Abstract
The human intestinal tract harbors a complex microbial ecosystem which plays a key role in nutrition and health. Although this microbiota has been studied in great detail by culture techniques, microscopic counts on human feces suggest that 60 to 80% of the observable bacteria cannot be cultivated. Using comparative analysis of cloned 16S rRNA gene (rDNA) sequences, we have investigated the bacterial diversity (both cultivated and noncultivated bacteria) within an adult-male fecal sample. The 284 clones obtained from 10-cycle PCR were classified into 82 molecular species (at least 98% similarity). Three phylogenetic groups contained 95% of the clones: the Bacteroides group, the Clostridium coccoides group, and the Clostridium leptum subgroup. The remaining clones were distributed among a variety of phylogenetic clusters. Only 24% of the molecular species recovered corresponded to described organisms (those whose sequences were available in public databases), and all of these were established members of the dominant human fecal flora (e.g., Bacteroides thetaiotaomicron, Fusobacterium prausnitzii, and Eubacterium rectale). However, the majority of generated rDNA sequences (76%) did not correspond to known organisms and clearly derived from hitherto unknown species within this human gut microflora.
There are up to 1014 total bacteria in the human intestinal tract, which is 10 to 20 times the total number of tissue cells in the entire body. The composition and activity of this flora have a profound influence on health and disease through their involvement in the nutrition, pathogenesis, and immune function of the host (10, 13). The gut microflora is an intricate ecosystem which has been investigated in great detail by anaerobic culture techniques (11, 27). The predominant genera in the large bowel are reported to be Bacteroides, Eubacterium, Clostridium, Ruminococcus, Peptococcus, Peptostreptococcus, Bifidobacterium, and Fusobacterium. Despite these intensive investigations, however, there is much concern that culture-based methods provide an incomplete picture of the diversity of the predominant organisms of the gut flora. Indeed, molecular genetic tools have indicated that 60 to 80% of the organisms in the total human microflora have not been cultivated (24). The phylogenetic analysis of bacterial 16S rRNA genes (rDNAs), amplified directly from complex communities, provides an efficient strategy for exploring the biodiversity of a particular biota. This PCR cloning strategy has been successfully applied to various ecosystems such as marine sediments (8, 17), marine picoplankton habitats (29), hot springs (33), soils (4), anaerobic digestor habitats (15), termite guts (28), the rumen (34), and the human colon (35). In order to derive a detailed phylogenetic inventory of the human gut flora, we have analyzed 284 cloned bacterial 16S rDNAs extracted from a fecal sample of a healthy 40-year-old male. This method has facilitated access to both cultivated and noncultivated microorganisms. Comparative sequence analysis revealed that only 24% of molecular species corresponded to known organisms, indicating that the vast majority of the dominant organisms of the human gut flora have so far eluded scientific description.
MATERIALS AND METHODS
Anaerobic culture.
A 1-g aliquot of a fecal sample (from a healthy 40-year-old man) was put immediately after collection into an anaerobic chamber (85% N2, 10% CO2, and 5% H2 gas phase) at 37°C and then serially 10-fold diluted in liquid casein yeast extract, which consists of Trypticase (2 g liter−1), NaCl (5 g liter−1), KH2PO4 (1 g liter−1), and yeast extract (2 g liter−1) (pH 7). Five 200-μl aliquots of a 10−8 dilution and three 100-μl aliquots of a 10−7 dilution were plated on brain heart infusion agar (Difco Laboratories, Detroit, Mich.) supplemented with hemin (5 mg liter−1) and yeast extract (5 g liter−1) and incubated anaerobically for 5 days at 37°C. The volunteer had not been prescribed antibiotics for at least 3 months prior to the study commencement and had no history of gastrointestinal disorder.
Microscopic counts.
An aliquot from the same fecal sample was serially diluted under anaerobic conditions. A 100-μl aliquot of a 10−6 dilution and 800 μl of 1× phosphate-buffered saline (PBS; 2.7 mM KCl, 138 mM NaCl, 1.5 mM KH2PO4, 20.4 mM NaH2PO4 [pH 7.2]) were adsorbed onto a 0.6-μm-pore-size polycarbonate filter (well diameter, 8 mm) with a membrane filter tower (Biocom, Les Ullis, France) under a pressure of 2 × 105 Pa, washed with 1 ml of PBS, and fixed for 1 h at 4°C with 4% (wt/vol) paraformaldehyde. The membrane was then washed with 1 ml of PBS and then with 300 μl of lysozyme buffer (50 mM EDTA, 100 mM Tris Cl [pH 8], 1% lysozyme). Four hundred microliters of lysozyme buffer was then added, and the mixture was incubated for 20 min at 42°C. The cells were washed with PBS and dehydrated by successive 3-min incubations in 50, 80, and 100% (vol/vol) ethanol. The membrane was incubated for 2 h at 46°C with 36 μl of hybridization buffer (0.9 M NaCl, 20 mM Tris Cl [pH 8], 15% [wt/vol] formamide, 0.01% [wt/vol] sodium dodecyl sulfate [SDS]) and 1.5 μl (138 ng) of a fluorescein-5-isothiocyanate (FITC)-labeled oligonucleotide probe which targets the domain Bacteria, S-D-Bact-0338-a-A-18 (5′ GCT GCC TCC CGT AGG AGT 3′ [2]), according to the nomenclature of the Oligonucleotide Probe Database (1). Cells were washed at 48°C with 0.2 M NaCl–20 mM Tris Cl (pH 8)–5 mM EDTA–0.01% (wt/vol) SDS. After the cells were dried, 40 μl of 0.001% (wt/vol) DAPI (4′,6-diamidino-2-phenylindole; Sigma, St. Louis, Mo.) was added and the mixture was incubated for 5 min in the dark. Glycerol-phosphate-buffered saline mounting fluid (Citifluor Ltd., Canterbury, United Kingdom) was added, and the membrane was analyzed. DAPI-positive and FITC-positive cells were visualized with an Optiphot2 epifluorescence microscope (Nikon, Champigny sur Marne, France) equipped with a diameter 25 series and Nikon EX 465-495 filter set (for FITC) or a Nikon EX 38/10 filter set (for DAPI). Positive cells were counted with the image analysis system Visiolab 1000 (Biocom) as previously detailed (3).
Extraction and purification of total DNA.
Immediately after collection, total DNA was extracted from a 125-mg fecal sample aliquot and purified as described by Godon et al. (15). The DNA concentration (225 ng μl−1) and its integrity (size, >21 kb) were estimated by agarose gel electrophoresis (with 1.5% [wt/vol] agarose–1× Tris-borate-EDTA–1 ng of ethidium bromide ml−1).
