Abstract
Teichoic acid-associated N-acetylglucosamine and rhamnose have been shown to serve as phage receptors in Listeria monocytogenes serotype 1/2a. We generated and characterized two single-copy Tn916ΔE mutants which were resistant to phage A118 and several other serotype 1/2a-specific phages. In one mutant the insertion was immediately upstream of the recently identified ptsHI locus, which encodes two proteins of the phosphoenolpyruvate-dependent carbohydrate uptake system, whereas in the other the insertion was immediately upstream of an operon whose most distal gene was clpC, involved in stress responses and virulence. Transduction experiments confirmed the association of the phage-resistant phenotype of these mutants with the transposon insertion. Phage A118 resistance of the mutants could be attributed to inability of the phage to adsorb onto the mutant cells, and biochemical analysis of cell wall composition showed that the teichoic acids of both mutants were deficient in N-acetylglucosamine. Rhamnose and other teichoic acid and cell wall components were not affected.
Structurally and antigenically, the anionic polymer teichoic acid is an important cell wall component of the gram-positive pathogen Listeria monocytogenes. It consists of a ribitol phosphate backbone with glycosidic substitutions that vary characteristically among the different serotypes of the pathogen (4, 17). The sugar substituents on teichoic acid are major antigenic determinants (7, 18). These substituents may be involved in different aspects of the interaction of the bacterium with its environment, including interactions with host cells during pathogenesis or with phages in the natural environment of the microorganism. Indeed, biochemical evidence suggests that N-acetylglucosamine and rhamnose substituents on teichoic acid serve as phage receptors (19).
Although a number of different serotypes of L. monocytogenes have been identified, most human infections are caused by strains of just three serotypes (serotypes 1/2a, 1/2b, and 4b) (3, 5, 15). The genetic and pathogenesis-related aspects of teichoic acid glycosylation in these clinically important serotypes are poorly understood. In the case of serotype 4b, such studies have been facilitated by the availability of highly specific monoclonal antibodies (9). Galactose and glucose serve as serotype-specific substituents on the teichoic acid of serotype 4b bacteria (4, 17), and the absence of either substituent markedly affects reactivity with the serotype-specific monoclonal antibodies (10, 12). Recently, we characterized a serogroup 4-specific gene, gtcA, which was essential for glycosylation of the teichoic acid of serotype 4b strains with galactose and glucose (12). In addition, a genomic region unique to serotype 4b, 4d, and 4e L. monocytogenes strains has been identified by using mutants lacking reactivity with the serotype-specific monoclonal antibodies (11). Similar genetic studies of teichoic acid glycosylation of serotype 1/2a or 1/2b strains, however, have not been performed. These strains lack galactose or glucose from teichoic acid and, instead, contain N-acetylglucosamine and rhamnose as sugar substituents (4, 7, 17). Since these substituents have been reported to function as phage receptors for serotype 1/2-specific phage A118 (19), we hypothesized that mutations that confer phage A118 resistance may be used to identify genes involved in the relevant glycosylation of teichoic acid. Here we describe construction of phage-resistant mutants of L. monocytogenes serotype 1/2a, identification of the insertionally inactivated loci, and the impact of the mutations on teichoic acid composition.
MATERIALS AND METHODS
Bacterial strains, phages, and growth conditions.
The bacterial and phage strains used in this work are listed in Table 1. Streptomycin-resistant strain 1/2a3 was previously designated Mack-strr (8). Bacteria were grown in brain heart infusion (BHI) broth or on tryptic soy agar supplemented with 0.7% yeast extract. When indicated, NaCl was added to a final concentration of 5% (wt/vol). All phages used in this work, including transducing phage LMUP35, have been shown to react with serotype 1/2a strains but not with serotype 4b strains (5a). The phages were propagated in strain 1/2a3. Infection assays and PFU determinations were performed as described previously (20). When appropriate, the antibiotics ampicillin (100 μg/ml), streptomycin (1,200 μg/ml), and erythromycin (10 μg/ml) were used.
TABLE 1.
