Abstract
Heterologous protein production can be doubled by increasing the copy number of the corresponding heterologous gene. We constructed a host-vector system in the yeast Kluyveromyces lactis that was able to induce copy number amplification of pKD1 plasmid-based vectors upon expression of an integrated copy of the plasmid recombinase gene. We increased the production and secretion of two heterologous proteins, glucoamylase from the yeast Arxula adeninivorans and mammalian interleukin-1β, following gene dosage amplification when the heterologous genes were carried by pKD1-based vectors. The choice of the promoters for expression of the integrated recombinase gene and of the episomal heterologous genes are critical for the mitotic stability of the host-vector system.
Circular DNA plasmids are present in some yeast species. The most extensively studied is the 2μm circular plasmid of Saccharomyces cerevisiae, which is considered a model for yeast plasmids (23, 36). Circular plasmids have been found in some Zygosaccharomyces osmotolerant yeasts (28, 29, 31), Torulaspora delbrueckii (8), and Kluyveromyces waltii and K. drosophilarum (15, 18). These plasmids do not confer any evident phenotype on the host cell. They do not have sequence homology and are species specific with respect to maintenance and replication (1, 3, 14). Nevertheless, they are approximately the same size, have similar structural organization, and share intramolecular dimorphism.
The plasmids are present in the host cells at high copy number (50 to 100 per cell) and in two equivalent molecular forms resulting from intramolecular recombination at inverted-repeated (IR) sequences. A model for copy number amplification, which involves site-specific recombination events during plasmid replication, has been proposed for the 2μm circle (22, 35). Expression of the plasmid recombinase gene (FLP) is repressed by the product of the plasmid-partitioning genes REP1 and REP2 (27, 34). A fourth gene (D) on the 2μm circular plasmid counteracts the regulatory function of the Rep1 and Rep2 proteins (24). A mathematical model for plasmid maintenance has been successfully applied in computer simulations and is supported by experimental data (33).
The pKD1 plasmid of K. drosophilarum can replicate and is stably maintained in Kluyveromyces lactis (3). This plasmid (Fig. 1A) has been isolated and sequenced (12, 18). pKD1 carries a replication origin, the two B and C genes involved in partitioning, a cis-acting partitioning locus (CSL) (5), a gene encoding a site-specific plasmid recombinase (A), and recombinase target sites in the IRs. Site-specific recombination in pKD1 requires the B and C gene products (6). Homologous recombination occurs frequently in K. lactis among plasmid circles (4). Inactivation of the recombinase gene decreases the plasmid copy number (6), suggesting that pKD1 plasmid amplification requires site-specific recombination. The absence of a fourth gene in pKD1 indicates that the regulation of the A gene might be different, and possibly simpler, than in the 2 μm plasmid.
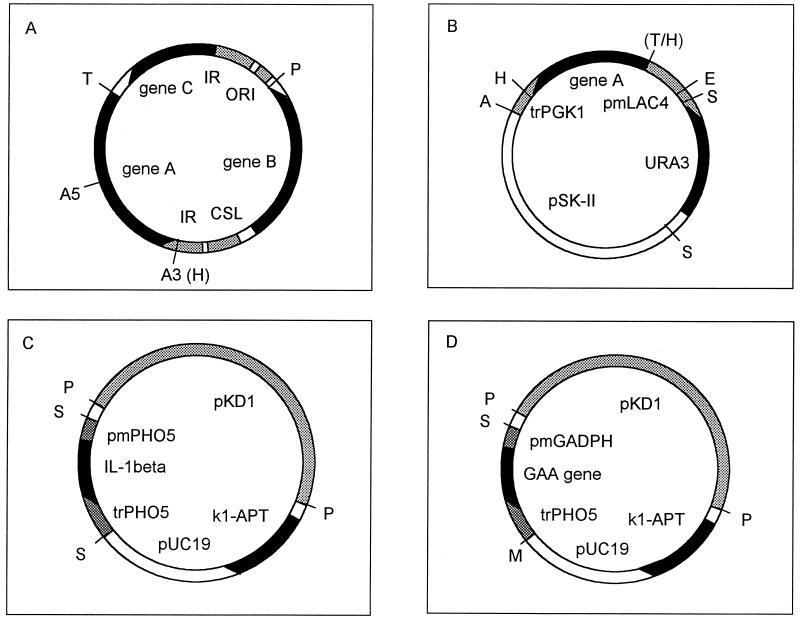
FIG. 1.
Structures of plasmid pKD1 and of vectors. Relevant restriction sites are indicated by capital letters as follows: A, SacI; E, EcoRI; H, HindIII; M, SmaI; P, SphI; S, SalI; T, StuI. (A) Plasmid pKD1. The three genes A, B, and C are indicated by solid boxes. The replication origin (ORI), the cis-acting stability locus (CSL), and the inverted repeats (IR) are dotted light gray patterns. Localization of the A3 and A5 primers is also indicated. (B) Integrative vector pLAU16. The A gene of pKD1 and the yeast marker URA3 are indicated by solid black boxes. The LAC4 promoter (pmLAC4) and the PGK1 terminator (trPGK1) are marked with stippling. pSK BluescriptII is placed between the SacI and SalI sites. (C and D) Expression vectors pGM-IL and pGM-GAM. The heterologous IL-1β and GAM genes and the yeast marker k1-APT fusion are marked in black. In dotted gray are shown the PHO5 and GAPDH promoters (pmPHO5 and pmGPDH) and PHO5 terminator (trPHO5). The entire and fully functional sequence of pKD1 contained in the vectors, and the pUC19 sequences are shaded in dotted light gray.