Amplification, cloning, and screening.
The forward primer S-D-Bact-0008-a-S-20 (5′ AGA GTT TGA TCC TGG CTC AG 3′ [18]), which targets the domain Bacteria, and the reverse primer S-*-Univ-1492-b-A-21 (5′ ACG GCT ACC TTG TTA CGA CTT 3′ [21]), which targets all living organisms, were used to amplify bacterial 16S rDNAs by PCR (9). Reaction tubes contained 225 ng (1 μl) of fecal DNA, 1.25 U of Taq DNA polymerase (AmpliTaq Gold; Perkin-Elmer Corporation, Foster City, Calif.), 1× AmpliTaq Gold reaction buffer, 2.5 mM MgCl2, 200 μM each deoxyribonucleotide triphosphate, and 0.44 μM each primer in a final volume of 50 μl. Initial DNA denaturation and enzyme activation steps were performed at 94°C for 10 min in a PTC 150 thermocycler (MJ Research, Inc., Watertown, Mass.), followed by 10 cycles of denaturation at 92°C for 1 min, annealing at 48°C for 1 min, and elongation at 72°C for 1 min 30 s, which was followed by a final elongation at 72°C for 15 min. PCR products were purified and concentrated with a QIAquick spin PCR purification kit (Qiagen, S.A., Courtaboeuf, France). Six reaction mixtures were pooled in order to increase the DNA yield, and amplicons were eluted with 30 μl of 10 mM Tris Cl (pH 8). Their concentration (22 ng μl−1) and size (1.5 kb) were estimated by electrophoresis as previously described. The purified products were ligated into the pUAg vector (R&D Systems Europe Ltd., Abingdon, United Kingdom) as specified by the manufacturer. Competent Escherichia coli DHI cells were transformed with ligation products by heat shock (40 s at 42°C). Recombinant cells were selected on Luria-Bertani medium with kanamycin (50 μg ml−1) and also with IPTG (isopropyl-β-d-thiogalactopyranoside) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Colonies which contained a plasmid with an insert could not produce β-galactosidase and degrade X-Gal; consequently, they were white. Possible redundancy among 80 clones was estimated by restriction fragment length polymorphism analysis. Two plasmid-targeted primers, the pUAg SEQ 5′ primer (5′ GCT ATG ACC ATG ATT ACG CCA AG 3′) and the pUAg SEQ 3′ primer (5′ TGT AAA ACG ACG GCC AGT GAA 3′) were used to amplify each insert by 35-cycle colony PCR. Products were digested with the restriction endonucleases HaeIII, KpnI, and Sau3A. Because of the multiplicity of patterns obtained, indicating less than 10% redundancy, all the clones were subjected to detailed sequence analysis.
Plasmid extraction and sequencing.
DNA preparations for sequencing were made with a QIAprep spin plasmid kit (Qiagen, S.A.) as specified by the manufacturer. Plasmids were eluted with 50 μl of water, and the products were stored at −80°C. Sizes and concentrations of plasmids were checked by 0.8% (wt/vol) agarose gel electrophoresis. Plasmidic inserts were sequenced with an automated ABI Prism 377 DNA sequencer (Applied Biosystems, Perkin-Elmer Corporation). Sequencing reactions were performed with a dRhodamine Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq DNA polymerase FS (fluorescent sequencing) (Perkin-Elmer Corporation) in a GeneAmp PCR system 9600 thermocycler (Perkin-Elmer Cetus Corporation). Two primers were used: S-D-Bact-0008-a-S-20 (5′ AGA GTT TGA TCC TGG CTC AG 3′), which targets the domain Bacteria, and S-*-Univ-0536-a-A-18 (5′ GWA TTA CCG CGG CKG CTG 3′ [14]), which is universal. The average length of fragments was 500 bases. For almost complete sequence determination, two other universal primers were used: S-*-Univ-0915-a-A-16 (5′ GCC CCC GYC AAT TCC T 3′ [36]) and S-*-Univ-1390-a-A-18 (5′ GAC GGG CGG TGT GTA CAA 3′ [36]).
Sequence analysis.
Sequences were edited to exclude the PCR primer binding sites. Newly determined sequences were compared to those available in public databases (Ribosomal Database Project [RDP] [25] and GenBank) in order to ascertain their closest relatives. A Genetics Computer Group software package (EGCG version 8.00; Peter Rice, The Sanger Centre, Cambridge, United Kingdom) was used for sequence analysis. Sequences from both the fecal sample and public databases were aligned with the PILEUP program. These alignments were checked and manually adjusted to reduce errors before the sequences were further used. Phylogenetic analysis was performed with the PHYLIP phylogeny inference package (version 3.572c; J. Felsenstein, University of Washington, Seattle). Distances were calculated with both the SIMILARITY and DNADIST (Kimura’s method option) programs. Trees based on the neighbor-joining algorithm were constructed with the NEIGHBOR program. Unrooted trees were drawn with the DRAWGRAM program. The stability of branches was assessed by the bootstrap method with the SEQBOOT (500 replicates), DNADIST, NEIGHBOR, and CONSENSE programs. Chimeric sequences were detected with the RDP CHECK_CHIMERA program (25) and by comparison of trees derived from the 5′ and 3′ ends of the generated sequences. The retrieval of the same sequence from two independent PCRs (this study and unpublished data) was also considered evidence of a nonchimeric sequence. An Operational Taxonomic Unit (OTU) or molecular species, as employed here, consisted of all sequences (fecal clones and reference strains) with less than 2% divergence from 400 to 450 aligned homologous nucleotides. This threshold was based on the conclusion of Godon and colleagues (15) that “the sequence divergence of clones belonging to the same OTU [is] generally low (between 2 and 0%)” and was also generally consistent with the results of the comparison of 16S rRNA homology and DNA-DNA reassociation values (30). Coverage was calculated by Good’s method (16), according to which the percentage of coverage was calculated with the formula [1 − (n/N)] × 100, where n is the number of molecular species represented by one clone (single-clone OTUs) and N is the total number of sequences.
Reference strains and cloned sequences used in phylogenetic analysis.