Bacterial and phage strains used in this study
| Phage or strain | Remarks | Source or reference |
|---|---|---|
| Phages | ||
| A118, A006, A602, A620 | M. Loessner | |
| LMUP 11, LMUP 31a, LMUP 32a | Silage | D. A. Hodgson |
| LMUP 32b, LMUP 33, LMUP 35 | Silage | D. A. Hodgson |
| LMUP 322, LMUP 41, LMUP 42 | Silage | D. A. Hodgson |
| LMUP 61a, LMUP 61b | Silage | D. A. Hodgson |
| EF 1, EF 2, EF 3, EF 4 | Sewage effluent | |
| CUSI153/95, DP857 | Lysogens | D. A. Hodgson |
| L. monocytogenes strains | ||
| Mack (= SLCC 5764) | 8 | |
| 10403S | D. A. Portnoy | |
| 1/2a3 | Streptomycin-resistant derivative of Mack | 8 |
| HLT2 | A118-resistant mutant | This study |
| HLT8 | A118-resistant mutant | This study |
| HLT18 | A118-resistant mutant | This study |
| HLT22 | A118-resistant mutant | This study |
| HLT2/1 | HLT2 transductant | This study |
| HLT8/2 | HLT8 transductant | This study |
| E. coli strains | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 hsdR17 (rK− mK+) supE44 thi-1 gyrA relA1 | X. F. Gao |
| HLT2-SPCR | DH5α with HLT2 SSP-PCR product cloned into pCR1000 | This study |
| HLT8-SPCR | DH5α with HLT8 SSP-PCR product cloned into pCR1000 | This study |
Identification of phage-resistant mutants.
Tn916ΔE was mobilized from Enterococcus faecalis RH110 to strain 1/2a3 by filter membrane matings as described previously (8). Four independent mutant banks were constructed. A sample (5 μl) from each mutant bank was added to 10 ml of fresh BHI broth containing streptomycin and erythromycin and incubated at 37°C overnight to eliminate the donor cells and allow the Listeria transconjugants to grow. After five serial transfers in media containing streptomycin and erythromycin, 1 ml from each mutant bank was mixed with 1 ml (2.01 × 109 PFU) of phage A118 at 22°C for 30 min, and samples (20 and 50 μl) from each mixture were plated onto BHI agar supplemented with streptomycin and erythromycin and incubated at 37°C overnight. Several isolated colonies on each plate were selected for further screening for resistance to A118, and 22 phage-resistant derivatives (HLT1 to HLT22) were identified. Mutants HLT2, HLT8, and HLT18 were derived from three separate mutant banks. Mutant banks of strain 10403S were constructed by using Tn917-LTV3 (1) and were kindly provided by D. A. Portnoy (University of California, Berkeley). Mutants of 10403S were identified as described above, and each mutant bank was screened separately.
Generation of transductants.
To generate transductants from the Tn916ΔE-carrying phage-resistant mutants, 500 μl of an overnight culture of the mutant was mixed with 500 μl of transducing phage LMUP35, and the mixture was incubated at room temperature for 30 min. Phage lysates were prepared as described previously (20) and were filtered (pore size, 0.22 μm). One milliliter of the phage lysate from each mutant was mixed with 1 ml of an overnight culture of wild-type parent strain 1/2a3. The phage-bacterium mixture was incubated at 22°C for 30 min and plated onto BHI agar plates containing erythromycin and sodium citrate (10 mM). Putative transductant colonies were obtained following 48 h of incubation at 35°C.
DNA manipulations and analyses.
Genomic DNA was extracted from L. monocytogenes and Tn916ΔE copy number was determined as described previously (11). Single specific primer PCR (SSP-PCR) (16) was used to amplify Tn916ΔE-flanking fragments as described previously (11); the primers used were primer OTL (5′CGGAATTCCGTGAAGTATCTTCCTACAG3′), which was derived from one of the termini of the transposon, and primer M13F (Pharmacia). The PCR fragments were purified (GeneClean) and cloned into pCR1000 (TA cloning; Invitrogen). Plasmids were purified with Wizard miniprep columns (Promega). Southern blotting was performed as described previously (11) by using DNA probes labeled with digoxigenin (Genius kit; Boehringer Mannheim). Sequences of the cloned fragments were determined and analyzed as described previously (12).
For PCR amplification within the transposon-targeted region of HLT2 we used primers HLT2F (5′GCGAAATAAGGGTGCTCTACA3′) and HLT2R (5′ACGTGAACGCCGTCTTTAGA3′). Primer HLT2F was designed on the basis of the SSP-PCR product sequence, whereas HLT2R was derived from the known sequence of ptsI (accession no. AFO30824). For PCR amplification in the transposon-targeted region of HLT8 we used transposon-terminal primer OTR (5′ACTTATCACACTTTATCAAGGTCA3′) and primer HLT8R (5′TATCTCGTCCTGCTTCTCTT3′), which was derived from the known sequence of ORF3 in the clpC locus (accession no. U40604). For PCR we used Taq polymerase (Promega) and previously described conditions (11).
Nucleotide sequence accession numbers.
Nucleotide sequences of the transposon-flanking regions in HLT2 and HLT8 have been deposited in the GenBank database under accession no. AF160962 and AF160963, respectively.
