Abstract
Phantoms are essential tools for assessing and verifying performance in computed tomography (CT). Realistic patient-based lung phantoms that accurately represent textures and densities are essential in developing and evaluating novel CT hardware and software. This study introduces PixelPrint, a 3D-printing solution to create patient-specific lung phantoms with accurate contrast and textures. PixelPrint converts patient images directly into printer instructions, where density is modeled as the ratio of filament to voxel volume to emulate local attenuation values. For evaluation of PixelPrint, phantoms based on four COVID-19 pneumonia patients were manufactured and scanned with the original (clinical) CT scanners and protocols. Density and geometrical accuracies between phantom and patient images were evaluated for various anatomical features in the lung, and a radiomic feature comparison was performed for mild, moderate, and severe COVID-19 pneumonia patient-based phantoms. Qualitatively, CT images of the patient-based phantoms closely resemble the original CT images, both in texture and contrast levels, with clearly visible vascular and parenchymal structures. Regions-of-interest (ROIs) comparing attenuation demonstrated differences below 15 HU. Manual size measurements performed by an experienced thoracic radiologist revealed a high degree of geometrical correlation between identical patient and phantom features, with differences smaller than the intrinsic spatial resolution of the images. Radiomic feature analysis revealed high correspondence, with correlations of 0.95–0.99 between patient and phantom images. Our study demonstrates the feasibility of 3D-printed patient-based lung phantoms with accurate geometry, texture, and contrast that will enable protocol optimization, CT research and development advancements, and generation of ground-truth datasets for radiomic evaluations.
Keywords: Computed Tomography, 3D-printing, COVID-19, lung imaging, patient-specific phantoms, radiomics
1. INTRODUCTION
Anthropomorphic phantoms, geometric image quality phantoms, and mathematical phantoms are fundamental tools for developing, optimizing, and evaluating novel methods in computed tomography (CT) research and clinical practice. While many different phantoms are available commercially and in research laboratories, there is a lack of patient-based phantoms that fully represent attenuation profiles and textures seen in clinical CT acquisitions, for example, for healthy and diseased lungs. Additionally, the academic and clinical CT community would benefit from a rapid and inexpensive manufacturing process compared to current commercial solutions. Over the last decade, fused deposition modeling (FDM)-based three-dimensional (3D) printing of various tissue-mimicking phantoms has been widely explored for validation and evaluation of CT imaging technology1–4. Studies have focused on several areas, including manufacturing geometrically correct models of organs5–9, generating realistic texture samples10,11, and creating accurate attenuation profiles12–15. The general procedure to 3D-print an anthropomorphic phantom from CT image data includes: (i) segmentation of regions/organs of interest in CT images, (ii) conversion of selected regions from volumetric data to triangulated surface geometry models (e.g., STL or SLA files), and (iii) use of printer-specific slicing software to apply proper parameters (e.g., extrusion rate, print speed, infill ratios, etc.) to generate instructions for printers to create 3D products. While this approach produces phantoms that resemble true anatomical structures, it still has shortcomings. First, spatial resolution is largely lost due to segmentation of regions and conversion to surface models. Second, for each region/surface model, the slicer software assigns unique infill and exterior walls (or perimeter), creating abrupt, unrealistic transitions between regions of different densities in the final product. Third, due to its reliance on segmentation, this method is susceptible to boundary placement errors.
A promising alternative is to directly translate DICOM image data into printer instructions16,17, usually referred to as geometric code or G-code. To generate different densities in 3D-printed CT phantoms, these methods utilize a pixel-by-pixel change in the filament extrusion rate, while maintaining a constant printing speed. Although this approach enables generation of sophisticated phantoms with realistic attenuation profiles, it falls short when printing high-resolution features. The reduction in spatial resolution is an important concern when generating natural image textures. Therefore, there is an unmet need for a rapid and inexpensive process for generating patient-based phantoms with accurate organ geometry, image texture, and attenuation profiles. We propose a 3D-printing solution that can achieve accurate organ geometry, image texture, and attenuation profiles, while eliminating the complexities and limitations of previous methods. Our solution is a one-step method for translating CT images into printer instructions (G-code) that can be used by any FDM 3D-printer. It combines varying printer speeds with a constant filament extrusion rate to control the density of each printed voxel.
