13 × 100 mm borosilicate tubes and evaporated to dryness at 50°C under a gentle stream of nitrogen. The dried extract was re-suspended with 500 μL of mobile phase (20:80 methanol: 10 mM ammonium formate pH 3) and transferred to an injection vial for analysis. The analysis was performed using a Thermo Scientific TSQ Quantiva triple quadrupole mass spectrometer interfaced with a Thermo Scientific Ultimate 3000XRS UHPLC system using a heated electrospray ion source (HESI). MS detection was performed in positive ion mode, using multiple reaction monitoring (MRM). The precursor-ion reactions for VE607 and IS were set to 393.4 → [98.2 + 142.1] and 441.2 → 263.1, respectively. Chromatographic separation was achieved using gradient elution with a Phenomenex Phenyl-Hexyl analytical column (150 × 2.0 mm I.D., 5 μm) operating at 50°C. The initial mobile phase condition consisted of methanol and 10 mM ammonium formate, pH 3 at a ratio of 20:80, respectively, and this ratio was maintained for 1 min. From 1 to 3 min a linear gradient was applied up to a ratio of 90:10 and maintained for 2 min. At 5.1 min, the mobile phase composition was reverted to 20:80 and the column was allowed to equilibrate for 8.9 min for a total run time of 14 min. The flow rate was fixed at 0.3 mL/min and VE607 and IS eluted at 2.7 and 4.9 min, respectively. Data acquisition and analysis were performed using Thermo Scientific Xcalibur 4.2.47. Calibration curves were calculated from the equation y = ax + b, as determined by weighted (1/x) linear regression of the calibration lines constructed from the peak-area ratio of VE607 to IS. The analytical range was set from 25 to 50,000 ng/mL and the sample concentrations were interpolated from the standard curve. The precision and accuracy of the method met generally accepted performance criteria in bioanalytical chemistry
SARS-CoV-2 infection and treatment conditions
For all in vivo experiments, the 6 to 8 weeks male and female mice were intranasally challenged with 1 × 105 FFU SARS-CoV-2_WA1_nLuc, in 25-30 μL volume under anesthesia (0.5 - 5% isoflurane delivered using precision Dräger vaporizer with oxygen flow rate of 1 L/min). VE607 was administered intraperitoneally (i.p. 25 mg/kg), 24 h prior to infection and daily until day 4 following infection. Lethargic and moribund mice or mice that had lost more than 20% of their body weight were sacrificed and considered to have succumbed to infection for Kaplan-Meier survival plots.
Bioluminescence imaging (BLI) of SARS-CoV-2 infection
All standard operating procedures and protocols for IVIS imaging of SARS-CoV-2 infected animals under ABSL-3 conditions were approved by IACUC, IBSCYU and YARC. All the imaging was carried out using IVIS Spectrum® (PerkinElmer) in XIC-3 animal isolation chamber (PerkinElmer) that provided biological isolation of anesthetized mice or individual organs during the imaging procedure. All mice were anesthetized via isoflurane inhalation (3–5% isoflurane, oxygen flow rate of 1.5 L/min) prior and during BLI using the XGI-8 Gas Anesthesia System. Prior to imaging, 100 μL of nanoluciferase substrate, furimazine (NanoGloTM, Promega, Madison, WI) diluted 1:40 in endotoxin-free PBS was retroorbitally administered to mice under anesthesia. The mice were then placed into XIC-3 animal isolation chamber (PerkinElmer) pre-saturated with isothesia and oxygen mix. The mice were imaged in both dorsal and ventral position at indicated days post infection. The animals were then imaged again after euthanasia and necropsy by spreading additional 200 μL of substrate on to exposed intact organs. Infected areas identified by carrying out whole-body imaging after necropsy were isolated, washed in PBS to remove residual blood and placed onto a clear plastic plate. Additional droplets of furimazine in PBS (1:40) were added to organs and soaked in substrate for 1–2 min before BLI.