RESULTS
Generation of L. monocytogenes mutants resistant to serogroup 1/2-specific phages.
Screening of four independent Tn916ΔE mutant banks of strain 1/2a3 and nine independent Tn917-LTV3 mutant banks of strain 10403S for resistance to serogroup 1/2-specific phage A118 led to identification of several resistant mutants. To determine whether the mutants were resistant to other serotype 1/2-specific phages as well, we screened six A118-resistant mutants of 1/2a3 and 18 resistant mutants of 10403S (two from each bank) with a panel of 20 additional phages which readily infected the parental strains, as described above. The 1/2a3-derived phage A118-resistant mutants formed plaques only when they were infected by phage LMPU35 (a transducing phage) and phage EGD857 but were resistant to the remaining 18 phages. The 10403S-derived phage A118-resistant mutants formed plaques only when they were infected by phage EGD857 and were resistant to all other phages, including LMPU35. These results suggested that mutants identified on the basis of their resistance to phage A118 were also resistant to several other phages. The reasons for the differential sensitivity of the 1/2a3-derived and 10403S-derived mutants to LMUP35 are not known, but they likely involve strain-specific differences in expression of the phage receptor or other aspects of the LMUP35-bacterial host interaction.
Transduction of the phage resistance phenotype.
Southern blots obtained with digoxigenin-labeled pAM120 as Tn916 probe showed that Tn916ΔE mutants HLT2, HLT8, HLT18, and HLT22 (derived from strain 1/2a3) carried a single copy of Tn916ΔE (data not shown). To confirm that phage resistance was associated with insertion of the transposon, we employed transducing phage LMUP35. LMUP35-mediated transduction of the erythromycin resistance marker of Tn916ΔE was accomplished with mutants HLT2, HLT8, HLT18, and HLT22, and the resulting transductants were screened for A118 resistance. The putative transductants of HLT2, HLT8, and HLT18 were all resistant to A118, suggesting that the phage resistance phenotype of these mutants was indeed associated with the transposon insertion. In contrast, all of the putative transductants of HLT22 that were examined proved to be sensitive to A118, suggesting that the phenotype of this mutant was not associated with the transposon insertion and was instead due to an unidentified spontaneous mutation. Mutant HLT22 was not studied further.
Unfortunately, transduction could not be implemented successfully with any of the 10403S-derived mutants, since, as mentioned above, these mutants were resistant to phage LMPU35 and no other transducing phages that could infect these strains were available. In the absence of sufficient evidence that the transposon insertion was indeed associated with the phage-resistant phenotype, the 10403S mutants were not studied further.
Localization of the transposon insertions of HLT2 and HLT8 in the ptsHI and clpC genomic regions, respectively, of L. monocytogenes.
SSP-PCR was successfully used to amplify transposon-flanking fragments from mutants HLT2 and HLT8. SSP-PCR fragments could not be obtained readily from mutant HLT18, and the insertionally inactivated locus in this mutant was not characterized further in this study. The SSP-PCR fragments from HLT2 (ca. 0.2 kb) and HLT8 (ca. 2.3 kb) were cloned in pCR1000 and used as probes in Southern blots of DNA from the wild type, mutants, and transductant derivatives. These Southern blots confirmed that the appropriate fragment had been amplified from each mutant and that the transductants harbored the transposon in the same restriction fragment as the corresponding mutants.
In the case of HLT2, the transposon was localized in a ca. 5-kb EcoRI genomic fragment in the mutant and the transductant derived from this mutant, HLT2/1 (Fig. 1).
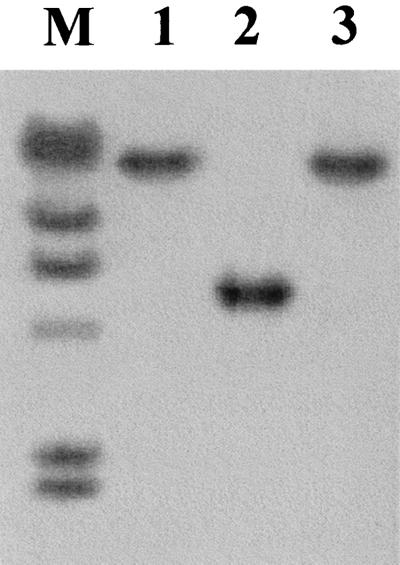
FIG. 1.