2. METHODS
2.1. PixelPrint
Conventional 3D-printing utilizes slicing software to convert 3D-models to printer instructions written in G-code, a widely used machine language defining 3D-printing parameters (e.g., layer height, retraction, print speed, etc.). We present a solution that converts volumetric CT DICOM images directly into G-code without segmentation or intermediate 3D-models. Applied to common FDM 3D-printers, PixelPrint produces multiple 2D layers, one layer at a time, to create phantoms. Each printed layer is mapped from a corresponding DICOM slice, with the physical scale controlled to ensure that the resulting phantom has the same dimensions as the scanned patient. For each printed layer, PixelPrint generates an array of spaced parallel filament lines at fixed spacing but of varying widths, creating a partial volume effect to form varying densities. PixelPrint computes the density of the input image at closely spaced intervals along each line and maps it into appropriate extrusion and printhead speeds over each interval. It then records one G-code command that defines a starting point, an end point, the filament extrusion, and speed for that interval. This process is repeated for every interval over every line, in every layer, until the entire volume is encoded in a G-code file. Since layers are deposited in alternating directions, the varying line widths create a matrix of high- and low-density regions that correlate with the original input image volume. The matrices are shifted in angle and location at each layer, and the printed layer height is much smaller than the typical CT slice thickness. Each CT slice of the printed phantom therefore contains multiple shifted layers, ensuring that reslicing of the CT image data does not result in sampling or moiré patterns. In our experiments, we found that altering the line width by varying the extrusion rate alone does not provide sufficient spatial resolution due to the inherently slow response time of the extrusion process. Instead, we maintain a constant filament flow rate while changing the speed of the printhead to control the extrusion width.
2.2. Patient-based phantoms
Phantoms were printed on a fused-filament 3D-printer (Lulzbot TAZ 6 with M175 tool head, Fargo Additive Manufacturing Equipment 3D, LLC Fargo, ND, USA) using a 0.25 mm brass nozzle. Polylactic Acid (PLA) filament with a diameter of 1.75 mm (MakeShaper, Keene Village Plastics, Cleveland, OH, USA) was extruded at a nozzle temperature of 210 °C. To improve adhesion, the build plate was heated to 50 °C. Printing speed varied from 3.0 to 30 mm/s, producing line widths from 1.0 to 0.1 mm. An institutional review board (IRB) approved this retrospective study. CT images of four patients who had been diagnosed with COVID-19 pneumonia and acute respiratory distress syndrome (ARDS) were selected from the PACS system at the Hospital of the University of Pennsylvania and anonymized. Depending on the disease severity, the images demonstrate isolated or wide-ranging regions with extensive fibro-proliferative changes, with both interstitial and alveolar components within the lung parenchyma. A 20 cm diameter ring surrounding the lung was added for better positioning of the phantom within the bore of an oval phantom representing a medium-sized patient (see details below). HU values were converted into filament line widths using a HU-to-density mapping calculated from a calibration phantom. A lower cut-off value of 10% and an upper cap of 100% material density were applied.
2.3. Data Acquisition & Analysis
For imaging, printed phantoms were placed inside the 20 cm bore of a technical phantom (Gammex MECT) to mimic attenuation profiles of an average-sized patient (300 × 400 mm2). For each patient-specific phantom, imaging was performed with the same CT scanner and protocol as the clinical acquisition (GE Revolution CT, Siemens Sensation-64/Edge/Drive). The resulting images were registered to the original patient image using simple-ITK. For three patients who were diagnosed with findings that are attributed to mild, moderate, and severe COVID-19 pneumonia, we extracted radiomics features from manually placed ROIs using the pyradiomics library18. Using the calculated radiomic feature vectors, we performed patient-phantom comparisons of textural and statical descriptors. In addition, regions-of-interest (ROIs) of different sizes in varied locations were manually placed in vessels and parenchyma of images from a fourth patient by an experienced thoracic radiologist (L.R., four years of experience) for mean HU values and standard deviations comparisons. Finally, size measurements of three small oval structures (two bronchi and one pulmonary artery) were performed by the radiologist on both patient and phantom images.
3. RESULTS
Printing each patient-based phantom required approximately 24 hours of printing time. A CT image of a phantom, shown in zoomed-in regions in the lower panels in Figure 1, closely resembles the original CT image in both texture and contrast levels, with clearly visible vascular and bronchial structures. Figure 2 shows the identical regions in patient and phantom data selected for density measurements. Although the patient image appears noisier than the phantom image, due to higher attenuation from the patient body, five ROIs show very similar mean values, with differences that are below 15 HU. Figure 3 presents three manually measured anatomical features. Manual size measurements performed by an experienced radiologist illustrate a high degree of geometrical correlation of details between the patient image and the phantom images, with differences that are smaller than the intrinsic spatial resolution of the scans.
Figure 1.