Our objectives in this study were (i) to construct a K. lactis strain containing a chromosomal copy of the recombinase A gene of pKD1 under the control of an inducible promoter and (ii) to use this strain for copy number amplification of pKD1-based vectors harboring expression cassettes for heterologous genes. Our results showed that copy number amplification could at least double heterologous protein production.
MATERIALS AND METHODS
Strains and media.
All the cloning steps were performed in Escherichia coli DH5α [φ80dlacZΔM15 Δ(lacZYA-argF)U169 deo rec-1 end-1 sup-44 λ thi-1 gyrA96 relA1]. The yeast strains used were MW98-8C (MATα uraA1-1 lysA argA pKD10) and PM6-7A (MATa uraA1-1 adeT-600 pKD1+). Yeast-peptone medium contained 2% peptone, 1% yeast extract, and 2% glucose (YPD) or 2% galactose (YPG); geneticin was added at 100 mg/liter when needed. SD medium contained 0.67% yeast nitrogen base (Difco, Detroit, Mich.), 20 mg of lysine and 20 mg of arginine per liter, and 2% glucose. In the low-phosphate medium LPi (1% peptone, 0.25% yeast extract, 2% glucose), the inorganic phosphate was eliminated by Mg2+ precipitation as previously described (13). Solid media contained 2% agar. Yeast cultures were inoculated at an optical density at 600 nm of ≤0.05 in Erlenmeyer flasks or aerated test tubes and incubated at 28°C on an orbital shaker at 175 rpm. Growth rates were determined as the rate of increase of optical density. Cell density at stationary phase was determined from cell counts in a Buerker chamber.
Construction of the integrative vector.
The 3′-terminal region of the pKD1 A gene was amplified from a pKD1 DNA preparation by PCR with the following oligonucleotides: A5 primer, (5′)GGG TCT AGA TCC AAG GAC TTT TGA GAT CTA CAA C(3′); A3 primer, (5′)GGG CCT GAG GAA GCT TGC CCC ATC ATG CCA CCA CCG TCC GCT GTG ATC GC(3′). The underlined nucleotides correspond to a HindIII site introduced by amplification and the recombinase gene stop codon. The positions of the primers are indicated in Fig. 1A. The amplification product, confirmed by sequencing, was fused in frame to the wild-type 5′ portion of the A gene and cloned into pSK-Bluescript II. The 1,373-bp StuI-HindIII fragment containing the A gene sequence was then placed into the HindIII site of the LAC4 promoter/PGK1 terminator cassette from vector pYG105, which has been used previously for the construction of the lactose- and galactose-inducible expression vector pYG107 (21). The A gene expression cassette was cloned into pSK-Bluescript II, and the URA3 marker gene from S. cerevisiae, which complements the uraA1-1 mutation of K. lactis (16), was subsequently inserted into the SalI site of the resulting construct to give pLAU16 (Fig. 1B).
Construction of expression vectors.
We constructed three pKD1-based expression vectors containing heterologous genes. In all cases, the expression cassette and the genetic markers were inserted into the SphI site of pKD1, which is located in a region without plasmid functions and permits the construction of very stable vectors (6).
The vector pKan707 (21) was digested with EcoRI, and the fragment harboring the pUC19 bacterial origin of replication and marker was circularized. This new vector, pKan007, also contained the K. lactis k1 promoter of the killer plasmid fused in frame with the aminoglycoside phosphotransferase (APT) gene of the bacterial transposon Tn903. The k1-APT fusion confers resistance to the antibiotic Geneticin. pKan007 was inserted into the SphI site of pKD1 to generate the stable replicative vector p3K31. The XbaI-HindIII fragment of vector pYG81 (20), containing the S. cerevisiae PHO5 promoter, the interleukin-1β (IL-1β) gene fused in frame to the K. lactis killer toxin secretory signal, and the PHO5 terminator, was cloned into pSK-Bluescript II. From the resulting vector, the IL-1β expression cassette was isolated as a SalI fragment and was cloned into the single SalI site of vector p3K31. The vector was called pGM-IL (Fig. 1C), and the expression of the heterologous gene was inducible in low-phosphate media (20). Vector pGMA-IL was identical to pGM-IL, except that the pKD1 moiety contained a 4-bp insertion in the A gene. This mutation inactivated the recombinase gene, giving rise to a vector with a lower basal copy number (6). The pGM-GAM expression vector was obtained by cloning the 2,776-bp SalI-SmaI fragment, containing the S. cerevisiae GAPDH promoter, the complete sequence of the Arxula adeninivorans glucoamylase gene including the secretory signal, and the PHO5 terminator, from vector pTS32-TAA (10) into the SalI and SmaI sites of p3K31 (Fig. 1D). In pGM-GAM the glucoamylase expression was constitutive in K. lactis. All molecular cloning procedures were performed by following protocols suggested by the suppliers of the products or by standard laboratory manuals (26).
Yeast transformation and stability of the transformants.