Southern blot detection of the genomic fragment harboring the Tn916ΔE insertion in HLT2. Genomic DNAs from HLT2 (lane 1), the wild-type parental strain (lane 2), and HLT2 transductant HTL2/1 (lane 3) were digested with EcoRI and hybridized with a digoxigenin-labeled SSP-PCR fragment flanking the transposon. Lane M contained lambda DNA digested with HindIII, which was labeled with digoxigenin and used as markers in the same blot (the fragment sizes are, from top to bottom, 23, 9.4, 6.5, 4.3, 2.3, and 2.0 kb).
In the case of HLT8, use of the SSP-PCR fragment as a probe revealed six hybridizing EcoRI fragments in parental strain 1/2a3. In HLT8 and in the phage-resistant HLT8 transductant HLT8/2, one of the closely spaced bands found in the wild type was missing, and a novel, large EcoRI fragment (ca. 21 kb) was present (Fig. 2). The hybridization patterns suggested that the probe contained sequences homologous to a repeated sequence in the genome of the bacteria (possibly a rRNA operon).
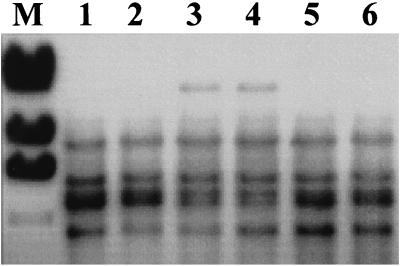
FIG. 2.
Southern blot detection of the genomic fragment harboring the Tn916ΔE insertion in HLT8. Genomic DNAs from the wild-type parental strain (lane 1), HLT8 (lane 3), transductant HTL8/2 (lane 4), and three other phage-resistant 1/2a3 mutants which harbored Tn916ΔE in loci other than the locus inactivated in HLT8 (lanes 2, 5, and 6) were digested with EcoRI and probed with the digoxigenin-labeled cloned SSP-PCR fragment flanking the transposon. Lane M contained the size markers described in the legend to Fig. 1 (only the largest four fragments are shown).
Sequence analysis of the cloned transposon-flanking fragments revealed that the insertions in HLT2 and HLT8 were either immediately upstream of (HLT2) or internal to (HLT8) sequences deposited in the database previously. In HLT2, the Tn916ΔE insertion was 63 nucleotides upstream of the promoter of the recently identified ptsH gene, which encodes the HPr protein of the phosphoenolpyruvate-dependent phosphotransferase system involved in carbohydrate uptake (2). In L. monocytogenes, ptsH was found to be cotranscribed with the downstream gene ptsI, which encodes enzyme I of the phosphotransferase system, and expression of ptsHI was induced by glucose (2). In HLT8, the insertion was at position 106 of the sequence whose accession number is U40604, 100 nucleotides upstream of the putative start codon of the first open reading frame (ORF1) in this sequence. ORF1 is the first member of a four-gene operon, the fourth gene of which encodes the ClpC ATPase, a protein involved in responses to several stress signals and in virulence (13, 14). Expression of the genes in this operon has been shown to be induced at 42°C (13). Although clpC has been extensively studied, the putative functions of the three preceding open reading frames of this operon remain unclear. Sequence motifs suggest that ORF1 and ORF2 may encode DNA-binding proteins, and the product of ORF3 has an arginine kinase-specific motif (13). The locations of the insertions in HLT2 and HLT8 and the genomic organizations of the corresponding regions are shown in Fig. 3.
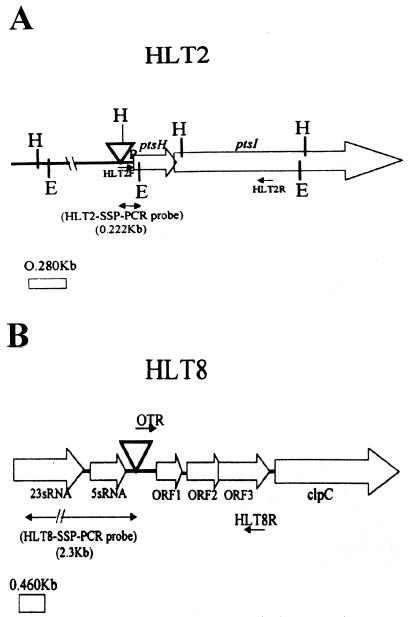
FIG. 3.
Genomic regions harboring the Tn916ΔE insertions in the phage-resistant mutants HLT2 (A) and HLT8 (B). The arrows indicate the direction of transcription, based on sequence analysis and on previously published descriptions of the loci (2, 13). Abbreviations: E, EcoRI; H, HindIII; ▿, Tn916ΔE. HLT2F, HLT2R, OTR, and HLT8R represent primers, as described in the text.