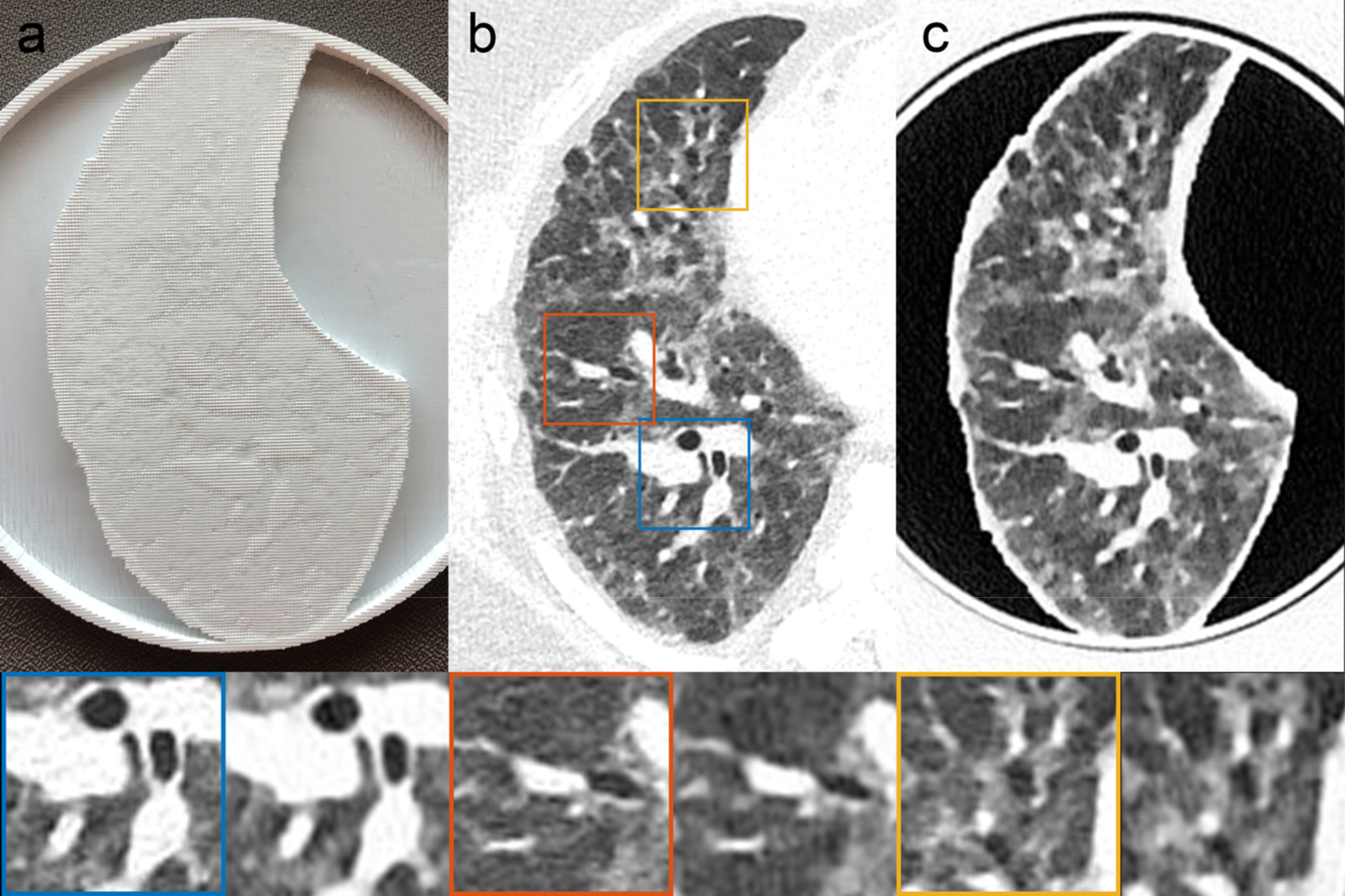
A patient-based lung 3D-printed phantom, with a high level of visual resemblance between the original CT images and the phantom CT images, both in texture and contrast levels. (a) Photograph of the printed patient-based phantom. (b) CT image of patient lung. (c) CT image of patient-based phantom. Blue, orange, and yellow boxes indicate zoomed-in regions of the patient DICOM image. Both CT images were acquired on the same scanner using the same (clinical) protocol. Window level/width: −500/1000 HU.
Figure 2.
