Abstract
Background
TP53 is rarely mutated in paediatric neuroblastoma. The prognosis of TP53 and TP53-associated genes in paediatric neuroblastoma is unclear. The objectives of the study were to analyse datasets of 2477 paediatric neuroblastoma patients from eight independent cohorts to reveal the prognosis of TP53 and TP53-associated genes.
Results
High TP53 mRNA expression was associated with shortened event-free survival and overall survival in paediatric neuroblastoma. Moreover, a higher enrichment score of the TP53 signalling pathway was associated with worse clinical outcomes of paediatric neuroblastoma. Among the genes associated with TP53, CCNE1, CDK2 and CHEK2 were correlated with unfavourable clinical outcomes, while SESN1 was correlated with favourable clinical outcomes of paediatric neuroblastoma in the eight independent neuroblastoma cohorts. TP53, CCNE1, CDK2 and CHEK2 were overexpressed in neuroblastoma patients with MYCN amplification, while SESN1 was downregulated in neuroblastoma patients with MYCN amplification. CCNE1, SESN1, MYCN amplification and age at diagnosis were independent prognostic markers of neuroblastoma. CCNE1 was also highly expressed in paediatric neuroblastoma patients with an age at diagnosis ≥ 18 months, while SESN1 was downregulated in paediatric neuroblastoma patients with an age at diagnosis ≥ 18 months. Combinations of CCNE1 with age at diagnosis or combinations of SESN1 with age at diagnosis achieved superior prognostic effects in paediatric neuroblastoma. Finally, we constructed a nomogram risk model of paediatric neuroblastoma based on age and TP53, CCNE1, CDK2, CHEK2 and SESN1 expression. The nomogram model could predict the overall survival of paediatric neuroblastoma and MYCN nonamplified paediatric neuroblastoma with high specificity and sensitivity.
Conclusions
TP53 and TP53-associated genes CCNE1, CDK2, CHEK2 and SESN1 were significantly associated with the clinical outcomes of paediatric neuroblastoma.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12863-022-01059-5.
Keywords: Paediatric neuroblastoma, TP53, CCNE1, CDK2, CHEK2, SESN1
Introduction
Paediatric neuroblastoma is the most common extracranial malignant tumour type in children [1, 2]. The aggressiveness and clinical outcomes of children with neuroblastoma are significantly different [3]. In 2009, the International Neuroblastoma Risk Group identified 13 prognostic factors of neuroblastoma through a large-scale cohort study, including age at diagnosis, tumour stage, histological type, degree of differentiation and MYCN amplification [4]. MYCN genetic amplification occurs in 25% of neuroblastoma [5] and is an unfavourable prognostic factor of neuroblastoma [6]. Paediatric neuroblastoma patients with an age at diagnosis ≥ 18 months have a poor prognosis [7]. The high clinical stage has adverse effects on the prognosis of children with neuroblastoma [8]. These risk stratifications provide a reference basis for the prognosis and choice of treatment of neuroblastoma. However, additional molecular biomarkers are still needed to provide better classifications and prognoses of paediatric neuroblastoma.
TP53 is referred to as the "guardian of the genome", and almost all key cellular activities are related to TP53 functions, such as apoptosis, cell cycle regulation, DNA repair and cell metabolism [9]. As a critical tumour suppressor, TP53 mutations have been identified in half of adult cancer patients [10]. Mutated TP53 induces tumour angiogenesis and promotes tumour metastasis [11, 12]. TP53 mutation is a poor prognostic factor for adult tumour patients [13]. Approximately 50% of relapsed adult tumours are correlated with TP53 loss of function [14, 15]. Restoration of mutant TP53 to a normal state represents a potential therapeutic concept for the treatment of adult tumours [16]. However, TP53 is rarely mutated in paediatric neuroblastoma [17, 18], and the functions of TP53 in paediatric neuroblastoma are largely unclear.
In addition to genetic mutations, TP53 may be involved in paediatric neuroblastoma through other mechanisms. Some variants of TP53 are associated with susceptibility to neuroblastoma [19]. TP53 protein frequently accumulates in neuroblastoma cells [20]. Moreover, TP53 is a target of MYCN. MYCN can directly bind to the TP53 promoter regions and upregulate TP53 mRNA expression [21]. Loss of TP53 function induces radioresistance in neuroblastoma by regulating metabolism [22]. In addition, the TP53 partner gene MDM2 is also a MYCN transcriptional target and is implicated in neuroblastoma pathogenesis [23, 24]. MDM2 overexpression maintains MYCN stabilization and translation in paediatric neuroblastoma cells [24] and represents an unfavourable prognostic factor of neuroblastoma [25]. Inhibition of MDM2 suppresses the progression of MYCN-dependent neuroblastoma [26]. MDM2-TP53 antagonists represent potential therapeutic drugs for paediatric neuroblastoma [27, 28]. Except for MDM2, TP53 is correlated with multiple other genes [29, 30]. However, in paediatric neuroblastoma, the prognosis of TP53 and its associated genes is still unclear.
Using the public paediatric neuroblastoma cohorts in Therapeutically Applicable Research to Generate Effective Treatments (TARGET), European Bioinformatics Institute (EMBL-EBI) and Gene Expression Omnibus (GEO) datasets, we analysed the prognosis of TP53 and TP53-associated genes in neuroblastoma. Our results suggested that TP53 and its associated genes CCNE1, CDK2, CHEK2, and SESN1 were significantly associated with the clinical event-free survival and overall survival of paediatric neuroblastoma. Combinations of CCNE1 expression and SESN1 expression with age at diagnosis achieved a better prognosis of neuroblastoma. Finally, we showed that a nomogram risk model based on age, TP53, CCNE1, CDK2, CHEK2 and SESN1 expression could predict the overall survival of paediatric neuroblastoma with high specificity and sensitivity.
Results
Higher TP53 mRNA expression is associated with worse clinical outcomes of paediatric neuroblastoma
We collected public paediatric neuroblastoma cohorts and designed a study process to determine the prognostic effects of TP53 and TP53-associated genes in paediatric neuroblastoma (Supplementary Fig. 1). First, the prognostic value of TP53 expression, the TP53 pathway and TP53-associated genes was determined using the TARGET, EMBL-EBI and GEO datasets. The independent prognostic factors in paediatric neuroblastoma were also determined. Finally, we also constructed a nomogram model to predict the overall survival of paediatric neuroblastoma based on age and TP53-associated genes.
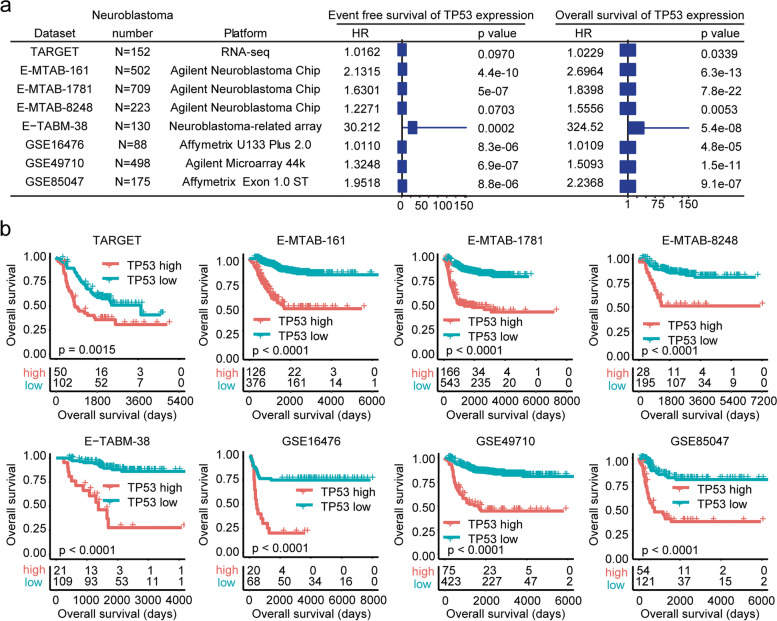
In total, 152 paediatric neuroblastoma patients from the TARGET dataset along with 2325 paediatric neuroblastoma patients from the EMBL-EBI and GEO datasets, including the E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets, were collected (Fig. 1a). The number of paediatric neuroblastoma patients in each dataset and the gene microarray platform of each dataset are shown in Fig. 1a.