The K. lactis strains were transformed by electroporation (7). Stability was measured by growing the transformants for 10 to 12 generations (about 24 h) on liquid YP medium to 1 × 108 to 3 × 108 cells/ml. The cultures were then diluted, plated on YPD at a density of more than 100 colonies per plate, and replica plated onto selective media. Stability was determined as the percentage of colonies maintaining the marker phenotype. For other generation time determinations, small aliquots of the grown cultures were reinoculated in fresh medium for successive rounds of growth.
Southern and Northern analysis of nucleic acids.
DNA samples were prepared from cell cultures by a standard procedure (11): cell lysis was obtained after Zymolyase incubation (Seikagaku-Koygo, Tokyo, Japan) and sodium dodecyl sulfate (SDS) treatment. Cellular debris were pelleted by centrifugation (10 min at 14,000 × g), and the aqueous phase was extracted with phenol and precipitated with ethanol. Resuspended DNA was incubated with RNase. RNA samples were prepared from late-logarithmic-phase cultures (108 cells/ml) after Zymolyase incubation and cell lysis by resuspension in hypotonic solution (0.5 M sorbitol, 0.1 M Tris-HCl [pH 7.5]). The aqueous phase was extracted with buffered phenol, and total RNA was precipitated with ethanol. DNA and RNA were fractionated by electrophoresis on agarose and agarose-formaldehyde gels, respectively. They were transferred to nylon membrane filters (Hybond-N; Amersham International, Little Chalfont, United Kingdom) and hybridized with radiolabelled probes as suggested by the supplier. Probes were labeled with the random primed DNA labeling kit (Boehringer, Mannheim, Germany). Double-stranded DNAs used as probes included the 1,373-bp StuI-HindIII fragment from the A gene, the 1.3-kbp HindIII fragment from the K. lactis actin gene (17), the 412-bp SacI-HindIII fragment from the PGK1 terminator, the 1,201-bp HindIII-SalI fragment from the LAC4 promoter, and the 1.3-kbp SalI fragment from transposon Tn903.
Interleukin detection.
The amount of biologically active IL-1β was determined by an immunochemical assay (h-Interleukin-1β ELISA; Boehringer). The enzyme-linked immunosorbent assay (ELISA) was performed both on the supernatants and on crude extracts of the transformed cells ground with glass beads (diameter, 0.5 mm). For crude-extract preparation, the cells were washed with water and resuspended in 0.9 M NaCl before being subjected to mechanical breaking. The total amount of extracted proteins was determined as described by Bradford (9). The number of broken cells was determined by estimating an average of 5 pg of protein per cell. No immunoactive material was found in samples from untransformed cells or in SDS-treated samples. The production of secreted IL-1β also was evaluated by Coomassie blue R-250 or silver staining of the supernatants from the cultures after gel electrophoresis. Aliquots of the cultures were collected and centrifuged (5 min at 14,000 × g). Supernatants were mixed 1:1 with loading buffer (0.1 M Tris-HCl [pH 7.4], 20% glycerol, 4% SDS, 5% β-mercaptoethanol, 0.02% bromphenol blue) and run on SDS-polyacrylamide gel electrophoresis (PAGE). A signal corresponding in size to IL-1β could be detected only in supernatants from LPi medium. The stained SDS-PAGE gels were also analyzed densitometrically (see below).
Glucoamylase detection.
Glucoamylase was determined by measurement of hydrolytic activity on starch. Starch-bound iodine has a peak of absorbance at 580 nm. Starch hydrolysis was assayed by measuring ΔA580/Δt as follows. A 75-μl volume of 3 M sodium acetate (pH 5.2) and 100 μl of 1% starch (soluble potato starch; Sigma, St. Louis, Mo.) were added to 5-ml volumes of supernatants from centrifuged cultures. The reaction mixtures were placed at room temperature (25°C). Samples were taken at regular intervals and cooled on ice for 2 min, and cold 0.1 N I2 in 0.12 M KI was added to a final concentration of 0.05 N I2. The A580 was measured immediately after iodine addition. One unit of glucoamylase activity was defined as the amount of enzyme allowing a decrease of 1 U of A580 per min of enzyme-substrate reaction. No glucoamylase activity could be detected in samples from untransformed BT16 cultures.
Densitometric analysis of specific RNA, plasmids, and proteins.
Plasmid DNA and transcript levels were measured relative to their levels in glucose-grown cells. The images of stained gels and autoradiographs were digitally acquired (ABEL-CAT 1.1.5; ABEL Science Ware srl, Pomezia, Italy) for the measurements of mRNA, plasmid DNA, and proteins. Sample lanes were scanned densitometrically with an image analyzer (Phoretix 1D; Non Linear Dynamics Ltd., Newcastle upon Tyne, United Kingdom). The volume of the signal, i.e., the number of pixels multiplied by the areas of the peaks, was considered proportional to the number of molecules detected by the staining or hybridization procedure. Calibration with standards was performed to permit interpolation of results. The total amount of mRNA loaded on a gel was determined by probing the filters with the K. lactis actin gene (17). The amount of DNA was determined both by measuring A260 and by performing densitometric analysis of the ethidium bromide-stained gels before capillary blotting.
RESULTS
Construction of a K. lactis strain harboring a chromosomally integrated inducible copy of the pKD1 recombinase gene.