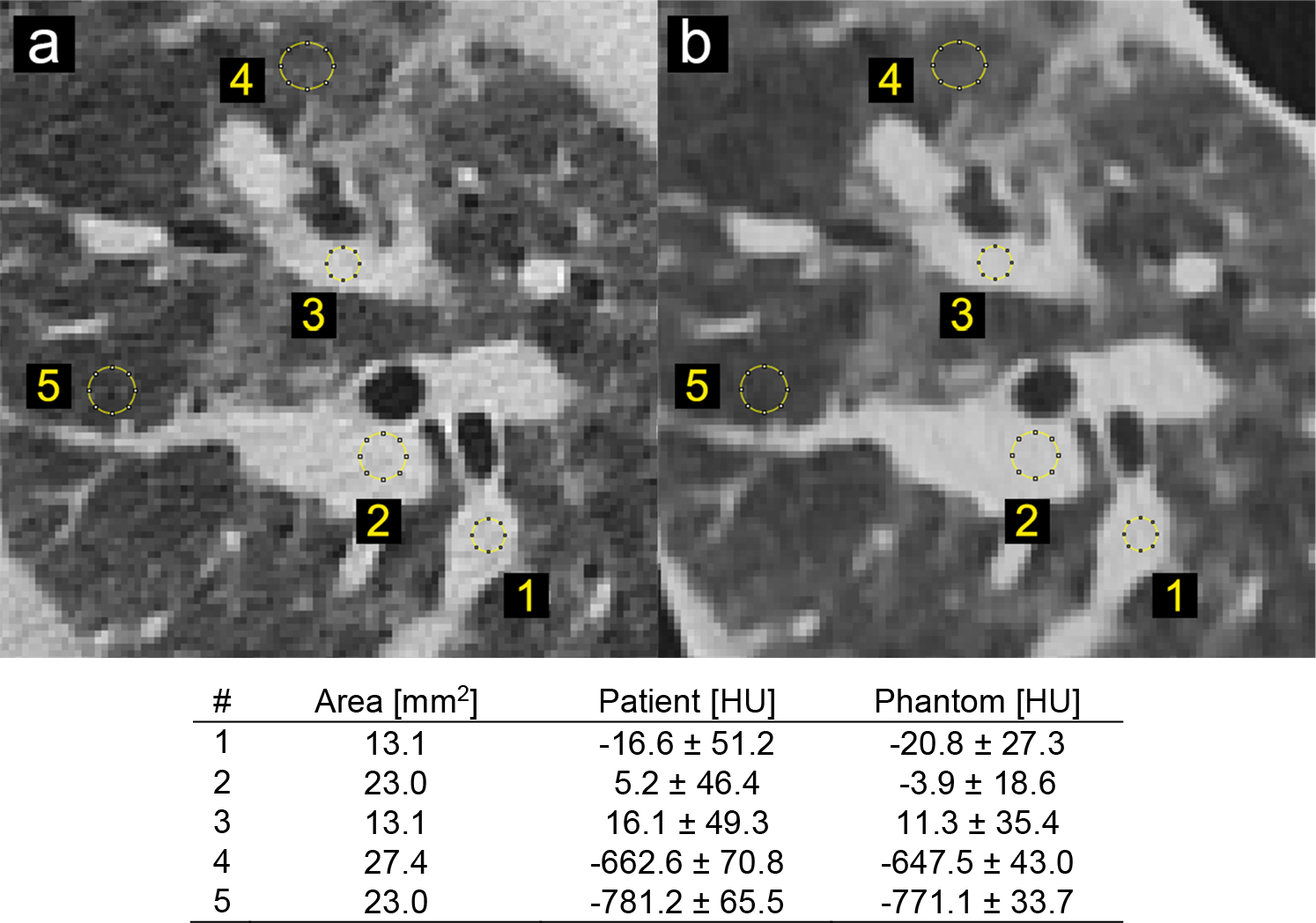
Locations and size of selected regions of interest for density measurements in patient and phantom images. (a) CT image of the original patient scan (b) CT image of the patient-based phantom. Window level/width: −500/1000 HU.
Figure 3.
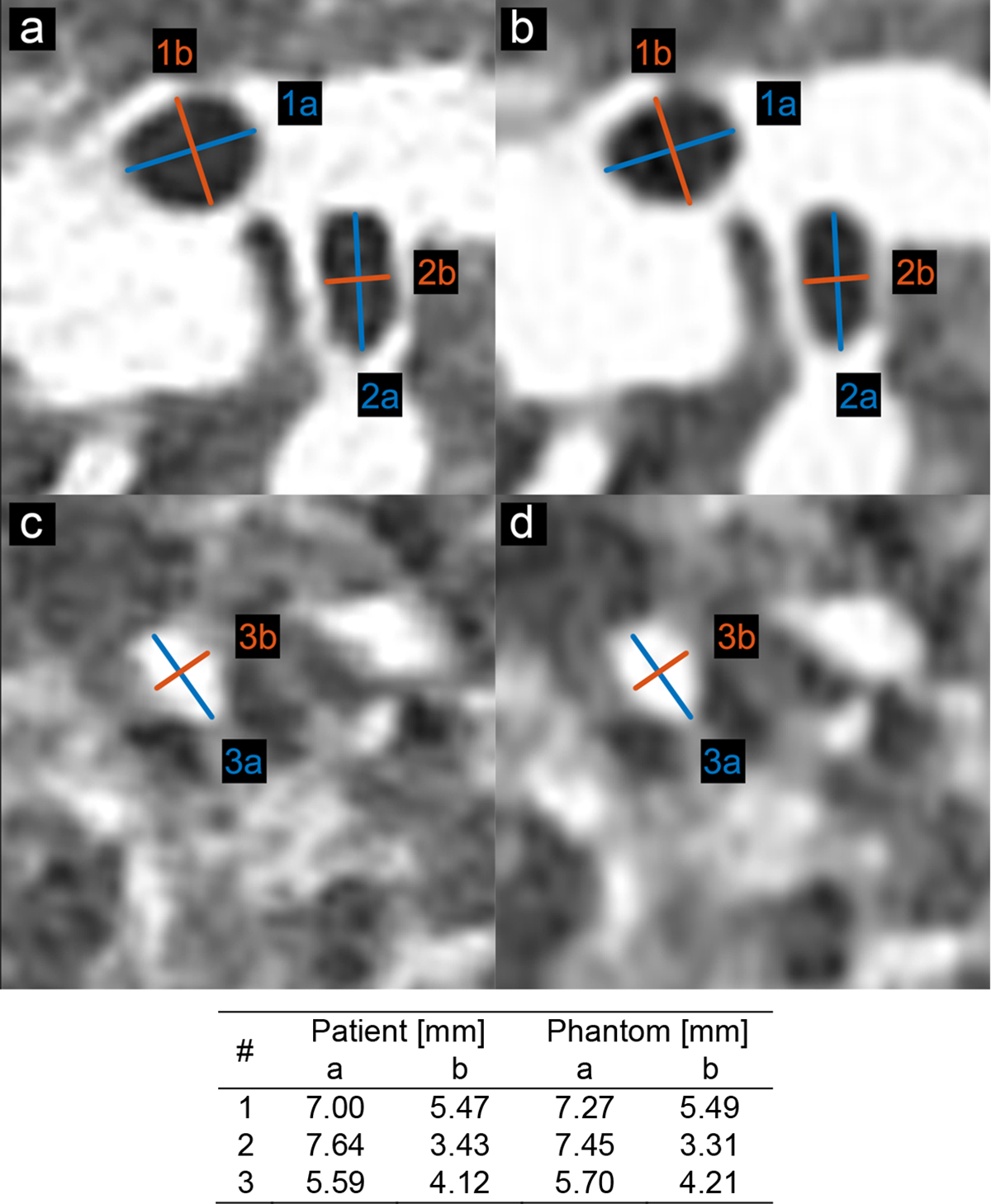
Locations and size measurements of selected anatomical features for size evaluations in patient and phantom data. (a) and (c) CT image of original (clinical) patient lung. (b) and (d) CT image of patient-based 3D-printed phantom. Window level/width: −500/1000 HU.
Radiomic features were calculated from manually selected volumetric ROIs of mild, moderate, and severe COVID-19 pneumonia patient images (Figure 4). To account for the large range of radiomic feature magnitudes (10−3–1010), the