Fig. 1.
Higher TP53 mRNA expression is associated with worse clinical outcomes of paediatric neuroblastoma. a Forest plots showing the associations of TP53 mRNA expression with neuroblastoma event-free survival and overall survival in the TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets. The number of paediatric neuroblastoma patients in each dataset and the gene microarray platform of each dataset are shown. b Different overall survival of paediatric neuroblastoma patients with higher or lower TP53 expression in the TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets
First, the prognostic effects of TP53 mRNA expression in paediatric neuroblastoma were determined. Univariate Cox regression analysis suggested that TP53 expression was associated with the event-free survival of paediatric neuroblastoma in the E-MTAB-161, E-MTAB-1781, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets (Fig. 1a). However, in the TARGET and E-MTAB-8248 datasets, TP53 mRNA expression was not significantly associated with the event-free survival of paediatric neuroblastoma (Fig. 1a). Moreover, TP53 expression was associated with the overall survival of paediatric neuroblastoma in all eight independent datasets: TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 (Fig. 1a).
Furthermore, paediatric neuroblastoma patients in each dataset were divided into TP53 higher and lower subgroups based on the mRNA expression levels of TP53. The different clinical outcomes of TP53 higher and lower subgroups were determined by Kaplan–Meier survival analysis. Lower TP53 expression was associated with prolonged event-free survival and overall survival of paediatric neuroblastoma in the TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets (Fig. 1b and Supplementary Fig. 2). These results suggested that TP53 mRNA expression was a critical prognostic factor of paediatric neuroblastoma.
A higher enrichment score of the TP53 signalling pathway is associated with worse clinical outcomes of paediatric neuroblastoma
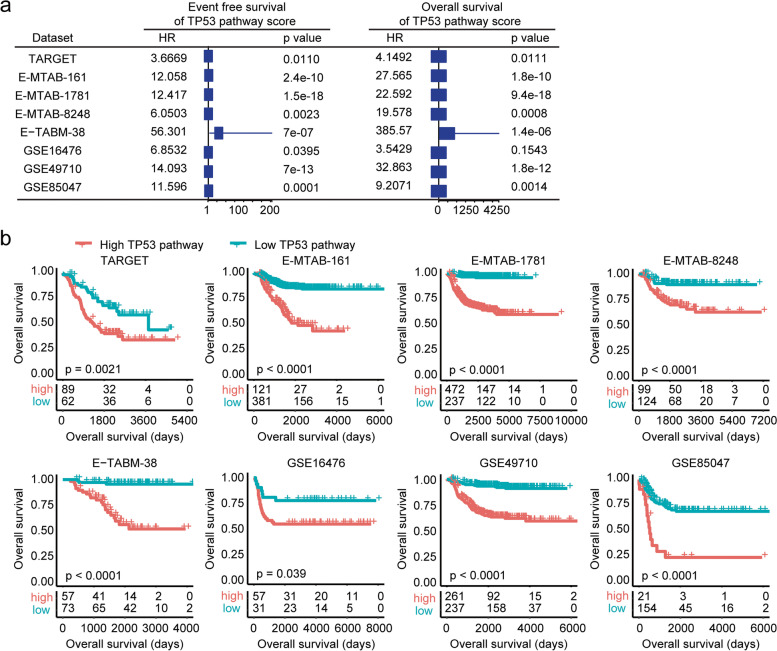
Using the ssGSEA, we determined the enrichment score of the TP53 signalling pathway in each paediatric neuroblastoma patient. We found that the enrichment score of the TP53 signalling pathway was associated with the event-free survival and overall survival of paediatric neuroblastoma in the TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets (Fig. 2a). Moreover, paediatric neuroblastoma patients with lower enrichment scores of the TP53 signalling pathway had prolonged event-free survival and overall survival in the eight independent datasets (Fig. 2b and Supplementary Fig. 3).
Fig. 2.
A higher enrichment score of the TP53 signalling pathway is associated with worse clinical outcomes of paediatric neuroblastoma. a Forest plots showing the associations of the TP53 signalling pathway with neuroblastoma event-free survival and overall survival in the TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets. b Different overall survival of paediatric neuroblastoma patients with higher TP53 signalling pathway enrichment scores or lower TP53 signalling pathway enrichment scores in the TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets
The TP53-associated genes CCNE1, CDK2, CHEK2 and SESN1 are correlated with the clinical outcomes of paediatric neuroblastoma
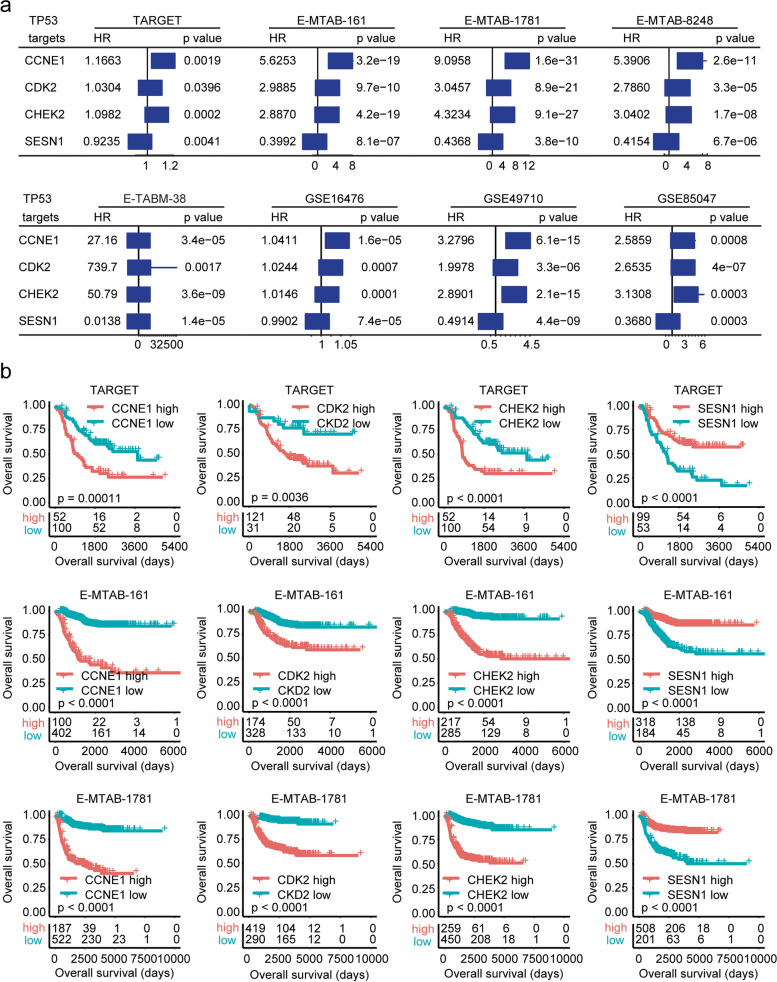
In the GSEA dataset, the TP53 signalling pathway lists 68 TP53-associated genes. We then determined the prognosis of the TP53-associated genes in the TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets. The prognosis of these 68 genes is shown in the supplementary table. Four TP53-associated genes, CCNE1, CDK2, CHEK2 and SESN1, were all detected and were associated with the overall survival of paediatric neuroblastoma in the eight independent datasets (Fig. 3a). However, the prognostic effects of CCNE1, CDK2, CHEK2 and SESN1 were different in paediatric neuroblastoma. Higher expression of CCNE1, CDK2 or CHEK2 was an unfavourable prognostic factor, while higher expression of SESN1 was a favourable prognostic factor in paediatric neuroblastoma (Fig. 3a).