The integrative vector pLAU16 was introduced into the MW98-8C (pKD10) K. lactis strain by electroporation. Several Ura+ transformants were isolated, and chromosomal integrants were selected by the stability of the Ura+ phenotype and by Southern analysis (results not shown). One transformant, BT16, showed 100% stability of the Ura+ phenotype after growth for 50 generations on YPD and YPG and showed hybridization of the gene A probe to chromosomal DNA (results not shown). The growth rate of strain BT16 on YPD and YPG was not affected by the pLAU16 integration compared to that of the parental strain MW98-8C. The number of cells at stationary phase was three- to fourfold smaller on YPG than on YPD. The correct arrangement of the LAC4 promoter, the recombinase gene, and the terminator in the integrant strain was verified by chromosomal DNA digestion with different restriction endonucleases and by Southern analysis (results not shown).
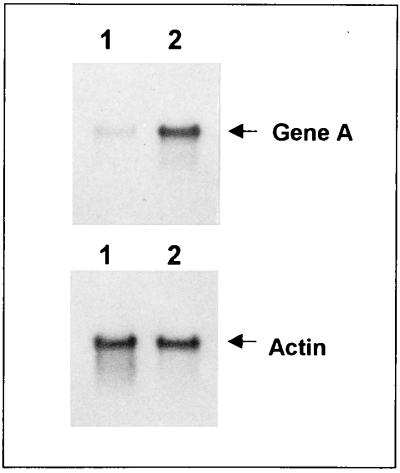
RNA was extracted from BT16 after growth on YPD and YPG and was fractionated on a denaturing agarose gel for Northern analysis. Hybridization with the A gene probe revealed a 10-fold induction of the recombinase gene expression on galactose (Fig. 2). The activity of the recombinase in strain BT16 was confirmed by its ability to isomerize a pKD1-derived vector harboring an inactive A gene (results not shown).
FIG. 2.
Northern analysis of the recombinase gene transcripts in the integrant strain BT16. Total RNA extracted from BT16 cells grown on YPD (lanes 1) and on YPG (lanes 2) was analyzed by hybridization with the A gene probe (top) and with the actin gene probe (bottom). No A gene transcript could be detected in RNA preparations from the parental strain MW98-8C (data not shown).
Stability and copy number of BT16 transformants.
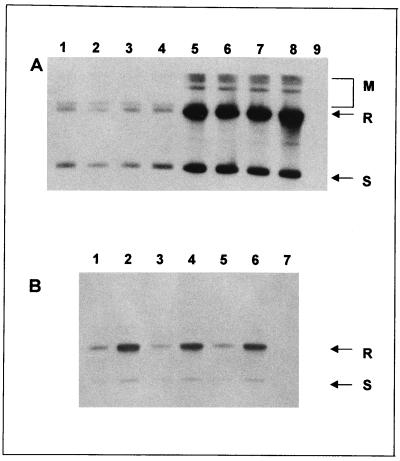
The expression vectors pGM-IL, pGMA-IL, and pGM-GAM were introduced into strains BT16 and MW98-8C by electroporation, and the Geneticin-resistant clones were selected. The stability of the transformants was measured after 24 h of growth on YPD and YPG (Table 1). All the vectors were very stable on both carbon sources. The basal copy number of vectors pGM-IL and pGM-GAM was estimated at 20 copies per cell, that of pGMA-IL, which carries an inactive copy of the recombinase gene, was 6 to 7 copies (6, 6a). Analysis of pGMA-IL transformants allowed us to identify effects of the chromosomal copy of the recombinase when the plasmid-borne recombinase inactive. Total DNA was extracted from the transformants and analyzed by the Southern procedure. Autoradiographs (Fig. 3) show increased levels of plasmid DNA in pGM-IL and pGM-GAM transformants after growth under inducing conditions, this was confirmed by densitometric analysis (Table 1). We observed a 6- to 7-fold increase of pGM-IL and pGM-GAM and an 11-fold increase of pGMA-IL copy number in BT16 transformants grown on YPG over those grown on YPD. Similar results were obtained when lactose instead of galactose was used as the inducer and carbon source. No significant increase in copy number could be observed in MW98-8C transformants.
TABLE 1.
Stability and copy number in pGM-IL, pGMA-IL, and pGM-GAM transformantsa
| Vector | Strain | Carbon source | Stability of integrated sequencesb (%) | Vector stabilityc (%) | Copy number increased |
|---|---|---|---|---|---|
| pGM-IL | BT16 | Galactose | 99 ± 1.5 | 95 ± 4.4 | 6.6 ± 1.6 |
| Glucose | 100 ± 0.0 | 100 ± 0.0 | |||
| pGMA-IL | BT16 | Galactose | 99 ± 1.9 | 78 ± 6.4 | 11 ± 3.3 |
| Glucose | 100 ± 0.0 | 89 ± 6.2 | |||
| pGM-GAM | BT16 | Galactose | 100 ± 0.0 | 97 ± 2.5 | 6.7 ± 1.4 |
| Glucose | 100 ± 0.0 | 100 ± 0.0 | |||
| pGM-IL | MW98-8C | Galactose | 99 ± 1 | 1.4 ± 0.4 | |
| Glucose | 99 ± 1 | ||||
| pGMA-IL | MW98-8C | Galactose | 98 ± 1.7 | 1.5 ± 0.2 | |
| Glucose | 96 ± 4.3 |
Results are means and standard deviations for four independent transformants.
Percentage of Ura+ cells after 24 h of growth on the indicated carbon source.
Percentage of G418r cells after 24 h of growth on the indicated carbon source.