Reference bacteria used for phylogenetic analysis are listed below with their GenBank accession numbers or Deutsche Sammlung von Mikroorganismen numbers (for sequences present only in the RDP database): Acholeplasma palmae (L33734), Bacteroides caccae (X83951), Bacteroides distasonis (M86695, M25249), Bacteroides eggerthii (L16485), Bacteroides forsythus (X73962), Bacteroides fragilis (X83943), Bacteroides merdae (X83954), Bacteroides ovatus (X83952), Bacteroides putredinis (L16497), Bacteroides splanchnicus (L16496), Bacteroides stercoris (X83953), Bacteroides thetaiotaomicron (M58763), Bacteroides uniformis (L16486), Bacteroides vulgatus (M58762), Butyrivibrio crossotus (X89981), Butyrivibrio fibrisolvens (U41167), Chlamydia pneumoniae (Z49873), Chlamydia psittaci (AB001778), Clostridium aminophilum (L04165), Clostridium aminovalericum (X73436), Clostridium celerecrescens (X71848), Clostridium clostridiiforme (M59089), Clostridium coccoides (M59090), Clostridium herbivorans (L34418), Clostridium leptum (M59095), Clostridium neopropionicum (X76746), Clostridium nexile (X73443), Clostridium oroticum (M59109), Clostridium polysaccharolyticum (X77839), Clostridium populeti (X71853), Clostridium sporosphaeroides (X66002), Clostridium symbiosum (M59112), Clostridium viride (X81125), Clostridium xylanolyticum (X76739), Coprococcus eutactus (D14148), Cytophaga fermentans (M58766), Escherichia coli (Z83205), Eubacterium brachy (U13038, Z36272), Eubacterium contortum (L34615), Eubacterium desmolans (L34618), Eubacterium eligens (L34420), Eubacterium formicigenerans (L34619), Eubacterium fossor (L34620), Eubacterium hadrum (DSM 3319), Eubacterium halii (L34621), Eubacterium lentum (DSM 2243), Eubacterium plautii (DSM 4000), Eubacterium ramulus (L34623), Eubacterium rectale (L34627), Eubacterium siraeum (L34625), Eubacterium sp. strain OS (Octopus Spring) type K (L04711), Eubacterium ventriosum (L34421), Eubacterium xylanophilum (L34628), Fusobacterium prausnitzii (X85022), Gemmata obscuriglobus (X56305), Geothrix fermentans (U41563), Holophaga foetida (X77215), Isosphaera pallida (X64372), Lachnospira pectinoschiza (L14675), Nitrospira gracilis (L35503), Oscillatoria williamsii (U83259), Phascolarctobacterium faecium (X72867), Pirellula marina (X62912), Planctomyces limnophilus (X62911), Porphyromonas cangingivalis (X76259), Porphyromonas catoniae (X82823), Porphyromonas endodontalis (L16491), Porphyromonas gingivalis (L16492), Porphyromonas salivosa (L26103), Prevotella denticola (L16467), Prevotella heparinolytica (L16487), Prevotella oralis (L16480), Prevotella oris (L16474), Prevotella oulora (L16472), Prevotella pallens (Y13106), Prevotella ruminicola (L16482), Prevotella veroralis (L16473), Prosthecobacter fusiformis (U60015), PVB_OTU_9A (U15118), Rikenella microfusus (L16498), Roseburia cecicola (L14676), Ruminococcus bromii (L76600), Ruminococcus flavefaciens ATCC 49949 (Af030449), Ruminococcus flavefaciens NJ (Af030446), Ruminococcus gnavus (L76597), Ruminococcus hansenii (M59114), Ruminococcus obeum (L76601), Ruminococcus productus (L76595), Ruminococcus torques (L76604), soil DNA clone EA25 (U51864), soil DNA clone MC17 (X64381), soil DNA clone MC18 (X64374), soil DNA clone MC31 (X64380), Spiroplasma apis (M23937), Streptococcus parasanguinis (Af003933), Streptococcus salivarius (M58839), Termitobacter aceticus (Z49863), unidentified bacterium (UB) A03 (Af052408), UB A07 (Af052409), UB A09 (Af052410), UB A10 (Af052411), UB A11 (Af052412), UB A12 (Af052413), UB A13 (Af052414), UB A14 (Af052415), UB A19 (Af052416), UB A20 (Af052417), UB A21 (Af052418), UB A22 (Af052419), UB A27 (Af052420), UB A54 (Af052421), UB A57 (Af052422), UB A71 (Af052423), unidentified rumen bacterium (URB) 30-05 (Af018503), URB JW7 (Af018440), URB JW21 (Af018551), URB RC23 (Af001713), URB RFN7 (AB009217), and Verrucomicrobium spinosum (X90515).
Nucleotide sequences accession numbers.
One representative of each OTU (except some OTUs with a single representative) was almost fully sequenced and deposited in GenBank with the acronym adhufec for adult human feces (accession no. AF132232 to AF132286).
RESULTS
Microscopic counts.
The fecal specimen moisture content was 26%. According to DAPI staining, there were 2.72 × 1011 ± 0.10 × 1011 cells per g (wet weight) of feces in the sample (means ± standard deviations for the same sample from randomly chosen microscopic fields) or 10.6 × 1011 ± 0.4 × 1011 cells per g (dry weight) of feces in the sample. According to oligonucleotide probe hybridization (with a probe which targets the domain Bacteria), there were 7.1 × 1011 ± 0.2 × 1011 bacterial cells per g (dry weight) of feces. The number of cultivated bacteria in the anaerobic chamber, with brain heart infusion medium, was 0.56 × 1011 ± 0.05 × 1011 CFU per g (wet weight) of feces or 2.2 × 1011 ± 0.2 × 1011 CFU per g (dry weight). These results indicated that organisms which grew anaerobically on a nonselective medium corresponded to 21% of bacteria counted through DAPI staining and 32% of those counted by oligonucleotide probe hybridization.
Overall phylogenetic analysis.
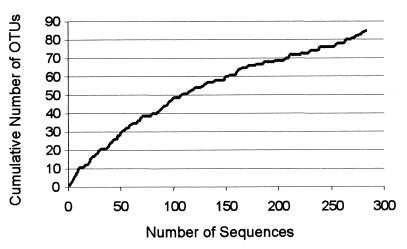
The average determined length of the rDNA sequences was 500 bases, and phylogenetic analysis was based on 400 to 450 aligned homologous nucleotides (E. coli positions 50 to 450 to 500). Phylogenetic trees based on partial sequences have the same topologies as those based on complete sequences: the established groups are identical, but some deep branches may slightly differ (23, 29). In this study, global phylogenetic inferences of the clones were aimed and partial sequences were well adapted (almost complete sequences were obtained and submitted to GenBank so that they could be used for further comparative analyses). Trees obtained with the SIMILARITY and NEIGHBOR programs were compared to those based on Kimura’s method and the NEIGHBOR program, and results were generally highly consistent. All clones were obtained by the same 10-cycle PCR with a bacterial primer (S-D-Bact-0008-a-S-20) and a universal primer (S-*-Univ-1492-b-A-21) by using the same ligation product but two transformations. The first transformation provided 120 clones, and the second transformation provided 400. Among the 284 clones analyzed, there were 82 distinct OTUs and the vast majority (95%) were distributed among three major monophyletic groups: the Bacteroides group, the Clostridium coccoides group, and the Clostridium leptum subgroup. In the three phylogenetic trees (Fig. 1 to 3) depicting these major phylogenetic lineages, each OTU has been represented by a single fecal clone. This clone was used as the reference sequence for calculating the phylogenetic distances with other sequences. The coverage (16) calculated for the 284 clones was 85%. In other words, the probability of the next cloned sequence falling in a novel OTU (not yet observed) was 15%. This value gives an estimation of how well the clones analyzed account for the biodiversity within the original sample, by the present methodology (i.e., with our PCR conditions and primer set). Another estimation (15) was obtained by plotting the cumulative number of OTUs as a function of the number of clones sequenced (Fig. 4). This second evaluation confirmed that this study nearly embraced the whole dominant biodiversity.