Sequence analysis showed that the region immediately upstream of the transposon insertion in HLT8 exhibited 97% identity with 5S and 23S ribosomal DNA sequences of Listeria ivanovii and L. monocytogenes (accession no. Y07639 and X64533, respectively). This suggests that in L. monocytogenes the clpC-containing operon is immediately downstream of an rrn operon, as mentioned by Rouquette et al. (13). This finding also agreed with our Southern blot hybridization data (Fig. 2) which suggested that the transposon-flanking fragment in HLT8 hybridized with a repeated sequence in the genome of L. monocytogenes.
The locations of the transposon insertions in HLT2 and HLT8 were confirmed by PCR by using primers located at the positions shown in Fig. 3. In the case of HLT2, the expected 1,243-bp PCR product was produced by using primer HLT2F, located between the transposon insertion and the beginning of the known ptsHI sequence, and primer HLT2R, located inside ptsI (Fig. 4). In the case of HLT8, the expected 2,044-bp PCR product was obtained by using primer OTR, located in the terminus of Tn916ΔE, and primer HLT8R, located at the 3′ end of ORF3 in the clpC operon (Fig. 4). These findings confirmed that the insertions were in the locations suggested by the analyses of the sequences of the transposon-flanking fragments.
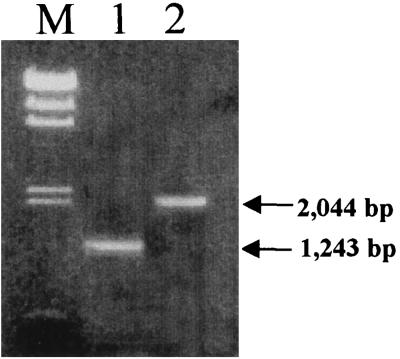
FIG. 4.
Confirmation of transposon insertion location. Primers from the regions indicated in Fig. 3 were used in PCR as described in the text. Lane M, lambda DNA digested with HindIII, used as molecular size markers (the fragment sizes are listed in the legend to Fig. 1; a 0.56-kb fragment is also included); lane 2, PCR product (1,243 bp) obtained by using primers HLT2F and HLT2R and genomic DNA of HLT2 as the template; lane 3, PCR product (2,044 bp) obtained by using primers OTR and HLT8R and genomic DNA of HLT8 as the template.
Inactivation of any one of at least three loci can confer phage A118 resistance.
Southern blot hybridization of eight other phage-resistant mutants (four mutants derived from 1/2a3 and four derived from 10403S) in which we used the HLT2 and HLT8 transposon-flanking fragments as probes showed that all other mutants, including HLT18, produced wild-type hybridization patterns with these probes (data not shown). Since our transduction data indicated that the phage resistance of HLT18 was associated with the single transposon insertion present in this mutant, we concluded that the genome of serotype 1/2a bacteria harbors at least three loci (i.e., the loci targeted in mutants HLT2, HLT8, and HLT18), any one of which, if insertionally inactivated, renders serotype 1/2a bacteria resistant to phage A118.
Absence of N-acetylglucosamine in the wall teichoic acid of the phage-resistant mutants.
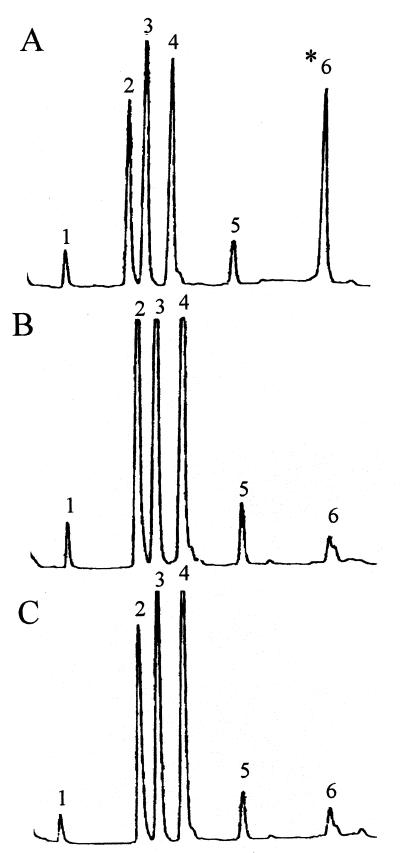
In view of the previously reported role of wall teichoic acid-associated N-acetylglucosamine and rhamnose as phage receptors in L. monocytogenes serotype 1/2a (19), we examined the cell wall composition of HLT2 and HLT8. The results of the biochemical analyses showed that the mutants had normal amounts of rhamnose in their teichoic acids but were markedly deficient in N-acetylglucosamine, which was present in only trace amounts (Fig. 5). The trace amounts of N-acetylglucosamine were probably derived from peptidoglycan, as indicated by the occurrence of minute quantities of muramic acid in the hydrofluoric acid hydrolysates.