Relative increase (YPG/YPD) of vector copy number per G418r cell.
FIG. 3.
Southern analysis of pGM-IL and pGM-GAM transformants of strain BT16. (A) Equal amounts of DNA from four independent pGM-IL transformants grown on YPD (lanes 1 to 4) and YPG (lanes 5 to 8) were analyzed by hybridization with the Tn903 probe. DNA from the untransformed BT16 strain was loaded in lane 9. (B) Equal amounts of DNA from three independent pGM-GAM transformants grown on YPD (lanes 1, 3, and 5) and YPG (lanes 2, 4, and 6) were analyzed by hybridization with the Tn903 probe. DNA from untransformed BT16 strain was loaded in lane 7. In both panels, the more intense signals detected by the probe corresponded to the supercoiled and the relaxed forms of the monomeric vectors, indicated by S and R, respectively. Various multimeric forms of pGM-IL, indicated by M in panel A, could also be detected in galactose-grown transformants.
To determine if the copy number increase also could be induced on the natural plasmid, pKD1 was genetically transmitted to a strain containing the chromosomal pLAU16 integration by crossing strains BT16 (pKD10) and PM6-7A (pKD1+) and subjecting them to sporulation. The haploid segregant GBM1 (MATα uraA1-1 ADET ARGA LYSA pKD1+) had both pKD1 and the chromosomal A gene integration, selected by using the corresponding Ura+ phenotype. pKD1 was detected by ethidium bromide staining of DNA samples fractionated on agarose gels. When grown on YPG, strain GBM1 exhibited a three- to fourfold increase in pKD1 copy number over that for YPD-grown cells (data not shown).
Glucoamylase production by BT16(pGM-GAM) transformants.
To determine if increased heterologous protein production resulted from increased vector copy number, four BT16(pGM-GAM) transformants, in which the glucoamylase gene is constitutively expressed, were inoculated in YPG and YPD. Samples were taken from late-logarithmic- and stationary-phase cultures, i.e., at 24, 48, and 72 h after inoculation. Stability and copy number of pGM-GAM and glucoamylase activity in supernatants were measured for each sample (Table 2). No activity was detected after 24 h. After 48 h, glucoamylase activity could be detected only in YPG supernatants. After 72 h, activity in YPG samples was eightfold higher than that in YPD samples. Crude extracts from transformed cells corresponding to the analyzed supernatants never showed glucoamylase activity. The copy number was stably maintained and higher in YPG-grown cells. Vector stability decreased only slightly in YPG, while the Ura+ phenotype, linked to the integrated copy of the A gene, showed a low stability after long incubation times on galactose. Plasmid loss and loss of the integrated sequences were always mutually exclusive events in the progeny of the transformant clones analyzed so far. This finding suggested that simultaneous overexpression of the heterologous gene and of the recombinase gene might be unfavorable.
TABLE 2.
Glucoamylase productiona
| Time (h) | Carbon source | Glucoamylase activityb (mU/ml) | Vector copy number increasec | Vector stabilityd (%) | Stability of integrated sequencese (%) |
|---|---|---|---|---|---|
| 24 | Galactose | 0 | 6.7 ± 1.4 | 100 ± 3 | 92 ± 7 |
| Glucose | 0 | 99 ± 3 | 98 ± 3 | ||
| 48 | Galactose | 59 ± 12 | 4.5 ± 0.6 | 90 ± 9 | 38 ± 25 |
| Glucose | 0 | 99 ± 1 | 99 ± 1 | ||
| 72 | Galactose | 113 ± 36 | 6.5 ± 0.8 | 82 ± 18 | 31 ± 22 |
| Glucose | 14 ± 6 | 100 ± 0 | 99 ± 1 |
Data are means and standard deviations for four independent transformants. Samples were collected at 24, 48 and 72 hours after inoculation.
Glucoamylase activity recovered in the growth medium of BT16 [pGM-GAM] transformants. The activity was determined as the rate of iodine release from starch.
Ratio of vector copy numbers in galactose-glucose-grown cells.
Percentages of Geneticin-resistant cells.
Percentages of Ura+ cells.
IL-1β production in BT16(pGM-IL) transformants.
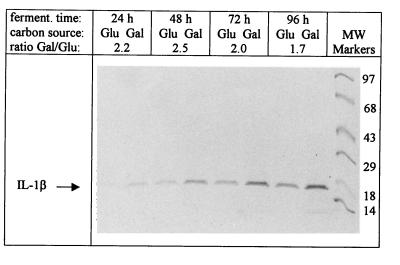
The integrated sequences and the expression vectors were unstable only when the heterologous gene and the integrated recombinase were simultaneously induced. To overcome this problem, IL-1β was produced by preculturing BT16(pGM-IL) transformants on YP medium supplemented with Geneticin and glucose or galactose as the carbon source to the stationary phase. In these media, IL-1β gene expression was repressed. The transformants were stable on both carbon sources, and the vector copy number increased following galactose induction of the A gene. The cells were collected and washed with water, inoculated at a density of 0.5 × 108 to 1.5 × 108 cells/ml in LPi medium, and further incubated for heterologous protein production. Only cells precultured on glucose could undergo two or three further duplications during the IL-1β production phase. Aliquots of the LPi supernatants were collected and loaded on an SDS-polyacrylamide gel for SDS-PAGE (Fig. 4). IL-1β accumulated during the fermentation, and production increased 1.7- to 2.5-fold when the cells were pregrown on galactose compared to glucose. The immunoactive IL-1β produced after 24 h was analyzed by ELISA (Table 3). The average amount of produced IL-1β was 42 ± 2.4 mg/ml in YPD samples and 89 ± 13 mg/ml in YPG samples. These results parallel those obtained by the gel-staining procedure. However, when the low cell density of the transformants pregrown on galactose was considered, the increase in interleukin production per cell on this carbon source was 7- to 11-fold, which approximates the increase in plasmid copy number. Based on IL-1β levels in both the supernatant and the cell extract, we conclude that the heterologous protein is largely secreted into the medium.