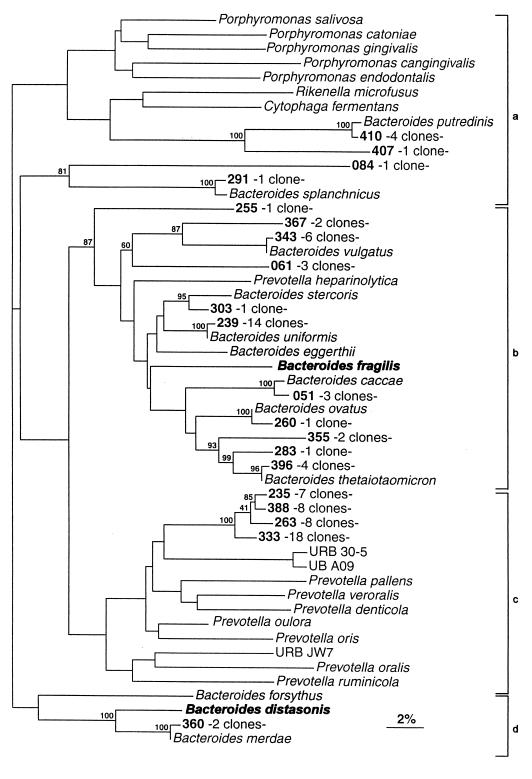
FIG. 1.
Phylogenetic tree derived from partial 16S rDNA sequence data for members of the Bacteroides group. Bar represents 2% sequence divergence. Designations of clones and key organisms used to name the subgroups are in boldface type. The tree was constructed with the SIMILARITY and NEIGHBOR programs. Bootstrap values are based on 500 replications. Vertical bars correspond to the Anaeroflexus assemblage (a) Bacteroides fragilis subgroup (b), Prevotella subgroup (c), and Bacteroides distasonis subgroup (d).
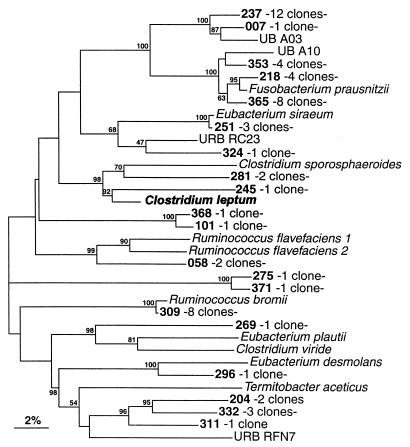
FIG. 3.
Phylogenetic tree derived from partial 16S rDNA sequence data for members of the Clostridium leptum subgroup. Bar represents 2% sequence divergence. Designations of clones and the key organism used to name the group are in boldface type. The tree was constructed with the SIMILARITY and NEIGHBOR programs. Bootstrap values are based on 500 replications.
FIG. 4.
Estimation of the biodiversity which was obtained by direct community analysis of a fecal sample. The cumulative number of OTUs is given as a function of the number of clones sequenced. Clones were randomly used.
Bacteroides group.
Phylogenetic distribution of the clones within the four subgroups of the Bacteroides group (RDP registration no. 2.7.1.1) are shown in Fig. 1. Aligned bases corresponding to E. coli positions 42 to 483 were used to construct this tree with the SIMILARITY and NEIGHBOR programs. Eighty-eight clones (20 OTUs) fell into the four subgroups. Seven clones were included in the Anaeroflexus assemblage (RDP registration no. 2.7.1.1.1), 2 clones were included in the Bacteroides distasonis subgroup (2.7.1.1.2, including Bacteroides merdae), 38 clones were included in the Bacteroides fragilis subgroup (2.7.1.1.3, including Bacteroides caccae and Bacteroides stercoris), and 41 clones were included in the Prevotella subgroup (2.7.1.1.4). The eight previously known OTUs were recovered within three of the four subgroups (the Prevotella subgroup being the exception). Among recognized organisms, the Bacteroides uniformis OTU was represented by 14 clones, the Bacteroides vulgatus OTU was represented by 6 clones, the Bacteroides putredinis and Bacteroides thetaiotaomicron OTUs were represented by 4 clones each, the Bacteroides caccae OTU was represented by 3 clones, the Bacteroides merdae OTU was represented by 2 clones, and the Bacteroides ovatus and the Bacteroides splanchnicus OTUs were represented by a single clone each. Thirteen OTUs (62%) had no closely related sequence (at least 98% similarity) in public databases and were derived from potentially new species. The 38 Prevotella-like clones (13% of total clone population and 43% of Bacteroides group clones) fell into four distinct groups. Sequence divergence ranged from 1.2 to 3.2% (GAP program with complete sequences), indicating that the four OTUs almost certainly derived from separate species. Interestingly, the four unknown OTUs were included in a robust cluster (global bootstrap value of 100%), which demonstrated that all these clones had a common ancestor within the Prevotella subgroup. From sequence divergence and tree topology considerations it was evident that the aforementioned unknown OTUs did not possess a specific affinity with any currently defined Prevotella species (sequence divergence values of >7.9% with recognized species). There were only seven one-clone OTUs within the Bacteroides group; the percentage of coverage in this group was hence high (91%). In other words, if we were to continue sequencing, 9 of 10 clones in this group would fall within an already delimited OTU.
Clostridium coccoides group.