FIG. 5.
Teichoic acid composition of wild-type parent strain 1/2a3 (A), mutant HLT2 (B), and mutant HLT8 (C). Teichoic acids were prepared and analyzed as described previously (4, 12). Peaks 1, glycerol; peak 2, anhydroribitol; peak 3, rhamnose; peak 4, ribitol; peak 5, glucose (trace levels); peak 6, glucosamine (indicated by asterisk). The small peak following peak 6 represents muramic acid (trace levels).
Phage A118 resistance is accompanied by failure of the phage to adsorb onto the mutant cells.
To determine whether the phage-resistant phenotype of the mutants was due to failure of the phage to adsorb and not to inhibition of other aspects of phage infection, we compared phage A118 adsorption onto mutant and wild-type cells. Significantly more phage particles were found in the supernatants when HLT2 and HLT8 were used as hosts than when wild-type strain 1/2a3 was used (Table 2). These results indicate that phage A118 failed to adsorb onto HLT2 or HLT8 cells and suggest that loss of a phage receptor accounted for the phage resistance of mutants HLT2 and HLT8.
TABLE 2.
Adsorption deficiency of phage-resistant mutants and their transductants
| Organism | Phage A118 adsorption (PFU ml−1)a |
|---|---|
| Parent strain 1/2a3 | 7 × 104 |
| Mutant HLT2 | 1.2 × 108 |
| Transductant HLT2/1 | >1.2 × 108 |
| Mutant HLT8 | 2.2 × 107 |
| Transductant HLT8/2 | 5.7 × 107 |
Adsorption of A118 was measured by determining the number of PFU remaining in the supernatant of a mixture containing A118 and the indicated strain, as described in the text. HLT2/1 and HLT8/2 are transductants obtained from HLT2 and HLT8, respectively. Similar results were obtained with two other transductants from each mutant.
Heat and salt sensitivity of mutant HLT8.
Mutants HLT2 and HLT8 grew normally at 4, 22, and 35°C in liquid or solid media. A higher temperature (42°C) and the presence of 5% NaCl at either 35 or at 4°C appeared to inhibit the growth of HLT8 (but not HLT2); the extent of inhibition was moderate in liquid cultures (data not shown) and more pronounced on solid media. Following 48 h of growth at 42°C, the average colony size of HLT8 was 0.75 mm, which was one-half the colony size of either the wild type or HLT2 (1.5 mm). HLT8 grew as well as HLT2 and the wild type at 22°C in the presence of 5% NaCl (data not shown). Following 36 h of growth at 35°C in the presence of 5% NaCl, however, the colony size of HLT8 was noticeably reduced (0.3 mm) compared to the colony size of the wild type or HLT2 (1.0 mm). A 75% reduction in the colony size of HLT8 compared to the wild-type or HLT2 colony size was observed during growth at 4°C in the presence of 5% NaCl, as determined over a 10-week period.
Colony formation at 42°C in the presence of 5% NaCl was too limited for both wild-type and mutant strains, and effective comparisons could not be made (data not shown).
DISCUSSION
The bacterial determinants which control phage sensitivity of L. monocytogenes are of interest ecologically and in terms of the application of phage typing schemes. Ecologically, phage sensitivity is likely to be crucial to the ability of the bacteria to persist and multiply in nature and in a food-processing environment. In previous work we found that strains representing one of the major epidemic clonal lineages of the pathogen had a unique cytosine modification of GATC sites in their genomes, which may have been associated with restriction activity against phages (20). Genetically, however, determinants of L. monocytogenes that may be involved either in phage restriction or in adsorption of phage have not been characterized yet.
Our findings suggest that phage A118 resistance of transposon-induced mutants HLT2 and HLT8 was associated with a lack of N-acetylglucosamine in the teichoic acid structure and with inability of the phage to adsorb onto the mutant bacteria. A previous report indicated that in L. monocytogenes serotype 1/2a teichoic acid-associated N-acetylglucosamine and rhamnose serve as phage receptors (19). Since HLT2 and HLT8 had normal rhamnose levels in their teichoic acids, we concluded that rhamnose is not sufficient as the only receptor, although it may still be essential as a component of the receptor complex. The role of rhamnose will be addressed more precisely by isolating mutants that lack this substituent in their teichoic acids.