Images were acquired and analyzed with Living Image v4.7.3 in vivo software package (Perkin Elmer Inc). Image acquisition exposures were set to auto, with imaging parameter preferences set in order of exposure time, binning, and f/stop, respectively. Images were acquired with luminescent f/stop of 2, photographic f/stop of 8. Binning was set to medium. Comparative images were compiled and batch-processed using the image browser with collective luminescent scales. Photon flux was measured as luminescent radiance (p/sec/cm2/sr). During luminescent threshold selection for image display, luminescent signals were regarded as background when minimum threshold setting resulted in displayed radiance above non-tissue-containing or known uninfected regions.
Focus forming assay
Titers of virus stocks was determined by standard plaque assay. Briefly, the 4 × 105 Vero-E6 cells were seeded on 12-well plate. 24 h later, the cells were infected with 200 μL of serially diluted virus stock. After 1 h, the cells were overlayed with 1 mL of pre-warmed 0.6% Avicel (RC-581 FMC BioPolymer) made in complete RPMI medium. Plaques were resolved at 48 h post infection by fixing in 10% paraformaldehyde for 15 min followed by staining for 1 h with 0.2% crystal violet made in 20% ethanol. Plates were rinsed in water to visualize plaques.
Measurement of viral burden
Indicated organs (nasal cavity, brain, lungs) from infected or uninfected mice were collected, weighed, and homogenized in 1 mL of serum free RPMI media containing penicillin-streptomycin and homogenized in 2 mL tube containing 1.5 mm Zirconium beads with BeadBug 6 homogenizer (Benchmark Scientific, TEquipment Inc). Virus titers were measured using three highly correlative methods. Frist, the total RNA was extracted from homogenized tissues using RNeasy plus Mini kit (Qiagen Cat # 74136), reverse transcribed with iScript advanced cDNA kit (Bio-Rad Cat #1725036) followed by a SYBR Green Real-time PCR assay for determining copies of SARS-CoV-2 N gene RNA using primers SARS-CoV-2 N F: 5′-ATGCTGCAATCGTGCTACAA-3′ and SARS-CoV-2 N R: 5′-GACTGCCGCCTCTGCTC-3′.
Quantification and statistical analysis
Data were analyzed and plotted using GraphPad Prism software (La Jolla, CA, USA). Statistical significance for pairwise comparisons were derived by applying non-parametric Mann-Whitney test (two-tailed). p values < 0.05 were considered significant; significance values are indicated as ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Acknowledgments
The authors thank the CRCHUM BSL3 and Flow Cytometry Platforms for technical assistance. The authors thank Hughes Charest and the LSPQ for the authentic SARS-CoV-2 virus. This work was supported by “Ministère de l’Économie et de l’Innovation du Québec, Programme de soutien aux organismes de recherche et d’innovation”, by the Fondation du CHUM, by a Canadian Institutes of Health Research (CIHR) foundation grant #352417 and by an Exceptional Fund COVID-19 from the Canada Foundation for Innovation (CFI) #41027 to A.F, and by R01 AI163395 from NIH NIAID to W.M. Work on the presented variants was also supported by the Sentinelle COVID Quebec network led by the LSPQ in collaboration with Fonds de Recherche du Québec Santé (FRQS) to A.F. The work was also supported by the Université de Montréal Faculty of Medicine fund “Combattre la COVID-19: de la prevention au controle” to C.B. and A.F.; A.F. is the recipient of a Canada Research Chair on Retroviral Entry # RCHS0235 950-232424. G.B.B. is the recipient of a Fonds de Recherche Québec— Santé (FRQS) PhD fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The graphical abstract was created with BioRender.com.The views expressed in this manuscript are those of the authors and do not reflect the official policy or position of the Uniformed Services University, the US Army, the Department of Defense, or the US Government.
Author contributions
A.F., C.B., and P.D.U. designed the studies. S.D., I.U., S.Y.G., J.G., M.M., D.V., G.B.B., V.T.V., G.G., F.G., J.R., D.Y., A.B.S., M.P., M.C., and C.A. performed the experiments. P.K., P.D.U., W.M., A.F., and C.B., funding acquisition and/or interpretation of results. Y.C., A.B.S., M.P., and M.C. contributed with unique reagents. A.F. S.D., P.D.U., and C.B. wrote the manuscript with inputs from others. Every author has read, edited, and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: July 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104528.