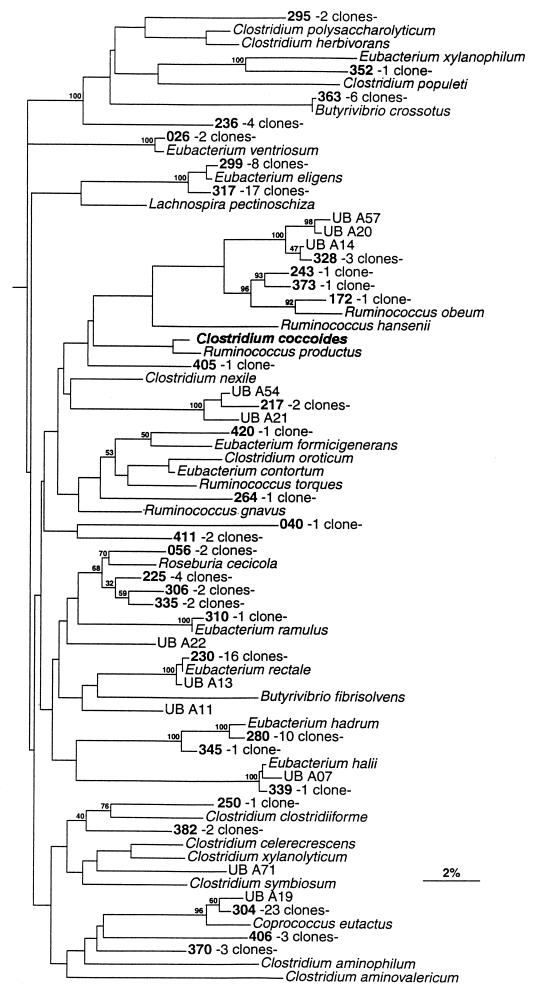
Figure 2 shows phylogenetic inferences among clones of the Clostridium coccoides group (RDP registration no. 2.16.4.1). This group corresponds to Clostridium rRNA subcluster XIVa (5). Aligned bases corresponding to E. coli positions 60 to 440 were used to construct this tree with the SIMILARITY and NEIGHBOR programs. There were 125 clones in this group, which were distributed within 31 OTUs, of which 24 (77%) were new. Only seven OTUs were previously known. The members of the Eubacterium eligens OTU were the most numerous, with 25 representatives. The Eubacterium rectale OTU was represented by 16 clones, the Eubacterium hadrum OTU was represented by 10 clones, the Butyrivibrio crossotus OTU was represented by 6 clones, and the Eubacterium ventriosum OTU was represented by 2 clones. The Eubacterium halii and the Eubacterium ramulus OTUs contained a single representative each. Although there were 14 OTUs with just a single clone, the coverage was high (89%) because 10 OTUs were composed of at least three clones. Four OTUs contained a high number of clones (at least 10) from the aforementioned Eubacterium eligens, Eubacterium rectale, and Eubacterium hadrum OTUs and OTU 304 (23 clones related to Coprococcus eutactus). Two distinct groups of clones were actually related to Eubacterium eligens. The first embraced eight clones and displayed approximately 0.4% sequence divergence from Eubacterium eligens. The second group, composed of 17 clones, exhibited 0.8% divergence from Eubacterium eligens. In addition, there was a 1.9% divergence between these two groups. Bootstrap values were high for each group, and the value was maximal (100%) for the whole cluster. In conclusion, these two clonal groups represented two distinct populations (sub-OTUs) within the Eubacterium eligens OTU.
FIG. 2.
Phylogenetic tree derived from partial 16S rDNA sequence data for members of the Clostridium coccoides group. Bar represents 2% sequence divergence. Designations of clones and the key organism used to name the group are in boldface type. The tree was constructed with the SIMILARITY and NEIGHBOR programs. Bootstrap values are based on 500 replications.
Clostridium leptum subgroup.
Figure 3 depicts the phylogenetic interrelationships of clones which belong to the Clostridium leptum subgroup (RDP registration no. 2.16.10.1.2). Termitobacter aceticus, URB RFN7, and URB RC23 were added to this group, which equates to Clostridium rRNA cluster IV (5). Aligned bases corresponding to E. coli positions 45 to 449 were used to construct the tree with the SIMILARITY and NEIGHBOR programs. The 57 clones related to this monophyletic group were classified in 20 OTUs. Only three OTUs were already known. Eight clones belonged to the Ruminococcus bromii OTU. A second OTU included Eubacterium siraeum and four fecal clones, whereas the remaining known OTU included Fusobacterium prausnitzii and four clones. The OTU exemplified by clone 218 showed 1.2% sequence divergence from Fusobacterium prausnitzii. In addition to the established Fusobacterium prausnitzii OTU, there were four other groups of clones closely related to this species. There were two distinct clusters: the first group included Fusobacterium prausnitzii and OTUs 218, 353, and 365 (intragroup sequence similarity, ≥95.7%), and the second group consisted of OTUs 007 and 237 (intragroup sequence similarity, 96.2%). The latter group displayed between 9 and 10% sequence divergence from Fusobacterium prausnitzii and related OTUs. Fusobacterium prausnitzii and those 28 clones (9.9% of total clone population) were confidently clustered, with a bootstrap value of 100%. Noticeably, OTU 237 with 12 clones represented 4.2% of the whole clone population of the sample. Seventeen cloned sequences (85%) from the Clostridium leptum subgroup corresponded to new molecular species; 11 OTUs were represented by only one clone, and the coverage was 81%.
Disparate clusters.
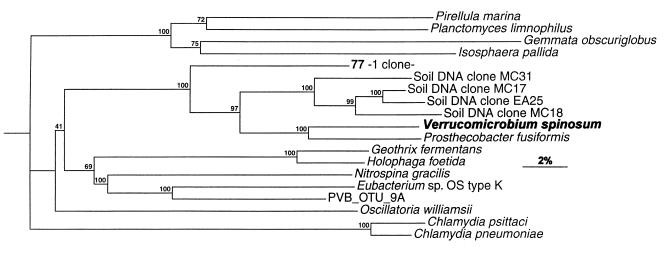
Several clones branched rather deeply in other phylogenetic groups. Two clones belonged to known OTUs: Streptococcus salivarius and Streptococcus parasanguinis (less than 1% sequence divergence). The 11 other sequences potentially represented new molecular species. Three clones (202, 262, and 279) had the URB RF39 rDNA clone as their closest relative, but phylogenetic distances ranged from 12 to 14%. They formed deep branches among Mycoplasma spp. and relatives (RDP registration no. 2.16.9). Of these three clones, clones 262 and 279 formed a single OTU whereas clone 202 represented a separate molecular species (divergence, 17%). Clone 173 also branched very deeply within Mycoplasma and relatives but was not associated with clones 202, 262, and 279. Two clones (035 and 395 in the same OTU) belonged to the Phascolarctobacterium faecium subgroup (RDP registration no., 2.16.3.1.4), within the Sporomusa group, and branched with Phascolarctobacterium faecium (originally isolated from a koala fecal sample [7]). A sequence divergence value of 10% between OTU 035 or 395 and Phascolarctobacterium faecium, however, showed that the relatedness between these organisms was not particularly high and that they represent genomically separate species. Clone 358 belonged to the Clostridium thermocellum subgroup (RDP registration no., 2.16.10.1.1), its closest neighbor (17% sequence divergence) being URB JW21, which itself currently has no close relative (34). Clone 376 branched with the Clostridium propionicum subgroup (RDP registration no., 2.16.4.2.1), and its closest relative was Clostridium neopropionicum (8.6% sequence divergence). Clone 323 belonged to the Atopobium group (RDP registration no., 2.16.1.3). Its closest relatives were Eubacterium fossor and Eubacterium lentum (<12% sequence divergence). Recently, the 16S rRNA of Collinsella aerofaciens (previously Eubacterium aerofaciens) was sequenced (20). It also belongs to this group, but our clone was not very close (>12% sequence divergence). Clone 429 belonged to the Eubacterium group (RDP registration no., 2.16.4.4). The phylogenetic distance between this clone and Eubacterium brachy was 15%. The remaining clone, 077, was linked to the prosthecate bacterium Verrucomicrobium spinosum (17% divergence). Phylogenetic inference of this clone within the Verrucomicrobium subdivision (35) is shown in Fig. 5.