Recently, we described gtcA, a novel, serotype-specific gene of L. monocytogenes serotype 4b; we found that mutations in this gene resulted in loss of galactose and marked reductions in the levels of glucose in the teichoic acid of this serotype (12). Similar studies with other serotypes have not been reported, and to our knowledge, HLT2 and HLT8 are the first genetically defined serotype 1/2a mutants that have been shown to be phage resistant and altered with respect to teichoic acid glycosylation. Interestingly, in both HLT2 and HLT8 the mutations were localized in previously identified genomic regions which were not known to be involved in teichoic acid glycosylation and phage adsorption.
In the case of HLT2, the transposon insertion upstream of the ptsHI promoter may have affected carbohydrate uptake and may have (directly or indirectly) inhibited incorporation of N-acetylglucosamine into teichoic acid, without apparently affecting incorporation of rhamnose. It is also possible, however, that depending on the transcriptional organization of the upstream region, the insertion may affect expression of a yet-to-be-identified gene(s) upstream of the insertion. Further characterization of the ptsHI locus and its upstream genomic region is needed to elucidate the impact of the HLT2 transposon insertion on incorporation of N-acetylglucosamine into the teichoic acid of L. monocytogenes serotype 1/2a. Previously, the ptsHI locus was characterized in a serotype 4b strain, and mutations in this locus were not described (2).
In the case of HLT8, the region upstream of the transposon insertion harbors an rrn operon transcribed in the same direction as the clpC operon. The transposon insertion was localized in the intergenic space between this rrn operon and the clpC operon and is expected, therefore, to have an effect on transcription of the latter. ClpC is a stress response protein which has recently been shown to be required for cell-to-cell spread of the bacteria during infection (13, 14). Therefore, it will be interesting to determine whether HLT8 has similar virulence-related deficiencies and to identify (by performing complementation and additional mutational studies) which of the genes in this operon is essential for incorporation of N-acetylglucosamine into the teichoic acid of serotype 1/2a strains. The reported difficulty in expressing some of these genes in Escherichia coli (14) may, however, complicate such genetic complementation studies. The high-temperature- and salt-sensitive phenotype of HLT8 (impaired growth in rich media at 42°C without NaCl, as well as at 4 and 35°C in the presence of 5% NaCl) suggests that this mutant is impaired with respect to ClpC-related responses. Interestingly, this phenotype was clearly evident in complex media (BHI), unlike the previously described clpC mutants, which grew normally in complex media and exhibited impaired growth under stress conditions only in synthetic media (13). Possible differences in the adaptive physiology of the parental strains used in these studies (LO28 versus 1/2a3) may account for the differences.
The localization of the HLT2 and HLT8 transposon insertions in regions related to important physiological and (in the case of HLT8, at least) pathogenesis functions suggests that the molecular basis underlying the expression of phage receptors and teichoic acid glycosylation may have coevolved with pathogenesis in L. monocytogenes serotype 1/2a. In addition to providing ligands that serve as phage receptors, proper teichoic acid glycosylation may be essential for surface anchoring and disposition of other surface components, including proteins, such as ActA and the surface-associated internalin proteins (internalins A and B), which have been implicated in the host cell-pathogen interaction (6). Additional molecular characterization of the mutants and the corresponding genomic regions will be needed to determine the involvement of these loci in phage-host cell interactions, teichoic acid glycosylation, and possibly other aspects relevant to the ecology and pathogenesis of L. monocytogenes.
ACKNOWLEDGMENTS
We are grateful to M. Loessner (Technical University of Munich, Munich, Germany) for providing phages A118, A006, A502, and A620 and to D. A. Portnoy (University of California, Berkeley) for providing strain 10403S and the 10403S mutant banks. We thank members of our laboratories for valuable feedback and support throughout this work.
This research was supported in part by U.S. Department of Agriculture National Research Initiative AAFS grant 95-37201-2031 and by ILSI-North America.
REFERENCES
- 1.Camilli A, Portnoy D A, Youngman P. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J Bacteriol. 1990;172:3738–3744. doi: 10.1128/jb.172.7.3738-3744.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christensen D P, Benson A K, Hutkins R W. Cloning and expression of the Listeria monocytogenes Scott A ptsH and ptsI genes, coding for Hpr and enzyme I, respectively, of the phosphotransferase system. Appl Environ Microbiol. 1998;64:3147–3152. doi: 10.1128/aem.64.9.3147-3152.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farber J M, Peterkin P I. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiedler F, Seger J, Schrettenbrunner A, Seeliger H P R. The biochemistry of murein and cell wall teichoic acids in the genus Listeria. Syst Appl Microbiol. 1984;5:360–376. [Google Scholar]
- 5.Gellin B G, Broome C V. Listeriosis. JAMA. 1989;261:1313–1320. [PubMed] [Google Scholar]
- 5a.Hodgson, D. A. Unpublished data.