Fig. 3.
TP53-associated genes CCNE1, CDK2, CHEK2 and SESN1 are correlated with the clinical outcomes of paediatric neuroblastoma. a Forest plots showing TP53-associated genes CCNE1, CDK2, CHEK2 and SESN1 in the prediction of the clinical event-free survival or overall survival of paediatric neuroblastoma in the TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets. b Kaplan–Meier curves showing the prognosis of the TP53-associated genes CCNE1, CDK2, CHEK2 and SESN1 in the TARGET, E-MTAB-161 and E-MTAB-1781 datasets
The Kaplan–Meier survival analysis further showed that overall survival was decreased in paediatric neuroblastoma patients with high CCNE1, CDK2 or CHEK2 expression (Fig. 3b and Supplementary Fig. 4). However, overall survival was increased in paediatric neuroblastoma patients with high SESN1 expression in the TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets (Fig. 3b and Supplementary Fig. 4).
TP53, CCNE1, CDK2, CHEK2 and SESN1 expression is correlated with MYCN amplification in neuroblastoma
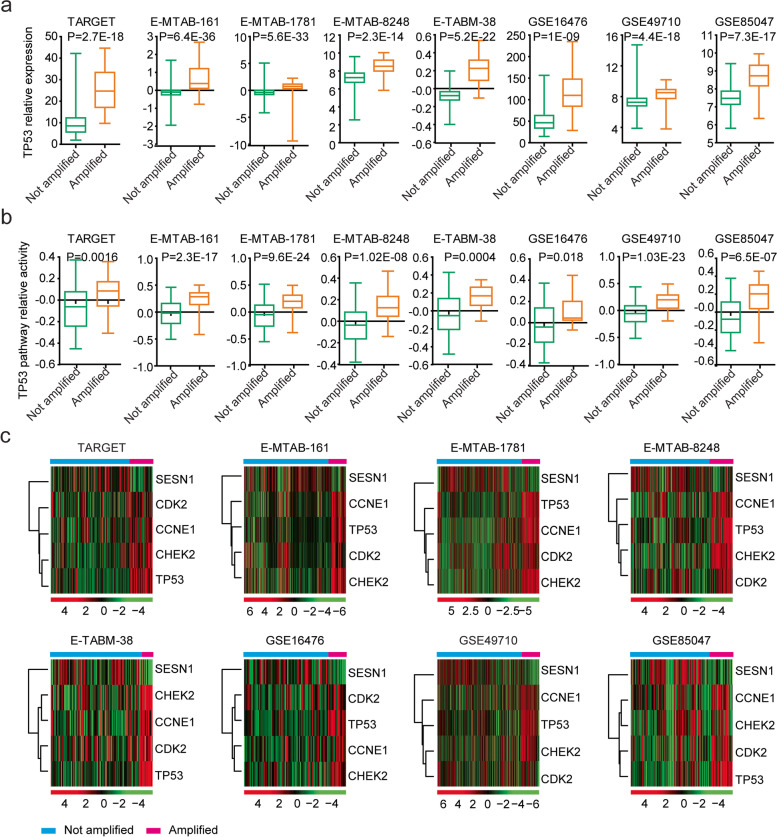
Previous results showed that TP53 was a direct target of MYCN in paediatric neuroblastoma [21]. Therefore, we determined the associations of TP53 expression with MYCN amplification in paediatric neuroblastoma cohorts. We found that TP53 mRNA levels were upregulated in MYCN-amplified paediatric neuroblastoma patients in the TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets (Fig. 4a). Moreover, the enrichment score of the TP53 signalling pathway was also associated with MYCN amplification in paediatric neuroblastoma patients (Fig. 4b).
Fig. 4.
TP53 and its associated genes are upregulated in MYCN-amplified paediatric neuroblastoma patients. a Box plots showing the relative TP53 expression levels in paediatric neuroblastoma patients with or without MYCN amplification. b Box plots show the relative TP53 signalling pathway enrichment score in paediatric neuroblastoma patients with or without MYCN amplification. c Heatmaps demonstrating the different expression levels of TP53, CCNE1, CDK2, CHEK2 and SESN1 in paediatric neuroblastoma patients with or without MYCN amplification in the TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets
The mRNA expression levels of CCNE1, CDK2, CHEK2 and SESN1 in paediatric neuroblastoma patients with or without MYCN amplification were also investigated. Similar to TP53, CCNE1, CDK2 and CHEK2 expression levels were all upregulated in paediatric neuroblastoma patients with MYCN amplification (Fig. 4c). In contrast, SESN1 expression was downregulated in paediatric neuroblastoma patients with MYCN amplification in the TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 independent paediatric neuroblastoma cohorts (Fig. 4c). The results were consistent with overexpression of CCNE1, CDK2 or CHEK2 being worse prognostic factors, while increased regulation of SESN1 was a better prognostic factor in paediatric neuroblastoma.