extracted features were normalized using a n additional ROI which was positioned on the mild COVID-19 pneumonia patient images and served as a standard radiomic feature vector. Figure 5 presents the resulting mean absolute relative differences of radiomic features (relative to the standard feature vector) for each phantom or patient ROI. It demonstrates a good correspondence between 3D-printed phantom images and clinical images for the mild and moderate patients, and a clear separation between disease severity for all phantom or patient ROIs. Correlations of radiomic feature relative differences between clinical and 3D-printed were found to be 0.9468 for the mild patient, 0.9806 for the moderate patient, and 0.9979 for the severe patient, demonstrating the high degree of contrast and textural correspondence between clinical images and 3D-printed phantoms using PixelPrint.
Figure 4.
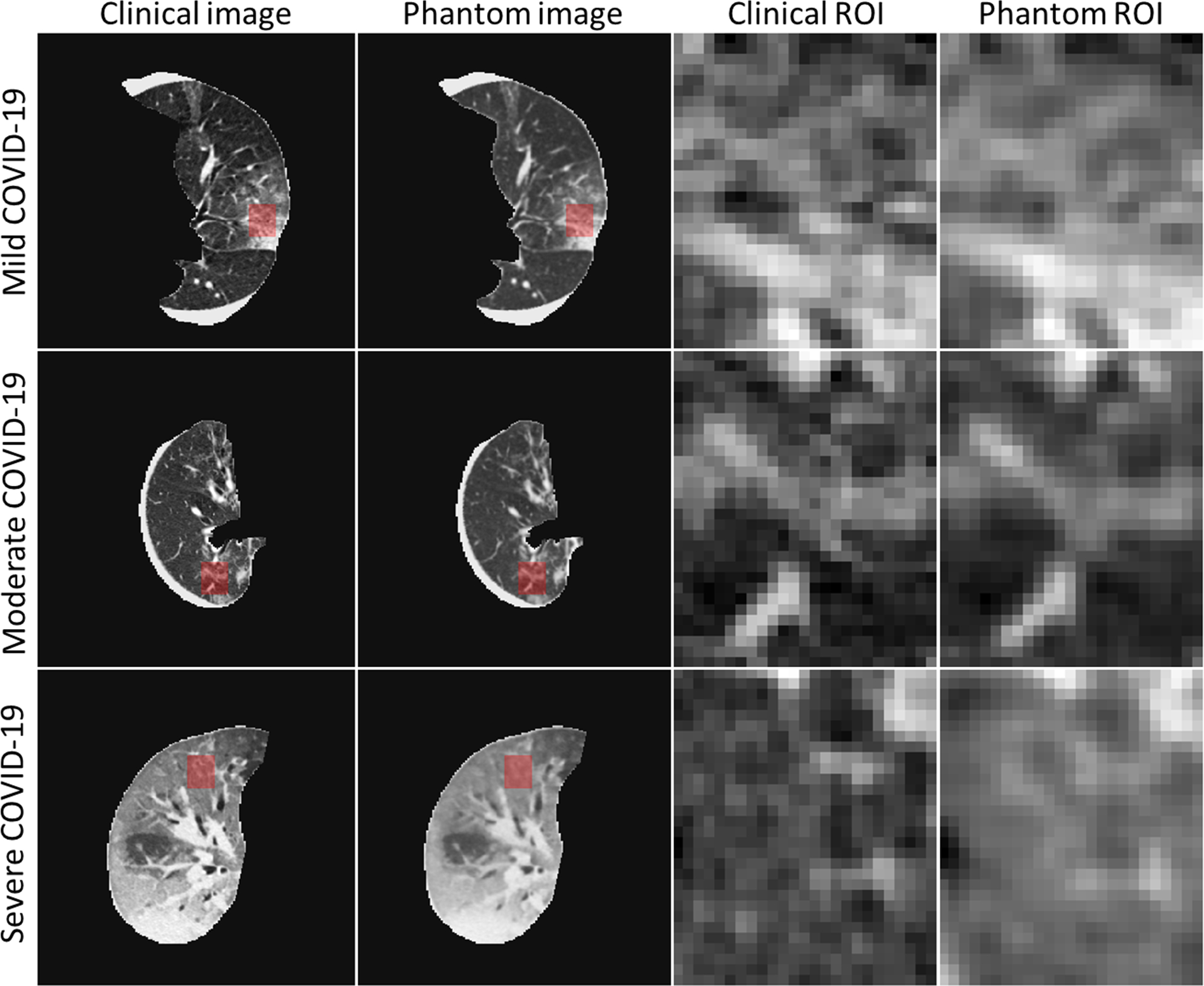
CT images of three patient and patient-specific 3D-printed phantom scans, together with the locations and sizes of the selected regions of interest (ROIs) for radiomic feature comparisons. Each ROI is spanned over five consecutive slices (only central slice presented here) and contains a total of 3750 pixels (5 × 25 × 30). Window level/width: −500/1200 HU.
Figure 5.
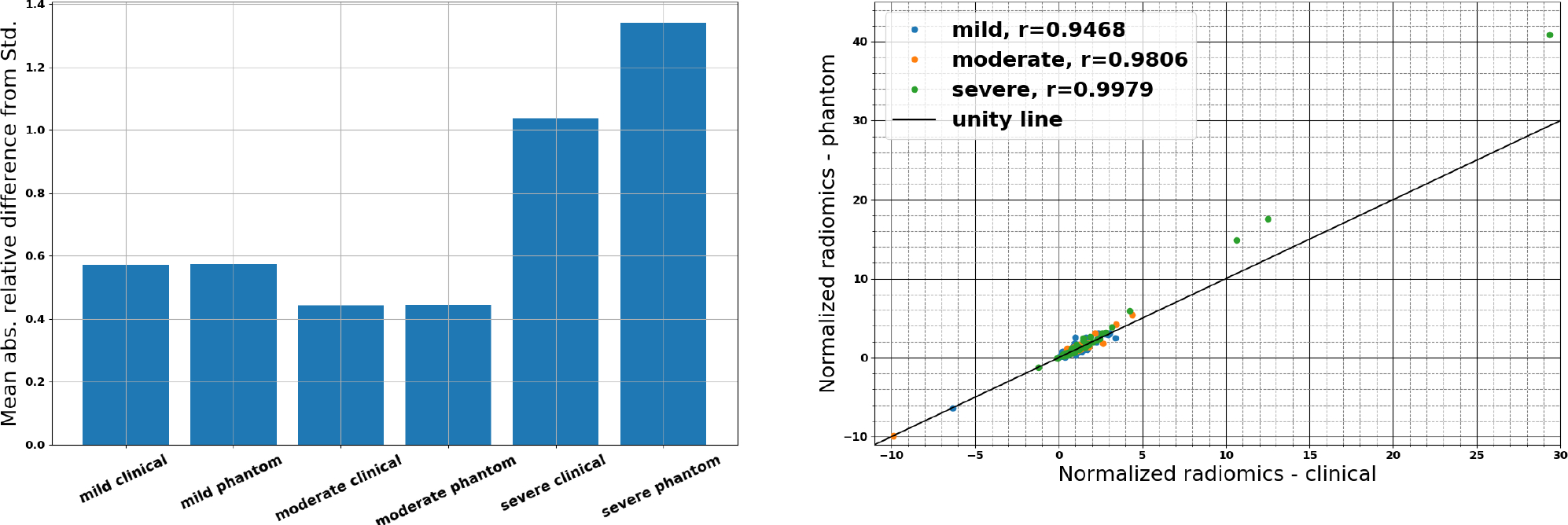
Comparison of mean absolute relative differences of radiomic features from the selected ROIs shown in Figure 4, demonstrating a correspondence between 3D-printed phantom and clinical images, and parametric response mappings demonstrating high patient-phantom correlations.