FIG. 4.
Interleukin detection in the supernatants of a typical BT16(pGM-IL) transformant by the gel-staining procedure. The time of sampling from the shift of cells to the production medium and the carbon source of the preculture are indicated in lines 1 and 2, respectively. In all lanes, 10 μl of the supernatant was loaded and electrophoresed. The gel was subsequently stained with Coomassie blue R-250 and analyzed densitometrically. Line 3 shows the results of this analysis, as the ratio of the intensities of the signals corresponding to the IL-1β produced by galactose-grown cells with respect to those for glucose-grown cells. This transformant secreted more than 400 mg of IL-1β per liter after 96 h in YPG. MW, molecular weight (in thousands).
TABLE 3.
Production of immunoactive IL-1βa
| Transformant | Carbon sourceb | Total amt of IL-1β producedc
|
Secreted IL-1β (%) | |
|---|---|---|---|---|
| μg/ml | pg/cell | |||
| 1 | Galactose | 101 | 1.7 | 87 |
| Glucose | 40 | 0.15 | 90 | |
| 2 | Galactose | 69 | 1.0 | 75 |
| Glucose | 40 | 0.12 | 76 | |
| 3 | Galactose | 97 | 0.9 | 80 |
| Glucose | 45 | 0.12 | 78 | |
The amount of immunoactive IL-1β was determined by ELISA on both culture medium and cell extracts. Three independent BT16(pGA-IL) transformants were analyzed after 24 h in production medium (LPi).
Carbon source in the precultivation medium.
Total amount of IL-1β (supernatant and cell extracts) produced per milliliter or per cell.
DISCUSSION
The lactose-assimilating yeast K. lactis has a versatile secretory system, which facilitates the secretion of heterologous proteins directed by various signal peptides (10, 19, 21, 25, 30, 32, 37). Heterologous genes can be introduced into this yeast on integrative or replicative vectors. In heterologous-protein production based on replicative plasmids, control of vector stability and copy number is essential in determining the total amount of product synthesized.
We found that the mechanisms of pKD1 plasmid copy number control and amplification are similar to those of the 2μm circular plasmid of S. cerevisiae, in that in both yeasts the level of expression of the plasmid recombinase is involved. Differences in the details of these mechanisms have been observed (6), but the present results show that in K. lactis the overexpression of a chromosomally integrated copy of the pKD1 recombinase gene results in a significant increase in plasmid copy number.
The copy number of the native pKD1 plasmid is in the range of 60 to 80 per cell (18) but is reduced to about 20 copies per cell for pKD1-based vectors bearing heterologous genes. A further threefold decrease in copy number is observed when the recombinase gene is inactivated in the recombinant vectors (6). Induction of the chromosomally integrated copy of the recombinase gene increased the copy number in all cases, although a saturation effect is suggested by the lower relative copy number increase obtained when basal copy number was higher.
We also found that the induction of amplification of a heterologous gene on a pKD1-based vector is followed by a corresponding increase in the heterologous-protein production. The product is largely secreted in the medium. The signal peptide of the K. lactis killer toxin drives the secretion of mature and bioactive IL-1β in both K. lactis and S. cerevisiae (2, 20). This secretory pathway was not saturated, as a result of the increased production of the secreted IL-1β. In S. cerevisiae the amount of produced and secreted IL-1β was as small as 1-2 mg/liter (2). With K. lactis as the host and pKD1-based multicopy expression vectors, production was increased to approximately 100 mg/liter (20). Our results demonstrated that production can be further increased, to 400 mg of secreted IL-1β per liter, by using the regulatory system of pKD1 to increase the plasmid copy number.
We had two major problems while increasing heterologous-protein production. The first was that the poor growth of the transformed strains on galactose, which is the inducer of the integrated recombinase gene expression, limited the increase in production. We can overcome this problem with a different promoter, which is inducible under conditions more favorable for cell growth. The second problem was that the simultaneous induction of the integrated recombinase gene and the heterologous gene on the vector resulted in the loss either of the plasmid or of the integrated sequences. We think that overproduction of the heterologous protein by multiple plasmid-borne genes exposes the cells to high-stress conditions and selects for cells that have lost either the heterologous gene or the chromosomal recombinase sequence that artificially increases plasmid copy number. We separated induction of the recombinase gene from the induced expression of heterologous genes and increased production of human IL-1β. Appropriate choice of the promoters and the conditions under which they are expressed is critical to maximize production of the heterologous protein.
ACKNOWLEDGMENTS
This work was supported by C.E.C. grant BIO4-CT96-0003 and by C.N.R. Target Project on Biotechnology grant 97.01162.49.