FIG. 5.
Phylogenetic tree derived from partial 16S rDNA sequence data for members of the Verrucomicrobium subdivision. The bar represents 2% sequence divergence. Designations of a clone and the key organism used to name the cluster are in boldface type. The tree was constructed with the SIMILARITY and NEIGHBOR programs. Bootstrap values are given for 500 replications.
Chimeras.
Of 295 sequences analyzed, 11 chimeric 16S rDNA sequences were detected. The chimeric sequences branched differently in trees based on 200 aligned bases at the 5′ and at the 3′ ends; the CHECK_CHIMERA program (RDP) confirmed this, and no identical sequences were found from an independent PCR (unpublished data). One chimera occurred between clone 333 and clone 263, two rDNA sequences which were very close and belonged to the Prevotella cluster. The first OTU had 18 representatives, and the second OTU had 8 representatives; the chimeric sequence was represented by 10 clones. This is consistent with the observation of Godon and colleagues that chimeric events have a higher tendency to occur between highly represented sequences than between poorly represented sequences (15). The second chimeric sequence also belonged to the Prevotella cluster. This sequence was longer than the others, which provided further evidence of its chimeric nature.
DISCUSSION
Total bacterial counts similar to that reported here (2.71 × 1011 ± 0.26 × 1011 bacterial cells per g [wet weight] of feces) were obtained by Langendijk et al. (24) from 10 fecal samples by DAPI staining. Moore and Holdeman (27) by direct microscopic clump counting found a greater range of values (5 × 1011 ± 4 × 1011 bacterial cells per g [dry weight] of feces) and an average moisture content of 21.5%. Our values were generally consistent with these results. Moore and Holdeman (27) counted 4.8 × 1011 ± 2.5 × 1011 CFU per g (dry weight). Finegold et al. (11) found between 5 × 1010 and 2.5 × 1014 CFU per g (dry weight) of feces and an average value of 1.6 × 1012. Mitsuoka et al. (26) estimated that the cultivated flora was 1.3 × 1011 CFU per g (dry weight), and Langendijk et al. recently reported a value of 3.87 × 1010 ± 0.47 × 1010 CFU per g (wet weight) of feces (24). Our value (0.56 × 1011 ± 0.05 × 1011 CFU per g [wet weight] of feces) was in general agreement with these results.
Percentages of cultivated flora according to Moore and Holdeman (27) ranged between 37 and 467%, with a mean of 94%. Wilson and Blitchington (35) cultivated 58% of fecal bacteria, while Langendijk et al. (24) cultivated 14% of fecal bacteria based on results of DAPI staining and 37% based on results of probe hybridization. This considerable variation may be explicable in terms of different culture methods (roll tubes and anaerobic chamber), media, and also counting methods (direct microscopic clump counting with Gram staining and Petroff-Hausser chamber counting with DAPI staining) used. The method used by Langendijk and colleagues (24) and that used in our study were comparable, and the results were in close agreement. An estimation of 68 to 79% of noncultured organisms reinforced the relevance of the present study, i.e., the development of a culture-independent strategy for assessing human gut microflora diversity.
It has been documented (27) that 400 to 500 species inhabit the human intestinal tract, but this value originated from statistical analyses: only 113 species were isolated (27) from 20 different samples. Moore and Holdeman (27) isolated between 13 and 30 different species from each fecal sample analyzed. Among our 82 molecular species, 20 corresponded to known cultivated bacteria. Of the OTUs recovered in this study, which were attributable to recognized flora, all corresponded to species known to reside in the human gut and were in good agreement with the results of the study of Moore and Holdeman (27). We also retrieved 62 new sequences which clearly derive from unknown species. Consequently, the species diversity revealed in our study was far greater than that anticipated based on culture data, particularly for gram-positive bacteria (i.e., Clostridium coccoides and Clostridium leptum groups). As indicated earlier, all known OTUs recovered in our study corresponded to established members of the indigenous human gut flora (11), e.g., Bacteroides thetaiotaomicron, Bacteroides vulgatus, Fusobacterium prausnitzii, Eubacterium eligens, Eubacterium siraeum, Eubacterium rectale, and Streptococcus salivarius. Some dominant species were, however, not detected in the analysis: Bifidobacterium spp., Collinsella aerofaciens, Ruminococcus productus, and Eubacterium biforme. Since the fecal sample analyzed contained genetic material of bifidobacteria (based on RNA dot blot hybridizations [data not shown]), the lack of bifidobacterial sequences in our study may be attributable to mismatches between the sequence of the 5′ PCR primer (S-D-Bact-0008-a-S-20) and bifidobacterial sequences. However, newly designed primers (data not shown) have failed to generate 16S rDNA clones of these organisms. It is pertinent to note that Wilson and Blitchington (35) also failed to find any bifidobacteria even though the PCR amplification primers used perfectly matched their reported sequences. It is possible, however, that denaturation during PCR was not effective for DNAs with the highest G+C contents to allow amplification of bifidobacterial sequences. This problem is being further investigated. We did not retrieve sequences attributable either to Collinsella aerofaciens or to Ruminococcus productus. By culture methods, these species have, however, been recovered only from 42 and 18% of fecal samples of Western subjects (11). Another organism not retrieved in the present inventory was Eubacterium biforme. This species belongs with Mycoplasma spp. and walled relatives and is isolated even less frequently than Collinsella aerofaciens or Ruminococcus productus (11). Interestingly, its 16S rDNA was not amplified by specific PCR with DNA from adult human feces (32). On the other hand, some of our known OTUs appeared to be more numerous than those obtained by culture techniques, even if we consider that the exact proportions were not necessarily conserved by the PCR step. The Coprococcus eutactus OTU represented 8% of the total clone population, whereas the proportion of this species in the study of Moore and Holdeman (27) was negligible and it was not reported in the study of Finegold et al. (11). Similarly, the Bacteroides uniformis OTU represented 5% of the total clone population in our investigation but was not reported in the Moore and Holdeman (27) or Finegold et al. (11) study. Bacteroides uniformis has, however, been retrieved from some fecal samples (22). Since phenotypic discrimination between Bacteroides uniformis and Bacteroides ovatus can be ambiguous (19), some isolates were conceivably previously misidentified.