CCNE1 and SESN1 are independent prognostic markers of neuroblastoma
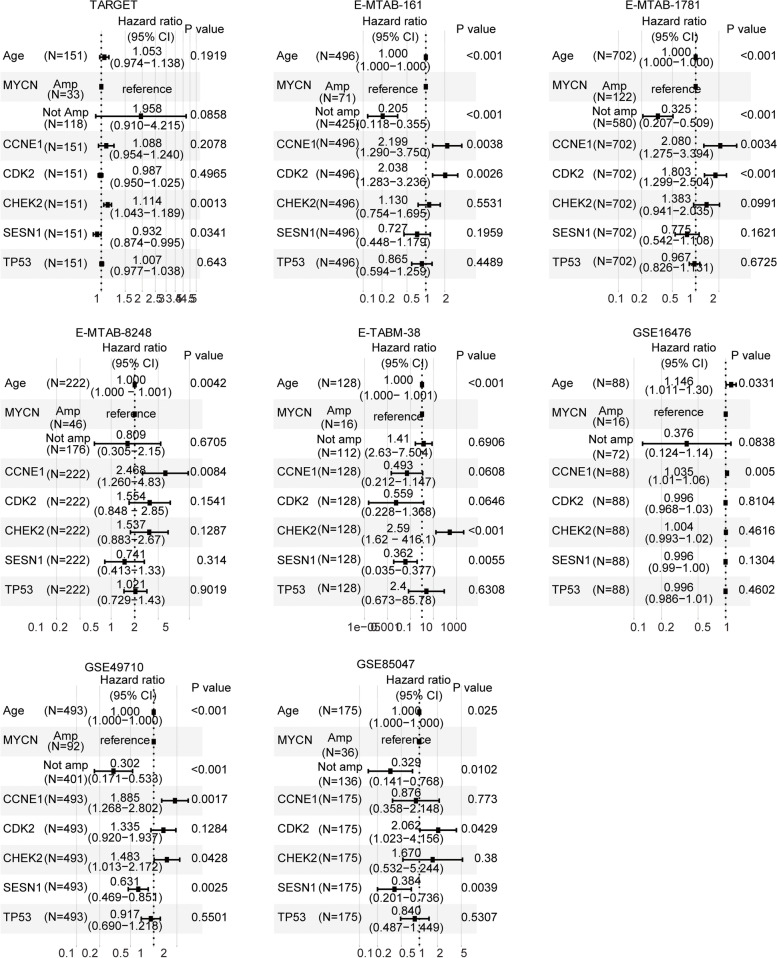
Age at diagnosis and MYCN amplification are critical determinants of the clinical outcomes of paediatric neuroblastoma [5]. We then assessed the associations of age at diagnosis, MYCN amplification, TP53, CCNE1, CDK2, CHEK2 and SESN1 in the prediction of the overall survival of neuroblastoma using a multivariate Cox regression assay. We found that age at diagnosis was an independent prognostic factor of paediatric neuroblastoma in the E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets (Fig. 5). MYCN amplification was also an independent prognostic factor in the E-MTAB-161, E-MTAB-1781, GSE49710 and GSE85047 datasets (Fig. 5). However, TP53 was not an independent prognostic factor of paediatric neuroblastoma in any of the eight independent paediatric neuroblastoma cohorts (Fig. 5).
Fig. 5.
CCNE1 and SESN1 are independent prognostic markers of neuroblastoma. Forest plots showing the associations of age, MYCN amplification, TP53, CCNE1, CDK2, CHEK2 and SESN1 expression with the clinical overall survival of paediatric neuroblastoma in the TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets
Moreover, CCNE1 was a prognostic factor of paediatric neuroblastoma in the E-MTAB-161, E-MTAB-1781, E-MTAB-8248, GSE16476 and GSE49710 datasets, independent of MYCN amplification and age at diagnosis (Fig. 5). CDK2 was also an independent prognostic factor of paediatric neuroblastoma in the E-MTAB-161, E-MTAB-1781 and GSE85047 datasets (Fig. 5). CHEK2 was an independent prognostic factor of paediatric neuroblastoma in the TARGET, E-TABM-38 and GSE49710 datasets (Fig. 5). SESN1 was an independent prognostic factor of paediatric neuroblastoma in the TARGET, E-TABM-38, GSE49710 and GSE85047 datasets (Fig. 5). Overall, age at diagnosis, MYCN amplification, CCNE1 and SESN1 were independent prognostic factors in at least four independent paediatric neuroblastoma cohorts.
Synergistic prognostic effects of CCNE1 with age at diagnosis in neuroblastoma
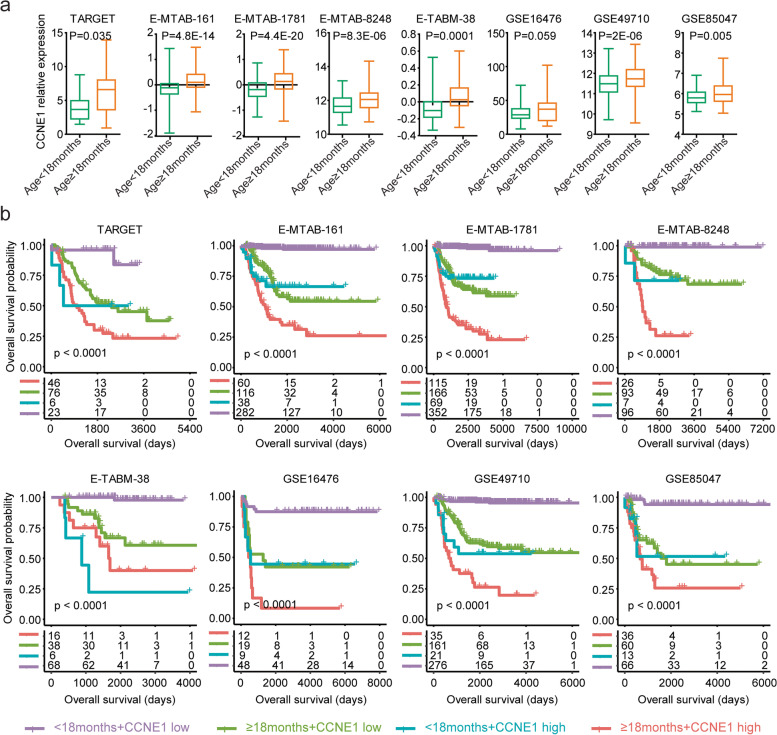
Since CCNE1 was a prognostic maker of neuroblastoma independent of age at diagnosis, the combinations of CCNE1 with age at diagnosis could achieve better prognostic effects in patients with paediatric neuroblastoma. First, in the TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE49710 and GSE85047 datasets, the expression levels of CCNE1 were higher in paediatric neuroblastoma patients with an age at diagnosis ≥ 18 months than in paediatric neuroblastoma patients with an age at diagnosis < 18 months (Fig. 6a).
Fig. 6.
Synergistic prognostic effects of CCNE1 with age at diagnosis in neuroblastoma. a Box plots showing the relative CCNE1 mRNA expression levels in paediatric neuroblastoma patients with an age at diagnosis ≥ 18 months or < 18 months in the TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets. b Paediatric neuroblastoma patients were divided into four subgroups based on the expression levels of CCNE1 and age of diagnosis. Different overall survival rates of paediatric neuroblastoma patients in each subgroup were determined
Furthermore, based on the expression levels of CCNE1 and age at diagnosis, paediatric neuroblastoma patients in each dataset were divided into four subgroups. Paediatric neuroblastoma patients with an age at diagnosis < 18 months and with CCNE1 lower expression levels had significantly better prognosis and mostly survived in the following timeframe (Fig. 6b). In contrast, paediatric neuroblastoma patients with an age at diagnosis ≥ 18 months and with higher CCNE1 expression levels had a significantly worse prognosis (Fig. 6b). Paediatric neuroblastoma patients with an age at diagnosis ≥ 18 months and with CCNE1 lower expression levels had medium overall survival risks (Fig. 6b). Paediatric neuroblastoma patients with an age at diagnosis < 18 months and with CCNE1 higher expression levels had the most diverse prognosis than other subgroups (Fig. 6b).