4. CONCLUSIONS
Over the last decade, several approaches have been proposed to produce clinically applicable CT phantoms. Kairn et al. introduced a method to generate a patient-based lung phantom19. They segmented CT images of the lung into three different regions and produced a tissue equivalent lung phantom. However, their approach is unable to meet the resolution requirements to represent structures in the lung parenchyma. Giron et al. and Joemai et al. developed a printed lung for image quality assessment in CT; their prints contain vascular structures with limited realistic lung textures8,20. Okkalidis et al. proposed a pixel-by-pixel algorithm17,21, translating DICOM images to printer instructions and printed patient-specific skull and chest phantoms. Results showed a reliable match in HU; however, detailed structures and textures within the lung are not visible. Jahnke et al. also introduced an alternative approach22 of stacking radiopaque 2D prints to form patient-based 3D-phantoms.
Our method enables the creation of realistic phantoms from clinical CT data, that can readily serve as ground-truth datasets, opening opportunities in the clinical and research arena. For day-to-day operations, our phantom concept allows optimization of CT protocols with a focus on specific clinical tasks, for example, introduction of advanced non-linear reconstruction algorithms23 can be challenging due to the limited clinical value of technical phantoms and ethical difficulties of scanning patients twice for this purpose. With our phantoms, an ample parameter space can be evaluated to determine the optimal solution with respect to radiation exposure and diagnostic image quality. A positive effect could be achieved for CT research and development by accelerating clinical evaluations with patient-based phantoms. Predominantly novel data-driven developments in artificial intelligence and radiomics can gain significantly from early access to realistic clinical data. One open challenge is the effect of differences in CT protocols and inter-vendor variabilities on radiomic features24–26. With a representative group of patient-based phantoms manufactured with PixelPrint, one would be able to evaluate this effect fully and determine a robust and rigorous operating space for radiomic feature extraction. Further, the same group of phantoms may assist as a tool to evaluate and validate harmonization strategies. In conclusion, the present study illustrates the possibility of creating 3D-printed patient-based lung phantoms with accurate organ geometry, image texture, and attenuation profiles. This may lead to a paradigm change for the development of novel CT hardware and software by enabling accelerated evaluation and validation with realistic patient-based data. Ultimately this will shape the clinical day-to-day routine and benefit patients with novel and standardized CT imaging.
5. ACKNOWLEDGMENTS
We acknowledge support through the National Institutes of Health (R01CA249538 and R01EB030494).
REFERENCES
- 1.Mitsouras D, Liacouras PC, Wake N & Rybicki FJ RadioGraphics Update: Medical 3D Printing for the Radiologist. RadioGraphics 40, E21–E23 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Filippou V & Tsoumpas C Recent advances on the development of phantoms using 3D printing for imaging with CT, MRI, PET, SPECT, and ultrasound. Medical Physics 45, e740–e760 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rengier F, Mehndiratta A, Von Tengg-Kobligk H, Zechmann CM, Unterhinninghofen R, Kauczor HU & Giesel FL 3D printing based on imaging data: Review of medical applications. International Journal of Computer Assisted Radiology and Surgery 5, 335–341 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Shahrubudin N, Lee TC & Ramlan R An overview on 3D printing technology: Technological, materials, and applications. Procedia Manufacturing 35, 1286–1296 (2019). [Google Scholar]
- 5.Dangelmaier J, Bar-Ness D, Daerr H, Muenzel D, Si-Mohamed S, Ehn S, Fingerle AA, Kimm MA, Kopp FK, Boussel L, Roessl E, Pfeiffer F, Rummeny EJ, Proksa R, Douek P & Noël PB Experimental feasibility of spectral photon-counting computed tomography with two contrast agents for the detection of endoleaks following endovascular aortic repair. European Radiology 1–8 (2018). doi: 10.1007/s00330-017-5252-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kopp FK, Daerr H, Si-Mohamed S, Sauter AP, Ehn S, Fingerle AA, Brendel B, Pfeiffer F, Roessl E, Rummeny EJ, Pfeiffer D, Proksa R, Douek P & Noël PB Evaluation of a preclinical photon-counting CT prototype for pulmonary imaging. Scientific Reports 8, 17386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muenzel D, Bar-Ness D, Roessl E, Blevis I, Bartels M, Fingerle AA, Ruschke S, Coulon P, Daerr H, Kopp FK, Brendel B, Thran A, Rokni M, Herzen J, Boussel L, Pfeiffer F, Proksa R, … Noël PB Spectral Photon-counting cT: Initial Experience with Dual-Contrast Agent K-Edge Colonography. Radiology (2017). doi: 10.1148/radiol.2016160890 [DOI] [PubMed] [Google Scholar]