We thank H. Fukuhara for useful discussions and F. Castelli for technical assistance.
REFERENCES
- 1.Araki H, Jearnpipatkul A, Tatsumi H, Sakurai T, Ushio K, Muta T, Oshima Y. Molecular and functional organization of the yeast plasmid pSR1. J Mol Biol. 1985;182:191–203. doi: 10.1016/0022-2836(85)90338-9. [DOI] [PubMed] [Google Scholar]
- 2.Baldari C, Murray J A H, Ghiara P, Cesareni G, Galeotti C L. A novel leader peptide which allows efficient secretion of a fragment of human interleukin 1β in Saccharomyces cerevisiae. EMBO J. 1987;6:229–234. doi: 10.1002/j.1460-2075.1987.tb04743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianchi M M, Falcone C, Chen X J, Wesolowski-Louvel M, Frontali L, Fukuhara H. Transformation of the yeast Kluyveromyces lactis by new vectors derived from the 1.6 μm circular plasmid pKD1. Curr Genet. 1987;12:185–192. [Google Scholar]
- 4.Bianchi M M, Frontali L, Fukuhara H. Active recombination of pKD1 derived vectors with resident pKD1 in Kluyveromyces lactis transformation. Curr Genet. 1989;15:253–260. [Google Scholar]
- 5.Bianchi M M, Santarelli R, Frontali L. Plasmid functions involved in the stable propagation of the pKD1 circular plasmid in Kluyveromyces lactis. Curr Genet. 1991;19:155–161. doi: 10.1007/BF00336481. [DOI] [PubMed] [Google Scholar]
- 6.Bianchi M M. Site-specific recombination of the circular 2μm-like plasmid pKD1 requires the integrity of the recombinase gene A and of the partitioning genes B and C. J Bacteriol. 1992;174:6703–6706. doi: 10.1128/jb.174.20.6703-6706.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Bianchi, M. M. Unpublished data.
- 7.Bianchi M M, Tizzani L, Destruelle M, Frontali L, Wesolowski-Louvel M. The ‘petite negative’ yeast Kluyveromyces lactis has a single gene expressing pyruvate decarboxylase activity. Mol Microbiol. 1996;19:27–36. doi: 10.1046/j.1365-2958.1996.346875.x. [DOI] [PubMed] [Google Scholar]
- 8.Blaisonneau J, Sor F, Cheret G, Yarrow D, Fukuhara H. A circular plasmid from the yeast Torulaspora delbrueckii. Plasmid. 1997;38:202–209. doi: 10.1006/plas.1997.1315. [DOI] [PubMed] [Google Scholar]
- 9.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein. Anal Biochem. 1977;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 10.Bui D M, Kunze I, Horstmann C, Schmidt T, Breunig K D, Kunze G. Expression of the Arxula adeninivorans glucoamylase gene in Kluyveromyces lactis. Appl Microbiol Biotechnol. 1996;45:102–106. doi: 10.1007/s002530050655. [DOI] [PubMed] [Google Scholar]
- 11.Cameron J R, Philippsen P, Davis R W. Analysis of chromosomal integration and deletions of yeast plasmids. Nucleic Acids Res. 1977;4:1429–1448. doi: 10.1093/nar/4.5.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X J, Saliola M, Falcone C, Bianchi M M, Fukuhara H. Sequence organization of the circular plasmid pKD1 from the yeast Kluyveromyces drosophilarum. Nucleic Acids Res. 1986;14:4471–4481. doi: 10.1093/nar/14.11.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X J, Fukuhara H. A gene fusion system using the aminoglycoside 3′-phopsphotransferase gene of the kanamycin-resistance transposon Tn903: use in the yeasts Kluyveromyces lactis and Saccharomyces cerevisiae. Gene. 1988;69:181–192. doi: 10.1016/0378-1119(88)90429-5. [DOI] [PubMed] [Google Scholar]
- 14.Chen X J, Bianchi M M, Suda K, Fukuhara H. Host range of the pKD1-derived plasmids in yeast. Curr Genet. 1989;16:95–98. doi: 10.1007/BF00393401. [DOI] [PubMed] [Google Scholar]
- 15.Chen X J, Cong Y S, Wesolowski-Louvel M, Li Y Y, Fukuhara H. Characterization of a circular plasmid from the yeast Kluyveromyces waltii. J Gen Microbiol. 1992;138:337–345. doi: 10.1099/00221287-138-2-337. [DOI] [PubMed] [Google Scholar]
- 16.De Louvencourt L, Fukuhara H, Heslot H, Wesolowski M. Transformation of Kluyveromyces lactis by killer plasmid DNA. J Bacteriol. 1983;154:737–742. doi: 10.1128/jb.154.2.737-742.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deshler J O, Larson G P, Rossi J J. Kluyveromyces lactis maintains Saccharomyces cerevisiae intron-encoded splicing signals. Mol Cell Biol. 1989;9:2208–2213. doi: 10.1128/mcb.9.5.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Falcone C, Saliola M, Chen X J, Frontali L, Fukuhara H. Analysis of a 1.6-μm circular plasmid from the yeast Kluyveromyces drosophilarum: structure and molecular dimorphism. Plasmid. 1986;15:248–252. doi: 10.1016/0147-619x(86)90044-2. [DOI] [PubMed] [Google Scholar]