Synergistic prognostic effects of SESN1 with age at diagnosis in neuroblastoma
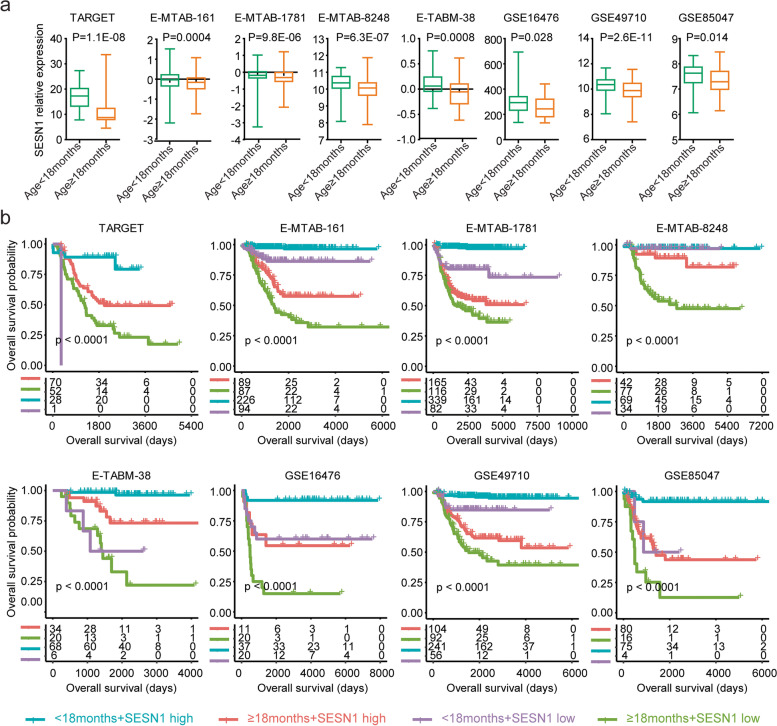
Consistent with the previous results that overexpression of SESN1 was a favourable prognostic factor of paediatric neuroblastoma, the expression levels of SESN1 were lower in paediatric neuroblastoma patients with an age at diagnosis ≥ 18 months in the TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets (Fig. 7a).
Fig. 7.
Synergistic prognostic effects of SESN1 with age at diagnosis in neuroblastoma. a Box plots show the relative SESN1 expression levels in paediatric neuroblastoma patients with an age at diagnosis ≥ 18 months or < 18 months. b Paediatric neuroblastoma patients were divided into four subgroups based on the expression levels of SESN1 and age at diagnosis. Different overall survival rates of paediatric neuroblastoma patients in each subgroup were determined
We also observed superior prognostic effects in the combinations of SESN1 with age at diagnosis in paediatric neuroblastoma. Paediatric neuroblastoma patients with an age at diagnosis < 18 months and SESN1 higher expression levels had a significantly better prognosis (Fig. 7b). In contrast, paediatric neuroblastoma patients with an age at diagnosis ≥ 18 months and SESN1 lower expression levels had a significantly worse prognosis (Fig. 7b). Paediatric neuroblastoma patients with an age at diagnosis ≥ 18 months and SESN1 higher expression levels had medium overall survival risks (Fig. 7b). Paediatric neuroblastoma patients with an age at diagnosis < 18 months and SESN1 lower expression levels had the most diverse prognosis than other subgroups (Fig. 7b).
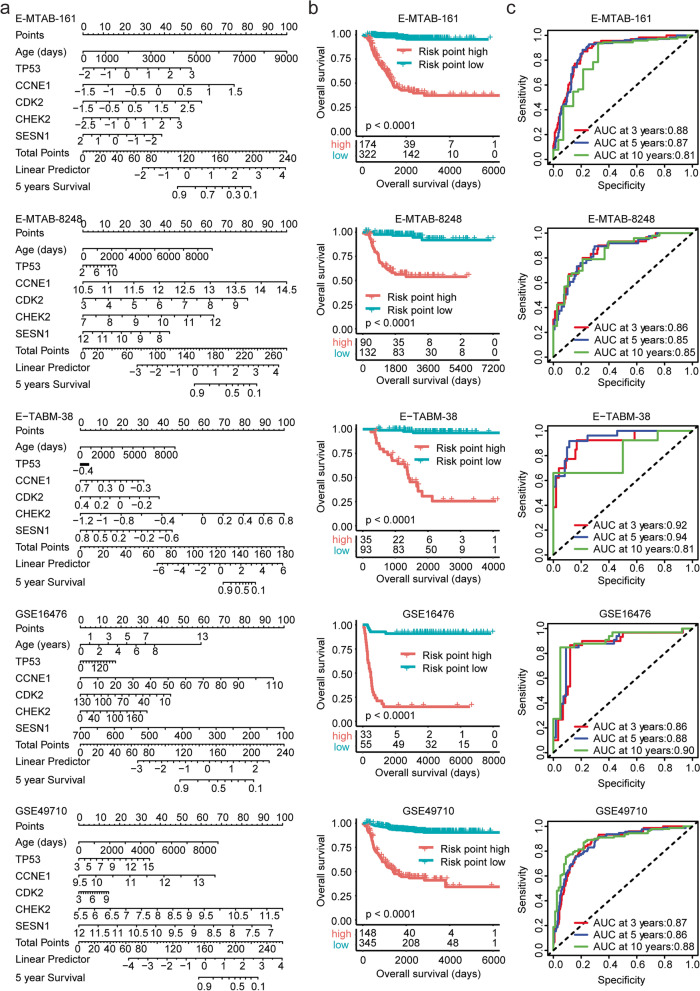
Construction of a nomogram model to predict the overall survival of paediatric neuroblastoma based on age and TP53, CCNE1, CDK2, CHEK2 and SESN1 expression
Our results suggested that TP53 and its associated genes CCNE1, CDK2, CHEK2 and SESN1 were all associated with the overall survival of paediatric neuroblastoma. We then constructed a nomogram model based on age at diagnosis and TP53, CCNE1, CDK2, CHEK2 and SESN1 expression features to predict the clinical overall survival of paediatric neuroblastoma (Fig. 8a). The risk point of each paediatric neuroblastoma patient in the E-MTAB-161, E-MTAB-8248, E-TABM-38, GSE16476 and GSE49710 datasets was obtained in the nomogram model. Paediatric neuroblastoma in the lower risk subgroup had significantly longer overall survival (Fig. 8b). Moreover, the ROC analysis in the E-MTAB-161, E-MTAB-8248, E-TABM-38, GSE16476 and GSE49710 datasets indicated that the nomogram model could predict the three-year, five-year or ten-year overall survival of paediatric neuroblastoma with high specificity and sensitivity (Fig. 8c).
Fig. 8.
Construction of a nomogram risk model of paediatric neuroblastoma based on age and TP53 CCNE1, CDK2, CHEK2 and SESN1 expression. a The nomogram model based on age and TP53 CCNE1, CDK2, CHEK2 and SESN1 expression levels in the E-MTAB-161, E-MTAB-8248, E-TABM-38, GSE16476 and GSE49710 datasets. b Kaplan–Meier curves showing the different overall survival rates of paediatric neuroblastoma patients between the low-risk subgroup and the high-risk subgroup. c The ROC curves showed the predictive specificity and sensitivity of the nomogram model. The AUC was calculated in the prediction of the three-year, five-year or ten-year overall survival of paediatric neuroblastoma
The nomogram model could predict the overall survival of MYCN nonamplified paediatric neuroblastoma
We previously showed that the expression levels of TP53, CCNE1, CDK2, CHEK2 and SESN1 were correlated with MYCN amplification in neuroblastoma patients. We further tested whether the TP53-based nomogram model could discriminate paediatric neuroblastoma cases with unfavourable outcomes among MYCN nonamplified patients. MYCN nonamplified patients from the TARGET, E-MTAB-161, E-MTAB-8248, E-TABM-38, GSE16476 and GSE49710 datasets were selected for investigation. MYCN nonamplified paediatric neuroblastoma patients with higher risk points had significantly shortened overall survival (Supplementary Fig. 5). Moreover, the ROC analysis in the TARGET, E-MTAB-161, E-MTAB-8248, E-TABM-38, GSE16476 and GSE49710 datasets showed that the nomogram model could predict the overall survival of MYCN nonamplified paediatric neuroblastoma (Supplementary Fig. 5).