Contributor Information
Andrés Finzi, Email: andres.finzi@umontreal.ca.
Christian Baron, Email: christian.baron@umontreal.ca.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact (andres.finzi@umontreal.ca) upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (andres.finzi@umontreal.ca) upon request.
References
- Adedeji A.O., Severson W., Jonsson C., Singh K., Weiss S.R., Sarafianos S.G. Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. J. Virol. 2013;87:8017–8028. doi: 10.1128/jvi.00998-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand S.P., Chen Y., Prevost J., Gasser R., Beaudoin-Bussieres G., Abrams C.F., Pazgier M., Finzi A. Interaction of human ACE2 to membrane-bound SARS-CoV-1 and SARS-CoV-2 S glycoproteins. Viruses. 2020;12:1104. doi: 10.3390/v12101104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball P. The lightning-fast quest for COVID vaccines - and what it means for other diseases. Nature. 2021;589:16–18. doi: 10.1038/d41586-020-03626-1. [DOI] [PubMed] [Google Scholar]
- Beaudoin-Bussieres G., Laumaea A., Anand S.P., Prevost J., Gasser R., Goyette G., Medjahed H., Perreault J., Tremblay T., Lewin A., et al. Decline of humoral responses against SARS-CoV-2 spike in convalescent individuals. mBio. 2020;11 doi: 10.1128/mBio.02590-20. e02590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Jr., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee D., Tauzin A., Marchitto L., Gong S.Y., Boutin M., Bourassa C., Beaudoin-Bussières G., Bo Y., Ding S., Laumaea A., et al. SARS-CoV-2 Omicron Spike recognition by plasma from individuals receiving BNT162b2 mRNA vaccination with a 16-weeks interval between doses. Cell Rep. 2021;38:110429. doi: 10.1016/j.celrep.2022.110429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. The dream vaccine. Science. 2021;372:227–231. doi: 10.1126/science.372.6539.227. [DOI] [PubMed] [Google Scholar]
- Dai W., Zhang B., Jiang X.M., Li J., Zhao Y., Xie X., Jin Z., Liu F., Peng J., Li Y., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Adam D., Beaudoin-Bussières G., Tauzin A., Gong S.Y., Gasser R., Laumaea A., Anand S.P., Privé A., Bourassa C., et al. SARS-CoV-2 spike expression at the surface of infected primary human airway epithelial cells. Viruses. 2022;14:5. doi: 10.3390/v14010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emi N., Friedmann T., Yee J.K. Pseudotype formation of murine leukemia virus with the G protein of vesicular stomatitis virus. J. Virol. 1991;65:1202–1207. doi: 10.1128/jvi.65.3.1202-1207.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K., et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Gong S.Y., Chatterjee D., Richard J., Prévost J., Tauzin A., Gasser R., Bo Y., Vézina D., Goyette G., Gendron-Lepage G., et al. Contribution of single mutations to selected SARS-CoV-2 emerging variants spike antigenicity. Virology. 2021;563:134–145. doi: 10.1016/j.virol.2021.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgren T. New method for fast and accurate binding-site identification and analysis. Chem. Biol. Drug Des. 2007;69:146–148. doi: 10.1111/j.1747-0285.2007.00483.x. [DOI] [PubMed] [Google Scholar]
- Halgren T.A. Identifying and characterizing binding sites and assessing druggability. J. Chem. Inf. Model. 2009;49:377–389. doi: 10.1021/ci800324m. [DOI] [PubMed] [Google Scholar]
- Halgren T.A., Murphy R.B., Friesner R.A., Beard H.S., Frye L.L., Pollard W.T., Banks J.L. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Harrison A.G., Lin T., Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41:1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Müller M.A., Drexler J.F., Glende J., Erdt M., Gützkow T., Losemann C., Binger T., Deng H., Schwegmann-Weßels C., et al. Differential sensitivity of bat cells to infection by enveloped RNA viruses: coronaviruses, paramyxoviruses, filoviruses, and influenza viruses. PLoS One. 2013;8:e72942. doi: 10.1371/journal.pone.0072942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pohlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennewein M.F., MacCamy A.J., Akins N.R., Feng J., Homad L.J., Hurlburt N.K., Seydoux E., Wan Y.H., Stuart A.B., Edara V.V., et al. Isolation and characterization of cross-neutralizing coronavirus antibodies from COVID-19+ subjects. Cell Rep. 2021;36:109353. doi: 10.1016/j.celrep.2021.109353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao R.Y., Tsui W.H., Lee T.S., Tanner J.A., Watt R.M., Huang J.D., Hu L., Chen G., Chen Z., Zhang L., et al. Identification of novel small-molecule inhibitors of severe acute respiratory syndrome-associated coronavirus by chemical genetics. Chem. Biol. 2004;11:1293–1299. doi: 10.1016/j.chembiol.2004.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokic G., Hillen H.S., Tegunov D., Dienemann C., Seitz F., Schmitzova J., Farnung L., Siewert A., Höbartner C., Cramer P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021;12:279. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Li J., Lai S., Gao G.F., Shi W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature. 2021;600:408–418. doi: 10.1038/s41586-021-04188-6. [DOI] [PubMed] [Google Scholar]
- Li W., Chen Y., Prévost J., Ullah I., Lu M., Gong S.Y., Tauzin A., Gasser R., Vézina D., Anand S.P., et al. Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern. Cell Rep. 2022;38:110210. doi: 10.1016/j.celrep.2021.110210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Uchil P.D., Li W., Zheng D., Terry D.S., Gorman J., Shi W., Zhang B., Zhou T., Ding S., et al. Real-time conformational dynamics of SARS-CoV-2 spikes on virus particles. Cell Host Microbe. 2020;28:880–891.e8. doi: 10.1016/j.chom.2020.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannar D., Saville J.W., Zhu X., Srivastava S.S., Berezuk A.M., Zhou S., Tuttle K.S., Kim A., Li W., Dimitrov D.S., Subramaniam S. Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding. Cell Rep. 2021;37:110156. doi: 10.1016/j.celrep.2021.110156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashalidis E.H., Śledź P., Lang S., Abell C. A three-stage biophysical screening cascade for fragment-based drug discovery. Nat. Protoc. 2013;8:2309–2324. doi: 10.1038/nprot.2013.130. [DOI] [PubMed] [Google Scholar]
- Nabel K.G., Clark S.A., Shankar S., Pan J., Clark L.E., Yang P., Coscia A., McKay L.G.A., Varnum H.H., Brusic V., et al. Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain. Science. 2021;375:eabl6251. doi: 10.1126/science.abl6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M.H.M., Søndergaard C.R., Rostkowski M., Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theor. Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., et al. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- Prévost J., Finzi A. The great escape? SARS-CoV-2 variants evading neutralizing responses. Cell Host Microbe. 2021;29:322–324. doi: 10.1016/j.chom.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost J., Gasser R., Beaudoin-Bussières G., Richard J., Duerr R., Laumaea A., Anand S.P., Goyette G., Benlarbi M., Ding S., et al. Cross-sectional evaluation of humoral responses against SARS-CoV-2 spike. Cell Rep. Med. 2020;1:100126. doi: 10.1016/j.xcrm.2020.100126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost J., Richard J., Gasser R., Ding S., Fage C., Anand S.P., Adam D., Gupta Vergara N., Tauzin A., Benlarbi M., et al. Impact of temperature on the affinity of SARS-CoV-2 Spike glycoprotein for host ACE2. J. Biol. Chem. 2021;297:101151. doi: 10.1016/j.jbc.2021.101151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappazzo C.G., Tse L.V., Kaku C.I., Wrapp D., Sakharkar M., Huang D., Deveau L.M., Yockachonis T.J., Herbert A.S., Battles M.B., et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science. 2021;371:823–829. doi: 10.1126/science.abf4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos K., Wu C., Damm W., Reboul M., Stevenson J.M., Lu C., Dahlgren M.K., Mondal S., Chen W., Wang L., et al. OPLS3e: extending Force Field Coverage for Drug-Like Small Molecules. J. Chem. Theor. Comput. 2019;15:1863–1874. doi: 10.1021/acs.jctc.8b01026. [DOI] [PubMed] [Google Scholar]