- 6.Ireton K, Cossart P. Host-pathogen interactions during entry and actin-based movement of Listeria monocytogenes. Annu Rev Genet. 1997;31:113–138. doi: 10.1146/annurev.genet.31.1.113. [DOI] [PubMed] [Google Scholar]
- 7.Kamisango K, Fujii H, Okumura H, Saiki I, Araki Y, Yamamura Y, Azuma I. Structural and immunochemical studies of teichoic acid of Listeria monocytogenes. J Biochem. 1983;93:1401–1409. doi: 10.1093/oxfordjournals.jbchem.a134275. [DOI] [PubMed] [Google Scholar]
- 8.Kathariou S, Metz P, Hof H, Goebel W. Tn916-induced mutations in the hemolysin determinant affecting virulence of Listeria monocytogenes. J Bacteriol. 1987;169:1291–1297. doi: 10.1128/jb.169.3.1291-1297.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kathariou S, Mizumoto C, Allen R D, Fok A K, Benedict A A. Monoclonal antibodies with a high degree of specificity for Listeria monocytogenes serotype 4b. Appl Environ Microbiol. 1994;60:3548–3552. doi: 10.1128/aem.60.10.3548-3552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei X-H. Molecular studies of surface antigens specific for serotype 4b Listeria monocytogenes. Ph.D. thesis. Honolulu: University of Hawaii; 1997. [Google Scholar]
- 11.Lei X-H, Promadej N, Kathariou S. DNA fragments from regions involved in surface antigen expression specifically identify Listeria monocytogenes serovar 4 and a subset thereof: cluster IIB (serotypes 4b, 4d, and 4e) Appl Environ Microbiol. 1997;63:1077–1082. doi: 10.1128/aem.63.3.1077-1082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Promadej N, Fiedler F, Cossart P, Dramsi S, Kathariou S. Cell wall teichoic acid glycosylation in Listeria monocytogenes serotype 4b requires gtcA, a novel, serogroup-specific gene. J Bacteriol. 1999;181:418–425. doi: 10.1128/jb.181.2.418-425.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rouquette C, Ripio M-T, Pellegrini E, Bolla J-M, Tascon R I, Vazquez-Boland J-A, Berche P. Identification of a ClpC ATPase required for stress tolerance and in vivo survival of Listeria monocytogenes. Mol Microbiol. 1996;21:977–987. doi: 10.1046/j.1365-2958.1996.641432.x. [DOI] [PubMed] [Google Scholar]
- 14.Rouquette C, de Chastelier C, Nair S, Berche P. The ClpC ATPase of Listeria monocytogenes is a general stress protein required for virulence and promoting early bacterial escape from the phagosome of macrophages. Mol Microbiol. 1998;27:1235–1245. doi: 10.1046/j.1365-2958.1998.00775.x. [DOI] [PubMed] [Google Scholar]
- 15.Schuchat A, Swaminathan B, Broome C V. Epidemiology of human listeriosis. Clin Microbiol Rev. 1991;4:169–183. doi: 10.1128/cmr.4.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shyamala V, Ferro-Luzzi Ames G. Genome walking by single-specific primer polymerase chain reaction: SSP-PCR. Gene. 1989;84:1–8. doi: 10.1016/0378-1119(89)90132-7. [DOI] [PubMed] [Google Scholar]
- 17.Uchikawa K, Sekikawa J, Azuma I. Structural studies on teichoic acids in cell walls of several serotypes of Listeria monocytogenes. J Biochem. 1986;99:315–327. doi: 10.1093/oxfordjournals.jbchem.a135486. [DOI] [PubMed] [Google Scholar]
- 18.Ullman W W, Cameron J A. Immunochemistry of the cell walls of Listeria monocytogenes. J Bacteriol. 1969;98:486–493. doi: 10.1128/jb.98.2.486-493.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wendlinger G, Loessner M J, Scherer S. Bacteriophage receptors on Listeria monocytogenes cells are the N-acetylglucosamine and rhamnose substituents of teichoic acids or the peptidoglycan itself. Microbiology. 1996;142:985–992. doi: 10.1099/00221287-142-4-985. [DOI] [PubMed] [Google Scholar]
- 20.Zheng W, Kathariou S. Host-mediated modification of Sau3AI restriction in Listeria monocytogenes: prevalence in epidemic-associated strains. Appl Environ Microbiol. 1997;63:3085–3089. doi: 10.1128/aem.63.8.3085-3089.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]