Discussion
Loss of TP53 function is detected in 50% of adult tumour patients and confers high metastasis and poor clinical outcomes [31]. However, in paediatric neuroblastoma, we showed that overexpression of TP53 was associated with worse clinical event-free survival and overall survival. Moreover, a higher enrichment score of the TP53 signalling pathway was also associated with worse clinical outcomes of paediatric neuroblastoma. Furthermore, TP53-associated genes CCNE1, CDK2, CHEK2 and SESN1 were all prognostic markers of paediatric neuroblastoma. A nomogram model based on age at diagnosis, TP53, CCNE1, CDK2, CHEK2 and SESN1 expression features predicted the clinical overall survival of paediatric neuroblastoma. Our results highlighted the critical functions of TP53 and its associated genes in the regulation of the progression of paediatric neuroblastoma.
Previous results have shown that CCNE1 is a target of MYCN [32] and an unfavourable prognostic marker of neuroblastoma [33]. CCNE1 kinase inhibitors are potential drugs for MYCN-dependent neuroblastoma [34]. Our results were consistent with these observations that CCNE1 was upregulated in MYCN-amplified paediatric neuroblastoma patients, and CCNE1 overexpression was associated with worse clinical outcomes of paediatric neuroblastoma. Furthermore, our results showed that CCNE1 was also overexpressed in paediatric neuroblastoma patients with an age at diagnosis ≥ 18 months. CCNE1 was an independent prognostic factor, and combinations of CCNE1 expression with age at diagnosis achieved better prognostic effects in neuroblastoma. CDK2 was also suggested to be an unfavourable prognostic factor for paediatric neuroblastoma [35]. CDK2 inhibition suppressed the progression of MYCN-amplified neuroblastoma [36], and CDK2 antagonists represent potential therapeutic drugs in MYCN-driven neuroblastoma [37, 38]. However, the prognostic value of CHEK2 and SESN1 in paediatric neuroblastoma has never been reported.
As a TP53 target gene, SESN1 is associated with the DNA damage response and mTOR signalling pathway [39]. In contrast to CCNE1, CDK2 and CHEK2, SESN1 had opposite expression levels and prognostic effects in paediatric neuroblastoma. SESN1 was downregulated in MYCN-amplified paediatric neuroblastoma patients, and SESN1 overexpression was associated with better clinical outcomes of paediatric neuroblastoma. Furthermore, our results showed that SESN1 was a prognostic factor independent of age and MYCN amplification. SESN1 was also overexpressed in paediatric neuroblastoma patients with an age at diagnosis ≥ 18 months, and combinations of SESN1 expression with age at diagnosis achieved better prognostic effects in neuroblastoma. Our results highlighted the new prognostic roles of SESN1 in paediatric neuroblastoma.
Paediatric neuroblastoma is extremely heterogeneous. Integrated analysis from different cohorts based on different gene expression technologies may provide more robust results [6]. Integrated analysis of eight independent datasets, TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047, suggested that TP53 and its associated genes CCNE1, CDK2, CHEK2 and SESN1 were significantly correlated with the clinical outcomes of paediatric neuroblastoma. However, those conclusions were generated from published datasets and lacked validation using additional experiments. Therefore, the functions of TP53 and its associated genes should be further studied in neuroblastoma cells.
Conclusions
TP53, CCNE1, CDK2, CHEK2 and SESN1 were significantly associated with the clinical event-free survival and overall survival of paediatric neuroblastoma. Combinations of CCNE1 and SESN1 with age at diagnosis achieved superior prognosis of neuroblastoma. A nomogram risk model based on age, TP53, CCNE1, CDK2, CHEK2 and SESN1 expression predicted the overall survival of paediatric neuroblastoma with high specificity and sensitivity.
Methods
Data collection
TARGET paediatric pancancer studies were collected from St Jude Children’s Research Hospital (https://ocg.cancer.gov/) [40]. The E-MTAB-161 [41–43], E-MTAB-1781 [44], E-MTAB-8248 [45] and E-TABM-38 [46–48] datasets were downloaded from EMBL-EBI (https://www.ebi.ac.uk/arrayexpress/). The GSE16476 [49–51], GSE49710 [52] and GSE85047 [53] datasets were collected from the GEO website (www.ncbi.nlm.nih.gov/geo).
Univariate and multivariable Cox regression
Univariate and multivariable Cox regression analyses were carried out using the “survival” and “survminer” packages in R software. The forest plots were generated using the “forestplot” and “ggforest” packages in R software. The hazard ratio (HR) and p values were determined using Cox regression.
Kaplan–Meier survival analysis
Kaplan–Meier plots were created using the “survival” and “survminer” packages in R software. Paediatric neuroblastoma patients were divided into “high” or “low” subgroups based on the best cut-off points using the “survminer” package. P values were determined using the log-rank test.
Single-sample Gene Set Enrichment Analysis (ssGSEA)
The TP53 signalling pathway-associated gene set (c2.cp.kegg.v7.2.symbols) was downloaded from the GSEA website (www.broad.mit.edu/gsea/index.html). The enrichment scores of the TP53 signalling pathway were determined using the “GSVA” package in R software. “GSVA” in the ssGSEA was an unsupervised method evaluating the enrichment score of the TP53 pathway in each sample based on the expression of 68 TP53-associated genes.
Heatmap presentation
The expression of TP53 and its datasets CCNE1, CDK2, CHEK2 and SESN1 in neuroblastoma patients with or without MYCN amplification was clustered using the “pheatmap” package in R software.
Nomogram model
The nomogram models were generated using the “rms” and “ggplot2” packages in R software. The risk score was calculated using the “nomogramFormula” package.
TimeROC curves
The TimeROC curves were generated using the “timeROC” package in R software. The area under the ROC curve (AUC) was calculated by the “survival” package.
Statistical analysis
The box plots were generated from GraphPad Prism software. Statistical analysis was performed using the two-tailed paired Student’s t test. A P value less than 0.05 was chosen to indicate a statistically significant difference.
Supplementary Information
Additional file 1: Figure S1. Working process to analyze the prognosis of TP53 and its associated genes in paediatric neuroblastoma. Figure S2. Kaplan-Meier curves showed the different event free survival of paediatric neuroblastoma patients with TP53 higher expressions or lower expressions in TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets. Figure S3. Kaplan-Meier curves showed the different event free survival of paediatric neuroblastoma patients with higher TP53 signaling pathway enrichment score or lower TP53 signaling pathway enrichment score in TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets. Figure S4. Kaplan-Meier curves showed the prognosis of TP53 associated genes CCNE1, CDK2, CHEK2 and SESN1 in E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets. Figure S5. The nomogram model could predict the overall survival of MYCN non-amplified paediatric neuroblastoma patients. (a) Kaplan-Meier curves showed the different overall survival of MYCN nonamplified paediatric neuroblastoma patients between low-risk sub-group and high-risk sub-group in TARGT, E-MTAB-161, E-MTAB-8248, E-TABM-38, GSE16476 and GSE49710 datasets. (c) The ROC curves showed the prediction of the three years, five years or ten years overall survival of MYCN non-amplified paediatric neuroblastoma.
Acknowledgements
Not applicable.