Among 50 bacterial 16S rDNA clones from a human fecal sample (35), 34% were related to the Bacteroides group, 10% were related to the Clostridium coccoides group, and 50% were related to the Clostridium leptum subgroup. The percentages of clones belonging to the Bacteroides group were similar in our study, but we found four times more clones in the Clostridium coccoides group and 2.5 times fewer clones in the Clostridium leptum subgroup. We recovered 16 clones in the Eubacterium rectale OTU. While Wilson and Blitchington (35) did not retrieve any sequence close to Eubacterium rectale, they cultivated this species from the fecal sample studied. One clone, designated low G+C #2, represented 24% of the total population in the study of Wilson and Blitchington (35) and was very close to Fusobacterium prausnitzii. Those authors also obtained one clone close to E. coli, one related to the Planctomyces group, and one designated β-proteobacterium 1, none of which were retrieved in our study. This difference may be due to interindividual differences, to the DNA extraction method, or to the PCR amplification procedure (i.e., we did not use the same primers).
More recently, 78 bacterial clones were obtained from a human fecal sample (37), but only those which corresponded to dominant bands in the reverse transcription-PCR-derived temperature gradient gel electrophoresis profiles were analyzed. In the Bacteroides group, only clones in the Prevotella subgroup were represented (37). These clones were very close in sequence to URB 30-05 (34) but branched from our Prevotella clones rather deeply (11% sequence divergence). Since many of our OTUs in the Bacteroides group had only a few representatives, the corresponding bands in the temperature gradient gel electrophoresis profile would have been nondominant. Only three sequences in the Clostridium coccoides group (37) were not related to the OTUs observed in this study. The others were related to Eubacterium rectale, Eubacterium halii, OTU 304 (Coprococcus eutactus related), OTU 217, and OTU 328, with 0.5 to 1.5% sequence divergence and high bootstrap values. Two OTUs (UB A03 and UB A10) belonged to the Clostridium leptum subgroup. They were clearly related to the two clusters related to the Fusobacterium prausnitzii OTU but represented separate OTUs. Fusobacterium prausnitzii was the second most frequently recovered organism in the study of Moore and Holdeman (23). It is now evident from two independent molecularly based analyses (35) that several genomically distinct OTUs proximal to Fusobacterium prausnitzii exist as dominant members of the human gut flora. The end products of metabolism of Fusobacterium prausnitzii (production of acetate, butyrate, and formate) may be shared by the whole cluster of Fusobacterium prausnitzii and related OTUs, thereby representing an important contributor to intestinal fermentations.
In an rDNA study of the rumen ecosystem of dairy cows (34), 133 cloned bacterial 16S rDNA sequences were analyzed. Of these, 49 were related to Prevotella ruminicola. The Prevotella clones obtained in our study belong to a quite different subcluster. It is clear, however, that in that study (34), an important proportion of clones belonged to the Prevotella subgroup and represented potentially new species. A possible bias should be investigated by probe hybridization methodologies. Whitford et al. (34) also reported seven clones within the Bacteroides group (Prevotella subgroup excluded), and three clones also related to this group fell outside of the Bacteroides group. The majority of the remaining clones belonged to Clostridium subcluster XIVa (Clostridium coccoides group) and cluster IV (Clostridium leptum subgroup) (5). However, as was also found in our study, some of the clones did not cluster within the main phylogenetic groups. Interestingly, these clones fell into the same disparate clusters as ours but branched rather deeply from these groups: Clostridium cluster III (including the Clostridium thermocellum subgroup), cluster IX (including the Phascolarctobacterium faecium subgroup), and subcluster XIVb (including the Clostridium propionicum subgroup) (see reference 5 for cluster designations).
It is now recognized that, due to limitations of culture-based techniques, knowledge of the human gut microbiota composition is far from complete. In particular, it is now generally, if not universally, believed that a significant proportion of the gut microbial diversity has not been cultivated. The autochthonous bacterial population plays a decisive role in the health and physiological function of the host (6, 10, 12). A thorough knowledge of the predominant bacteria within the human gut is, for example, critical to understanding how the normal flora may provide colonization resistance and stimulate immune responses to pathogenic bacteria. Similarly, to implement effective dietary strategies (e.g., use of probiotics and prebiotics) aimed at modulating the normal flora for improved health (13, 31), accurate methods for monitoring bacterial changes are absolutely essential.
In this study, we have conducted a molecular genetic analysis of rDNA amplicons generated directly from a single human fecal sample. The results have shown that 95% of this flora could be assigned to three major phylogenetic lineages (viz., the Bacteroides, Clostridium coccoides, and Clostridium leptum groups). Significantly, an in-depth phylogenetic analysis revealed that the great majority of the observed rDNA diversity was attributable to hitherto unknown dominant microorganisms within the human gut. The detailed molecular diversity inventory derived from our direct fecal analysis will form a baseline and comparative framework for other age-related, diet, gut disorder, and geography-based studies. Such extended studies (of another adult, one elderly person, and several babies) are already planned. In addition, the generated sequences should facilitate the isolation of corresponding extant organisms (i.e., molecular species related to Fusobacterium prausnitzii and the new cluster within the Prevotella subgroup) and also aid in the design of new species- and group-specific gene probes, offering powerful tools with which to study gut microbial ecology. The novel molecular species, retrieved from a single adult, will be quantified by dot blotting and in situ hybridizations of several fecal samples to assess their contribution to the total flora without the bias resulting from PCR amplification.
ACKNOWLEDGMENTS
We thank Patrick Tailliez and Violaine Rochet for the microscopic counts; Françoise Marcille for the culture counts; and Roger Hutson, Rakia Chouari, and Emmanuelle Zumstein for help in sequencing.
This study was supported by grants from the Bureau des Ressources Génétiques and the European Union for research project FLAIR CT97-3035.