- 8.Hernandez-Giron I, den Harder JM, Streekstra GJ, Geleijns J & Veldkamp WJH. Development of a 3D printed anthropomorphic lung phantom for image quality assessment in CT. Physica Medica 57, 47–57 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Abdullah KA, McEntee MF, Reed W & Kench PL Development of an organ-specific insert phantom generated using a 3D printer for investigations of cardiac computed tomography protocols. Journal of Medical Radiation Sciences 65, 175–183 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Gang G, Brehler M, Shi H & Stayman J 3D-Printed Textured Phantoms for Assessment of High Resolution CT. in Medical Physics E209–E210 (2019). [Google Scholar]
- 11.Solomon J, Ba A, Bochud F & Samei E Comparison of low-contrast detectability between two CT reconstruction algorithms using voxel-based 3D printed textured phantoms. Medical Physics 43, 6497–6506 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Ardila Pardo GL, Conzelmann J, Genske U, Hamm B, Scheel M & Jahnke P 3D printing of anatomically realistic phantoms with detection tasks to assess the diagnostic performance of CT images. European Radiology 30, 4557–4563 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pegues H, Knudsen J, Tong H, Gehm ME, Wiley BJ, Samei E & Lo J Using inkjet 3D printing to create contrast-enhanced textured physical phantoms for CT. in 10948, 181 (SPIE-Intl Soc Optical Eng, 2019). [Google Scholar]
- 14.Okkalidis N A novel 3D printing method for accurate anatomy replication in patient-specific phantoms. Medical Physics 45, 4600–4606 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Okkalidis N & Marinakis G Technical Note: Accurate replication of soft and bone tissues with 3D printing. Medical Physics 47, 2206–2211 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Okkalidis N & Marinakis G Technical Note: Accurate replication of soft and bone tissues with 3D printing. Medical Physics 47, 2206–2211 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Okkalidis N A novel 3D printing method for accurate anatomy replication in patient-specific phantoms. Medical Physics 45, 4600–4606 (2018). [DOI] [PubMed] [Google Scholar]
- 18.JJM van G., A F, C P, A H, N A, V N, RGH B-T, JC F-R, S P & HJWL A Computational Radiomics System to Decode the Radiographic Phenotype. Cancer research 77, e104–e107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kairn T, Zahrani M, Cassim N, Livingstone AG, Charles PH & Crowe SB Quasi-simultaneous 3D printing of muscle-, lung- and bone-equivalent media: a proof-of-concept study. Physical and Engineering Sciences in Medicine 43, 701–710 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Joemai RMS & Geleijns J Assessment of structural similarity in CT using filtered backprojection and iterative reconstruction: A phantom study with 3D printed lung vessels. British Journal of Radiology 90, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Solomon J, Ba A, Bochud F & Samei E Comparison of low-contrast detectability between two CT reconstruction algorithms using voxel-based 3D printed textured phantoms. Medical Physics 43, 6497–6506 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Jahnke P, Schwarz S, Ziegert M, Schwarz FB, Hamm B & Scheel M Paper-based 3D printing of anthropomorphic CT phantoms: Feasibility of two construction techniques. European Radiology 29, 1384–1390 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Willemink MJ & Noël PB The evolution of image reconstruction for CT—from filtered back projection to artificial intelligence. European Radiology 29, 2185–2195 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ger RB, Zhou S, Chi PCM, Lee HJ, Layman RR, Jones AK, Goff DL, Fuller CD, Howell RM, Li H, Stafford RJ, Court LE & Mackin DS Comprehensive Investigation on Controlling for CT Imaging Variabilities in Radiomics Studies. Scientific Reports 8, 13047 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackin D, Fave X, Zhang L, Fried D, Yang J, Brian Taylor, Rodriguez-Rivera E, Dodge C, Jones AK & Court L Measuring computed tomography scanner variability of radiomics features. Investigative Radiology 50, 757–765 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larue RTHM, van Timmeren JE, de Jong EEC, Feliciani G, Leijenaar RTH, Schreurs WMJ, Sosef MN, Raat FHPJ, van der Zande FHR, Das M, van Elmpt W & Lambin P Influence of gray level discretization on radiomic feature stability for different CT scanners, tube currents and slice thicknesses: a comprehensive phantom study. Acta Oncologica 56, 1544–1553 (2017). [DOI] [PubMed] [Google Scholar]