Abbreviations
- TARGET
Therapeutically Applicable Research to Generate Effective Treatments
- EMBL-EBI
European Bioinformatics Institute
- GEO
Gene Expression Omnibus
- HR
Hazard ratio
- ssGSEA
Single sample Gene Set Enrichment Analysis
- AUC
Area under the ROC curve
Authors’ contributions
HW designed the study and wrote the manuscript. HW, XW and LX performed the data analysis. JZ supervised the work.
Funding
The present study was sponsored by the Fujian Provincial Health Technology Project (grant no: 2021GGA049).
Availability of data and materials
The datasets generated and/or analysed during the current study are available from the TARGET (https://ocg.cancer.gov/), EMBL-EBI (https://www.ebi.ac.uk/arrayexpress/) and the GEO websites (www.ncbi.nlm.nih.gov/geo).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Haiwei Wang and Xinrui Wang equally contributed to this work.
Contributor Information
Haiwei Wang, Email: hwwang@sibs.ac.cn.
Ji Zhang, Email: Zj11222@rjh.com.cn.
References
- 1.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma Lancet. 2007;369(9579):2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 2.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362(23):2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3(3):203–216. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 4.Cohn SL, Pearson AD, London WB, Monclair T, Ambros PF, Brodeur GM, et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol. 2009;27(2):289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell K, Gastier-Foster JM, Mann M, Naranjo AH, Van Ryn C, Bagatell R, et al. Association of MYCN copy number with clinical features, tumor biology, and outcomes in neuroblastoma: A report from the Children’s Oncology Group. Cancer. 2017;123(21):4224–4235. doi: 10.1002/cncr.30873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Wang X, Xu L, Zhang J, Cao H. Prognostic significance of MYCN related genes in pediatric neuroblastoma: a study based on TARGET and GEO datasets. BMC Pediatr. 2020;20(1):314. doi: 10.1186/s12887-020-02219-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang H, Wang X, Xu L, Zhang J, Cao H. Age related gene DST represents an independent prognostic factor for MYCN non-amplified neuroblastoma. BMC Pediatr. 2021;21(1):272. doi: 10.1186/s12887-021-02753-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park JR, Eggert A, Caron H. Neuroblastoma: biology, prognosis, and treatment. Hematol Oncol Clin North Am. 2010;24(1):65–86. doi: 10.1016/j.hoc.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8(4):275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 10.Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, et al. TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum Mutat. 2016;37(9):865–876. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- 11.Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137(1):87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 12.Weissmueller S, Manchado E, Saborowski M, Morris JPt, Wagenblast E, Davis CA, et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling. Cell. 2014;157(2):382–94. doi: 10.1016/j.cell.2014.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Wang X, Xu L, Lin Y, Zhang J, Cao H. Identification of genomic alterations and associated transcriptomic profiling reveal the prognostic significance of MMP14 and PKM2 in patients with pancreatic cancer. Aging (Albany NY) 2020;12(18):18676–18692. doi: 10.18632/aging.103958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yogev O, Barker K, Sikka A, Almeida GS, Hallsworth A, Smith LM, et al. p53 Loss in MYC-Driven Neuroblastoma Leads to Metabolic Adaptations Supporting Radioresistance. Cancer Res. 2016;76(10):3025–3035. doi: 10.1158/0008-5472.CAN-15-1939. [DOI] [PubMed] [Google Scholar]
- 15.Carr-Wilkinson J, O’Toole K, Wood KM, Challen CC, Baker AG, Board JR, et al. High Frequency of p53/MDM2/p14ARF Pathway Abnormalities in Relapsed Neuroblastoma. Clin Cancer Res. 2010;16(4):1108–1118. doi: 10.1158/1078-0432.CCR-09-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9(12):862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 17.Tweddle DA, Malcolm AJ, Bown N, Pearson AD, Lunec J. Evidence for the development of p53 mutations after cytotoxic therapy in a neuroblastoma cell line. Cancer Res. 2001;61(1):8–13. [PubMed] [Google Scholar]
- 18.Vogan K, Bernstein M, Leclerc JM, Brisson L, Brossard J, Brodeur GM, et al. Absence of p53 gene mutations in primary neuroblastomas. Cancer Res. 1993;53(21):5269–5273. [PubMed] [Google Scholar]
- 19.Diskin SJ, Capasso M, Diamond M, Oldridge DA, Conkrite K, Bosse KR, et al. Rare variants in TP53 and susceptibility to neuroblastoma. J Natl Cancer Inst. 2014;106(4):dju047. doi: 10.1093/jnci/dju047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tweddle DA, Pearson AD, Haber M, Norris MD, Xue C, Flemming C, et al. The p53 pathway and its inactivation in neuroblastoma. Cancer Lett. 2003;197(1–2):93–98. doi: 10.1016/s0304-3835(03)00088-0. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, Iraci N, Gherardi S, Gamble LD, Wood KM, Perini G, et al. p53 is a direct transcriptional target of MYCN in neuroblastoma. Cancer Res. 2010;70(4):1377–1388. doi: 10.1158/0008-5472.CAN-09-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slack A, Chen Z, Tonelli R, Pule M, Hunt L, Pession A, et al. The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proc Natl Acad Sci U S A. 2005;102(3):731–736. doi: 10.1073/pnas.0405495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slack A, Lozano G, Shohet JM. MDM2 as MYCN transcriptional target: implications for neuroblastoma pathogenesis. Cancer Lett. 2005;228(1–2):21–27. doi: 10.1016/j.canlet.2005.01.050. [DOI] [PubMed] [Google Scholar]
- 24.Gu L, Zhang H, He J, Li J, Huang M, Zhou M. MDM2 regulates MYCN mRNA stabilization and translation in human neuroblastoma cells. Oncogene. 2012;31(11):1342–1353. doi: 10.1038/onc.2011.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inomistova MV, Svergun NM, Khranovska NM, Skachkova OV, Gorbach OI, Klymnyuk GI. Prognostic significance of MDM2 gene expression in childhood neuroblastoma. Exp Oncol. 2015;37(2):111–115. doi: 10.31768/2312-8852.2015.37(2):111-115. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z, Lin Y, Barbieri E, Burlingame S, Hicks J, Ludwig A, et al. MDM2 deficiency suppresses MYCN-Driven neuroblastoma tumorigenesis in vivo. Neoplasia. 2009;11(8):753–762. doi: 10.1593/neo.09466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zafar A, Wang W, Liu G, Xian W, McKeon F, Zhou J, et al. Targeting the p53-MDM2 pathway for neuroblastoma therapy: Rays of hope. Cancer Lett. 2021;496:16–29. doi: 10.1016/j.canlet.2020.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Maerken T, Rihani A, Dreidax D, De Clercq S, Yigit N, Marine JC, et al. Functional analysis of the p53 pathway in neuroblastoma cells using the small-molecule MDM2 antagonist nutlin-3. Mol Cancer Ther. 2011;10(6):983–993. doi: 10.1158/1535-7163.MCT-10-1090. [DOI] [PubMed] [Google Scholar]
- 29.Mirza A, Wu Q, Wang L, McClanahan T, Bishop WR, Gheyas F, et al. Global transcriptional program of p53 target genes during the process of apoptosis and cell cycle progression. Oncogene. 2003;22(23):3645–3654. doi: 10.1038/sj.onc.1206477. [DOI] [PubMed] [Google Scholar]