REFERENCES
- 1.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The Oligonucleotide Probe Database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bidnenko E, Mercier C, Tremblay J, Tailliez P, Kulakauskas S. Estimation of the state of the bacterial cell wall by fluorescent in situ hybridization. Appl Environ Microbiol. 1998;64:3059–3062. doi: 10.1128/aem.64.8.3059-3062.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A. The phylogeny of the genus Clostridium: proposal of five new general and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 6.Cummings J H, Macfarlane G T. The control and consequences of bacterial fermentations in the human colon. J Appl Bacteriol. 1991;70:443–459. doi: 10.1111/j.1365-2672.1991.tb02739.x. [DOI] [PubMed] [Google Scholar]
- 7.Del Dot T, Osawa R, Stackebrandt E. Phascolarctobacterium faecium gen. nov., spec. nov., a novel taxon of the Sporosuma group of Bacteria. Syst Appl Microbiol. 1993;16:380–384. [Google Scholar]
- 8.Devereux R, Mundfrom G W. A phylogenetic tree of 16S rRNA sequences from sulfate-reducing bacteria in a sandy marine sediment. Appl Environ Microbiol. 1994;60:3437–3439. doi: 10.1128/aem.60.9.3437-3439.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dore J, Sghir A, Hannequart-Gramet G, Corthier G, Pochart P. Design and evaluation of a 16S rRNA-targeted oligonucleotide probe for specific detection and quantitation of human faecal Bacteroides populations. Syst Appl Microbiol. 1998;21:65–71. doi: 10.1016/S0723-2020(98)80009-X. [DOI] [PubMed] [Google Scholar]
- 10.Ducluzeau R. Role of experimental microbial ecology in gastroenterology. In: Bergone-Berezin E, editor. Microbial ecology and intestinal infections. Berlin, Germany: Springer-Verlag; 1988. pp. 7–26. [Google Scholar]
- 11.Finegold S M, Sutter V L, Mathisen G E. Normal indigenous flora. In: Hentges D J, editor. Human intestinal microflora in health and disease. New York, N.Y: Academic Press; 1983. pp. 3–31. [Google Scholar]
- 12.Gibson, G. R., and M. B. Roberfroid (ed.). Colonic microbiota, nutrition and health, in press. Kluwer Academic Publishers, Dodrecht, The Netherlands.
- 13.Gibson G R, Roberfroid M B. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr. 1995;125:1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 14.Giovannoni S J, DeLong E F, Olsen G J, Pace N R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988;170:720–726. doi: 10.1128/jb.170.2.720-726.1988. . (Erratum, 170:2418.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Godon J J, Zumstein E, Dabert P, Habouzit F, Moletta R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol. 1997;63:2802–2813. doi: 10.1128/aem.63.7.2802-2813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Good I J. The population frequencies of species and the estimation of population parameters. Biometrica. 1953;40:237–264. [Google Scholar]
- 17.Gray J P, Herwig R P. Phylogenetic analysis of the bacterial communities in marine sediments. Appl Environ Microbiol. 1996;62:4049–4059. doi: 10.1128/aem.62.11.4049-4059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hicks R E, Amann R I, Stahl D A. Dual staining of natural bacterioplankton with 4′,6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992;58:2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holdeman L V, Kelley R W, Moore W E C. Bacteroides. In: Krieg N R, Holt G J, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams & Wilkins Co.; 1984. pp. 604–631. [Google Scholar]
- 20.Kageyama A, Benno Y, Nakase T. Phylogenetic and phenotypic evidence for the transfer of Eubacterium aerofaciens to the genus Collinsella as Collinsella aerofaciens gen. nov., comb. nov. Int J Syst Bacteriol. 1999;49:557–565. doi: 10.1099/00207713-49-2-557. [DOI] [PubMed] [Google Scholar]
- 21.Kane M D, Poulsen L K, Stahl D A. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl Environ Microbiol. 1993;59:682–686. doi: 10.1128/aem.59.3.682-686.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koornhof H J, Richardson N J, Wall D M, Moore W E. Fecal bacteria in South African rural blacks and other population groups. Isr J Med Sci. 1979;15:335–340. [PubMed] [Google Scholar]
- 23.Lane D J, Pace B, Olsen G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langendijk P S, Schut F, Jansen G J, Raangs G C, Kamphuis G R, Wilkinson M H, Welling G W. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol. 1995;61:3069–3075. doi: 10.1128/aem.61.8.3069-3075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitsuoka T, Ohno K, Benno Y, Suzuki K, Namba K. The fecal flora of man. IV. Communication: comparison of the newly developed method with the old conventional method for the analysis of intestinal flora. Zentbl Bakteriol Orig A. 1976;234:219–233. [PubMed] [Google Scholar]
- 27.Moore W E, Holdeman L V. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl Microbiol. 1974;27:961–979. doi: 10.1128/am.27.5.961-979.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paster B J, Dewhirst F E, Cooke S M, Fussing V, Poulsen L K, Breznak J A. Phylogeny of not-yet-cultured spirochetes from termite guts. Appl Environ Microbiol. 1996;62:347–352. doi: 10.1128/aem.62.2.347-352.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt T M, DeLong E F, Pace N R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 31.Tannock G W. Probiotic properties of lactic-acid bacteria: plenty of scope for fundamental R & D. Trends Biotechnol. 1997;15:270–274. doi: 10.1016/s0167-7799(97)01056-1. [DOI] [PubMed] [Google Scholar]
- 32.Wang R F, Cao W W, Cerniglia C E. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl Environ Microbiol. 1996;62:1242–1247. doi: 10.1128/aem.62.4.1242-1247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weller R, Weller J W, Ward D M. 16S rRNA sequences of uncultivated hot spring cyanobacterial mat inhabitants retrieved as randomly primed cDNA. Appl Environ Microbiol. 1991;57:1146–1151. doi: 10.1128/aem.57.4.1146-1151.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitford M F, Forster R F, Beard C E, Gong J, Teather R M. Phylogenetic analysis of rumen bacteria by comparative sequence analysis of cloned 16S rRNA genes. Anaerobe. 1998;4:153–163. doi: 10.1006/anae.1998.0155. [DOI] [PubMed] [Google Scholar]
- 35.Wilson K H, Blitchington R B. Human colonic biota studied by ribosomal DNA sequence analysis. Appl Environ Microbiol. 1996;62:2273–2278. doi: 10.1128/aem.62.7.2273-2278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng D, Alm E W, Stahl D A, Raskin L. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol. 1996;62:4504–4513. doi: 10.1128/aem.62.12.4504-4513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zoetendal E G, Akkermans A D, De Vos W M. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854–3859. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]