- Madhavi Sastry G., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- Schrödinger . LLC; 2020. Release 2020-4. Induced Fit Docking Protocol. Glide. [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifahmadian M., Arya T., Bessette B., Lecoq L., Ruediger E., Omichinski J.G., Baron C. Monomer-to-dimer transition of Brucella suis type IV secretion system component VirB8 induces conformational changes. FEBS J. 2017;284:1218–1232. doi: 10.1111/febs.14049. [DOI] [PubMed] [Google Scholar]
- Tauzin A., Gong S.Y., Beaudoin-Bussières G., Vézina D., Gasser R., Nault L., Marchitto L., Benlarbi M., Chatterjee D., Nayrac M., et al. Strong humoral immune responses against SARS-CoV-2 Spike after BNT162b2 mRNA vaccination with a 16-week interval between doses. Cell Host Microbe. 2022;30:97–109.e5. doi: 10.1016/j.chom.2021.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauzin A., Nayrac M., Benlarbi M., Gong S.Y., Gasser R., Beaudoin-Bussières G., Brassard N., Laumaea A., Vézina D., Prévost J., et al. A single dose of the SARS-CoV-2 vaccine BNT162b2 elicits Fc-mediated antibody effector functions and T cell responses. Cell Host Microbe. 2021;29:1137–1150.e6. doi: 10.1016/j.chom.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong T.R. Therapies for coronaviruses. Part I of II – viral entry inhibitors. Expert Opin. Ther. Patents. 2009;19:357–367. doi: 10.1517/13543770802609384. [DOI] [PubMed] [Google Scholar]
- Ullah I., Prévost J., Ladinsky M.S., Stone H., Lu M., Anand S.P., Beaudoin-Bussières G., Symmes K., Benlarbi M., Ding S., et al. Live imaging of SARS-CoV-2 infection in mice reveals that neutralizing antibodies require Fc function for optimal efficacy. Immunity. 2021;54:2143–2158.e15. doi: 10.1016/j.immuni.2021.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Yang M.L., Duan Z.L., Liu F.L., Jin L., Long C.B., Zhang M., Tang X.P., Xu L., Li Y.C., et al. Dalbavancin binds ACE2 to block its interaction with SARS-CoV-2 spike protein and is effective in inhibiting SARS-CoV-2 infection in animal models. Cell Res. 2021;31:17–24. doi: 10.1038/s41422-020-00450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.V., et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848.e3. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Muruato A.E., Zhang X., Lokugamage K.G., Fontes-Garfias C.R., Zou J., Liu J., Ren P., Balakrishnan M., Cihlar T., et al. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat. Commun. 2020;11:5214. doi: 10.1038/s41467-020-19055-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Wang Y., Liu C., Zhang C., Han W., Hong X., Wang Y., Hong Q., Wang S., Zhao Q., et al. Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM. Sci. Adv. 2021;7:eabe5575. doi: 10.1126/sciadv.abe5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Rao Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 2021;19:685–700. doi: 10.1038/s41579-021-00630-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Han Y., Ding S., Finzi A., Mothes W., Lu M. SARS-CoV-2 variants exhibit increased kinetic stability of open spike conformations as an evolutionary strategy. mBio. 2021;13:e0322721. doi: 10.1128/mbio.03227-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Luan X., Li Z., Zhou Z., Wang Q., Gao M., Wang X., Zhou F., Shi J., You E., et al. Structural basis for inhibition of the SARS-CoV-2 RNA polymerase by suramin. Nat. Struct. Mol. Biol. 2021;28:319–325. doi: 10.1038/s41594-021-00570-0. [DOI] [PubMed] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z.L., Qiu X.D., Wu S., Liu Y.T., Zhao T., Sun Z.H., Li Z.R., Shan G.Z. Blocking effect of demethylzeylasteral on the interaction between human ACE2 protein and SARS-CoV-2 RBD protein discovered using SPR technology. Molecules. 2020;26:57. doi: 10.3390/molecules26010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact (andres.finzi@umontreal.ca) upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (andres.finzi@umontreal.ca) upon request.