- 19.Ferminan E, Dominguez A. Heterologous protein secretion directed by a repressible acid phosphatase system of Kluyveromyces lactis: characterization of upstream region-activating sequences in the KlPHO5 gene. Appl Environ Microbiol. 1998;64:2403–2408. doi: 10.1128/aem.64.7.2403-2408.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleer R, Chen X J, Amellal N, Yeh P, Fournier A, Guinet F, Gault N, Faucher D, Folliard F, Fukuhara H, Mayaux J-F. High-level of secretion of correctly processed recombinant human interleukin-1β in Kluyveromyces lactis. Gene. 1991;107:285–295. doi: 10.1016/0378-1119(91)90329-a. [DOI] [PubMed] [Google Scholar]
- 21.Fleer R, Yeh P, Amellal N, Maury I, Fournier A, Bacchetta F, Baduel P, Jung G, L’Hote H, Becquart J, Fukuhara H, Mayaux J-F. Stable multicopy vectors for high-level secretion of recombinant human serum albumin by Kluyveromyces yeasts. Bio/Technology. 1991;9:968–974. doi: 10.1038/nbt1091-968. [DOI] [PubMed] [Google Scholar]
- 22.Futcher A B. Copy number amplification of the 2μm circle plasmid of Saccharomyces cerevisiae. J Theor Biol. 1986;119:197–204. doi: 10.1016/s0022-5193(86)80074-1. [DOI] [PubMed] [Google Scholar]
- 23.Futcher A B. The 2μm circle plasmid of Saccharomyces cerevisiae. Yeast. 1988;4:27–40. doi: 10.1002/yea.320040104. [DOI] [PubMed] [Google Scholar]
- 24.Murray J A H, Scarpa M, Rossi N, Cesareni G. Antagonistic controls regulate copy number of the yeast 2μ plasmid. EMBO J. 1987;6:4205–4212. doi: 10.1002/j.1460-2075.1987.tb02768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rocha T L, Paterson G, Crimmins K, Boyd A, Sawyer L, Fothergill-Gilmore L A. Expression and secretion of recombinant ovine β-lactoglobulin in Saccharomyces cerevisiae and Kluyveromyces lactis. Biochem J. 1996;313:927–932. doi: 10.1042/bj3130927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Som T, Armstrong K A, Volkert F C, Broach J R. Autoregulation of 2μm circle gene expression provides a model for maintenance of stable plasmid copy levels. Cell. 1988;52:27–37. doi: 10.1016/0092-8674(88)90528-4. [DOI] [PubMed] [Google Scholar]
- 28.Toh-e A, Tada S, Oshima Y. 2-μm DNA-like plasmid in the osmophilic haploid yeast Saccharomyces rouxii. J Bacteriol. 1982;151:1380–1390. doi: 10.1128/jb.151.3.1380-1390.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toh-e A, Araki H, Utatsu I, Oshima Y. Plasmid resembling 2-micron DNA in the osmotolerant yeasts Saccharomyces bailii and Saccharomyces bisporus. J Gen Microbiol. 1984;130:2527–2534. doi: 10.1099/00221287-130-10-2527. [DOI] [PubMed] [Google Scholar]
- 30.Tokunaga M, Ishibashi M, Tatsuda D, Tokunaga H. Secretion of mouse alpha-amylase from Kluyveromyces lactis. Yeast. 1997;13:699–706. doi: 10.1002/(SICI)1097-0061(19970630)13:8<699::AID-YEA124>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 31.Utatsu I, Sakamoto S, Imura T, Toh-e A. Yeast plasmid resembling 2μm DNA: regional similarities and diversities at the molecular level. J Bacteriol. 1987;169:5537–5545. doi: 10.1128/jb.169.12.5537-5545.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Den Berg J A, van der Laken K J, van Ooyen J J, Renniers T C H M, Rietveld K, Schaap A, Brake A J, Bishop R J, Schultz K, Moyer D, Richman M, Shuster J R. Kluyveromyces as a host for heterologous gene expression: expression and secretion of prochymosin. Bio/Technology. 1990;8:135–139. doi: 10.1038/nbt0290-135. [DOI] [PubMed] [Google Scholar]
- 33.Van Der Sand S T, Greenhalf W, Gardner D C J, Oliver S G. The maintenance of self-replicating plasmids in Saccharomyces cerevisiae: mathematical modelling, computer simulations and experimental tests. Yeast. 1995;11:641–658. doi: 10.1002/yea.320110705. [DOI] [PubMed] [Google Scholar]
- 34.Veit B E, Fangman W L. Copy number and partition of the Saccharomyces cerevisiae 2μm plasmid controlled by transcription regulators. Mol Cell Biol. 1988;8:4949–4957. doi: 10.1128/mcb.8.11.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volkert F C, Broach J R. Site-specific recombination promotes plasmid amplification in yeast. Cell. 1986;46:541–550. doi: 10.1016/0092-8674(86)90879-2. [DOI] [PubMed] [Google Scholar]
- 36.Volkert F C, Wilson D W, Broach J R. Deoxyribonucleic acid plasmids in yeast. Microbiol Rev. 1986;53:299–317. doi: 10.1128/mr.53.3.299-317.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walsh D J, Bergquist P L. Expression and secretion of a thermostable bacterial xylanase in Kluyveromyces lactis. Appl Environ Microbiol. 1997;63:3297–3300. doi: 10.1128/aem.63.8.3297-3300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]