- 30.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9(5):402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 31.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 32.Mao L, Ding J, Perdue A, Yang L, Zha Y, Ren M, et al. Cyclin E1 is a common target of BMI1 and MYCN and a prognostic marker for neuroblastoma progression. Oncogene. 2012;31(33):3785–3795. doi: 10.1038/onc.2011.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taran K, Owecka A, Kobos J. Prognostic importance of cyclin E1 expression in neuroblastic tumors in children. Pol J Pathol. 2013;64(2):149–152. doi: 10.5114/pjp.2013.36016. [DOI] [PubMed] [Google Scholar]
- 34.Dolman ME, Poon E, Ebus ME, den Hartog IJ, van Noesel CJ, Jamin Y, et al. Cyclin-Dependent Kinase Inhibitor AT7519 as a Potential Drug for MYCN-Dependent Neuroblastoma. Clin Cancer Res. 2015;21(22):5100–5109. doi: 10.1158/1078-0432.CCR-15-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bo L, Wei B, Wang Z, Kong D, Gao Z, Miao Z. Bioinformatics analysis of the CDK2 functions in neuroblastoma. Mol Med Rep. 2018;17(3):3951–3959. doi: 10.3892/mmr.2017.8368. [DOI] [PubMed] [Google Scholar]
- 36.Molenaar JJ, Ebus ME, Geerts D, Koster J, Lamers F, Valentijn LJ, et al. Inactivation of CDK2 is synthetically lethal to MYCN over-expressing cancer cells. Proc Natl Acad Sci U S A. 2009;106(31):12968–12973. doi: 10.1073/pnas.0901418106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poon E, Liang T, Jamin Y, Walz S, Kwok C, Hakkert A, et al. Orally bioavailable CDK9/2 inhibitor shows mechanism-based therapeutic potential in MYCN-driven neuroblastoma. J Clin Invest. 2020;130(11):5875–5892. doi: 10.1172/JCI134132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Z, Wang Z, Pang JC, Yu Y, Bieerkehazhi S, Lu J, et al. Multiple CDK inhibitor dinaciclib suppresses neuroblastoma growth via inhibiting CDK2 and CDK9 activity. Sci Rep. 2016;6:29090. doi: 10.1038/srep29090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134(3):451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma X, Liu Y, Liu Y, Alexandrov LB, Edmonson MN, Gawad C, et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature. 2018;555(7696):371–376. doi: 10.1038/nature25795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oberthuer A, Juraeva D, Li L, Kahlert Y, Westermann F, Eils R, et al. Comparison of performance of one-color and two-color gene-expression analyses in predicting clinical endpoints of neuroblastoma patients. Pharmacogenomics J. 2010;10(4):258–266. doi: 10.1038/tpj.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duffy DJ, Krstic A, Halasz M, Schwarzl T, Fey D, Iljin K, et al. Integrative omics reveals MYCN as a global suppressor of cellular signalling and enables network-based therapeutic target discovery in neuroblastoma. Oncotarget. 2015;6(41):43182–43201. doi: 10.18632/oncotarget.6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu L, Chu P, Lingeman R, McDaniel H, Kechichian S, Hickey RJ, et al. The Mechanism by Which MYCN Amplification Confers an Enhanced Sensitivity to a PCNA-Derived Cell Permeable Peptide in Neuroblastoma Cells. EBioMedicine. 2015;2(12):1923–1931. doi: 10.1016/j.ebiom.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oberthuer A, Juraeva D, Hero B, Volland R, Sterz C, Schmidt R, et al. Revised risk estimation and treatment stratification of low- and intermediate-risk neuroblastoma patients by integrating clinical and molecular prognostic markers. Clin Cancer Res. 2015;21(8):1904–1915. doi: 10.1158/1078-0432.CCR-14-0817. [DOI] [PubMed] [Google Scholar]
- 45.Koneru B, Lopez G, Farooqi A, Conkrite KL, Nguyen TH, Macha SJ, et al. Telomere Maintenance Mechanisms Define Clinical Outcome in High-Risk Neuroblastoma. Cancer Res. 2020;80(12):2663–2675. doi: 10.1158/0008-5472.CAN-19-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westermann F, Muth D, Benner A, Bauer T, Henrich KO, Oberthuer A, et al. Distinct transcriptional MYCN/c-MYC activities are associated with spontaneous regression or malignant progression in neuroblastomas. Genome Biol. 2008;9(10):R150. doi: 10.1186/gb-2008-9-10-r150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oberthuer A, Berthold F, Warnat P, Hero B, Kahlert Y, Spitz R, et al. Customized oligonucleotide microarray gene expression-based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol. 2006;24(31):5070–5078. doi: 10.1200/JCO.2006.06.1879. [DOI] [PubMed] [Google Scholar]
- 48.Hoene V, Fischer M, Ivanova A, Wallach T, Berthold F, Dame C. GATA factors in human neuroblastoma: distinctive expression patterns in clinical subtypes. Br J Cancer. 2009;101(8):1481–1489. doi: 10.1038/sj.bjc.6605276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483(7391):589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 50.Molenaar JJ, Domingo-Fernandez R, Ebus ME, Lindner S, Koster J, Drabek K, et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat Genet. 2012;44(11):1199–1206. doi: 10.1038/ng.2436. [DOI] [PubMed] [Google Scholar]
- 51.Lamers F, Schild L, Koster J, Speleman F, Ora I, Westerhout EM, et al. Identification of BIRC6 as a novel intervention target for neuroblastoma therapy. BMC Cancer. 2012;12:285. doi: 10.1186/1471-2407-12-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang C, Gong B, Bushel PR, Thierry-Mieg J, Thierry-Mieg D, Xu J, et al. The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat Biotechnol. 2014;32(9):926–932. doi: 10.1038/nbt.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rajbhandari P, Lopez G, Capdevila C, Salvatori B, Yu J, Rodriguez-Barrueco R, et al. Cross-Cohort Analysis Identifies a TEAD4-MYCN Positive Feedback Loop as the Core Regulatory Element of High-Risk Neuroblastoma. Cancer Discov. 2018;8(5):582–599. doi: 10.1158/2159-8290.CD-16-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Working process to analyze the prognosis of TP53 and its associated genes in paediatric neuroblastoma. Figure S2. Kaplan-Meier curves showed the different event free survival of paediatric neuroblastoma patients with TP53 higher expressions or lower expressions in TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets. Figure S3. Kaplan-Meier curves showed the different event free survival of paediatric neuroblastoma patients with higher TP53 signaling pathway enrichment score or lower TP53 signaling pathway enrichment score in TARGET, E-MTAB-161, E-MTAB-1781, E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets. Figure S4. Kaplan-Meier curves showed the prognosis of TP53 associated genes CCNE1, CDK2, CHEK2 and SESN1 in E-MTAB-8248, E-TABM-38, GSE16476, GSE49710 and GSE85047 datasets. Figure S5. The nomogram model could predict the overall survival of MYCN non-amplified paediatric neuroblastoma patients. (a) Kaplan-Meier curves showed the different overall survival of MYCN nonamplified paediatric neuroblastoma patients between low-risk sub-group and high-risk sub-group in TARGT, E-MTAB-161, E-MTAB-8248, E-TABM-38, GSE16476 and GSE49710 datasets. (c) The ROC curves showed the prediction of the three years, five years or ten years overall survival of MYCN non-amplified paediatric neuroblastoma.
Data Availability Statement
The datasets generated and/or analysed during the current study are available from the TARGET (https://ocg.cancer.gov/), EMBL-EBI (https://www.ebi.ac.uk/arrayexpress/) and the GEO websites (www.ncbi.nlm.nih.gov/geo).